Abstract
Recent evidence suggests that a subset of hepatocellular carcinomas (HCCs) are derived from liver cancer stem cells (LCSCs). In order to isolate and characterize LCSCs, reliable markers that are specific to these cells are required. We evaluated the efficacy of a range of cancer stem cell (CSC) markers in isolating and characterizing LCSCs. We show that the most widely used CSC markers are not specific to LCSCs. By western analysis, protein expression of the common markers showed no significant difference between HCC tumor tissues and adjacent non-cancerous liver. Further, isolation of LCSCs from common HCC cell lines using FACScan and microbeads showed no consistent marker expression pattern. We also show that LCSCs have unique subtypes. Immunohistochemistry of HCC tissues showed that different HCCs express unique combinations of LCSC markers. Quantitative real-time polymerase chain reaction analysis showed that LCSCs isolated using different markers in the same HCC phenotype had different expression profiles. Likewise, LCSCs isolated from different HCC phenotypes with the same marker also had unique expression profiles and displayed varying resistance profiles to Sorafenib. Thus, using a range of commonly used CSC markers in HCCs and cell lines, we demonstrate that currently available markers are not specific for LCSCs. LCSCs have unique subtypes that express distinctive combinations of LCSC markers and altered drug resistance profiles, making their identification problematic.
Introduction
Hepatocellular carcinoma (HCC) is an aggressive cancer in which the majority of patients will die within a year of diagnosis [1]. It is currently the fifth most common cancer worldwide and the third most common cause of cancer mortality [2]. HCC is more prevalent in Asia and sub-Saharan Africa, where it accounts for 80% of cases diagnosed worldwide [2]. The vast majority of these tumors develop as a result of chronic liver injury caused by infections with hepatitis B virus (HBV) and hepatitis C virus (HCV), alcohol abuse, non-alcoholic steatohepatitis, and exposure to liver toxins such as aflatoxin and oral contraceptives. Among these, HBV and HCV infections are responsible for more than 80% of all HCC cases [1–3]. HCC has a bleak prognosis which is largely attributed to a poor understanding of the molecular mechanisms that control the initiation, progression, and treatment refractoriness of the tumor. Currently available options for the treatment of advanced liver cancer, including chemotherapy, internal radiation, local ablation, and anti-angiogenesis therapies, have shown limited efficacy.
It is now recognized that a small proportion of the cells within HCCs possess stem cell properties, including unlimited proliferative ability, a strong potential for self-renewal, and unlimited differentiation ability into cancer cell progeny [4]. These cells are termed cancer stem cells (CSCs) or tumor initiating cells. The role of liver cancer stem cells (LCSCs) in HCC has been verified in immunocompromised mice [5]. However, it is currently unknown how LCSCs originate.
HCC generally develops as a result of chronic liver inflammation that is induced most commonly by hepatitis B and C. In this context, LCSCs are highly active during liver inflammation and cirrhosis [6], and persistent liver injury activates the progenitor cell compartment, leading to their replication [7,8] and an increased ability for clonal expansion into tumors [9]. It has been reported that ∼55% of the small dysplastic foci that are the premalignant lesions of HCC consist of progenitor cells [5,6], and about 25%–50% of HCCs express markers of progenitor cells and possess progenitor cell compartments [6]. Liver progenitor cells and LCSCs are types of adult stem cells, which are the only cell type that persists in the tissue for a sufficient length of time to acquire genetic changes, leading to neoplastic development, suggesting that HCC is derived from oncogenic mutations in liver stem cells [10,11]. LCSCs may also be derived from the dedifferentiation of mature hepatocytes into a dedifferentiated state [12].
It is now also believed that CSCs may be responsible for treatment failure and relapse or metastasis, as these cells are resistant to most of the currently available chemotherapy and radiotherapy [10,11]. Therefore, elimination of CSCs has the potential to improve patient outcomes and survival.
Successful identification of LCSCs is a pre-requisite for a better understanding of the molecular mechanisms by which liver cancer initiates, progresses, evades treatment, relapses, and metastasizes. Currently, identification of CSCs is achieved through several approaches, including (1) flow cytometry separation using CSCs surface markers [13]; (2) detection of side population by the Hoechst 33342 exclusion assay [14]; (3) in vitro floating tumor sphere formation in serum-free medium [15] coupled with xenograft tumor formation in immune-deficient mice [16,17]; and (4) other assays such as aldehyde dehydrogenase (ALDH) activity assay [18] and immunohistochemistry analysis [15]. Several LCSC markers have been reported, including CD133, CD90, CD44, epithelial cell-adhesion molecule (EpCAM), CD13, OV6, and ALDH [19,20]. However, the reliability of each of these markers in identifying true LCSCs varies with LCSCs sorted from different laboratories showing high heterogeneity [21]. LCSCs isolated using different markers may show similar stemness. For example, CD90+ cells have been shown to be present in the majority of HCC but not in cirrhotic patients. These cells possessed stem cell properties and were able to stably form tumors in nude mice [20]. Similarly, CD133+ and CD44+ cells displayed stem cell properties and a greater tumorigenic ability compared with CD133− and CD44− cells [19,20]. So far, no universal markers for LCSCs have been identified, and isolating LCSC on the basis of single markers is problematic. Further, some stem cell markers may not be specific to LCSC, and they are generally not universally expressed in all LCSCs [20,22].
Given the problems of isolating LCSC using stem cell markers, there is a need for a comprehensive evaluation of the effectiveness of stem cell markers to characterize and isolate LCSC. Our study tested a range of LCSC markers in both human HCC and HCC cell lines to evaluate the effectiveness of these markers to characterize and isolate LCSCs.
Materials and Methods
HCC tissues
Fourteen cases of human HCC and matched adjacent non-cancerous tissues from patients who had undergone treatment at Westmead Hospital were collected. Tissues were collected from a single HCC lesion along with surrounding normal tissue. All HCCs were clinically and pathologically confirmed. Written consents were obtained from all patients before sample collection. The study was approved by the Human Ethics Committee of the Western Sydney Area Health Service.
Culture of HCC cell lines
The HCC cell line HuH7 was cultured in Dulbecco's modified Eagle's medium+10% fetal calf serum (FCS) at 37°C, 5% CO2. HCC cell lines PLC/PRF/5, SNU182, and SNU423 were cultured in RPMI+10% FCS at 37°C, 5% CO2.
Antibodies
Unconjugated antibodies against CD44, EpCAM, and alpha fetoprotein (AFP) were purchased from Cell Signaling. Unconjugated antibodies against CD133 and ALDH were purchased from Abcam. Unconjugated anti-CD90 was purchased from Epitomics. Microbead-conjugated and PE-conjugated antibodies against CD44, CD133, EpCAM, and CD90 were purchased from Miltenyi Biotec.
Protein extraction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and western blots
Tissues were lysed in protein extraction buffer [50 mM Tris-HCL (pH=7.4), 150 mM NaCl, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM ethylenediaminetetraaceticacid, 1 mM ethylene glycol tetraacetic acid, 1 mM dithiothreitol, 0.1 mM phenyl methyl sulfonyl fluoride, 1 mM sodium orthovanadate, 1% glycerol, 1% Triton X-100, and protease inhibitor cocktail] (Roche) using steel beads on a Tissue Lyser (Qiagen). Cell lysates were centrifuged at 14,000 rpm for 10 min at 4°C. The protein concentration in the supernatant was quantified using a DCA Protein Assay (BioRad) with bovine serum albumin (BSA) as the standard. An equal amount of protein (30 μg) for each sample was loaded onto an 8%–10% sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred to a polyvinylidene fluoride membrane (Hybond-P; Amersham Biosciences Piscataway). The membranes were blocked in tris-buffered saline +0.1% Tween 20 (TBST) +5% skim milk. The membranes were then incubated overnight in a 1:1,000 dilution of primary antibodies in TBST at 4°C. The membranes were washed thrice for 5 min in TBST, incubated in a 1:10,000 dilution of horseradish peroxidise (HRP)-conjugated anti-rabbit or anti-mouse secondary antibodies in TBST for 1 h, washed again in TBST, and developed using the Amersham ECL western blotting detection reagents (GE Healthcare). All membranes were stripped with a stripping buffer (2% SDS, 100 mM 2-mercaptoethanol, and 62.5 mM Tris-HCl (pH 6.7) at room temperature for 30 min with gentle shaking and re-probed with an antibody against β-actin (Sigma) as a control for protein loading.
Construction of a tissue microarray and immunohistochemistry
Formalin-fixed and paraffin-embedded donor HCC blocks were used to make a single tissue microarray (TMA) block. In each donor block, morphologically representative areas were identified and marked on the respective hematoxylin and eosin (H&E) slides. Duplicate cores (0.6 mm) were extracted from the chosen region and placed into the recipient paraffin block using the Beecher Manual Tissue Microarrayer Model MTA-1 system (Beecher Instruments). The differentiation stage, lymphovascular invasion (LVI), and tumor stage were determined for each HCC tumor from the H&E slides.
Immunohistochemistry
The TMA blocks constructed earlier were cut into sections of 4 μm thickness, dewaxed in xylene, and rehydrated in graded alcohols. For antigen retrieval, slides were placed in a 1% (w/v) zinc sulfate antigen retrieval solution and boiled for 30 min in a microwave. Specimens were then allowed to cool for 15 min followed by a 5-min wash in water. Before immunohistochemical staining, the slides were incubated with 3% H2O2 in phosphate-buffered saline (PBS) for 10 min to quench endogenous peroxidase activity, washed twice (5 min each) in TBS+0.025% Triton X-100, and blocked in TBS+1% BSA+10% FCS for 2 h. For immunohistochemical staining, consecutive slides were incubated overnight at 4°C with the respective primary antibodies (1:500). The slides were then washed twice in TBS+0.025% Triton X-100 and incubated for 1 h in corresponding HRP-conjugated secondary antibody (1:10,000) diluted in TBS+1% BSA. Slides were washed in TBST, developed with 2,3-diaminobenzidine tetrahydrochloride for 10 min, and counterstained with Hematoxylin QS (Vector Laboratories, Inc.) for 30 s. The slides were then washed, dehydrated in graded alcohol, mounted using Safety Mount No. 4 (Fronine Laboratories), and visualized on a Leica DMBL (Leica Microsystems) attached to a Spot RT KE (Spot Diagnostic Instruments). Images were analyzed using Spot Basic V4.1 (Spot Diagnostic Instruments). Immunostaining results were quantified using a TMA staining evaluation protocol. [23].
Magnetic bead separation
A total of 5×106 cells were harvested from each of the HCC cell lines. These cells were then labeled, respectively, with EpCAM, CD44, CD90, or CD133 antibodies conjugated to magnetic beads (Miltenyi Biotec) according to the manufacturer's instructions. Cells were then co-labeled with EpCAM, CD44, CD90, or CD133 conjugated to PE (Miltenyi Biotec) according to the manufacturer's instructions. Antibody positive and negative cells were then separated using MACS LS separation columns (Miltenyi Biotec) and identified as antibody-enriched or -depleted cell populations, respectively.
Flow cytometry
Cells were centrifuged at 1,500 g for 5 min. The supernatant was discarded, and the cell pellet was resuspended in PBS, then run on an FACS CantoII Flow Cytometer (Becton Dickenson). The data were analyzed using BD Diva Software (Becton Dickenson) to calculate the percentage of positive cells.
Tumor sphere assay
Cells were resuspended in tumor sphere assay media (1% w/v sodium pyruvate, 1% w/v minimum essential media non-essential amino acids, 1% v/v insulin transferrin selenium, 1 μM dexamethasone, 200 μM l-ascorbic acid 2-phosphate, 10 mM nicotinamide, 20 ng/mL epidermal growth factor, and 10 ng/mL fibroblast growth factor-2) at a concentration of 2×104 cells/mL. One milliliter of the cell suspension was seeded on a Costar® Ultra Low Attachment 6-well plate (Corning). The number of tumor spheres was counted under a phase-contrast microscope.
Quantitative real-time polymerase chain reaction analysis
RNA from EpCAM enriched HuH7 and PLC/PRF/5 cells were extracted using FavorPrep™ Tissue Total RNA Mini Kit (Favorgen) according to the manufacturer's instructions. cDNA was synthesized from the extracted RNA using M-MLV Reverse Transcriptase (Promega) according to the manufacturer's instructions. Gene expression was quantified by adding 5 ng of cDNA to 10 μL Fast SYBR® Green and 0.5 μM forward and reverse primers of the stemness genes (Oct3/4, TAT, CD90, CYP3A4, HNF3B, AFP, CD13, HNF1A, CD133, ALDH1A1, CK19, and CD45). Reaction volumes were adjusted to 20 μL with sterile water. Stemness gene expression was normalized to the expression of the housekeeping gene β-actin.
Cell proliferation
Cells were seeded at a density of 5×103 in a 96-well plate and cultured overnight. Cells were then treated with various concentrations of Sorafenib ranging from 0 to 50 μM and cultured for a further 24 h. After treatment, cell proliferation was determined by a Cell Proliferation ELISA, BrdU (Roche Applied Science) according to the manufacturer's instructions.
Statistical analysis
Differences between samples were determined using a two-sample t-test on Microsoft Excel 2010. Differences were deemed significant if a P-value was less than 0.05.
Results
Expression of LCSC markers in HCC
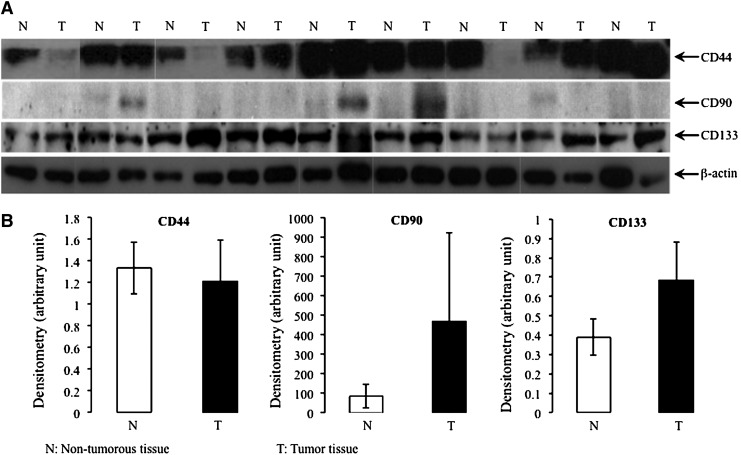
To investigate the specificity of currently used stem cell markers in detecting LCSCs, the expression of the three most commonly used CSC markers (CD44, CD90, and CD133) was tested by western blots in nine cases of human HCC. As shown in Fig. 1, there was no significant difference in the expression of the stem cell markers between tumor tissues and adjacent non-cancerous liver (P>0.05, two-way analysis of variance). The positivity of each of the three CSC markers varied to a great extent. For example, relative to the surrounding non-cancerous liver and normalized against β-actin in each compartment, it was evident that increased CD44 expression was found in 3 out of 9 tumors. In another three tumors, the expression of CD44 was decreased, and in the remaining cases, no change was found between adjacent non-cancerous liver and tumor. CD90 was barely detected in all non-cancerous liver tissues. Increased expression of CD90 was found in 3 out of 9 cases of HCC. For CD133, four HCCs (4/9) showed increased expression, two (2/9) showed decreased expression, and the remaining three showed no significant changes.
FIG. 1.
Western blot analysis of cancer stem cell (CSC) markers (CD44, CD90, and CD133) in human hepatocellular carcinoma (HCC) (A). The house keeping gene β-actin was used as the internal control. (B) Protein expression of the CSC markers was quantified by densitometry and normalized against the expression of β-actin.
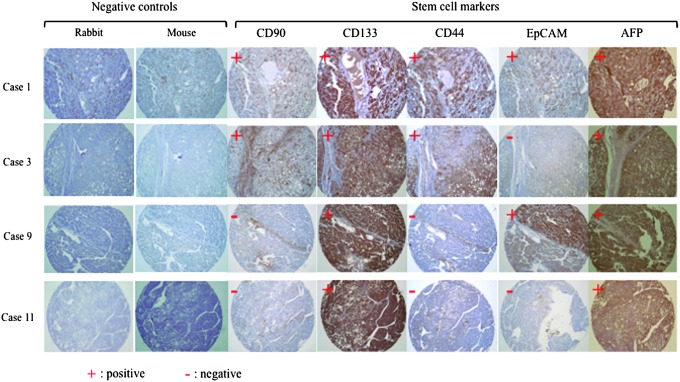
Immunohistochemical staining for CSC markers in human tumors is a widely used methodology for assessing CSCs in situ. It can also provide information on the origin, recurrence, and metastasis of the cancers, as well as on the possible correlation between the extent of CSC expression and patient survival [15]. To determine the accuracy of CSC markers to identify LCSC, we next examined the expression patterns of the well-characterized CSC markers CD44, CD90, CD133 and two other promising markers (EpCAM, and AFP) in 14 HCC tumor tissues by immunohistochemistry. As shown in Table 1, of the 14 HCCs tested, only two samples (HCC 1 and 2) were positive for all five stem cell markers, five samples (HCC 3–7) were positive for four markers, three samples (HCC 8–10) were positive for three markers, another three samples (HCC 11–13) were positive for two, and one sample (HCC14) was only positive for one marker. Typical immunostaining results (for HCC 1, 3, 9, and 11 only) are shown in Fig. 2. We then analyzed the co-expression patterns for these CSC markers in the 14 HCCs. As shown in Table 2, none of the CSC markers showed co-expression in all 14 HCCs tested. Co-expression of CD133 with AFP was found in 12 out of 14 HCC tissues. Co-expression of CD44 with CD133, CD44 with AFP, and EpCAM with CD133 was found in eight HCC cases, respectively. In 7 out of 14 HCC tissues, AFP co-expressed with EpCAM, and in 6 out of 14 cases, EpCAM co-expressed with CD44. Only 4 out of 14 HCC cases showed co-expression of CD133 with CD90 and AFP with CD90. Three out of 14 cases showed co-expression of CD44 with CD90, and 2 out of 14 cases showed co-expression of EpCAM with CD44.
Table 1.
Expression of Cancer Stem Cell Markers in Hepatocellular Carcinoma Tissues As Determined by Immunohistochemistry
| HCC case | CD44 | CD90 | CD133 | EpCAM | AFP | No. of positive markers/total case |
|---|---|---|---|---|---|---|
| 1 | + | + | + | + | + | 5/5 |
| 2 | + | + | + | + | + | 5/5 |
| 3 | + | + | + | − | + | 4/5 |
| 4 | + | − | + | + | + | 4/5 |
| 5 | + | − | + | + | + | 4/5 |
| 6 | + | − | + | + | + | 4/5 |
| 7 | + | − | + | + | + | 4/5 |
| 8 | − | + | + | − | + | 3/5 |
| 9 | − | − | + | + | + | 3/5 |
| 10 | + | − | + | − | + | 3/5 |
| 11 | − | − | + | − | + | 2/5 |
| 12 | − | − | + | − | + | 2/5 |
| 13 | − | − | + | + | − | 2/5 |
| 14 | − | − | + | − | − | 1/5 |
HCC, hepatocellular carcinoma; EpCAM, epithelial cell-adhesion molecule; AFP, alpha-fetoprotein.
FIG. 2.
Immunohistochemistry of human HCC. Serial sections from the paraffin-embedded tissue microarray blocks were used to stain for the CSC markers CD44, CD90, CD133, epithelial cell-adhesion molecule (EpCAM), and alpha-fetoprotein (AFP). Magnification:×100. Negative controls for rabbit monoclonal antibodies (CD90 and CD133) and mouse monoclonal antibodies (CD44, EpCAM, and AFP) are shown for each section.
Table 2.
Co-Expression of Cancer Stem Cell Markers in Hepatocellular Carcinoma Tissues As Determined by Immunohistochemistry
| Markers | CD44 | CD90 | CD133 | EpCAM | AFP |
|---|---|---|---|---|---|
| CD44 | — | — | — | — | — |
| CD90 | 3/14 | — | — | — | — |
| CD133 | 8/14 | 4/14 | — | — | — |
| EpCAM | 6/14 | 2/14 | 8/14 | — | — |
| AFP | 8/14 | 4/14 | 12/14 | 7/14 | — |
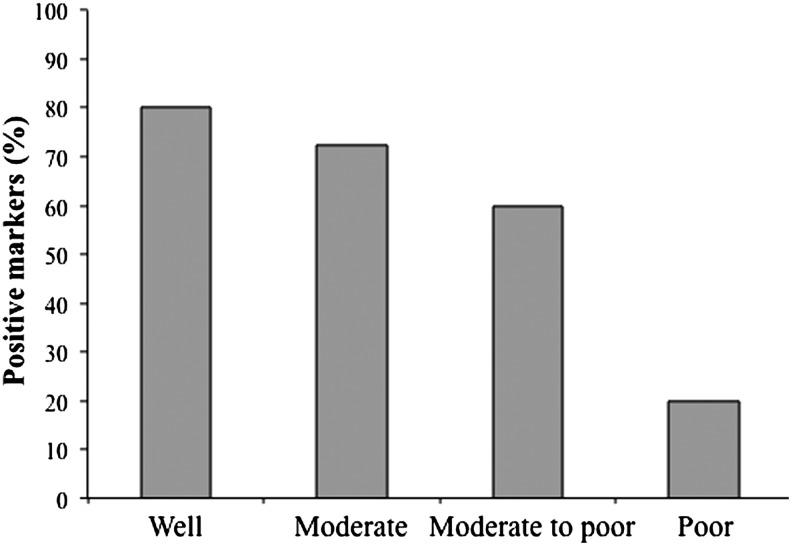
The relationship between HCC marker expression and disease state was also examined. Table 3 shows that the expression of LCSC markers did not relate to LVI (an indicator of metastasis) or tumor stage. However, the expression of LCSC markers appears to be positively related to the degree of differentiation with a greater percentage of LCSC markers being positive in tissues with greater differentiation (Fig. 3; Table 3).
Table 3.
Expression of Liver Cancer Stem Cell Markers and Their Correlation with Degree of Hepatocellular Carcinoma Tumor Differentiation, Lymphovascular Invasion, and Staging
| HCC case | Ratio of positive markers | Differentiation status | LVI | Tumor stage |
|---|---|---|---|---|
| 1 | 5/5 | Moderate | No | T1 |
| 2 | 5/5 | Moderate | No | T1 |
| 3 | 4/5 | Well | No | T2 |
| 4 | 4/5 | Moderate | No | T2 |
| 5 | 4/5 | Moderate | Yes | T2 |
| 6 | 4/5 | Moderate | No | T1 |
| 7 | 4/5 | Moderate to poor | Yes | T2 |
| 8 | 3/5 | Moderate to poor | Yes | T3b |
| 9 | 3/5 | Moderate to poor | Yes | T4 |
| 10 | 3/5 | Moderate | No | T1 |
| 11 | 2/5 | Moderate | Yes | T2 |
| 12 | 2/5 | Moderate to poor | Yes | T1 |
| 13 | 2/5 | Moderate | Yes | T2 |
| 14 | 1/5 | Poor | No | T1 |
Tumor staging: T1, solitary tumor without vascular invasion; T2, solitary tumor with vascular invasion or multiple tumors, none more than 5 cm; T3a, multiple tumors more than 5 cm; T3b, single tumor or multiple tumors of any size involving a major branch point of the portal vein or hepatic vein; T4, tumor (s) with direct invasion of adjacent organs other than the gallbladder or with perforation of visceral peritoneum.
LVI, lymphovascular invasion.
FIG. 3.
Percentage of liver cancer stem cell (LCSC) markers positive in HCCs with different differentiation status.
Isolation of LCSCs using stem cell markers
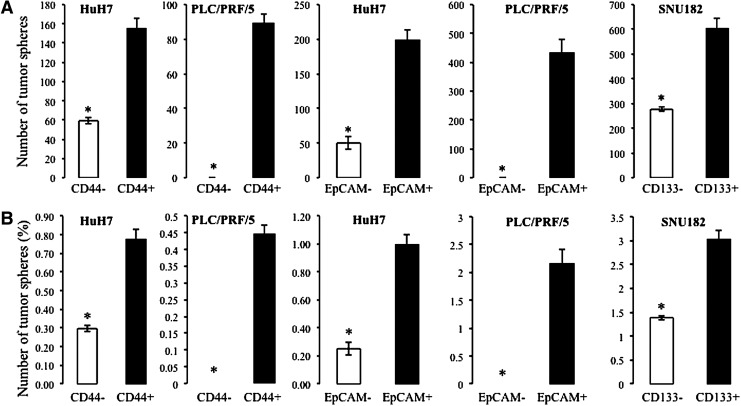
In order to confirm the specificity of the commonly used CSC markers in isolating LCSCs, four HCC cell lines (HuH7, SNU423, PLC/PRF/PRF/5, and SNU182) were segregated into enriched or depleted subpopulations based on the expression of the CSC markers CD44, CD90, CD133, and EpCAM, and the ability of these marker-enriched fractions to form spheres in appropriate culture conditions was tested. By flow cytometry analysis, the proportion of stem cells expressing the four CSC markers mentioned earlier varied considerably across the cell lines tested as shown in Table 4. The vast majority of SNU182 and SNU423 cells were positive for CD44 (93.4% and 97.8%, respectively), whereas only 3.7% of HuH7 cells and 0.1% of PLC/PRF/5 cells were positive for CD44. When the enriched CD44+ cells from HuH7 and PLC/PRF/5 cell lines were cultured in an appropriate culture condition, they formed significantly more tumor spheres than CD44− cells (two sample t-test, P<0.05) (Table 5; Fig. 4). The percentage of sphere formation in marker negative versus positive fractions is shown in Table 5.
Table 4.
Expression Percentage of Common Cancer Stem Cell Markers in Hepatocellular Carcinoma Cell Lines by Flow Cytometry
| Cell type | CD44 (%) | CD90 (%) | CD133 (%) | EpCAM (%) |
|---|---|---|---|---|
| HuH7 | 3.7 | 0 | 43.9 | 55.2 |
| SNU423 | 93.4 | 0 | 0 | 0 |
| PLC/PRF/5 | 0.1 | 0 | 36.9 | 0.3 |
| SNU182 | 97.8 | 0 | 13.2 | 61.5 |
Table 5.
Percentage of Cells that Formed Tumor Spheres in the Antibody-Enriched (+Fraction) Relative to Antibody-Depleted Fractions (−Fraction) from Hepatocellular Carcinoma Cells, As Determined by Tumor Sphere Assay
| |
CSC markers |
|||||||
|---|---|---|---|---|---|---|---|---|
| |
CD44 |
CD90 |
CD133 |
EpCAM |
||||
| Cell type | +Fraction | −Fraction | +Fraction | −Fraction | +Fraction | −Fraction | +Fraction | −Fraction |
| HuH7 | 0.78±0.06 | 0.30±0.06 | No expression | 0.69±0.07 | 0.71±0.80 | 1.00±0.07 | 0.25±0.05 | |
| SNU423 | High expression | No expression | No expression | No expression | ||||
| PLC/PRF/5 | 0.45±0.03 | 0.00±0.00 | No expression | 0.51±0.11 | 0.61±0.12 | 2.16±0.25 | 0.00±0.00 | |
| SNU182 | High expression | No expression | 3.02±0.19 | 1.39±0.05 | 3.20±0.35 | 3.08±0.26 | ||
No expression: No expression of CSC marker by flow cytometry.
High expression:>90% of cells were positive for CSC marker by flow cytometry.
Antibody-enriched fraction (+fraction) formed higher percentage of tumor spheres compared with antibody-depleted fraction (−fraction) (shown in bold, two sample t-test, P<0.05). The percentage of tumor spheres was determined as follows: [no. of spheres formed/total number of cells seeded]×100.
Data are expressed as mean±standard deviation (n=3).
CSC, cancer stem cell.
FIG. 4.
Number of tumor spheres (A) and overall percentage of tumor spheres (B) in antibody-enriched and antibody-depleted fractions of HCC cell lines (HuH7, PLC/PRF/5, and SNU182 cells). *Indicates significantly greater number of tumour spheres in antibody enriched compared to antibody depleted fractions. Significant differences were determined by a two sample t-test (p<0.05).
CD133 was positive in 43.9% of HuH7 cells, 36.9% of PLC/PRF/5 cells, and 13.2% of SNU182 cells; whereas no expression of CD133 was detected in SNU423 cells (Table 4). The CD133-enriched fraction of SNU182 cells showed significantly increased tumor sphere formation compared with their parental cells (two sample t-test, P<0.05) (Table 5; Fig. 4). However, the CD133-enriched fraction of HuH7 and PLC/PRF/5 cells did not exhibit increased tumor sphere formation (Table 5).
EpCAM was found to be positive in 55.2% of HuH7 cells and in 61.5% of SNU182 cells, whereas no expression was detected in SNU423 cells (Table 4). Approximately 0.3% of PLC/PRF/5 were positive for EpCAM. The EpCAM-enriched HuH7 and PLC/PRF/5 cells displayed increased tumor sphere formation compared with their respective parental cell populations (two sample t-test, P<0.05) (Table 5; Fig. 4). The EpCAM-enriched SNU182 cells did not show increased tumor sphere formation (Table 5).
No CD90 was detected in the four HCC cell lines tested (Table 4).
Expression of stemness markers in LCSCs
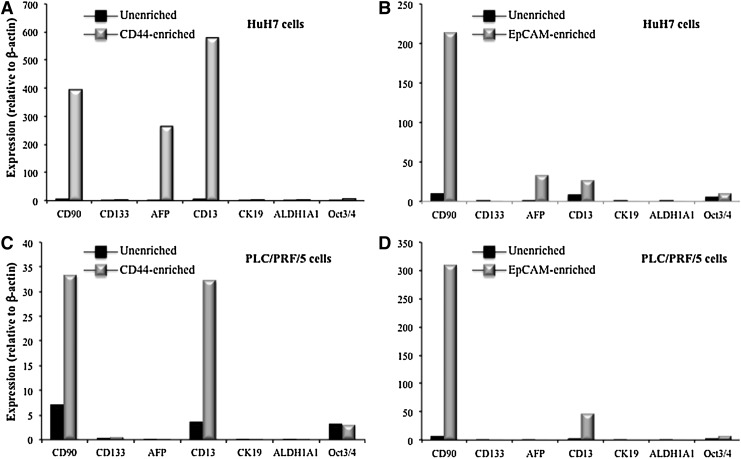
To determine the validity of using different LCSC markers to isolate unique LCSC subtypes, a quantitative real-time polymerase chain reaction array was undertaken to test the expression of a range of LCSC markers (CD90, CD133, AFP, CD13, CK19, and ALDH1A1) and the stemness marker (Oct3/4) in CD44 and EpCAM-enriched fractions of HuH7 cells. As shown in Fig. 5, in both CD44 and EpCAM-enriched fractions of HuH7 cells, three stem cell markers CD90, AFP, and CD13 were markedly increased compared with unenriched fractions (Fig. 5A, B). The stemnesss marker Oct3/4 had a greater increase in expression in EpCAM enriched populations compared with CD44-enriched counterparts. In contrast, in both CD44 and EpCAM-enriched fractions of PLC/PRF/5 cells, CD90 and CD13, and to a lesser extent, CD133 was markedly increased compared with non-enriched cell populations (Fig. 5C, D). AFP did not show a significant change between the marker-enriched and unenriched fractions. Similar to HuH7 cells, the stemnesss marker Oct3/4 showed increased expression in EpCAM-enriched but not in CD44-enriched PLC/PRF/5 cells populations.
FIG. 5.
Expression of CSC markers (CD90, CD133, AFP, CD13, CK19, and ALDH1A1) and the stemness marker (Oct/3/4) in CD44 (A, C) and PLC/PRF/5 (B, D)-enriched HuH7 cells (A, B) and PLC/PRF/5 cells (C, D). Expression was detected by quantitative real-time polymerase chain reaction, and normalized against the expression of β-actin.
Sorafenib resistance of LCSCs
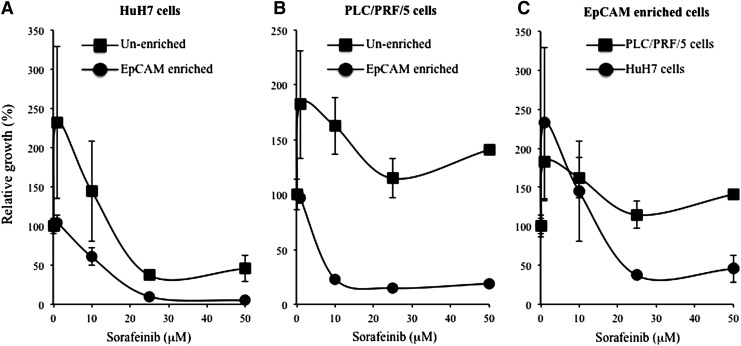
Resistance to chemotherapeutic agents is a clinically relevant feature of CSCs. LCSCs have been shown to be resistant to Sorafenib [24]. We, therefore, compared the effect of Sorafenib on the growth of EpCAM-enriched HCC cells. EpCAM-enriched HuH7 and PLC/PRF/5 cells, as well as their unenriched counterparts, were treated with various concentrations of Sorafenib for 24 h. As expected, at a concentration of 10 μM and above, EpCAM-enriched HuH7 and PLC/PRF/5 cells were more resistant to Sorafenib compared with the unenriched populations (Fig. 6A, B) (two-sample t-test, P<0.05). When cell growth was compared between EpCAM-enriched HuH7 cells and EpCAM-enriched PLC/PRFR/5 cells, EpCAM-enriched PLC/PRF/5 cells were more resistant to Sorafenib treatment than the EpCAM-enriched HuH7 cells (Fig. 6C) (two-sample t-test, P<0.05).
FIG. 6.
LCSCs (EpCAM+) cells are more resistant to Sorafenib treatment. EpCAM-enriched HuH7 (A) and PLC/PRF/5 (B) cells were treated with Sorafenib at concentrations of 0–50 μM for 24 h. Relative growth was measured by BrDU cell proliferation and expressed as a percentage of growth relative to Sorafenib 0 μM treatment. (C) EpCAM-enriched PLC/PRF/5 cells appear to be more resistant to Sorafenib than the EpCAM-enriched HuH7 cells. Data are expressed as mean±standard error (n=2).
Discussion
According to current theory, primary liver cancers originate from either differentiated liver cells (hepatocytes) or LCSCs [25]. Common causative agents for HCC such as HCV can induce liver cancer via activation of CSC pathways [25]. Further, currently available HCC therapies often fail because of their limited ability to kill CSCs. These CSCs generally remain quiescent but viable and retain the capability of regenerating new tumors [26]. Given the importance of CSCs in HCC, therefore, characterization of CSCs from the tumor bulk is an important pre-requisite for effective CSC targeting.
Many CSC markers have been used to isolate and characterize LCSCs. The most commonly used markers include CD133, CD44, CD90, CK19, Epcam, and ALDH [5,11]. In our study, we observed that these CSC markers were present in both tumors and adjacent non-cancerous liver, and that the positive percent and staining intensity vary considerably between cases. No consistent expression patterns were observed in the available cases. Furthermore, LCSCs isolated using different markers in the same cell line or the same markers in different cell lines had a unique expression profile. We believe that this finding reflects the strong heterogeneity of origin of liver cancer and possibly the varied etiology of HCC. Our findings that the studied CSC markers were expressed at various levels in different HCC cell lines further supports the heterogeneous nature of liver cancer cells.
Studies by others have also shown that CSC markers are not specific to LCSCs. In particular, CD133 was found to be expressed in both LCSCs and hepatocytes [22]. CD133 is the most abundant CSC marker for many solid tumors and hematopoetic malignancies. Its role as a marker in liver cancer is questionable, as the percentage of tumor cells expressing CD133 varied considerably across the commonly used HCC cell lines, ranging from <1% in SMMC-7721 cells, to 66% in HuH7 cells, and >90% in Hep3B cells [22]. In human liver, it was reported that CD133 is sporadically expressed in 1%–3% of HCC tissues and 0.025%–0.1% of non-tumorous liver [27]. CD133 has also been observed in ductular reactions in both acute and chronically damaged livers [28] as well as in normal biliary epithelium [29]. In our study, 43.9% of non-enriched HuH7 cells and 36.9% of non-enriched PLC/PRF/5 cells were positive for CD133. By immunoblotting and immunohistochemistry, all HCC tissues that we studied expressed CD133. However, when the well-established CSC markers CD44 and EpCAM were used to enrich the LCSC cell population, it was revealed that the expression of CD133 between the marker enriched and non-enriched population was minimal, and the expression level of CD133 was much less than that of the other markers such as CD90 and CD13. In addition, CD133 co-expressed with four other markers (CD44, CD90, EpCAM, and AFP) in only 2 out of 14 HCC cases. Clearly, caution should be exercised in using CD133 as an LCSC marker.
CD44, a cell adhesion molecule, is another well-characterized LCSC marker [30,31]. Aberrant expression of CD44 has been linked to the development of several human tumors such as prostate, colorectal, and breast cancer [31–33], and as a CSC marker for these tumors [34–36]. As previously reported, the tumorigenic properties of some CSCs isolated using other markers such as CD133 or CD90 were attributed to the expression of CD44 [20,22]. The expression of CD44 in HCC is also related to a higher frequency of extrahepatic metastasis and poor survival [37]. It has, thus, been suggested that the combination of CD44 with other markers such as CD90+/CD44+ and CD44+/CD133+ may be of more value in isolating LCSCs [21,22]. Our data shows high expression of CD44 in most HCC (8/14), and CD44 appears to co-express with many of the other CSC markers studied. The CD44+ fraction of HuH7 and PLC/PRF/5 showed increased numbers of tumor spheres compared with the CD44− fractions, and the CD44-enriched fractions of HuH7 and PLC/PRF/5 cells showed increased resistance to Sorafenib. However, the other two cell lines tested (SNU423 and SNU182) did not show increased tumor sphere formation in the CD44+-enriched fraction. These data demonstrate that CD44 may be a reliable LCSC marker for some but not all types of HCC cells, again reflecting the well-reported heterogeneity of liver cancer.
CD90 expression was shown in a previous study to be expressed in all HCCs and HCC cell lines, but with no expression in cirrhotic or adjacent noncancerous tissue [20,38]. In our study, CD90 was detectable only in a minority of HCC tissues, and no CD90 was detected in the four cell lines we tested. Thus, the efficacy of using CD90 as an LCSC marker is unclear.
Based on published data and our own studies, caution needs to be taken in isolating and characterizing LCSC using the currently available CSC markers. Combining more than one marker has been shown to increase the isolating efficiency [21,22]. However, in immunohistochemistry analysis, we could not reveal a ubiquitous co-expression pattern between any of the LCSC markers. CD90 and CD44 only co-expressed in a fraction of HCC tissues tested. The best co-expression pattern was between CD133 and AFP. However, AFP is a well-characterized marker that is used to detect all HCCs [39], and CD133 expression appears to be non-specific with no significant difference in expression between human HCC tumor tissues and adjacent noncancerous tissues. Furthermore, AFP appears to be markedly up-regulated in EpCAM-enriched HuH7 cells, but not in PLC/PRF/5 cells, suggesting a possible cell-specific phenotype.
Our findings suggest that LCSCs have unique subtypes. Using different markers to isolate LCSCs in the same HCC cell lines, or isolating LCSCs from different cell lines using the same markers may result in different phenotypes, each with a distinct expression profile of the LCSC markers and stemness markers. LCSCs isolated using the same markers from different cell lines may also have unique biological properties with different resistant levels to Sorafenib. Similar phenomena have been reported in breast cancer [40,41]. Therefore, a combination of at least two markers may provide a better efficiency in LCSC isolation and characterization, as was reported by others [22].
In summary, our current study has further confirmed the heterogeneity and complex nature of CSC biology. By using a range of commonly used LCSC markers on the same HCCs and cell lines, our study is the first to provide a comprehensive screen of LCSC markers in HCC. We have shown that the reported markers are not specific to LCSCs, and LCSC subtypes express different LCSC markers. A “universal” LCSC marker(s) is (are) unlikely present. We believe that in order to utilize LCSC markers to isolate LCSC, a wide range of markers should be examined for each HCC phenotype and linked with experimental evidence in immunodeficient mice (such as NOD-SCID mice) to confirm LCSC enrichment.
Acknowledgments
This work was supported by grants from the NSW Cancer Institute to LQ (ID: 08/FRL/1-04), by National Health and Medical Research Council grants to J.G. and L.Q. (ID: APP1006200, and APP1047417), and by the Robert W. Storr bequest to the Sydney Medical Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Yuen M-F. Hou J-L. Chutaputti A. Hepatocellular carcinoma in the Asia pacific region. J Gastroenterol Hepatol. 2009;24:346–353. doi: 10.1111/j.1440-1746.2009.05784.x. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A. Newell P. Hoshida Y. Inherited hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2010;24:725–734. doi: 10.1016/j.bpg.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Chiba T. Kita K. Zheng Y-W. Yokosuka O. Saisho H. Iwama A. Nakauchi H. Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 5.Ma S. Chan K-W. Hu L. Lee TK-W. Wo JY-H. Ng IO-L. Zheng BJ. Guan X-Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 7.Alison MR. Islam S. Lim S. Stem cells in liver regeneration, fibrosis and cancer: the good, the bad and the ugly. J Pathol. 2009;217:282–298. doi: 10.1002/path.2453. [DOI] [PubMed] [Google Scholar]
- 8.Zender L. Spector MS. Xue W. Flemming P. Cordon-Cardo C. Silke J. Fan S-T. Luk JM. Wigler M, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumble ML. Croager EJ. Yeoh GCT. Quail EA. Generation and characterization of p53 null transformed hepatic progenitor cells: oval cells give rise to hepatocellular carcinoma. Carcinogenesis. 2002;23:435–445. doi: 10.1093/carcin/23.3.435. [DOI] [PubMed] [Google Scholar]
- 10.Hermann PC. Bhaskar S. Cioffi M. Heeschen C. Cancer stem cells in solid tumors. Semin Cancer Biol. 2010;20:77–84. doi: 10.1016/j.semcancer.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Mikhail S. He AR. Liver cancer stem cells. Int J Hepatol. 2011:486954. doi: 10.4061/2011/486954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Y. Kitisin K. Jogunoori W. Li C. Deng C-X. Mueller SC. Ressom HW. Rashid A. He AR, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-B and IL-6 signaling. PNAS. 2008;105:2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tirino V. Desiderio V. Paino F. Pappacio G. De Rosa M. Methods for cancer stem cell detection and isolation. Methods Mol Biol. 2012;879:513–529. doi: 10.1007/978-1-61779-815-3_32. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda K. Saikawa Y. Ohashi M. Kumagai K. Kitajima M. Okano H. Matsuzaki Y. Kitagawa Y. Tumor initiating potential of side population cells in human gastric cancer. Int J Oncol. 2009;34:1201–1207. [PubMed] [Google Scholar]
- 15.Willan PM. Farnie G. Application of stem cell assays for the characterization of cancer stem cells. Stem Cell Biol Reg Med Part. 2011;4:259–282. [Google Scholar]
- 16.Meyerrose TE. Herrbrich P. Hess DA. Nolta JA. Immune-deficient mouse models for analysis of human stem cells. Biotechniques. 2003;35:1262–1272. doi: 10.2144/03356ss06. [DOI] [PubMed] [Google Scholar]
- 17.Hess DA. Mouse models for studying normal and cancer stem cells. Stem Cell Biol Reg Med Part 4. In: Allan AL, editor. Cancer Stem Cells in Solid Tumors. Humana Press; NY: 2011. pp. 311–325. [Google Scholar]
- 18.Ma I. Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 19.Ho DW. Yang ZF. Yi K. Lam CT. Ng MN. Yu WC. Lau J. Wan T. Wang X, et al. Gene expression profiling of liver cancer stem cells by RNA sequencing. PLoS One. 2012;7:e37159. doi: 10.1371/journal.pone.0037159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang ZF. Ngai P. Ho DW. Yu WC. Ng MNP. Lau CK. Li MIY. Tam KH. Lam CT. Poon RTP. Fan ST. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 21.Liu L-L. Fu D. Ma Y. Shen X-Z. The power and the promise of liver cancer stem cell markers. Stem Cells Dev. 2011;20:2023–2030. doi: 10.1089/scd.2011.0012. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Z. Hao X. Yan M. Yao M. Ge C. Gu J. Li J. Cancer stem/progenitor cells are highly enriched in CD133+ CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067–2078. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
- 23.Von Wasielewski R. Mengel M. Wiese B. Rudiger T. Muller-Hermelink HK. Keipe H. Tissue array technology for testing interlaboratory and interobserver reproducibility of immunohistochemical estrogen receptor analysis in a multicenter trial. Am J Clin Pathol. 2002;118:675–682. doi: 10.1309/URLK-6AVK-331U-0V5P. [DOI] [PubMed] [Google Scholar]
- 24.Xin H-W. Ambe CM. Hari DM. Wiegand GW. Miller TC. Chen J-Q. Anderson AJ. Ray S. Mullinax JE, et al. Label-retaining liver cancer cells are relatively resistant to sorafenib. Gut. 2013 doi: 10.1136/gatjnl-2012-303261. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rountree CB. Mishra L. Willenbring H. Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology. 2012;55:298–306. doi: 10.1002/hep.24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dean M. Fojo T. Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 27.Lee TK. Cheung VC. Ng IO. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.05.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Tsuchiya A. Kamimura H. Takamura M. Yamagiwa S. Matsuda Y. Sato Y. Nomoto M. Ichida T. Aoyagi Y. Clinicopathological analysis of CD133 and NCAM human hepatic stem/progenitor cells in damaged livers and hepatocellular carcinomas. Hepatol Res. 2009;39:1080–109O. doi: 10.1111/j.1872-034X.2009.00559.x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshikawa S. Zen Y. Fujii T. Sato Y. Ohta T. Aoyagi Y. Nakanuma Y. Characterization of CD133+ parenchymal cells in the liver: histology and culture. World J Gastroenterol. 2009;15:4896–4906. doi: 10.3748/wjg.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YH. Huang CJ. Chao JR. Chen ST. Lee SF. Yen JJ. Yang-Yen HF. Coupling of osteopontin and its cell surface recepto CD44 to the cell survival response elicited by interleukin-3 or granulocyte macrophage colony-stimulating factor. Mol Cell Biol. 2000;20:2734–2742. doi: 10.1128/mcb.20.8.2734-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison H. Sherman LS. Legg J. Banine F. Isacke C. Haipek CA. Gutmann DH. Ponta H. Herrlich P. The NF2 tumor supressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H. Yang XL. Rosada C. Hamilton SR. August JT. CD44 expression in colorectal adenomas is an early event occurring prior to K-ras and p53 gene mutation. Arch Biochem Biophys. 1994;310:504–507. doi: 10.1006/abbi.1994.1199. [DOI] [PubMed] [Google Scholar]
- 33.Nagabhushan M. Pretlow TG. Guo YJ. Amini SB. Pretlow TP. Sy MS. Altered expression of CD44 in human prostate cancer during progression. Am J Clin Pathol. 1996;106:647–651. doi: 10.1093/ajcp/106.5.647. [DOI] [PubMed] [Google Scholar]
- 34.Du L. Wang H. He L. Zhang J. Ni B. Wang X. Jin H. Cahuzac N. Mehrpour M. Lu Y. Chen Q. CD44 is of functional importance for colorectal cancer stem cells cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 35.Wright MH. Calcagno AM. Salcido CD. Carlson MD. Ambudkar SV. Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaishi S. Okumura T. Tu S. Wang SS. Shibata W. Vigneshwaran R. Gordon SA. Shimada Y. Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endo K. Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathological parameters, p53 expression, and patient survival. J Hepatol. 2000;32:78–84. doi: 10.1016/s0168-8278(00)80192-0. [DOI] [PubMed] [Google Scholar]
- 38.Yang ZF. Ho DW. Ng MN. Lau CK. Yu WC. Ngai P. Chu PWK. Lam CT. Poon RTP. Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Li D. Mallory T. Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta. 2001;313:15–19. doi: 10.1016/s0009-8981(01)00644-1. [DOI] [PubMed] [Google Scholar]
- 40.Dontu G. El-Ashry D. Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Park SY. Lee HE. Li H. Shipitsin M. Gelman R. Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16:876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]