Abstract
Synthetic biology aims to make genetic systems more amenable to engineering, which has naturally led to the development of Computer-Aided Design (CAD) tools. Experimentalists still primarily rely on project-specific ad-hoc workflows instead of domain-specific tools, suggesting that CAD tools are lagging behind the front line of the field. Here, we discuss the scientific hurdles that have limited the productivity gains anticipated from existing tools. We argue that the real value of efforts to develop CAD tools is the formalization of genetic design rules that determine the complex relationships between genotype and phenotype.
Computer-Aided Design tools for synthetic biology
Several groups have been developing Computer-Aided Design (CAD) solutions for synthetic biology [1–10] yet the transcriptional complexity of published artificial gene networks has been leveling off since 2005 [11]. After ten years of high-expectations and hype in synthetic biology, engineering biological systems has proved more challenging than anticipated [12]. The lack of sufficient tools in synthetic biology has spurred intense efforts to develop CAD software. Unfortunately, experimental synthetic biologists still rely largely on project-specific, ad-hoc development processes that combine construct assembly, data collection, data analysis, and mathematical modeling.
Five recent reviews have comprehensively covered the current state of computational tools for synthetic biology [13–17]. Thus, here we limit our description of these efforts to a brief, general overview. We also constrain the review to software specific to synthetic biology by excluding the many commercial software packages that are useful synthetic biologists, but intended for a broader user base. CAD tools for synthetic biology facilitate the design of larger systems from smaller genetic parts by providing users with visual, textual, or programming-language-like interfaces, or automatically generate designs from intended function. These tools assume that data such as sequence and description are attached to each part by the user or in some database. The aggregated parts sequences can then be leveraged to produce the corresponding physical DNA. Many tools include some level of functional modeling capabilities, which rely on the user providing the necessary equations and/or parameters (Figure 1). As a result most projects will require the use of multiple software tools that need to be integrated in software “stacks”. Box 1 discusses some of the technical and legal challenges that could hamper the integration of software tools.
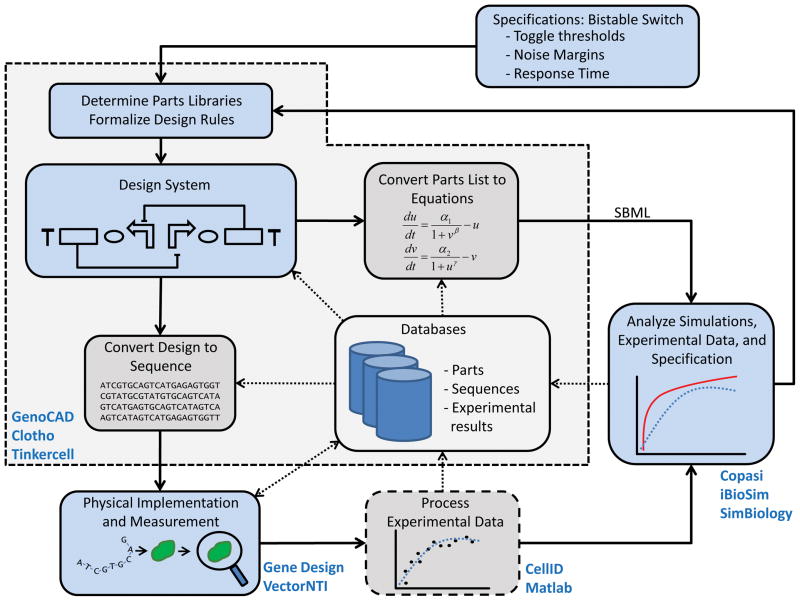
Figure 1.
GDA design flow. Synthetic biology projects typically rely on iterative workflows composed of different tasks. Emerging GDA tool chains rely on numerous software applications that support different phases of the project workflow. The development of a genetic switch [72] will start by expressing the design objective as a list of quantitative requirements: input toggle thresholds, noise margins, switching response time, etc. Once the objective is specified, it is possible to develop a list of genetic parts useable for the project. The choice of biological parts will involve factors such as use of the parts in prior projects, quality of the data characterizing the parts function, or intellectual property considerations. The formalization of design rules often takes place in parallel to the parts library development. Design rules may express rules such as whether it is acceptable to have polycistronic expression cassettes or if the design should be split between different plasmids. Only after parts have been selected and a strategy has been agreed upon is it possible to start designing constructs. In the fabrication phase, the construct is assembled usually by combining de novo gene synthesis and cloning of existing DNA sequences. Users use molecular biology software suites to facilitate assembly or order the sequence from a gene synthesis company. Experimentalists insert the synthetic DNA molecule into the host of choice and collect phenotypic data. Experimental data is then processed, for example by reducing microscopy images into time series of quantitative data. Performance is evaluated by considering simulations, experimental data, and the original specifications. At nearly every stage, software interacts with databases to reuse leverage past work or to store current work for future use. The shaded area delimited by dashes delineates stages facilitated by synthetic biology CAD software, while other stages are handled by more general purpose software. Text in blue indicates examples of software providing assisting at each stage.
Box 1. Software Integration.
A crucial evolution in EDA has been the integration of previously independent design flow steps as the assumptions allowing for isolation have eroded. Some examples have been alluded to previously: device physics are infringing upon the Boolean assumption, layout and function are intermingling, and testing is becoming more interlinked with design. Beginning with the development of centralized CAD frameworks in the early 1980’s, this progression has led to integrated CAD tools in EDA [41, 66] that have evolved to be modular and able to communicate with one another. In GDA, design flow stages are intrinsically interlaced, and as such tools should be designed for integration rather than forcing them apart. In that perspective, the importance of ongoing efforts to develop open source software frameworks and data exchange standards like the Synthetic Biology Open Language (SBOL) (Table 1) cannot be underestimated [67, 68].
As the field matures, many of the GDA applications will need to be integrated into custom software stacks as is happening in mainstream bioinformatics [69]. Different integration models have different legal implications. One model consists of integrating data by accessing specialized web services. This model is illustrated by the rapid development of scientific workflow systems that facilitate this data integration [70, 71]. The model is attractive because the tools only need to share a common language like SBOL, saving the end-user the effort of installing, maintaining, and integrating different software components. Yet, there are many potential difficulties with this integration model: (i) dependence on computational services provided on a volunteer basis by a third party creates potential vulnerability (ii) moving large amounts of data to web-hosted services can be inefficient, and (iii) sending sensitive data to a third-party server may be undesirable. The other approach to software integration consists of integrating different applications that complement each other but execute on the end-user computational resources. This integration model raises some software licensing issues. In order to prevent corporations from segmenting GDA markets into proprietary silos like in EDA, it is prudent to foster the emergence of a vibrant GDA software development community. In addition to avoiding market lockout resulting from proprietary software, it is crucial to ensure that the code base developed by the GDA community is free from hidden intellectual property claims and is licensed under permissive terms allowing academic and corporate stakeholders to reuse existing code bases.
Thus far in synthetic biology, relatively simple design goals such as “exhibits oscillations” [18] have advanced to increasingly sophisticated goals such as “fast, robust, tunable oscillators” [19] or “synchronized oscillators” [20]. As the field moves towards real-world applications [21], tools that can adequately predict functionality from design will be indispensible. Similarly, designers need to begin considering alternative design approaches and corresponding comparison metrics [22] to move beyond proof of concept designs. Recently announced design-to-spec competitions like CAGEN (Competitive Assessment of Genetically Engineered Networks) and GenoCon (International Rational Genome Design Contest) aim to help address this need (Table 1).
Table 1.
List of GDA-relevant websites
| Website | URL | Brief description |
|---|---|---|
| Bio-Design Automation | www.biodesignautomation.org | Community and annual event focused on Bio-Design Automation |
| CAGEN: Critical Assessment of Genetically Engineered Networks | openwetware.org/wiki/CAGEN | Competition aimed at more predictable genetic circuits |
| GenoCon International Rational-Genome-Design Contest | genocon.org | Competition aimed at practical genetic circuit engineering |
| iGEM Synthetic Biology based on standard parts | www.partsregistry.org | Library of genetic components |
| Joint BioEnergy Institute Registry | registry.jbei.org | Library of genetic components |
| biofab | www.biofab.org | Facility that provides standardized collections of genetic components |
| Synthetic Biology Open Language (SBOL) | www.sbolstandard.org | Community effort to develop data exchange standards |
The goal of this paper is to first explore the scientific hurdles that have limited the productivity gains anticipated from existing CAD tools, and to second argue that the real value of these efforts is not in promised productivity gains, but in the formalization of genetic design rules. Formalizations inherently test commonly held conceptions of how genetic systems work and consequently drive investigation into one of the most fundamental questions of genetics: How does phenotype arise from complicated networks of elements coded in the genotype? [23]
The slow maturation of EDA
CAD tools are ubiquitous in nearly all fields of engineering. They provide two primary functions: simplifying common tasks and making designs convenient for communication and evaluation. For example, blueprints are designed much faster on a computer than with traditional drafting. Moreover, CAD tools can generate layouts of each floor, 3D renderings of the building, or models of the building’s thermal performance.
In electronics, the development of consistent suites of CAD tools is called Electronic Design Automation (EDA). EDA utilizes iterative “design flows,” where key design processes are looped back on themselves until the design meets required specifications. Abstract representations of an electronic design in EDA can be organized into a hierarchy consisting of high-level description, logical description, and physical layout.
The spectacular success of EDA over the last 50 years [24] provides an inspiring model for synthetic biology. The synthetic biology counterpart of EDA is sometimes referred to as Bio-Design Automation (Table 1). However, the alternative name Genetic Design Automation (GDA) may better emphasize that synthetic biology focuses more on engineering DNA molecules than other biological objects [25]. Many have proposed that synthetic biologists leverage expertise in the design of electrical circuits, and in the same way GDA can draw from the development of EDA. In EDA, Hardware Description Languages (HDLs) are a special category of programming languages used to formally describe immensely complicated designs in a compact way. These languages rely extensively on the existing abstraction that digital circuits operate under the laws of Boolean algebra, which allows hugely complex circuits to be designed reliably.
This assumption does not exist for GDA. Exploratory work on an HDL for GDA has progressed under the assumption that similar enabling assumptions will emerge [10]. In parallel, efforts within the EDA community to describe analog and mixed analog-digital circuits with HDLs are ongoing. The challenges of extending HDLs to analog circuits are very similar to the challenges faced in GDA, and indeed some works have explored these similarities [26, 27].
The next sections describe three difficult problems that need to be solved before the level of automation in EDA can be achieved with GDA. There is mounting evidence that the first generation of GDA tools will not be able to ignore some of the most challenging problems currently faced by the EDA community.
Engineering fantasies: the scientific gaps facing GDA
Transitions between DNA sequence, model, and fabrication are currently hindered not so much by the implementation of CAD tools, but rather by three difficult scientific challenges: (i) predictability of components, (ii) decoupling of design and fabrication and (iii) experimental characterization methods. These outstanding questions are summarized in Box 2 and discussed in more details below.
Box 2. Outstanding questions.
Should parts be standardized or designed? Standardization of genetic parts relies on the assumption that parts function is context-independent. This hypothesis greatly simplifies many aspects of the design process. Evidence of context-dependencies affecting parts function may limit the success of standardization efforts. Designing custom parts for use in a particular context is an alternative to standardization.
Should design be orthogonal to fabrication? It is desirable to formalize manufacturing expertise as design rules that could be used to compare the cost of manufacturing functionally equivalent designs. Beyond the potential of substantial savings in manufacturing expenses, this effort is likely to uncover subtle relationships between the structure and functions of DNA sequences.
Should design be orthogonal to measurement? In order to ensure that a design can be validated it is important to integrate models of the measurement protocols in the design phase. Measurement strategies relying on fluorescent reporter genes introduce perturbations in the design that have been poorly characterized so far.
Can GDA facilitate collaboration across specialties? GDA can foster collaboration between specialists of different scientific and engineering domains but it is a social and intellectual challenge to develop languages and workflows that allow these specialists to communicate effectively.
Off-the-shelf Components
One of the popular visions for synthetic biology describes catalogues of clearly defined genetic parts that can be easily combined into larger genetic constructs with predictable biological function. This vision motivated the development of the BioBrick assembly standard and the Registry of Standardized Genetic Parts, a database of BioBrick compatible parts (Table 1). Tools that aggregate models of basic genetic components to form system-scale models are being developed [1, 2, 4, 28], but the lack of data sheets listing quantitative parameters characterizing the parts behavior has hampered the use of these tools for designing artificial gene networks [29]. Projects such as BioFAB (Table 1) are attempting to address this issue by characterizing large numbers of parts and standardizing data collection techniques [30].
Recent efforts to quantitatively characterize the effects of different parts on gene expression is revealing a complex landscape of context-dependencies that somewhat challenges the assumption that parts can be characterized in isolation. For instance, the RBS sequence was first assumed by many in the field to determine translation efficiency in prokaryotes independently from the downstream coding sequence. However, sequences around the translation start site can influence the secondary structure of the mRNA, which is long known to play a crucial role in the translation rate [31]. Tools utilizing thermodynamic models [32, 33] are now available to predict the translation initiation efficiency in prokaryotes using sequence both upstream and downstream of the translational start site. Coupling between translation and transcription elongation rates [34] also represents a challenge to the standardization of components, though the assumption that initiation, not elongation, is the rate limiting step in transcription may be a valid approximation. As a result, tools that can predict behavior based on sequence, thermodynamics, or other methods are emerging as increasingly attractive.
The above issues can be avoided by characterizing on a gene-by-gene or device-by-device level, a trend already apparent in the field [11]. Creation of device variants or automatically generated devices [35] should consider the many context dependencies that affect parts. Yet, even such low levels of granularity might prove to have unexpected context dependencies. Computational studies, inspired by impedance-matching in electronics, have demonstrated an effect termed “retroactivity” in which the performance of one genetic device is influenced by connecting a downstream device [36, 37]. Just as electronic circuit designers are currently running into major power limitations, synthetic biologists are almost certain to run into limits on the many ingredients necessary for gene expression. How the availability of resources within a cell impacts the performance of individual genetic components and devices will also become an important consideration.
Decoupling of design and fabrication
Historically, recombinant DNA technologies were so limited that fabrication constrained design to the point that software focused almost entirely on assisting cloning rather than design of function. The recent emergence of generic DNA fabrication methods, including standardized assembly of genetic parts or de novo gene synthesis [38] led to the emergence of DNA-sequence design as a new scientific problem [39, 40]. Because it is now possible to assume that generic DNA fabrication processes can assemble any sequence genetic engineers imagine, design and fabrication tend to be considered orthogonal engineering problems.
In EDA, the assumption of the Boolean abstraction has allowed fabrication to be considered mostly orthogonal to the rest of the design process. As circuit densities have rapidly increased, fabrication constraints have become more closely intertwined with other constraints such as timing delay and power consumption [41]. Consequently, increasingly integrated tools consider constraints from multiple design domains simultaneously. Genetic design is still in the process of moving away from fabrication technologies that constrain the design space to achieve complete independence between design and manufacture. For instance, BioBrick assembly standards have moved from the original standard precluding assembly of fusion proteins (BBF RFC 10) to proposed standards allowing fusions [42]or scarless assembly (BBR RFC 26, 39). Most recently, single-step assembly methods [43] have become popular.
Despite the simplification decoupling offers, there are distinct advantages to recoupling design and manufacturing during the design phase. Poor design strategies can create manufacturing problems. For example, repeated use of the same parts can cause sequence verification difficulty and structural instabilities [44]. Sequences with high GC content are notoriously difficult to amplify. Even though experienced gene synthesis companies will be able to synthesize most DNA sequences ordered by their customers, price and time to delivery vary greatly with sequence complexity. Ignoring such manufacturing constraints during the design phase will significantly increase the cost and duration of GDA cycles. Since many projects require the characterization of large numbers of design variants, the cost and time to fabricate these designs is still one of the bottlenecks of the GDA loop. Sophisticated design strategies are necessary to formalize manufacturing constraints [45] and optimize fabrication without serious detriment to function. For example, tools that can adjust codon bias or match non-unique sequences to function [32, 33, 35] could ameliorate manufacturing concerns. Sample tracking tools could suggest reuse of sequence segments resulting in shortened and cheaper assembly cycles. Algorithms to optimize fabrication processes [46] will have to be connected to design tools to give designers an immediate appreciation of the manufacturing cost of candidate designs.
Parts Characterization
The problem of part definitions extends to the ways experimental data are collected, used, and shared. Models vary from project to project and typically have only one or two fluorescent reporters as measured outputs. Since the models typically have many more parameters to estimate, finding parameter sets that are predictive from the many possible sets that match a given dataset is difficult. Combined with the range of measurement techniques used in the lab and the unknown impact of even small changes in experimental conditions, standardization and reuse of collected data are challenging.
Though not necessarily an explicit step in standard EDA design flows, Design for Test (DFT) is a background constraint throughout the process. As electronic circuits have become more complex, validation testing has become correspondingly more difficult. First, immense complexity makes exhaustive validation intractable, so intelligent minimization of test programs has evolved along with sophisticated testing equipment [47]. Second, the density of modern chips has made accessing nodes on internal layers without unintended performance effects a major problem. In living cells, the problem is much worse. Unknown cellular mechanisms, genetic instabilities, molecular noise, measurement accuracy, and inability to measure key components have prevented the verification of synthetic biology designs.
DFT in GDA starts by ensuring consistency between the design specification, experimental characterization methods, and mathematical paradigms used to model the behavior. For example, models based on deterministic equations can be supported by cell culture assays, whereas stochastic models call for single cell observations. Experimental design, common in some other engineering fields and recently emerging as a hot topic in systems biology [48], can guide experimentalists to optimal sets of measurements and avoid non-informative data collection. Depending on the system, this approach can also predict data sets that will determine all model parameters to within a given tolerance [49]. Similar techniques are frequently used in EDA to design intelligent testing programs, particularly for System-on-a-Chip designs where analog components commingle with digital circuits. Not considering the experimental characterization during the design phase may lead to designs that simply cannot be adequately tested.
The problem of collecting time-course data from inside living cells has been transformed by the emergence of fluorescent proteins. However, just like making an internal node in a chip accessible for test affects its performance, the maturation rate and protein half-life buffer fluorescence signals from the physiological events of interest [50]. Unfortunately, even parameters related to measurements such as the in vivo maturation rate of fluorescent proteins are difficult to determine accurately due to cell-to-cell variation and unknown dependencies on host strain, metabolic state, and environmental conditions. Also, the use of genes tagged with fluorescent domains is common, but the effects poorly understood. For example, how does a fluorescent tag affect the degradation rate of the fusion protein? Experimental work is needed to answer these questions.
New measurement devices will greatly facilitate the ability to determine in vivo parameter values and their dependencies on the continuously variable cellular environment. The emergence of time lapse fluorescent microscopy has allowed scientists to measure the dynamics of molecular mechanisms in individual cells [51]. This technique currently requires custom integration of optical equipment, image processing software, and microfluidic systems [52, 53]. Noise analysis connects the processed data to models of the underlying mechanisms [51]. Current image processing and analysis are typically postprocessing steps, but these prototypes prefigure a new generation of instruments that will acquire raw images, process them in real time, and implement data reduction algorithms to extract high-level statistics for comparison with design-phase models (Figure 2). Microfluidic systems will allow for complicated input control of environmental parameters that should open avenues to applying some advanced testing techniques seen in EDA DFT, such as frequency response analysis.
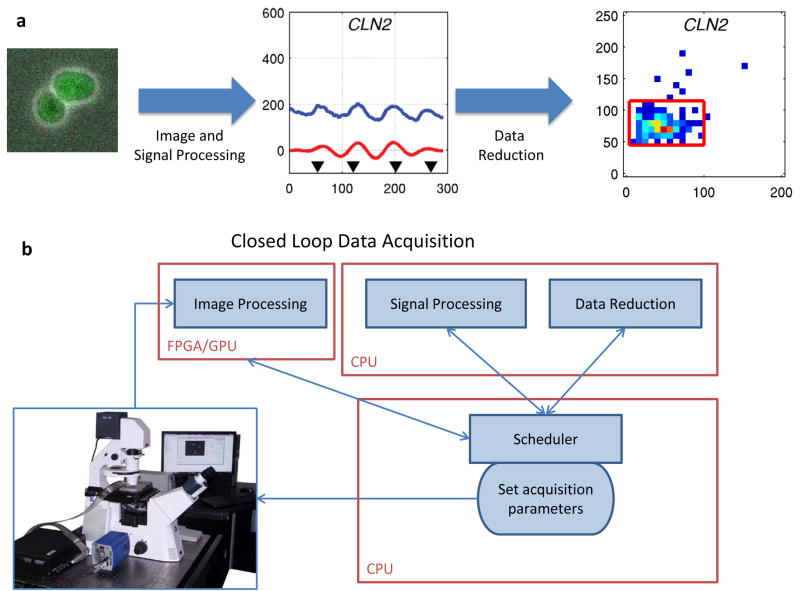
Figure 2.
New Instruments Connect Design and Experiment. (a) Using time-lapse microscopy for characterizing the dynamics of gene networks requires the development of custom suite of image and signal processing software along with data reduction algorithms. The mathematical models used to reduce movies into high-level statistics are necessarily related to the models used to design the gene network as ultimately experimental data need to be reconciled with model predictions. (b) Microscopy movies have traditionally been analyzed in a post-processing step. However, it is conceivable that in a near future the data analysis will be performed in real time by the computer controlling the microscope and the microfluidic system giving the user an experience similar to the use of a flow-cytometer. This information could also be used by the user to manually interact with the cells under observation. Alternatively, control algorithm could be developed to program the instrument to take specific actions such as changing the growth medium in response to specific behaviors of the cell populations.
A Scientific Renewal
Several authors recently argued that synthetic biology will lead to a better understanding of biology [54, 55]. In this spirit, we would like to question the assumption that the immediate value of GDA lies in its potential to accelerate the progress of experimental synthetic biologists. In the short term, efforts to develop GDA tools may be better justified as attempts to formalize genetic design principles. The assumption that models used in engineering can be extrapolated to biology can easily and rightfully be challenged by biologists. A more effective way of using GDA to engage the dialogue between engineers and life scientists might be to present GDA models as formal and compact representations of biological hypotheses. GDA then becomes a framework to express and test biological hypotheses, a form of scientific investigation common in the life sciences [56–58].
It is important to consider here two fundamental differences between EDA and GDA. The dynamics of genetic networks is largely determined by the interactions between large macromolecules confined to the small volume of a living cell. As a result, the dynamics of a genetic network are inherently stochastic in nature. There is even mounting evidence that many regulatory processes are based on molecular noise instead of merely attempting to mitigate its negative effects [59]. Electronic circuits use so many electrons that they behave deterministically. However, a consequence of the miniaturization and increasing power efficiency of silicon devices is a drastic reduction of the fluxes of electrons and a concomitant increase of the intrinsic electronic noise. It will be interesting to see if and how the EDA and GDA communities will work together to solve the problems associated with design automation of noisy systems.
Another important difference between GDA and EDA is that complexity in EDA was derived, while complexity in GDA was evolved. EDA has progressed by structured, rational improvements on the mathematical formalisms that express physical realities, incrementally allowing higher and higher complexity. As a result, the emergence of high-level function can always be traced to the lowest level components. On the other hand, since genetic systems evolved by random mutation, it is not clear that they follow rigorous design rules, and we cannot yet trace high-level function back to the low-level components in most cases. Synthetic biologists are walking in the footsteps of 50 years of effort by molecular geneticists to understand the design rules of genetic systems. Yet, the engineering mindset provides a new spark. The understanding of a genetic mechanism is truly put to the test when an engineer attempts to use the general principle to build something new. Formalization of these principles tests the theory and opens new areas of investigation when the theory is found lacking. A prime example of this is the aforementioned RBS calculator. Attempts to use “standard” ribosome binding sites failed and led to predictive thermodynamic models. The possibility to deoptimize the sequence of viral genes by taking advantage of codon pair bias [60, 61] is opening new research directions to better understand translation [62]. At a higher level of organization, the refactoring of the T7 genome resulted in reduced fitness that it is not completely understood [63].
In the current state, synthetic biology remains painfully slow, prohibitively expensive, and excessively labor-intensive. As an example, consider the progression from two 2002 theoretical papers on genetic oscillators [64, 65] to corresponding experimental publications in 2008 [19] and 2010 [20]. The unifying vision of a seamless GDA flow provides a collaborative framework for large interdisciplinary teams rather than relying on exceptional individual investigators familiar with all aspects of the design process. There is still a great deal of foundational work and biological discovery remaining before GDA materializes into suites of software tools facilitating design to specification entirely in silico. In the short term, closing some of the capability gaps will catalyze the emergence of more integrated teams that better handle the complex interdependencies between design, fabrication, and measurement. The tools these teams will generate may not have the elegance of an integrated solution, but they will provide new computational resources that should percolate beyond the confines of the synthetic biology community to benefit a larger population of life scientists.
Acknowledgments
This work is supported by NSF Awards 0850100 and 0963988 and by grant R01-GM078989 from the National Institutes of Health. MWL was supported by a fellowship from Science Application International Corporation and by a Department of Defense SMART Scholarship under OSD-T&E (Office of Secretary Defense-Test and Evaluation), Defense–Wide/PE0601120D8Z National Defense Education Program (NDEP)/BA-1, Basic Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hill AD, et al. SynBioSS: the synthetic biology modeling suite. Bioinformatics. 2008;24:2551–2553. doi: 10.1093/bioinformatics/btn468. [DOI] [PubMed] [Google Scholar]
- 2.Chandran D, et al. TinkerCell: modular CAD tool for synthetic biology. J Biol Eng. 2009;3:19. doi: 10.1186/1754-1611-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czar MJ, et al. Writing DNA with GenoCAD (TM) Nucleic Acids Research. 2009;37:W40–W47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchisio MA, Stelling J. Computational design of synthetic gene circuits with composable parts. Bioinformatics. 2008;24:1903–1910. doi: 10.1093/bioinformatics/btn330. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigo G, et al. Asmparts: assembly of biological model parts. Syst Synth Biol. 2007;1:167–170. doi: 10.1007/s11693-008-9013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers CJ, et al. iBioSim: a tool for the analysis and design of genetic circuits. Bioinformatics. 2009;25:2848–2849. doi: 10.1093/bioinformatics/btp457. [DOI] [PubMed] [Google Scholar]
- 7.Rialle S, et al. BioNetCAD: design, simulation and experimental validation of synthetic biochemical networks. Bioinformatics. 2010;26:2298–2304. doi: 10.1093/bioinformatics/btq409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilitchenko L, et al. Eugene - a domain specific language for specifying and constraining synthetic biological parts, devices, and systems. PLoS ONE. 2011;6:e18882. doi: 10.1371/journal.pone.0018882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia B, et al. Developer’s and User’s Guide to Clotho v2.0 A Software Platform for the Creation of Synthetic Biological Systems. Methods Enzymol. 2011;498:97–135. doi: 10.1016/B978-0-12-385120-8.00005-X. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen M, Phillips A. Towards programming languages for genetic engineering of living cells. J R Soc Interface. 2009;6(Suppl 4):S437–450. doi: 10.1098/rsif.2008.0516.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 12.Kwok R. Five hard truths for synthetic biology. Nature. 2010;463:288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- 13.Marchisio MA, Stelling J. Computational design tools for synthetic biology. Curr Opin Biotechnol. 2009;20:479–485. doi: 10.1016/j.copbio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Clancy K, Voigt CA. Programming cells: towards an automated ‘Genetic Compiler’. Current Opinion in Biotechnology. 2010;21:572–581. doi: 10.1016/j.copbio.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald JT, et al. Computational design approaches and tools for synthetic biology. Integr Biol. 2011;3:97–108. doi: 10.1039/c0ib00077a. [DOI] [PubMed] [Google Scholar]
- 16.Alterovitz G, et al. The challenges of informatics in synthetic biology: from biomolecular networks to artificial organisms. Briefings in bioinformatics. 2010;11:80–95. doi: 10.1093/bib/bbp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voigt CA, editor. Synthetic Biology Parts B: Computer Aided Design and DNA Assembly. Academic Press; 2011. [Google Scholar]
- 18.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 19.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danino T, et al. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ball DA, et al. Co-design in synthetic biology: a system-level analysis of the development of an environmental sensing device. Pac Symp Biocomput. 2010:385–396. [PubMed] [Google Scholar]
- 23.Benfey PN, Mitchell-Olds T. Perspective - From genotype to phenotype: Systems biology meets natural variation. Science. 2008;320:495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sangiovanni-Vincentelli A. The tides of EDA. Ieee Design & Test of Computers. 2003;20:59–75. [Google Scholar]
- 25.Myers CJ, et al. In: Roychowdhury J, editor. Genetic design automation; ICCAD ’09 Proceedings of the 2009 International Conference on Computer-Aided Design; ACM; 2009. pp. 713–716. [Google Scholar]
- 26.Gendrault Y, et al. Synthetic biology methodology and model refinement based on microelectronic modeling tools and languages. Biotechnol J. 2011;6:796–806. doi: 10.1002/biot.201100083. [DOI] [PubMed] [Google Scholar]
- 27.Pêcheux FMM, Lallement C. Is SystemC-AMS an appropriate “promoter” for the modeling and simulation of bio-compatible systems?. Proceedings of 2010 IEEE International Symposium on Circuits and Systems (ISCAS); 2010. pp. 1791–1794. [Google Scholar]
- 28.Cai Y, et al. Modeling structure-function relationships in synthetic DNA sequences using attribute grammars. PLoS Comput Biol. 2009;5:e1000529. doi: 10.1371/journal.pcbi.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canton B, et al. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 30.Kelly JR, et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salis HM, et al. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Na D, et al. Mathematical modeling of translation initiation for the estimation of its efficiency to computationally design mRNA sequences with desired expression levels in prokaryotes. BMC Syst Biol. 2010;4:71. doi: 10.1186/1752-0509-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proshkin S, et al. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamata I, et al. In: Deaton R, Suyama A, editors. Automatic design of DNA logic gates based on kinetic simulation; DNA Computing and Molecular Programming 15th International Conference, DNA; Springer; 2009. pp. 88–96. [Google Scholar]
- 36.Del Vecchio D, et al. Modular cell biology: retroactivity and insulation. Mol Syst Biol. 2008;4:161. doi: 10.1038/msb4100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KH, Sauro HM. Measuring retroactivity from noise in gene regulatory networks. Biophys J. 2011;100:1167–1177. doi: 10.1016/j.bpj.2010.12.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czar MJ, et al. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Goler JA, et al. Genetic design: rising above the sequence. Trends Biotechnol. 2008;26:538–544. doi: 10.1016/j.tibtech.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 41.Scheffer L, et al. EDA for IC implementation, circuit design, and process technology. CRC Taylor & Francis; 2006. [Google Scholar]
- 42.Anderson JC, et al. BglBricks: A flexible standard for biological part assembly. J Biol Eng. 2010;4:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira PH, et al. Structural instability of plasmid biopharmaceuticals: challenges and implications. Trends Biotechnol. 2009;27:503–511. doi: 10.1016/j.tibtech.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Cai Y, et al. GenoCAD for iGEM: a grammatical approach to the design of standard-compliant constructs. Nucleic Acids Res. 2010;38:2637–2644. doi: 10.1093/nar/gkq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Densmore D, et al. Algorithms for automated DNA assembly. Nucleic Acids Res. 2010;38:2607–2616. doi: 10.1093/nar/gkq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheffer LK, et al. EDA for IC system design, verification, and testing. CRC Taylor & Francis; 2006. [Google Scholar]
- 48.Kreutz C, Timmer J. Systems biology: experimental design. FEBS J. 2009;276:923–942. doi: 10.1111/j.1742-4658.2008.06843.x. [DOI] [PubMed] [Google Scholar]
- 49.Apgar JF, et al. Sloppy models, parameter uncertainty, and the role of experimental design. Mol Biosyst. 2010;6:1890–1900. doi: 10.1039/b918098b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, et al. Mathematical analysis and quantification of fluorescent proteins as transcriptional reporters. Biophysical Journal. 2008;94:2017–2026. doi: 10.1529/biophysj.107.122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Locke JCW, Elowitz MB. Using movies to analyse gene circuit dynamics in single cells. Nature Reviews Microbiology. 2009;7:383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett MR, Hasty J. Microfluidic devices for measuring gene network dynamics in single cells. Nat Rev Genet. 2009;10:628–638. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charvin G, et al. Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 2010;8:e1000284. doi: 10.1371/journal.pbio.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elowitz M, Lim WA. Build life to understand it. Nature. 2010;468:889–890. doi: 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bashor CJ, et al. Rewiring Cells: Synthetic Biology as a Tool to Interrogate the Organizational Principles of Living Systems. Annual Review of Biophysics. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kell DB, Oliver SG. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays. 2004;26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- 57.King RD, et al. Functional genomic hypothesis generation and experimentation by a robot scientist. Nature. 2004;427:247–252. doi: 10.1038/nature02236. [DOI] [PubMed] [Google Scholar]
- 58.Evans J, Rzhetsky A. Machine Science. Science. 2010;329:399–400. doi: 10.1126/science.1189416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balazsi G, et al. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28:723–U1729. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan LY, et al. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1:2005.0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hasty J, et al. Synthetic gene network for entraining and amplifying cellular oscillations. Phys Rev Lett. 2002;88:148101. doi: 10.1103/PhysRevLett.88.148101. [DOI] [PubMed] [Google Scholar]
- 65.McMillen D, et al. Synchronizing genetic relaxation oscillators by intercell signaling. Proc Natl Acad Sci U S A. 2002;99:679–684. doi: 10.1073/pnas.022642299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barnes TJ. Electronic CAD frameworks. Kluwer Academic Publishers; 1992. [Google Scholar]
- 67.Galdzicki M, et al. Standard biological parts knowledgebase. PLoS ONE. 2011;6:e17005. doi: 10.1371/journal.pone.0017005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peccoud J, et al. Essential information for synthetic DNA sequences. Nat Biotechnol. 2011;29:22. doi: 10.1038/nbt.1753. discussion 22–23. [DOI] [PubMed] [Google Scholar]
- 69.Stajich JE, Lapp H. Open source tools and toolkits for bioinformatics: significance, and where are we? Briefings in bioinformatics. 2006;7:287–296. doi: 10.1093/bib/bbl026. [DOI] [PubMed] [Google Scholar]
- 70.McPhillips T, et al. Scientific workflow design for mere mortals. Future Gener Comp Sy. 2009;25:541–551. [Google Scholar]
- 71.Shon J, et al. Scientific workflows as productivity tools for drug discovery. Current opinion in drug discovery & development. 2008;11:381–388. [PubMed] [Google Scholar]
- 72.Gardner TS, et al. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]