Abstract
Core-shell conjugated polymer nanoparticles (CPNs) were fabricated by complexing a semi-flexible, primary amine-containing conjugated polymer (CP) with hyaluronic acid (HA). Flexibility introduced in the rigid rod conjugated backbone allows backbone reorganization to increase π-π interaction under ionic complexation, resulting in core-shell nanoparticles with a hydrophobic CP core wrapped with a HA shell. The core-shell nanoparticles exhibited no cellular toxicity and high cancer cell specificity with minimal binding to normal cells.
INTRODUCTION
Recently, conjugated polymers (CPs) have attracted much attention for sensing,1–4 labeling,5–10 and delivery11–13 of biological substances owing to their excellent photophysical and biophysical properties. CPs are synthetic macromolecules containing fully conjugated π-electrons along the backbones, resulting in high luminescence brightness, high photostability, and excellent energy transfer.14,15 By incorporating hydrophilic polar side chains and functional entities such as sensing units or targeting ligands, various conjugated polyelectrolytes (CPEs) and conjugated polymer nanoparticles (CPNs) have been synthesized and used for biological applications in aqueous environments.16–18
The photophysical excellence and robustness of CP-based nanomaterials are typically useful for labeling target cells or tissues. Biophysically, amphiphilicity of the CPEs and CPNs is important for cellular interaction and subsequent cellular entry because the cell surface contains both negatively charged proteoglycans and hydrophobic membrane lipids. Especially, particles with high surface-to-volume ratios exhibit size, shape, and functional group-dependent cellular interactions and entry efficiency.19, 20 Therefore, functional modifications and structural modulations of CP-based nanomaterials are highly anticipated to obtain desired biophysical properties for improved cellular applications.
Previously, we fabricated positively charged CPNs by treating a non-aqueous soluble, primary amine-containing rigid rod CP with organic acids followed by dialysis.21, 22 The aggregation structures of CPNs, after complete removal of organic acids, were dependent on the nature of organic acids. Acetic acid treatment produced loose aggregation of rigid rod CP backbones, while increased π-π interactions among the backbones were observed when the same batch of polymer was treated with tartaric acid (i.e., dicarboxylic acid). From this observation, we hypothesized that if CPs containing flexible units along the backbones are treated with polymeric acids, π-π interaction among the backbones will be dramatically increased because the semi-flexibility of the non-aqueous soluble backbones will help backbone reorganization to maximize hydrophobic chain interaction. If complexation between the non-aqueous soluble CP and polyanion contributes to increasing aqueous solubility of CPs, random complex formations is expected with no significant spectral changes. To test this hypothesis and functionalize the CP with cancer cell-specific ligands, we employed a linear polysaccharide, hyaluronic acid (HA), which contains N-acetyl-D-glucosamine and D-glucuronic acid units. HA has specific binding with cell surface receptors such as CD44 and RHAMN, which are overexpressed in many cancer cells.23 Numerous cancer drugs and polymers have been modified with HA for targeted drug and gene delivery.24–28
In this note, we report that core-CP shell-HA nanoparticles is obtained when semi-flexible CPs are treated with polyanionic HA, while non-flexible CPs produce random complexes upon HA treatment. The core-shell nanoparticles are nontoxic to cells and exhibit high cancer cell specificity through the specific binding of HA to cancer cell surface receptors.
RESULTS AND DISCUSSION
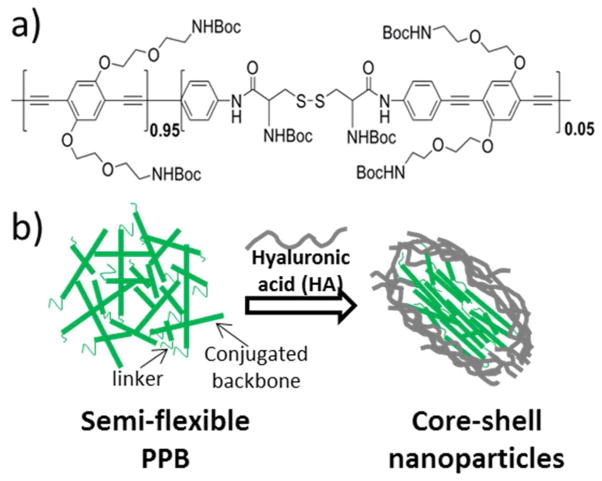
A semi-flexible poly(phenylenebutadiynylene) (PPB), which contains a small fraction of flexible non-conjugating units along the rigid conjugated backbones, was prepared by following our published method21 and used for the complexation with HA (Figure 1). The semi-flexible PPB was treated with organic acids, dialyzed against water, and complexed with HA.
Figure 1.
a) Chemical structure of the semi-flexible PPB. b) A schematic presentation of structural reorganization of the semi-flexible PPB upon HA complexation. The semi-flexible PPB in water was prepared by deprotection of Boc group using organic acid treatments followed by dialysis.
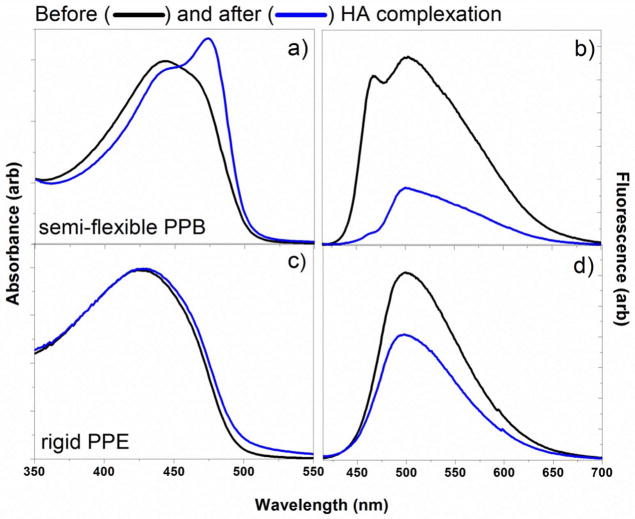
The semi-flexible PPB exhibits characteristic photophysical properties of the fully conjugated PPBs, because the amount of non-conjugated unit is limited (~5–10%) not to perturb the π-electron conjugation along the backbone. The photophysical and physical properties of the semi-flexible PPB in water were similar to the characteristics of loosely aggregated CPs with fully conjugated backbones. When the semi-flexible PPB was mixed with HA, the resulting complex exhibited dramatic changes in both absorption and emission spectra. As shown in Figure 2, a new strong absorption peak appeared at ~474 nm (30 nm red-shifted upon complexation) (Figure 2a), and the emission intensity of the complex decreased significantly (Figure 2b). We believe that unlike complexes formed between conjugated rigid oligomers and polysaccharides, which exhibit both red-shift and fluorescent increase through chain end-to-end alignment induced by linear polysaccharides,29,30 the loosely aggregated, non-aqueous soluble PPBs connected with flexible linkers were able to reorganize to increase interpolymer π-π interactions, similarly to the high molecular weight, fully conjugated poly(phenyleneethynylene) (PPEs) at the air-water interface under mechanical forces.31 As a control, a fully conjugated PPE containing the same amine side chains was treated with HA and the spectroscopic behaviors were monitored. We employed the structurally similar PPE as a control due to poor solubility of PPB without flexible linkers. Upon complexation with HA, the control PPE exhibited only a small increase in the absorption maximum (<5 nm) without a spectral shape change (Figure 2c), and the fluorescent intensity decreased slightly (Figure 2d). This implies that no significant structural reorganization of PPE occurred upon HA complexation, supporting that the semi-flexibility of PPB is responsible for the significant spectral changes upon ionic complexation.
Figure 2.
Absorbance and emission spectra of semi-flexible PPB (a and b) and rigid PPE (c and d) before and after HA complexation. A sharp absorption peak at 474 nm (a) and decreased emission intensity of the semi-flexible PPB after HA complexation supports the formation of ordered aggregation among the semi-flexible conjugated backbones. No significant changes were observed upon HA complexation of the control PPE (c and d).
While the hydrodynamic radius of the semi-flexible PPB at the low concentration (10 μM) could not be determined due to insufficient scattering intensity from the loose aggregations, the ionic complexes produced strong light scattering, implying the formation of dense particles. The hydrodynamic radius of the semi-flexible PPB/HA complex was successfully determined to be 51.35 ± 0.77 nm. Zeta potential value of the semi-flexible PPB/HA nanoparticles in water at neutral pH was −25.5 ± 11.7 mV (Supporting Information Figure S1). The hydrodynamic radius of the control PPE/HA complex was determined to be 79.49 ± 0.18 nm, with a zeta potential of −32.6 ± 5.6 mV (Supporting Information Figure S2). The control PPE/HA was almost 30 nm larger than the semi-flexible PPB/HA, implying that more loosely aggregated particles were formed.
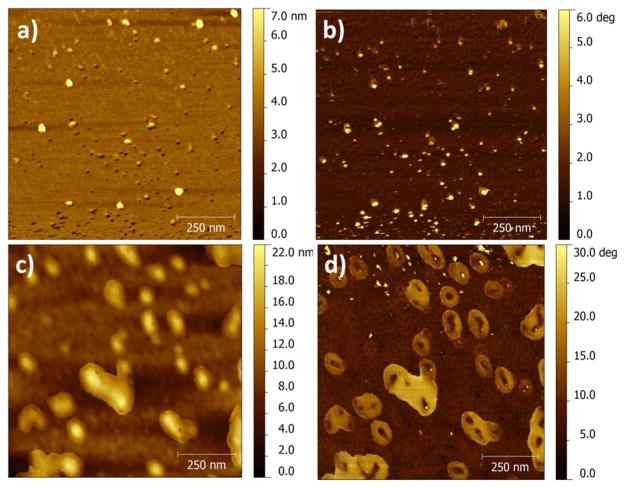
Atomic force microscopic (AFM) imaging of the semi-flexible PPB/HA nanoparticle supported the formation of elongated core-shell nanoparticles with average size of 58 ± 13 nm (by measuring lateral sizes), as shown in Figure 3c, while the semi-flexible PPB without HA complexation exhibited mixed particles with no specific shapes (Figure 3a). AFM phase image also showed that core-shell particles were favorably formed at a molar ratio of 1:3 of the semi-flexible PPB to HA. Both at the lower and higher ratios, no defined core-shell nanoparticles were observed (Supporting Information Figure S3). Higher density (represented as dark color) was observed in the center of the nanoparticles, while lower density (bright color) was observed in the shell. Since the particles were prepared on an aminosilanized mica surface, it is difficult to determine the shape and size of the intact nanoparticles in water by the AFM imaging. The AFM images are believed to be flattened during the sample preparation determined by the height analysis (Supporting Information Figure S3). The topographic and phase images of the control PPE/HA show circular particles with no core-shell shape, with an average size and height of 81±9 and 4±0.5 nm, respectively (Supporting Information Figure S4). The AFM imaging of the control PPE/HA further supports that HA does not cause structural reorganization of rigid rod CPs.
Figure 3.
AFM topographic (a and c) and phase (b and d) images of the semi-flexible PPB (a and b) and semi-flexible PPB/HA nanoparticles formed at 1:3 molar ratio (c and d). The semi-flexible PPB/HA exhibits elongated particles on a mica surface (c), and the phase image (d) reveals that the complexes are core-shell nanoparticles.
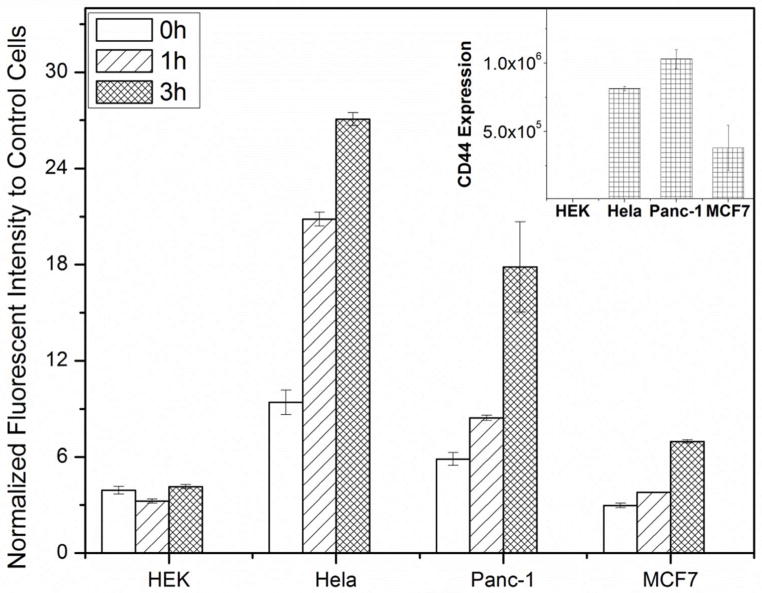
Cancer cell specificity of the core-shell nanoparticle was examined by incubating various cells including cancer and normal cells with the core-shell nanoparticles for different times, and the normalized mean fluorescent intensity ratios to the control cells were plotted by analyzing flow cytometry data (Figure 4). As shown in Figure 4, cancer cells overexpressing CD44 [i.e., human cervical carcinoma (HeLa) and human pancreatic carcinoma cell lines (Panc-1)] exhibited an intensity increase as the incubation time increased, while normal human embryonic kidney cells (HEK) and cancer cells with low CD44 expression exhibited low fluorescent intensity over the incubation times. The labeling specificity coincided well with the CD44 expression levels, which were measured by incubating fluorescently labeled antibody against CD44 with various cells (inset of Figure 4). As expected, the core-shell nanoparticles exhibit no toxicity measured by ATP consumption of the treated cells (Supporting Information Figure S5).
Figure 4.
Normalized fluorescent mean intensity ratios of various cells treated with the semi-flexible PPB/HA (1:3) up to 3 h. Cancer cells overexpressed CD44 (inset) exhibit higher fluorescent intensities than those from cells with low CD44 expression, supporting specific labeling of cancer cells via HA-CD44 interaction.
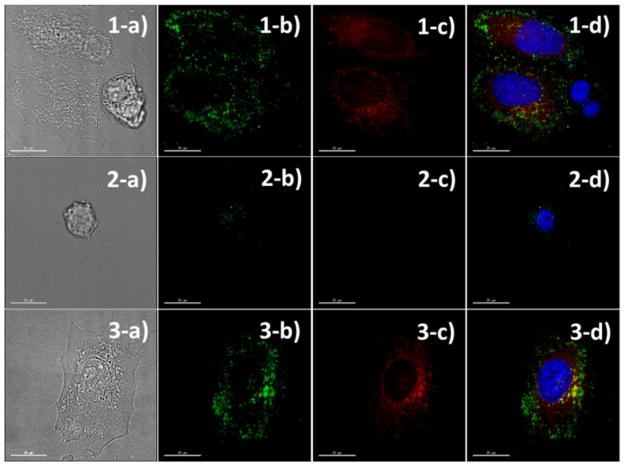
The specific labeling of cancer cells by the core-shell particles was further confirmed by fluorescence microscopic imaging of cancer cells co-cultured with normal HEK cells. Before co-culturing, HeLa cells were pre-labeled with a fluorescent dye (CellTracker Red) to fluorescently distinguish the cancer cells from normal cells. It is known that some fluorescent dyes have poor retention in the cell and can readily diffuse through the cell membrane. CellTracker Red was used because it is converted into an impermeable dye by forming covalent bonds inside cells. After incubating the cells for 1 h, the cells were rinsed, stained (for labeling of nucleus), and fixed for microscopic imaging. Because the cancer cells possibly influence the normal cell growth, each cell line was independently cultured and incubated with the core-shell nanoparticles under the same incubation condition. As shown in Figure 5a, strong green fluorescent signals were observed as scattered dots throughout the HeLa cells (pre-labeled with a red fluorescent dye), while much weaker green signals were observed from the normal HEK cells, indicating that the core-shell nanoparticles preferentially labeled HeLa cells through the specific HA-CD44 binding followed by endocytosis. For the control cells cultured independently, HeLa cells exhibited many green fluorescent dots throughout the cytosol, while the number and intensity of green dots observed from HEK were significantly low (Figure 5b).
Figure 5.
Fluorescent microscopic (b-d) and contrast (a) images of HeLa/HEK mixed cells (1), HEK cells (2), and HeLa cells (3) incubated with the semi-flexible PPB/HA for 1 h, respectively. HeLa cells were fluorescently pre-labeled with a red dye (column c) before the core-shell nanoparticle incubation. Core-shell nanoparticles were seen under the green channel (column b); the nuclei were stained with a blue dye; and merged images were seen in column d. The core-shell nanoparticles preferentially labeled HeLa cells while exhibiting low binding to normal HEK cell.
CONCLUSION
In summary, we have fabricated core-shell nanoparticles by complexing a semi-flexible PPB with a linear polysaccharide, HA. The resulting fluorescent core-shell nanoparticles exhibited high cancer cell specificity with low adsorption to normal cells. Since the size and shape of nanomaterials significantly influence labeling and delivery efficiency of biological substances, the concept of core-shell nanoparticle formation through controlled aggregation introduced in this work will contribute to novel biomaterials syntheses and fabrications.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health (SC1GM092778-01A1). We acknowledge the use of AFM in Professor Wenzhi Li’s lab.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information. General experimental and synthetic procedures, DLS, AFM, flow cytometry, and cellular toxicity protocols with corresponding figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pinto MR, Schanze KS. Proc Natl Acad Sci USA. 2004;101:7505–7510. doi: 10.1073/pnas.0402280101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaylord BS, Heeger AJ, Bazan GC. Proc Natl Acad Sci USA. 2002;99:10954–10957. doi: 10.1073/pnas.162375999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 4.Wang LH, Pu KY, Li J, Qi XY, Li H, Zhang H, Fan CH, Liu B. Adv Mater. 2011;23:4386–4391. doi: 10.1002/adma.201102227. [DOI] [PubMed] [Google Scholar]
- 5.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc Natl Acad Sci USA. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim IB, Shin H, Garcia AJ, Bunz UHF. Bioconjugate Chem. 2007;18:815–820. doi: 10.1021/bc0603440. [DOI] [PubMed] [Google Scholar]
- 7.Rahim NAA, McDaniel W, Bardon K, Srinivasan S, Vickerman V, So PTC, Moon JH. Adv Mater. 2009;21:3492–3496. [Google Scholar]
- 8.Moon JH, McDaniel W, MacLean P, Hancock LE. Angew Chem Int Ed. 2007;46:8223–8225. doi: 10.1002/anie.200701991. [DOI] [PubMed] [Google Scholar]
- 9.Yu JB, Wu CF, Zhang XJ, Ye FM, Gallina ME, Rong Y, Wu IC, Sun W, Chan YH, Chiu DT. Adv Mater. 2012;24:3498–3504. doi: 10.1002/adma.201201245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Lee J, Jeong EJ, Kronk A, Elenitoba-Johnson KSJ, Lim MS, Kim J. Adv Mater. 2012;24:2479–2484. doi: 10.1002/adma.201103895. [DOI] [PubMed] [Google Scholar]
- 11.Moon JH, Mendez E, Kim Y, Kaur A. Chem Commun. 2011;47:8370–8372. doi: 10.1039/c1cc10991j. [DOI] [PubMed] [Google Scholar]
- 12.Silva AT, Alien N, Ye CM, Verchot J, Moon JH. BMC Plant Biol. 2010;10:291. doi: 10.1186/1471-2229-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng X, Lv F, Liu L, Yang Q, Wang S, Bazan GC. Adv Mater. 2012;24:5428–5432. doi: 10.1002/adma.201202145. [DOI] [PubMed] [Google Scholar]
- 14.Bunz UHF. Acc Chem Res. 2001;34:998–1010. doi: 10.1021/ar010092c. [DOI] [PubMed] [Google Scholar]
- 15.Roncali J. Chem Rev. 1997;97:173–205. doi: 10.1021/cr950257t. [DOI] [PubMed] [Google Scholar]
- 16.Thomas SW, Joly GD, Swager TM. Chem Rev. 2007;107:1339–1386. doi: 10.1021/cr0501339. [DOI] [PubMed] [Google Scholar]
- 17.Zhu CL, Liu LB, Yang Q, Lv FT, Wang S. Chem Rev. 2012;112:4687–4735. doi: 10.1021/cr200263w. [DOI] [PubMed] [Google Scholar]
- 18.Pecher J, Mecking S. Chem Rev. 2010;110:6260–6279. doi: 10.1021/cr100132y. [DOI] [PubMed] [Google Scholar]
- 19.Tan SJ, Jana NR, Gao SJ, Patra PK, Ying JY. Chem Mater. 2010;22:2239–2247. [Google Scholar]
- 20.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 21.Vokatá T, Moon JH. Macromolecules. 2013;46:1253–1259. doi: 10.1021/ma3019975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko YJ, Mendez E, Moon JH. Macromolecules. 2011;44:5527–5530. doi: 10.1021/ma200661h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toole BP. Nat Rev Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 24.Needham CJ, Williams AK, Chew SA, Kasper FK, Mikos AG. Biomacromolecules. 2012;13:1429–1437. doi: 10.1021/bm300145q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh EJ, Kim KS, Kim YR, Jung HS, Beack S, Kong WH, Scarcelli G, Yun SH, Hahn SK. Biomacromolecules. 2012;13:2554–2561. doi: 10.1021/bm300796q. [DOI] [PubMed] [Google Scholar]
- 26.Choi KY, Yoon HY, Kim JH, Bae M, Park RW, Kang YM, Kim IS, Kwon IC, Choi K, Jeong SY, Kim K, Park JH. ACS Nano. 2011;5:8591–8599. doi: 10.1021/nn202070n. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Bae BC, Na K. Bioconjugate Chem. 2010;21:1312–1320. doi: 10.1021/bc100116v. [DOI] [PubMed] [Google Scholar]
- 28.Choi KY, Chung H, Min KH, Yoon HY, Kim K, Park JH, Kwon IC, Jeong SY. Biomaterials. 2010;31:106–114. doi: 10.1016/j.biomaterials.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 29.Tang YL, Hill EH, Zhou ZJ, Evans DG, Schanze KS, Whitten DG. Langmuir. 2011;27:4945–4955. doi: 10.1021/la1050173. [DOI] [PubMed] [Google Scholar]
- 30.Tang Y, Zhou Z, Ogawa K, Lopez GP, Schanze KS, Whitten DG. Langmuir. 2009;25:21–25. doi: 10.1021/la803111b. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Swager TM. Nature. 2001;411:1030–1034. doi: 10.1038/35082528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.