Abstract
Background
As fluorescent microscopy has developed, significant insights have been gained into the establishment of immune response within secondary lymphoid organs, particularly in draining lymph nodes. While established techniques such as confocal imaging and intravital multi-photon microscopy have proven invaluable, they provide limited insight into the architectural and structural context in which these responses occur. To interrogate the role of the lymph node environment in immune response effectively, a new set of imaging tools taking into account broader architectural context must be implemented into emerging immunological questions.
Methods and Results
Using two different methods of whole-organ imaging, optical clearing and three-dimensional reconstruction of serially sectioned lymph nodes, fluorescent representations of whole lymph nodes can be acquired at cellular resolution. Using freely available post-processing tools, images of unlimited size and depth can be assembled into cohesive, contextual snapshots of immunological response. Through the implementation of robust iterative analysis techniques, these highly complex three-dimensional images can be objectified into sortable object data sets. These data can then be used to interrogate complex questions at the cellular level within the broader context of lymph node biology.
Conclusions
By combining existing imaging technology with complex methods of sample preparation and capture, we have developed efficient systems for contextualizing immunological phenomena within lymphatic architecture. In combination with robust approaches to image analysis, these advances provide a path to integrating scientific understanding of basic lymphatic biology into the complex nature of immunological response.
Imaging Lymph Node Dynamics
As imaging techniques have become increasingly sophisticated, significant progress has been made in the imaging of cellular populations and architectural components within secondary lymphoid organs. While the earliest studies relied largely on light microscopy in conjunction with traditional histological techniques, the development of fluorescent technology has revolutionized our ability to interrogate specific cellular populations, and in conjunction with flow cytometry, has proven invaluable in establishing basic concepts of lymphoid activation within the context of the lymph node (LN) macro-environment.1–5 The further development of confocal microscopy into a widely available, basic immunological technique has allowed the inclusion of spatial context and cellular interactions into studies that had previously been limited to in vitro and biochemical investigation.
A major leap in the ability to address spatial questions within biological tissue came with the introduction of multiphoton microscopy (MPM), and eventually, intravital MPM (IV-MPM).6–13 Where traditional confocal analysis can image depths nearing 40 μm, current MPM allows for cellular resolution at depths approaching 250 μm. Equally important, the decreased illumination time of individual z planes has significantly reduced fluorophore bleaching, allowing longer image acquisition sessions, and opening the door for live, in vivo fluorescent imaging analysis. As a result, the last decade has seen an explosion in the understanding of cellular migration and interactions within draining lymph nodes and has provided a view of secondary lymphoid organs as dynamic, responsive environments.14–19
While the application of IV-MPM has been critical in establishing the kinetics of immune activation within draining lymph nodes, limitations in the technology still exist, and contextualizing cellular information within the greater lymph node environment has remained difficult. The use of transgenic murine reporter systems, in vivo labeling with monoclonal antibodies, and the adoptive transfer of fluorescent cells has extended live imaging capacity into complex three-dimensional environments. However, even with optimal imaging conditions and a skilled operator, the existing limits of MPM allow, at best, the capture of approximately one-fifth of an average murine popliteal LN (PLN; a common model for studies of LN dynamics). Because the resolution required to track migrating cells requires high magnification, global information about lymph node dynamics and structure is often lost in the pursuit of high definition images of specific cellular processes. New imaging techniques allowing the imaging and analysis of population-level dynamics have been required to further the understanding of the lymph node response to immunological challenge.
Fluorescent Whole-Mount Imaging
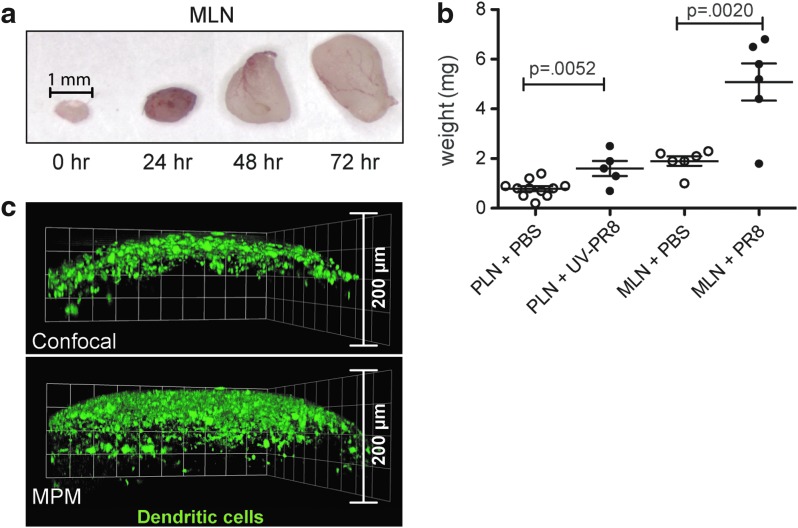
Although whole-mount imaging has been widely used,20–22 attempting to capture a complete set of data reflecting intact LNs is technically challenging and requires special consideration during sample preparation. A LN (such as the PLN or lung-draining mediastinal LN; MLN) from a naïve mouse has a diameter of approximately 1 mm, and this size can double or triple within hours of infection or vaccination (Fig. 1a). As a result, traditional whole-mount imaging by MPM, with an average penetration depth of 150 μm, is inadequate to provide large-scale contextual information (Fig. 1b). This fundamental challenge has three potential solutions: 1) generate new imaging technology; 2) alter sample preparation in ways that increase the imaging depth of the existing technology; or 3) separate the tissue into smaller components that can be imaged sequentially using the current technology. As the first option is usually outside the investigator's control, the second and third options are more feasible, although extensive development and optimization are still required.
FIG. 1.
Inflammation of murine LNs following administration of influenza. (a) Size increase in the lung-draining mediastinal lymph node (MLN) at 0, 24, 48, and 72 h following infection with influenza strain A/H1N1 Puerto Rico 8 (PR8). (b) Weight increases in popliteal LNs (PLNs) collected from mice injected SC in the footpad with PBS or UV-inactivated PR8 and in MLNs from mice administered PBS or PR8 intratracheally. Both PLNs and MLNs were collected at 72 h post-injection/infection. (c) Comparison of confocal and MP imaging of whole-mount LN tissue. PLNs from CD11c-eYFP reporter mice were isolated and imaged at depths from 0–200 μm to demonstrate comparative tissue penetration. A color version of this figure is available in the online article at www.liebertpub.com/lrb
One fundamental technical concern in biological imaging is the highly inconsistent refractive index within mammalian tissue. This inconsistency causes an extremely high degree of scattering as light passes through the tissue, limiting both the penetration depth of the laser and the resolution of light emitted from excited fluorophores. While the longer wavelengths used in MPM can increase the penetration depth substantially due to decreased excitation scattering, emission scattering is not addressed and still results in substantial imaging limitations. To bypass these issues, various methods of optically clearing whole-mount tissue have been developed.23 Fundamentally, the clearing process involves the fixation of the tissue, followed by the “replacement” of the cell cytoplasm with an optically neutral buffer, finally resulting in an intact, clarified organ.
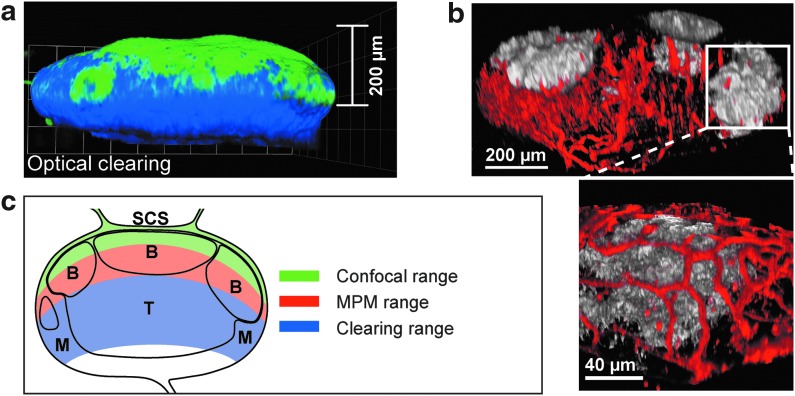
Using a recent technique described by Ertürk et al. (2012), LNs can be effectively cleared following in vivo labeling of the LN architecture. This method produces transparent tissue that can be effectively imaged at greatly increased depths due to reduced light scattering (Fig. 2a). By combining optical clearing with in vivo labeling and confocal microscopy, full LNs can be imaged, allowing striking resolution of vascular and lymphatic structures throughout the node (Fig. 2b). This approach has led to a better understanding of vascular and medullary complexity within the PLN, which is difficult to glean from existing literature, and has provided a clearer view of 3D LN structure. The schematic in Figure 2c illustrates the differences in tissue penetration depth using various imaging approaches.
FIG. 2.
Optical clearing of whole LNs. (a) Confocal micrograph of optically cleared PLN. mAb specific for the subcapsular sinus (anti-CD169, shown in green) and medulla (anti-F4/80; shown in blue) were injected SC 4h prior to LN harvest. (b) Confocal micrograph of optically cleared PLN, illustrating the complexity of the vascular and follicular architecture within the PLN. C57BL/6 mouse was injected IV with antibodies specific for blood vessels (anti-CD31; shown in red) 4h prior to LN harvest and was injected IV with antibodies specific for follicular dendritic cells (anti-8C12; shown in white) 24h prior to LN harvest. (c) Cartoon of approximate tissue penetration depths achievable through different imaging approaches. A color version of this figure is available in the online article at www.liebertpub.com/lrb
While optical clearing is advantageous in that it allows for the acquisition of whole LNs in a single z-stack, the clearing process can be time-consuming and may not be suitable for protein fluorophores (i.e., PE, APC) subject to rapid bleaching. Additionally, any fluorophore residing in the cytoplasm of the cell (i.e., not bound to a membrane) can be washed away during the process of cell permeabilization. These considerations make optical clearing extremely useful in the analysis of stable architectural components such as the vasculature, but render this method less feasible for high-definition analysis of dynamic populations requiring transgenic models. To address this issue, an alternative approach has been developed to obtain population-level data within the LN: whole-organ image reconstruction.
Whole Organ Image Reconstruction
Because optically clearing tissue does not always allow the tissue fluorescence to be preserved, this technique can be incompatible with experimental designs requiring transgenic reporter systems. Instead, by sectioning biological tissue into smaller components more amenable to imaging, and then stitching individual images together to create a single reconstructed image, fluorescence data can be acquired more easily, albeit with a loss of continuity in the z plane.
In attempting to clarify cellular dynamics, several groups have turned to 3D reconstruction to resolve complex interactions, although this approach is generally limited as implementation can be tedious and technically challenging.24,25 By serially cryosectioning LNs into thin sections (∼10–20 μm) that are then imaged by confocal microscopy, captured images can be combined effectively into serial stacks to give expanded views of 3D structure. Although this approach has been effective in approaching several questions,18,26 compiling enough images to reconstruct whole organs is a daunting task.
Alternatively, by replacing confocal capture with MPM, larger LNs can be broken down into thick sections (∼50–100 μm), which greatly increases the efficiency of imaging and processing. By serially cryosectioning PLNs into 50 μm slices and imaging the tissue by MPM, complete or partial LN reconstructions can be obtained relatively quickly and efficiently (Fig. 3a,b). Because of this increased efficiency, population and architectural questions can be addressed within the broader context of whole LNs from any tissue that is supportive of confocal imaging.
FIG. 3.
Serial imaging and whole LN reconstruction of the PLN. (a) Serial images used for reconstruction of in vivo labeled PLN. CD11c-eYFP mice were pre-injected SC with antibodies specific for CD169 (green) and CD35 (red), and PLNs were harvested 1 h after PBS injection into the footpad. PLN was sectioned (50 μm cryosections) and imaged for reconstruction. (b) Reconstructed image of a PLN from (a).
Automatic Acquisition of Tiled Stacks in the XY Plane and Post-Acquisition Stitching
While 3D reconstruction of full LNs has shown promise, a clear limitation has arisen in the size of LNs following vaccination or live infection. As shown in Figure 1b, there is a significant increase in the size of the lung-draining mediastinal LN (MLN) over the course of several days following infection. Manual imaging of multiple fields of view can be used to overcome these size increases, but two potential problems are introduced: 1) large sections of the LN can be missed, due to insufficient overlap between fields of view, and 2) the time required for imaging can quickly become prohibitive if the operator images unnecessarily large degrees of overlap between the fields of view.
As motorized imaging stages have developed, automated “tiling” options (the capture of multiple, adjacent xy tiles which can subsequently be stitched into a single image) has become available in imaging packages such as Fluoview by Olympus. Using various methods of automated xy acquisition, sequential tiled stacks can be captured and then recombined in the xy plane, followed by the z reconstruction of multiple stitched planes to form a single recombined image (Fig. 4). When using this approach, it is important to consider that the center of each section must be accurately identified, and that sufficient imaging overlap (usually 10–15%) is needed to ensure reliable stitching by available open source processing modules,27 although reconstruction alignment tools have begun to address these issues in high end imaging software.
FIG. 4.
Automatic tiling and whole LN reconstruction approach.Thick tissue sections, either labeled in vivo with antibodies injected prior to LN harvest or labeled post-sectioning, are imaged as individual fields of view, or “tiles.” The tiles are then stitched together, resulting in an xy panorama for each tissue section. The sections are then compiled in the z direction, resulting in a fully reconstructed LN, ready for subsequent examination and analysis. A color version of this figure is available in the online article at www.liebertpub.com/lrb
Whole LN Image Analysis
As imaging has been increasingly used to answer complex biological questions, robust image analysis and quantification strategies have become critical to achieve the significance and reproducibility demanded by other, more established techniques. Although imaging is often been viewed as a subjective approach to answering biological questions, advances in image analysis and processing (a field in its own right) available in various software formats have allowed for increasingly objective measurements of cellular behavior within the LN.28 Nonetheless, due to the sheer size of the data involved in whole-organ analysis, processing can quickly become cumbersome and overwhelming using traditional analysis techniques.
Since the establishment of IV-MPM, commercial software packages (i.e., Imaris and Volocity) have attempted to stay ahead of the imaging field by introducing comprehensive software suites designed for complex analysis of 3D systems. These packages are resource intensive, often requiring dedicated processing machines, and are frequently cost-prohibitive. While these analysis options are usually robust, they can have difficulty processing large numbers of events in three dimensions, as required by whole-organ analysis, and offer little flexibility outside of built-in functions. Where flexibility is required, open source software such as ImageJ is often utilized, although there is little functionality in the third dimension and iterative analysis/data assembly can be challenging without programming expertise.
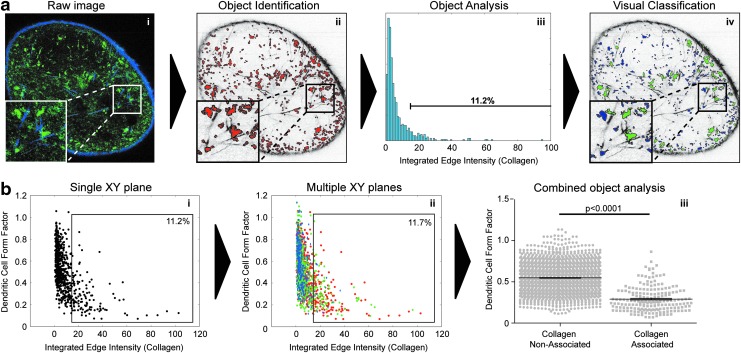
A third option, utilizing open source software designed for the development of large-scale imaging screens, has recently become available. Developed by the Broad Institute, CellProfiler29 is a particularly powerful 2D image analysis tool that offers almost unlimited flexibility in image analysis, highly functional iterative processing, and coherent data assembly. The use of CellProfiler and similar software has led to the emergence of histocytometry, the process of identifying individual objects within imaging data, which can then be sorted and analyzed much like flow cytometric data (Fig. 5a). Although this technique depends on high-quality imaging and 2D analysis, its ability to process large data sets quickly and provide extensive, flexible quantification makes it ideal for whole LN analysis on a cellular level. By isolating representative xy planes from reconstructions, and establishing parameters for object identification and analysis, multiple LNs can be analyzed efficiently, allowing for direct comparison between treatment groups at the cellular, image plane, and ultimately, whole LN levels (Fig. 5b).
FIG. 5.
Object identification and multiparameter analysis in the PLN. (a) Sample of a data analysis pipeline identifying DCs in contact with collagen within the PLN. (i) Raw image is loaded into the CellProfiler pipeline; (ii) Image is separated into its component channels, and DCs are identified using pre-defined algorithms; (iii) DCs are analyzed for multiple parameters, and are gated on co-localization of the DC perimeter with collagen signal; (iv) DCs are sorted based on the displayed gate, and are visually classified as collagen-associated DCs (green) or as nonassociated DCs (blue) superimposed on the original collagen channel image. (b) Sample of combinatorial analysis from multiple XY planes showing DC form factor (a measure of the spherical nature of each object) as a function of collagen association. (i) Scatter plot of the analysis for a single XY plane, showing DC form factor as a function of collagen association. (ii) Combined data from (3) XY planes, representing 150 μm of contiguous imaging data from (1) PLN gated on collagen associated DCs. (iii) DC form factor analysis from (ii) of collagen associated vs. nonassociated DCs. A color version of this figure is available in the online article at www.liebertpub.com/lrb
Application of Whole LN Imaging/Analysis
The rapid development of fluorescent imaging techniques has had a profound impact on the kinds of biological questions that can be asked. Investigating the interactions of different cell populations within the LN, the duration of time that such contacts last, and where these events occur, cannot be accomplished with traditional laboratory techniques such as flow cytometry. However, as we have expanded our knowledge of cellular events that occur in the LN, it has become increasingly clear that traditional, thin-section imaging is insufficient to address questions requiring a broader spatial context. Although biologists have obtained tantalizing glimpses of cell–cell interactions during early immune events within the LN,4,30–32 it has become equally important to determine in what architectural context these events occur.
The 3D representation of secondary lymphoid organs has initiated a new chapter in the arena of biological imaging. The ability to image whole LNs provides the considerable advantage of preserving the tissue integrity, thereby resulting in a clearer, more global picture of the LN structure and organization, and illustrating where specific cell types fit within this structural scaffold. By expanding the field of view to encompass half, it not more, of the total volume of the LN, regions of interest can be defined within an architectural context, and more informed questions can then be addressed through specific quantitative analysis (Fig. 6). As imaging and analysis software continues to become more flexible, exciting new questions can not only be addressed globally, but also objectively quantified.
FIG. 6.
Quantitative demonstration of lymphangiogenesis following influenza infection. Whole MLN reconstructions of mice administered PBS (left, 0 h) or influenza strain A/H1N1 Puerto Rico 8 (right, 72 hpi). The MLN tissue was thick-sectioned (50 μm), stained with antibodies specific for B220 (red), CD3 (green), and Lyve-1 (blue), imaged by confocal microscopy, and reconstructed by the approach shown in Figure 4. Lymphatic vessels (LVs) were then identified as objects within single xy planes and quantified across a total of (6) nonsequential xy planes for each reconstructed LN. XY planes were analyzed sequentially as outlined in Figure 5 to identify LVs within the T cell cortex. Quantitation of the numbers of identified LVs within each plane is shown with statistical analysis (Student's t test, n=6 XY planes). A color version of this figure is available in the online article at www.liebertpub.com/lrb
Some of the most intriguing questions that will be best addressed by these imaging techniques include: how does the structure of the LN change in the early aftermath of vaccination or infection? Where in the LN do the major players in the immune response localize, and for what purpose? What are the kinetics of cellular arrival to different compartments of the overall LN? And perhaps most importantly, how can our newfound knowledge of cell migration and localization be exploited to better inform vaccine design and disease treatment?
The use of whole-organ imaging is quickly proving invaluable to both confirm and challenge existing views of LN architecture and the role of LN-resident cell types. Future advances in both imaging and analysis techniques stand to illuminate the dynamic, exquisitely sensitive nature of secondary lymphoid organs and their positions as command centers for the initiation of immunity.
Author Disclosure Statement
This study is financially supported by NIH-NIAID (1 P01 AI078897 to M.C.C., 5 P01 AI07889705 to U.H.v.A.), NIH T32 Training Grant in Transplantation (T32 AI007498 to M.C.W.), and the American Lung Association (RT-224269-N to C.H.). Drs. Herndon and Carrol and M.C. Woodruff and B.A. Heesters disclosed on conflicts of interest or financial ties.
M.W., B.H., and C.H. performed all experiments and analysis described in text. M.W., C.H., and M.C. designed this study and developed the manuscript for publication.
References
- 1.Miller MJ. Safrina O. Cahalan MD. Imaging the single cell dynamics of DC4+ T cell activation by dendritic cells in lymph nodes. J Exp Med. 2004;200:847–856. doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mempel TR. Henrickson SE. Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 3.Bajenoff M. Egen JG. Koo LY. Laugier JP. Brau F. Glaichenhaus N. Germain RN. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrasco YR. Batista FD. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus in the lymph node. Immunity. 2007;27:160–171. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Allen CD. Okada T. Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reese AJM. The effect of hypoxia on liver secretion studied by intravital fluorescence microscopy. Br J Exp Pathol. 1960;41:527–535. [PMC free article] [PubMed] [Google Scholar]
- 7.Reese AJ. Rimington C. Biliary and urinary excretion of porphyrins in the rate studied by intravital fluorescence microscopy. Br J Exp Pathol. 1964;45:30–36. [PMC free article] [PubMed] [Google Scholar]
- 8.Atherton A. Born GVR. Quantitative investigation of the adhesiveness of circulating polymorphonuclear leucocytes to blood vessel walls. J Physiol. 1972;222:447–474. doi: 10.1113/jphysiol.1972.sp009808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denk W. Strickler JH. Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 10.Williams RM. Piston DW. Webb WW. Two-photon molecular excitation provides intrinsic 3-dimensional resolution for laser-based microscopy and microphotochemistry. FASEB J. 1994;8:804–813. doi: 10.1096/fasebj.8.11.8070629. [DOI] [PubMed] [Google Scholar]
- 11.Bargatze RF. Jutila MA. Butcher EC. Distinct roles of L-selectin and integrins alpha 4 beta 7 and LFA-1 in lymphocyte homing to Peyer's patch-HEV in situ: The multistep model confirmed and refined. Immunity. 1995;3:99–108. doi: 10.1016/1074-7613(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 12.Sμmen C. Mempel TR. Mazo IB. Von Andrian UH. Intravital microscopy; Visualizing immunity in context. Immunity. 2004;21:315–329. doi: 10.1016/j.immuni.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Kilarski WW. Guc E. Teo JC. Oliver SR. Lund AW. Swartz MA. Intravital immunofluorescence for visualizing the microcirculatory and immune microenvironments in the mouse ear dermis. PLos One. 2013:8. doi: 10.1371/journal.pone.0057135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Andrian UH. Intravital microscopy of the peripheral lymph node microcirculation in mice. Microcirculation. 1996;3:287–300. doi: 10.3109/10739689609148303. [DOI] [PubMed] [Google Scholar]
- 15.Stein JV. Cheng G. Stockton BM. Fors BP. Butcher EC. von Andrian UH. L-selectin-mediated leukocyte adhesion in vivo: Microvillous distribution determines tethering efficiency, but not rolling velocity. J Exp Med. 1999;189:37–50. doi: 10.1084/jem.189.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindquist RL. Shakhar G. Dudziak D, et al. Visualizing dendritic cell networks in vivo. Nat Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez SF. Lukacs-Kornek V. Kuligowski MP, et al. Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for hμmoral immunity in draining lymph nodes. Nat Immunol. 2010;11:427–434. doi: 10.1038/ni.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigorova IL. Panteleev M. Cyster JG. Lymph node cortical sinus organization and relationship to lymphocyte egress dynamics and antigen exposure. Proc Natl Acad Sci USA. 2010;107:20447–20452. doi: 10.1073/pnas.1009968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Textor J. Peixoto A. Henrickson SE. Sinn M. von Andrian UH. Westermann J. Defining the quantitative limits of intravital two-photon lymphocyte tracking. Proc Natl Acad Sci USA. 2011;108:12401–12406. doi: 10.1073/pnas.1102288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerber SA. Turner MJ. Lugade AA. Moran JP. Frelinger JG. Lord EM. Characterization of a lymph node within the mouse prostate: Detailed analysis using whole mount histology. Prostate. 2005;63:105–116. doi: 10.1002/pros.20169. [DOI] [PubMed] [Google Scholar]
- 21.He Y. Rajantie I. Pajusola K, et al. Vascular endothelial cell growth factor receptor 3-mediated activation of lymphatic endothelium is crucial for tumor cell entry and spread via lymphatic vessels. Cancer Res. 2005;65:4739–4746. doi: 10.1158/0008-5472.CAN-04-4576. [DOI] [PubMed] [Google Scholar]
- 22.Kotsμma M. Parashurama N. Smith BR. Wo J. Ito K. Gambhir SS. Nondestructive, serial in vivo imaging of a tissue-flap using a tissue adhesion barrier: Applications for IVM imaging in the mammary fat pad and lymph node. IntraVital. 2012;1:69–76. [Google Scholar]
- 23.Erturk A. Becker K. Jahrling N, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 24.Paish EC. Green AR. Rakha EA. Macmillan RD. Maddison JR. Ellis IO. Three-dimensional reconstruction of sentinel lymph nodes with metastatic breast cancer indicates three distinct patterns of tumour growth. J Clin Pathol. 2009;62:617–623. doi: 10.1136/jcp.2009.065219. [DOI] [PubMed] [Google Scholar]
- 25.Bin M. Lin Z. Winkelbach S. Lindenmaier W. Dittmar KEJ. Automatic registration of serial sections of mouse lymph node by using Image-Reg. Micron. 2008;39:387–396. doi: 10.1016/j.micron.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Miller MJ. Wei SH. Parker I. Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 27.Emmenlauer M. Ronneberger O. Ponti A, et al. XuvTools: Free, fast and reliable stitching of large 3D datasets. J Microscopy. 2009;233:42–60. doi: 10.1111/j.1365-2818.2008.03094.x. [DOI] [PubMed] [Google Scholar]
- 28.Gerner MY. Kastenmuller W. Ifrim I. Kabat J. Germain RN. Histocytometry: A method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambrecht MR. Sabatini DM. Carpenter AE. CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques. 2007;42:72–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 30.Murooka TT. Deruaz M. Marangoni F, et al. HIV-infected T cells are migratory vehicles for viral dissemination. Nature. 2012;490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrickson SE. Mempel TR. Mazo IB, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol. 2008;9:282–291. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray EE. Friend S. Suzuki K. Phan TG. Cyster JG. Subcapsular sinus macrophage fragmentation and CD169+ bleb acquisition by closely associated IL-17-committed innate-like lymphocytes. PLos One. 2012;7:e38258. doi: 10.1371/journal.pone.0038258. [DOI] [PMC free article] [PubMed] [Google Scholar]