Abstract
Pluripotent stem cell differentiation recapitulates aspects of embryonic development, including the regulation of morphogenesis and cell specification via precise spatiotemporal signaling. The assembly and reorganization of cadherins within multicellular aggregates may similarly influence β-catenin signaling dynamics and the associated cardiomyogenic differentiation of pluripotent embryonic stem cells (ESCs). In this study, dynamic changes in β-catenin expression and transcriptional activity were analyzed in response to altered cell adhesion kinetics during embryoid body (EB) formation and differentiation. Modulation of intercellular adhesion kinetics by rotary orbital mixing conditions led to temporal modulation of T-cell factor/lymphoid enhancer-binding factor activity, as well as changes in the spatial localization and phosphorylation state of β-catenin expression. Slower rotary speeds, which promoted accelerated ESC aggregation, resulted in the early accumulation of nuclear dephosphorylated β-catenin, which was followed by a decrease in β-catenin transcriptional activity and an increase in the gene expression of Wnt inhibitors such as Dkk-1. In addition, EBs that exhibited increased β-catenin transcriptional activity at early stages of differentiation subsequently demonstrated increased expression of genes related to cardiomyogenic phenotypes, and inhibition of the Wnt pathway during the initial 4 days of differentiation significantly decreased cardiomyogenic gene expression. Together, the results of this study indicate that the expression and transcriptional activity of β-catenin are temporally regulated by multicellular aggregation kinetics of pluripotent ESCs and influence mesoderm and cardiomyocyte differentiation.
Introduction
Pluripotent embryonic stem cells (ESCs) are a promising cell source for therapies that are aimed at treating degenerative and chronic diseases in which native tissues are damaged beyond their endogenous capacity for repair. Elucidating the mechanisms regulating ESC fate decisions will significantly aid in the development of directed differentiation approaches, in order to produce large quantities of differentiated cells for regenerative therapies. Differentiation of ESCs is commonly initiated by the spontaneous aggregation of cells via E-cadherin, a Ca2+-dependent homophilic adhesion molecule [1]. The resulting multicellular aggregates, termed “embryoid bodies” (EBs), recapitulate morphogenic events that are similar to those of preimplantation stage embryos, including differentiation into cell phenotypes comprising the three germ lineages (ectoderm, endoderm, and mesoderm) [2,3]. Although EBs often lack the spatial organization to directly mimic the regulation of tightly controlled spatiotemporal signaling exhibited in vivo, ESCs maintain the capacity to respond to similar molecular cues, thus motivating the analysis of developmentally relevant signaling pathways in the context of stem cell differentiation [4].
Transcriptional activation induced by the β-catenin signaling pathway controls cell fate decisions during early embryonic development, aiding in the regulation of embryonic patterning and axis formation [5], primitive streak formation [6], and mesoderm differentiation [7]. Similarly, in ESCs, β-catenin signaling is required for the maintenance of pluripotency [8,9], and mesoderm differentiation [10], as well as self-organization and axis formation within EBs [11]. Two main pools of β-catenin are present within cells [12]: (i) β-catenin sequestered at the cell membrane within adherens junctions, specifically as a mediator between cadherins and the actin cytoskeleton [13–15], and (ii) stabilized cytoplasmic β-catenin, which mediates transcription on destabilization and translocation to the nucleus [16,17]. Canonical Wnt signaling is a well-studied pathway that is involved in the initiation of β-catenin-regulated transcription [18]. In the absence of Wnt, cytoplasmic β-catenin is phosphorylated by the glycogen synthase kinase 3β (GSK3β)/adenomatous poliposis coli (APC)/Axin complex and is targeted for ubiquitination [5,19–21]. On Wnt binding to members of the seven-transmembrane Frizzled receptor family, the Frizzled receptor heterodimerizes with low-density lipoprotein receptor-related protein 5 and/or 6 (LRP5/6) [22], which, in turn, causes the disruption of the GSK3β/APC/Axin complex [23], resulting in the accumulation of stabilized, cytoplasmic β-catenin [24,25]. The cytoplasmic stabilization of dephosphorylated β-catenin permits the translocation of β-catenin to the nucleus, where it acts as a transcriptional co-activator with T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors [17] to regulate the transcription of target genes involved in a wide range of cellular processes, including cardiac specification and morphogenesis during embryonic development [26,27] and ESC differentiation [28–30]. Thus, β-catenin is a central component in cell–cell interactions and gene transcription, both of which are key factors regulating embryonic and ESC morphogenesis.
Although the presence of Wnt ligands and the destabilization of the GSK3β/APC/Axin complex are required to permit the transcriptional activity of β-catenin, the temporal onset and duration of signaling may also be modulated by the presence of E-cadherin adhesions between cells, which sequester β-catenin at the membrane [31,32]. Studies have illustrated the potential for cross-talk between β-catenin associated with E-cadherin and its availability to participate in signaling and transcriptional activities [33]; therefore, the intercellular adhesion events mitigating initial EB formation may also regulate downstream β-catenin-regulated transcription, thus potentially modulating the cardiomyogenic potential of the ESC population.
The formation and maintenance of EBs in rotary orbital suspension culture produces increased yields of homogeneous EB populations [34–37] and is a relatively facile technique to modulate ESC aggregation kinetics by simply varying the orbital mixing speed [36]. Interestingly, previous studies demonstrated that the extent of cardiomyogenic differentiation in rotary cultures was modulated by the orbital speed, with the conditions promoting accelerated EB formation kinetics also exhibiting increased cardiomyocyte differentiation [36]. Therefore, the control of ESC aggregation afforded by the rotary orbital suspension culture platform enables a systematic study of aggregation-induced modulation of β-catenin signaling kinetics and the associated regulation of cardiomyocyte differentiation.
The objective of the present study was to investigate the dynamics of β-catenin transcriptional activity in response to modulation of ESC aggregation kinetics during EB formation. β-catenin protein expression and localization were assessed via immunostaining for total and dephosphorylated β-catenin isoforms and immunoblotting of cytoplasmic and nuclear protein fractions, in conjunction with analysis of transcriptional activity using stably transduced luciferase TCF/LEF reporter mouse ESCs. In addition, gene expression of downstream targets of β-catenin signaling and cardiomyocyte differentiation were analyzed to determine the relationship between β-catenin protein expression, localization, Wnt/β-catenin pathway signaling dynamics, and differentiated phenotypes. The findings of this work demonstrate that the dynamics of ESC aggregation during multicellular assembly via cadherins modulates the β-catenin signaling pathway and alters cardiomyogenic gene transcription in ESCs.
Materials and Methods
ESC culture
Murine ESCs (D3 cell line) were cultured on 0.1% gelatin-coated tissue culture-treated plates (Corning). Culture media consisted of Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal bovine serum (Hyclone), 100 U/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL amphotericin (Mediatech), 2 mM L-glutamine (Mediatech), 1× MEM nonessential amino acid solution (Mediatech), 0.1 mM 2-mercaptoethanol (Fisher), and 103 U/mL of leukemia inhibitory factor (LIF; ESGRO, Chemicon). Cultures were re-fed with fresh media every other day, and passaged at approximately 70% confluence.
EB formation and culture
EBs were formed, as previously described, by inoculating a single suspension of ESCs at 2×105 cells/mL into 100 mm bacteriological grade polystyrene Petri dishes with 10 mL ESC media without LIF [34]. Dishes were incubated at 37°C with 5% CO2 either statically or placed on rotary orbital shakers (Lab-Line Lab Rotator, Barnstead International) at 25, 40, or 55 rpm to impart hydrodynamic conditions [36]. 90% of media was exchanged every 2 days by collecting the EBs via gravity sedimentation and re-suspending the cultures in fresh media.
Whole-mount EB immunostaining
EBs were collected by sedimentation at days 1, 2, 4, and 7 of differentiation, rinsed 3× with phosphate-buffered saline (PBS), and formalin (4% formaldehyde) fixed at room temperature for 45 min with rotation. EBs were then rinsed 3× (5 min with rotation) in EB wash/block buffer (2% BSA/0.1% Tween-20 in PBS) and permeabilized in 0.05% Triton X-100 and 2% BSA solution for 1 h at 4°C with rotation. EBs were blocked in wash/block buffer for 2 h at 4°C with rotation. After permeabilization and blocking, EBs were incubated with polyclonal rabbit anti-β-catenin (Millipore; 1:200) and monoclonal mouse anti-active β-catenin (Millipore; 1:50) specific to dephosphorylated Ser-33 and Thr-41 [19] (anti-aβC, Millipore; 1:50) or with monoclonal rat anti-E-cadherin (Sigma, 1:200) at 4°C overnight with rotation, rinsed in wash buffer, and incubated with secondary antibodies (Alex Fluor 488 anti-rabbit and Alexa Fluor 546 anti-mouse, Invitrogen; 1:200) for 4 h at 4°C with rotation. EBs were rinsed, counterstained with Hoechst (1:100), and imaged with a multiphoton laser scanning confocal microscope (Zeiss LSM 510 NLO).
Quantification of immunostaining
The relative intensity of expression of E-cadherin, β-catenin, and aβC was quantified with CellProfiler image analysis software [38] using a custom-written script. Briefly, the outlines of individual cells were determined based on primary identification of nuclei followed by expansion of the bounded region to determine cell boundaries. The intensity of staining for E-cadherin, β-catenin, and aβC was then determined on a per cell basis, with approximately 50–100 cells per EB. The total expression levels for each experimental condition were calculated from the average of at least three individual representative EBs.
Luciferase transduction
Lentiviral transduction of mESCs with a luciferase TCF/LEF reporter construct (Cignal™ TCF/LEF luciferase reporter; SA Biosciences) was performed to generate stably transduced puromycin resistant clones that expressed luciferase in response to β-catenin signaling. ESCs were plated at 50,000 cells/well on 0.1% gelatin-coated tissue culture treated six-well dishes (Corning) and cultured for 24 h before the introduction of Cignal Lenti vectors. Stable transduction of D3 ESCs was accomplished by incubating cells with Cignal™ Lenti TCF/LEF vectors at Multiplicity of Infection (MOI) levels (3, 10, or 25) for 24 h in the presence of 1 μg/mL polybrene (Sigma). ESCs were incubated with lentivirus particles for 24 h, and after an additional 72 h, transduced cells were selected using 3 μg/mL of puromycin (Sigma); concentration was determined from a kill curve using 0.5–3.0 μg/mL. After transduction and selection, individual colonies were chosen using cloning rings and 0.05% trypsin-EDTA, and plated onto 0.1% gelatin-coated tissue culture-treated 100 mm dishes. The selected clones were maintained in ESC media supplemented with 3 μg/mL puromycin for an additional 2 weeks and assessed for luciferase expression via anti-luciferase immunostaining, luciferase activity (with and without LiCl treatment), and alkaline phosphatase activity.
ESC immunostaining
Transduced clones and naïve D3s were cultured on 0.1% gelatin-adsorbed tissue culture-treated polystyrene six-well dishes. At ∼70% confluence, cells were rinsed 3× with PBS, formalin fixed in the wells for 10 min, and rinsed again with PBS. At ∼70% confluence, ESCs were fixed with formalin (4% formaldehyde), rinsed with PBS, permeabilized, and blocked with 0.05% Triton X-100/2% BSA/PBS solution for 1 h at room temperature. ESCs were then incubated overnight at 4°C with polyclonal rabbit anti-β-catenin (Millipore; 1:200), monoclonal rat anti-E-cadherin (Sigma; 1:200) [19], monoclonal mouse anti-luciferase (Santa Cruz Biotech; 1:50), or polyclonal goat anti Oct-4 (Santa Cruz Biotech; 1:100). Cells were rinsed with PBS 3×, and then incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen; 1:200) for 2 h at room, counterstained with Hoechst (10 μg/mL), mounted, and cover slipped. To assess alkaline phosphatase activity, ESCs were stained using the Vector Red alkaline Phosphatase Substrate kit (Vector Laboratories). Briefly, cells were incubated in Vector Red substrate in 100 mM Tris-HCl buffer to allow color development for 30 min at room temperature while protected from light. After color development, cells were rinsed with Tris-HCl buffer and water, mounted, and cover slipped. Images were acquired with a Nikon TE 2000 inverted microscope (Nikon, Inc.) with a SpotFlex camera (Diagnostic Instruments) or an EVOS fl inverted microscope (Advanced Microscopy Group).
Luciferase activity quantification
ESCs were cultured with or without the addition of LiCl (25 mM) for 24 h to disrupt the GSK3β/APC/Axin complex and inhibit the phosphorylation of β-catenin, ultimately resulting in increased TCF/LEF activity within transduced cells [39]. Luciferase activity was quantified using the Luciferase Assay System (Promega), according to manufacturer's instructions. Briefly, ESCs or EBs were rinsed 3× in PBS, lysed with rotation at 4°C for 10 min, vortexed for 5 s, and centrifuged at 10,000 rcf for 5 min. The supernatant was collected and transferred to a prechilled microcentrifuge tube. Twenty microliters of the cell lysate was added to 100 μL of Luciferase Assay Reagent, and luminescence was detected using a Femtomaster FB12 luminometer (Zylux Corporation). Relative light units were normalized to μg of DNA per sample as determined by Quant-It PicoGreen assay (Invitrogen).
Protein fractionation
EBs were collected at days 2 and 4 of differentiation for western blot analysis. The NE-PER cell fractionation kit (Pierce) was used to separate the cytoplasmic and nuclear fractions of undifferentiated cells and differentiating EBs. Briefly, cells and EBs were lysed using CER I reagent supplemented with 500× protease inhibitor cocktail (Calbiochem) and 50× phosphatase inhibitor cocktail (Calbiochem), followed by addition of the CER II reagent. After centrifugation at 16,000 rcf, the cytoplasmic supernatant fraction was collected. The remaining nuclear pellet was incubated in the NER reagent containing 500× protease and 50× phosphatase inhibitors, and centrifuged at 16,000 rcf for 10 min followed by collection of the supernatant containing the nuclear fraction.
Western blotting
Sample protein concentration was determined using the BCA Protein Quantification kit (Pierce); equal amounts of protein per sample (10 μg for cytoplasmic fractions; 35 μg for nuclear fractions) were mixed with loading buffer (0.1 M Tris-HCl containing sodium dodecyl sulfate (SDS), glycerol, biomophenol blue, and β-mercaptoethanol); incubated at 95°C for 5 min; and loaded in 4%–15% Mini-PROTEAN TGX gels (Bio-Rad). Vertical electrophoresis was performed using the Mini-PROTEAN Treta Cell (Bio-Rad) system with SDS/polyacrylamide gel electrophoresis (PAGE) running buffer (Tris base/glycine/SDS solution) at 200V for 30 min. A protein ladder (Precision Plus Protein Kaleidoscope, 10–250 kDa; Bio-Rad) was also loaded into each gel as a molecular weight reference.
After SDS/PAGE separation, protein was transferred to a nitrocellulose membrane (Bio-rad) via semi-dry transfer (Trans-Blot SD; Bio-Rad) at 25V for 20 min. Membranes were then blocked in near infrared blocking medium (Rockland Immunochemicals) for 1 h and subsequently incubated with primary antibodies for β-catenin (polyclonal rabbit anti-β-catenin; Millipore; 1:400) and loading controls for cytoplasmic (polyclonal rabbit anti-GAPDH; Pierce; 1:400) and nuclear (monoclonal mouse anti-TATA binding protein, TBP; Abcam; 1:2000) fractions overnight at 4°C. Primary antibody incubation was followed by washes in 0.01% Tween-20/PBS solution, and membranes were incubated with IR secondary antibodies (680 anti-mouse and anti-rabbit, LiCor Biosciences; 1:2000), followed by washes in PBS/0.1% Tween-20 solution. Blots were imaged using the Odyssey Infrared imager (LiCor Biosciences).
Quantitative PCR
Expression of Wnt pathway agonists and antagonists, as well as cardiogenic genes were assessed via quantitative PCR from EBs formed with different aggregation kinetics, as well as for those supplemented with Wnt inhibitors (5 μM Inhibitor of Wnt Production-4, IWP-4; Stemgent) during the first 4 days of differentiation [28]. RNA was extracted from undifferentiated ESCs and from EBs using the RNeasy Mini kit (Qiagen Incorporated). Reverse transcription for complementary DNA synthesis was performed from 1 μg RNA using the iScript cDNA synthesis kit (Bio-Rad), and quantitative PCR was performed with SYBR green technology on the MyiQ cycler (Bio-Rad). Primer sequences and annealing temperatures for Wnt-1, Wnt-3a, Dickkopf-1 (Dkk-1), Brachyury T (B-T), mesoderm posterior 1 (Mesp-1), myocyte enhancer factor-2c (Mef-2c), Nkx2.5, α-myosin heavy chain (α-MHC), myosin light chain-2 ventricle (MLC-2v), and 18S ribosome, are listed in Supplementary Table S1; (Supplementary Data are available online at www.liebertpub.com/scd). Each primer set was designed with either Beacon Designer software (Invitrogen) (B-T, Mesp-1, Mef-2c, α-MHC, and MLC-2v) or the Integrated DNA technologies INC design Website (www.idtdna.com) (Wnt11, Wnt3a, Dkk-1, and Nkx2.5) and validated with appropriate cell controls. In order to account for variability in expression of housekeeping genes, absolute gene expression concentrations were calculated against standard curves and represented per μg of total RNA.
Statistical analysis
Experimental conditions were examined with triplicate samples for a minimum of two independent experiments, and the data values presented reflect the mean value±standard error. One- or two-way analysis of variance was performed to determine statistical significance (P<0.05) between experimental groups and time points, and where significant, it was followed by post-hoc Tukey analysis to define statistical differences (P<0.05) between specific experimental variables.
Results
Spatial localization of E-cadherin and association with β-catenin in ESCs
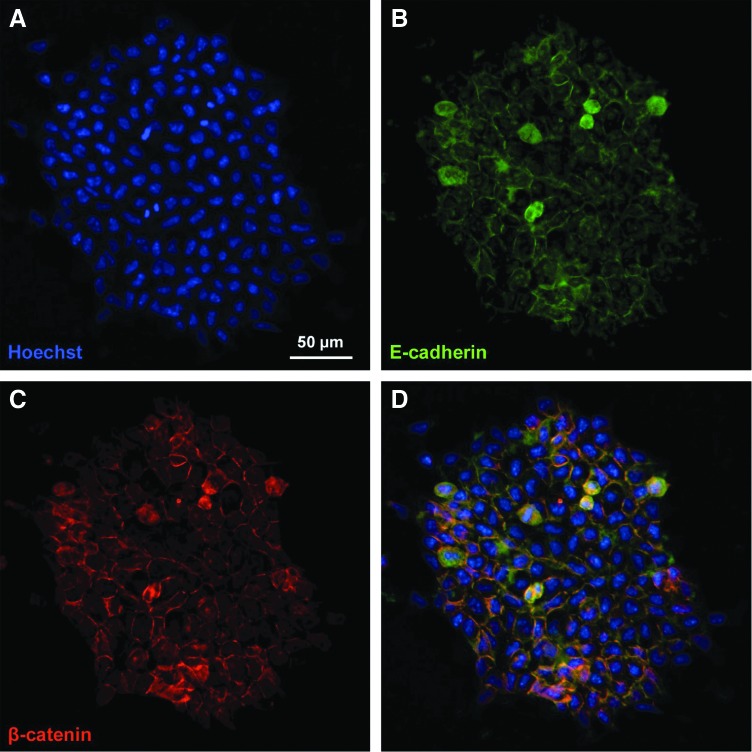
Previous studies have demonstrated that the Wnt signaling pathway is an important regulator of pluripotency, and that nuclear accumulation of β-catenin via treatment using a GSK-3 inhibitor aids in the maintenance of ESC self-renewal [8]. Consistent with published reports, undifferentiated mESC colonies expressed β-catenin, which was co-localized with E-cadherin at the plasma membrane, as well as expressed within the cytoplasm and nucleus (Fig. 1). The spatial distribution of both proteins was similar throughout the majority of cells in the colony, with the exception of cells exhibiting pyknotic nuclei, which demonstrated disrupted patterns of both β-catenin and E-cadherin. In addition, cells at the perimeter of colonies expressed decreased β-catenin at the membrane, which was concurrent with decreases in E-cadherin in regions lacking intercellular connections. Overall, the spatial localization of β-catenin and E-cadherin indicates the existence of both molecules during undifferentiated expansion before EB formation, as well as the localization of β-catenin both at the membrane (localized with E-cadherin) and in the cytoplasm and nucleus.
FIG. 1.
β-catenin and E-cadherin expression patterns within embryonic stem cells (ESCs). Localization of (A) nuclei, (B) E-cadherin, and (C) β-catenin in undifferentiated ESCs demonstrates expression primarily at the cell membrane between adjacent cells (D). Green=E-cadherin, red=β-catenin, blue=nuclei. Scale bar=50 μm. Color images available online at www.liebertpub.com/scd
Spatiotemporal expression of E-cadherin in response to EB formation kinetics
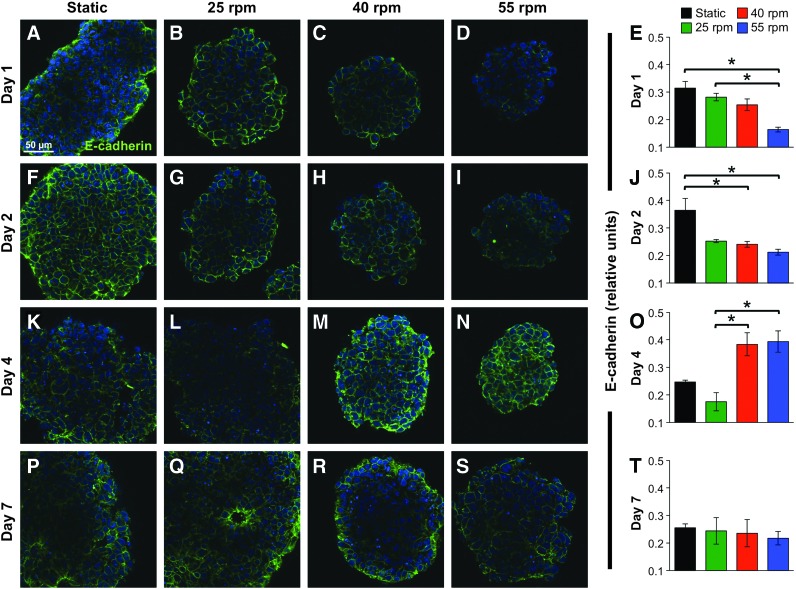
On aggregation and continued differentiation of EBs in rotary orbital suspension culture, E-cadherin was localized to the cell membrane at sites of intercellular connections, and was relatively homogeneously expressed throughout by most cells in the EB (Fig. 2). Altering the kinetics of cell–cell aggregation via changes in rotary orbital speed [36] suggested the modulation of E-cadherin expression during the course of differentiation. Specifically, the delayed aggregation kinetics of EBs formed at 55 rpm led to significantly decreased E-cadherin expression after 1 day (Fig. 2D), compared with 25 rpm rotary orbital conditions, which support more rapid EB formation (Fig. 2B). In addition, while the expression of E-cadherin appeared to decrease over 4 days of differentiation at 25 rpm (Fig. 2L), cultures maintained at 40 and 55 rpm (Fig. 2M, N) exhibited significantly increased expression of E-cadherin at the cell membrane. Taken together, β-catenin was co-localized with E-cadherin in undifferentiated ESCs, and on EB formation, E-cadherin localization was temporally modulated in response to altered cell association kinetics.
FIG. 2.
E-cadherin expression patterns during embryoid body (EB) differentiation. Static (A, F, K, P) and rotary EBs at 25 (B, G, L, Q), 40 (C, H, M, R), and 55 rpm (D, I, N, S) were stained for E-cadherin+ cells at days 1 (A–D), 2 (F–I), 4 (K–N), and 7 (P–S) of differentiation. Quantification of the staining patterns of E-cadherin (E, J, O, T), suggesting that E-cadherin expression within EBs was dynamically modulated during differentiation as a result of changes in ESC aggregation kinetics. Scale bar=50 μm. Green=E-cadherin, blue=nuclei. n=3, *P<0.05. Color images available online at www.liebertpub.com/scd
Spatiotemporal expression of β-catenin in response to EB formation kinetics
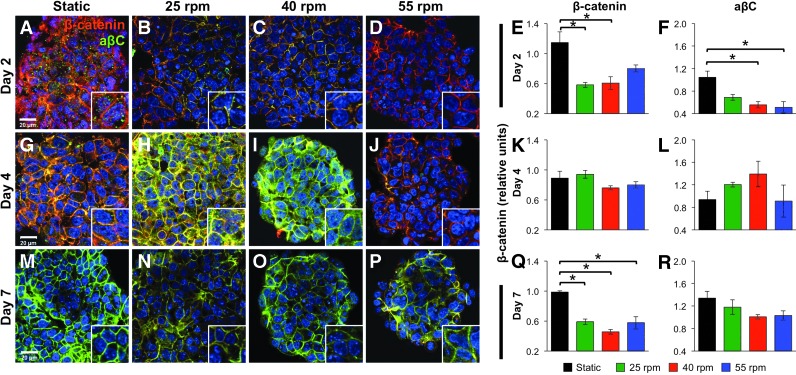
Based on the temporal modulation of E-cadherin and its established association with β-catenin, the spatial patterns of total (β-catenin+, red in Fig. 3) and dephosphorylated, transcriptionally active β-catenin (aβC+, green in Fig. 3) were assessed within EBs maintained in different culture conditions during the first 7 days of differentiation. Overall, static EBs exhibited significantly increased expression of β-catenin at early and late time points, compared with rotary conditions (Fig. 3A, E). Moreover, EBs maintained in static conditions exhibited nuclear expression of aβC at day 2 of differentiation (Fig. 3A, inset), accompanied by intercellular β-catenin expression at the cell membrane. Conversely, EBs from rotary orbital suspension cultures exhibited more homogeneous expression patterns of β-catenin and aβC over time, in which β-catenin was distinctly located either within the nucleus or at the cell membrane (Fig. 3B–D, H–J, N–P). After 2 days of differentiation, significantly decreased expression of β-catenin and aβC was observed within rotary EBs (Fig. 3B, C, inset), and aβC appeared to be sequestered at the cell membrane (Fig. 3B–D). However, at day 4 of differentiation, aβC expression appeared to increase within 25 and 40 rpm EBs, and altered localization of aβC was suggested by the appearance of aβC+nuclei, which was distinct from aβC previously observed at the cell membrane (Fig. 3H, I). In general, 55 rpm EBs exhibited little dephosphorylated β-catenin, and the majority of β-catenin expression was restricted to the cell membrane at days 2 and 4 of differentiation (Fig. 3D, J). After 7 days of differentiation, the expression of β-catenin was significantly decreased in all rotary conditions compared with static EBs, and little to no nuclear staining of β-catenin was observed in EBs from any of the culture conditions (Fig. 3M–P), suggesting that β-catenin transcriptional activity was diminished. Overall, patterns of β-catenin expression within differentiating EBs indicated that ESC aggregation modulates both the expression patterns and phosphorylation state of β-catenin.
FIG. 3.
Expression and phosphorylation state of β-catenin within EBs. Static (A, G, M) and rotary EBs at 25 (B, H, N), 40 (C, I, O), and 55 rpm (D, J, P) were stained for β-catenin+ and aβC+ cells at days 2 (A–D), 4 (G–J), and 7 (M–P) of differentiation. Quantification of the expression of β-catenin (E, K, Q) and aβC (F, L, R) indicated differences in β-catenin phosphorylation and location during differentiation. Scale bar=20 μm. Red=β-catenin, green=aβC, yellow=β-catenin/aβC overlay, blue=nuclei. n=3, *P<0.05. Color images available online at www.liebertpub.com/scd
Interestingly, a comparison of the dynamic expression patterns of E-cadherin (Fig. 2) and β-catenin (Fig. 3) over time revealed significant correlations, including a positive correlation (P<0.01) between E-cadherin and aβC after 2 days of differentiation and a negative correlation (P<0.03) between E-cadherin and total β-catenin expression after 4 days of differentiation (Table 1). Together, such results suggest an interplay between intercellular adhesions and β-catenin, with increased early expression of E-cadherin, followed by decreased E-cadherin levels, supporting increased expression of aβC and total β-catenin, respectively.
Table 1.
Correlation of E-Cadherin and B-Catenin Expression Dynamics During EB Differentiation
| Day of differentiation | Protein expression | Pearson's r | P-value | |
|---|---|---|---|---|
| 2 | E-cadherin | Total B-catenin | 0.80 | 0.20 |
| Active B-catenin | 0.99 | 0.01* | ||
| polyacrylamide gel electrophoresis | ||||
| 4 | E-cadherin | Total B-catenin | −0.97 | 0.03* |
| Active B-catenin | 0.03 | 0.97 | ||
| 7 | E-cadherin | Total B-catenin | 0.67 | 0.33 |
| Active B-catenin | 0.85 | 0.15 | ||
Values in bold denote correlations that are statistically significant. *P<0.05.
β-catenin cellular localization in response to EB formation kinetics
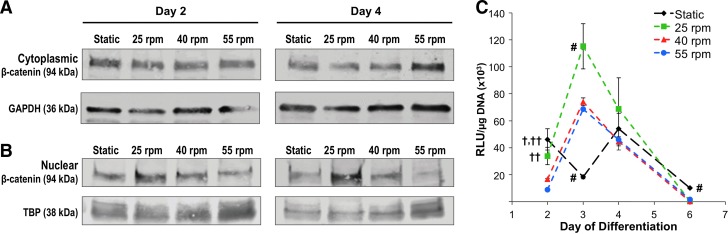
The intracellular localization of total β-catenin was further examined by western blot analysis of cytoplasmic (Fig. 4A) and nuclear (Fig. 4B) protein fractions from EBs cultured in static or rotary (25, 40, and 55 rpm) conditions at days 2 and 4 of differentiation. EBs from all conditions expressed total β-catenin in both the cytoplasm and nucleus at all time points examined (Fig. 4A, B). Overall, the expression of β-catenin in the cytoplasm remained relatively constant across all conditions at both time points examined (Fig. 4A). In contrast, nuclear fractions suggested differences in the β-catenin expression across different EB formation conditions. Specifically, EBs maintained at 25 rpm appeared to exhibit increased nuclear expression of total β-catenin at both days 2 and 4 of differentiation compared with other rotary conditions (Fig. 4B), which was consistent with the increased expression of aβC noted by immunofluorescence in 25 rpm EBs at day 4 of differentiation (Fig. 3F). Taken together, these data suggest the dynamic regulation of β-catenin protein expression, phosphorylation state, and localization during the initial stages of differentiation in response to intercellular adhesion kinetics.
FIG. 4.
β-catenin cellular localization and T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) activity in differentiating EBs. Although cytoplasmic (A) expression of β-catenin was similar among culture conditions, nuclear (B) β-catenin exhibited changes in expression among static and rotary (25, 40, 55 rpm) EBs after days 2 and 4 of differentiation. TCF/LEF-mediated luciferase activity was transiently increased by rotary orbital culture compared with static EBs, with a peak in expression after 3 days of differentiation (C); within rotary conditions, EBs maintained at 25 rpm yielded the largest increase in luciferase activity. P<0.05, #compared with all other conditions, †compared with 40 rpm, and ††compared with 55 rpm. Color images available online at www.liebertpub.com/scd
TCF/LEF activity in response to ESC aggregation kinetics
To quantitatively assess β-catenin transcriptional activity with high temporal resolution during the initial stages of EB differentiation, ESCs were transduced with a TCF/LEF-luciferase lentivirus. Transduced cells retained a typical undifferentiated morphological appearance at all MOI levels examined (Supplementary Fig. S1A–C); however, only ESCs transduced with 10 and 25 MOI survived after puromycin treatment (Supplementary Fig. S1D–F). Stably transduced clones were characterized based on morphological appearance, doubling time, pluripotency (alkaline phosphatase activity and Oct-4 protein expression), luciferase expression, and the capacity to form EBs (Supplementary Fig. S2), all of which supported the maintenance of the pluripotent ESC phenotype. The clones that exhibited the greatest dynamic range in luciferase activity in response to LiCl treatment were selected for additional analyses (Supplementary Fig. S2G). Importantly, transduced ESCs formed and maintained EBs that were similar in size and appearance to EBs formed from naïve ESCs [36] at various rotary speeds (25, 40 and 55 rpm) and in static conditions (Supplementary Fig. 2H), indicating the capacity to quantitatively assess β-catenin transcriptional activity in response to ESC aggregation kinetics.
EBs cultured under all conditions exhibited transient β-catenin transcriptional activity during the first 2–6 days of differentiation (Fig. 4C). After 2 days of EB differentiation, static and 25 rpm EBs exhibited the highest levels of luciferase activity, compared with 40 and 55 rpm (P<0.05). By day 3, EBs from all rotary conditions exhibited increased β-catenin transcriptional activity compared with static cultures (P<0.001, 25 rpm; P<0.01, 40 rpm; P<0.05, 55 rpm), with the β-catenin transcription in 25 rpm EBs statistically increased compared with the other two rotary conditions (P<0.05). Although EBs cultured in rotary orbital suspension increased transcription activity between days 2 and 3 of differentiation, statically cultured EBs exhibited decreased TCF/LEF activity over the same time course (Fig. 4C). Within 6 days of differentiation, all cultures exhibited decreased β-catenin transcription, below the initial levels at day 2, though static EBs maintained an increased level of TCF/LEF activity compared with any of the rotary orbital culture conditions (P<0.001) (Fig. 4C). Overall, the TCF/LEF transcriptional activity was most active within the first 3 days of EB differentiation and increasing the EB aggregation kinetics via rotary orbital conditions (25 rpm) enhanced the peak β-catenin transcriptional activity between days 2 and 4 of differentiation, consistent with immunofluorescence and western blot, which together reflected increased β-catenin protein expression and nuclear localization.
Temporal dynamics of β-catenin-target gene expression
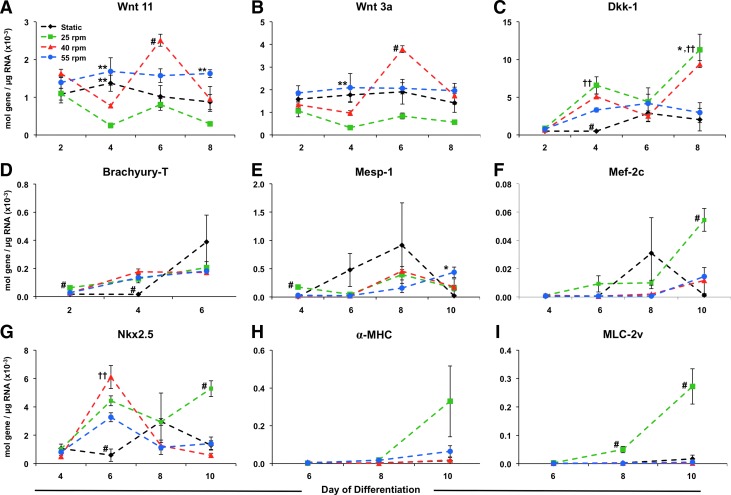
Since β-catenin expression, phosphorylation state, localization, and transcriptional activity were modulated by ESC aggregation kinetics, the downstream expression of β-catenin-target genes was examined. The expression patterns of canonical and noncanonical Wnt genes (Wnt3a and Wnt11, respectively) exhibited similar temporal expression (Fig. 5A, B). After 2 days of differentiation, EBs from all culture conditions expressed similar levels of both Wnt11 and Wnt3a; however, after 4 days of differentiation, 25 rpm EBs significantly decreased Wnt11 expression compared with static and 55 rpm (P<0.05, static; P<0.01, 55 rpm), accompanied by a decrease in Wnt3a expression compared with 55 rpm (P<0.05). At day 6 of differentiation, 40 rpm EBs exhibited increased Wnt11 and Wnt3a expression levels compared with all other conditions (P<0.05) (Fig. 5A, B). Interestingly, expression of Dkk-1, a canonical Wnt inhibitor, increased in 25 rpm EBs at days 4 and 8 compared with static and 55 rpm EBs (P<0.05) (Fig. 5A–C). Moreover, Dkk-1 expression increased in all rotary conditions compared with static culture at day 4 of differentiation (P<0.001, 25 rpm; P<0.01, 40 rpm; P<0.05, 55 rpm) (Fig. 5C).
FIG. 5.
Expression of β-catenin-target cardiomyogenic genes. qPCR analysis of RNA from static and rotary EBs (25, 40, 55 rpm) was performed at day 2, 4, 6, 8, and 10 of differentiation. Expression levels of noncanonical and canonical Wnts (A, B) were similar, but were modulated by culture condition, with 25 rpm rotary conditions exhibiting decreased levels of both Wnt11 and Wnt3a. In contrast, Wnt inhibitor Dkk-1 expression was increased within rotary EBs compared with static EBs (C), also corresponding to increased mesoderm-related gene expression within rotary conditions compared with static (D, E). In addition, early cardiomyogenic markers Mef-2c and Nkx2.5 were increased by rotary orbital culture (F, G), with 25 rpm conditions resulting in increased expression of sarcomeric muscle genes α-MHC and MLC-2v (H, I). n=3, P<0.05, #compared with all other conditions, *compared with static, **compared with 25 rpm, and ††compared with 55 rpm. Color images available online at www.liebertpub.com/scd
Expression of Brachyury-T (B-T), an early marker associated with primitive streak formation and mesoderm differentiation, was increased significantly after 2 days of differentiation within 25 rpm EBs compared with all other culture conditions (P<0.05), followed by increased expression at day 4 in all rotary conditions compared with static culture (P<0.01). Mesoderm posterior 1 (Mesp-I), a direct target of TCF/LEF regulated transcription [40], which is also implicated in early mesoderm differentiation within ESCs [10], exhibited increased expression in 25 rpm conditions at day 4 of differentiation compared with all other culture conditions (P<0.001).
Temporally controlled Wnt/β-catenin signaling has been implicated in the specification of mesoderm cells toward cardiac progenitors and subsequent cardiomyocyte maturation [27,41,42]. Nkx2.5, an early marker of cardiomyocyte progenitors, exhibited increased expression at early time points in all rotary orbital culture conditions compared with static cultures (Fig. 5G, H). After 6 days of differentiation, Nkx2.5 expression was greater in all rotary conditions compared with static culture (P<0.005, 25 rpm; P<0.001, 40 rpm; P<0.05, 55 rpm), and Nkx2.5 continued to exhibit increased expression in 25 rpm EBs compared with all other culture conditions after 10 days of differentiation (P<0.005). Likewise, 25 rpm EBs expressed increased levels of Myocyte enhancer 2c (Mef-2c) after 10 days of differentiation compared with all other conditions (P<0.005, static; P<0.01, 40 & 55 rpm).
Expression of sarcomeric muscle-related genes α-myosin heavy chain (α-MHC) and myosin light chain 2 ventricle (MLC-2v), phenotypic markers of more mature cardiomyocytes, were observed by day 10 of differentiation within EBs maintained at 25 rpm (Fig. 5H, I). MLC-2v expression was significantly increased within 25 rpm EBs compared with other culture conditions (P<0.05, static; P<0.01, 40 & 55 rpm) as early as day 8 of differentiation compared with other culture conditions, and continued to exhibit increased expression after 10 days of differentiation (P<0.005) (Fig. 5I). Together, the targets of the Wnt/β-catenin pathway (Mesp-1, Mef-2c) exhibited increased expression at early stages of differentiation (day 4), followed by increased expression of Wnt and β-catenin inhibitors (Dkk-1, Nkx2.5) and cardiomyogenic gene transcription in response to accelerated aggregation kinetics within 25 rpm rotary orbital cultures.
Regulation of cardiomyogenic gene transcription by Wnt/β-catenin signaling
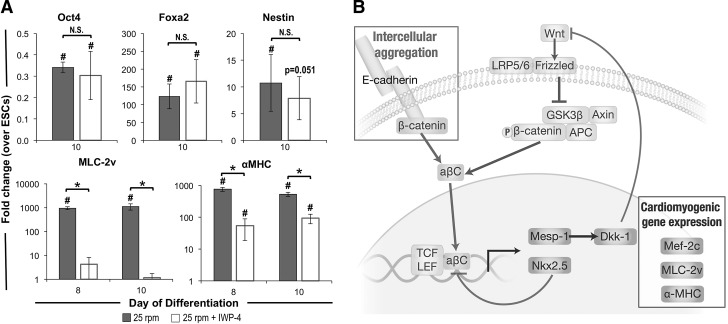
To establish a more mechanistic link between the observed regulation of β-catenin signaling and cardiomyogenic gene transcription, 25 rpm EBs were supplemented with inhibitors of canonical Wnt/β-catenin signaling (IWP-4; 5 μM) during the initial 4 days of differentiation (Fig. 6A). As expected, the pluripotency marker Oct-4 was significantly decreased compared with ESCs, and expression of genes related to endoderm (Foxa2) and ectoderm (Nestin) lineages was significantly increased compared with ESCs; however, Wnt inhibition did not significantly affect the expression of genes related to pluripotency, endoderm, or ectoderm differentiation. In contrast, inhibition of Wnt signaling significantly abrogated the previously observed increase in cardiomyogenic genes α-MHC and MLC-2v, thus indicating the role of canonical Wnt/β-catenin signaling in mediating the cardiomyogenic gene transcription downstream of intercellular aggregation at 25 rpm. The results of this study suggest that the dynamic interplay between E-cadherin-mediated cellular adhesion and Wnt signaling leads to nuclear accumulation and TCF/LEF transcriptional activation via dephosphorylated, β-catenin (aβC), thereby inducing downstream cardiomyogenic gene expression (Fig. 6B).
FIG. 6.
Influence of canonical Wnt/β-catenin signaling in aggregation-mediated cardiogenic gene transcription. Expression of genes related to pluripotency (Oct-4), endoderm (Foxa2), and ectoderm (Nestin) phenotypes was not altered by inhibition of Wnt signaling via treatment with IWP-4 during the first 4 days of differentiation at 25 rpm; however, cardiomyogenic markers α-MHC and MLC-2v were significantly decreased on Wnt inhibition (A). The proposed model depicting Wnt/β-catenin-mediated increase in cardiomyogenic gene transcription resulting from intercellular adhesion of ESCs (B). n=3, *P<0.05 *compared to matched treatment and #compared to undifferentiated ESCs.
Discussion
In this study, β-catenin expression, localization, and transcriptional activity, as well as cardiomyogenic gene transcription were assessed in response to changes in ESC aggregation kinetics. Overall, rotary orbital culture increased β-catenin-regulated transcription and cardiomyogenic gene expression compared with static culture conditions. The spatiotemporal location and phosphorylation state of β-catenin was modulated by rotary orbital speed, with slower rotary speeds (faster ESC aggregation kinetics), resulting in increased accumulation of nuclear, dephosphorylated β-catenin at early time points (Figs. 3 and 4). In addition, gene transcription downstream of β-catenin was increased initially by slower rotary speeds, evidenced by increased TCF/LEF activity and the expression of Mesp-1 and Mef-2c (Fig. 5). Moreover, inhibition of Wnt signaling significantly decreased cardiomyogenic gene transcription. Overall, this study illustrates that β-catenin signaling and downstream cardiomyogenic gene transcription are enhanced within culture conditions that promote increased multicellular aggregation kinetics.
Recent studies have illustrated that activation of the β-catenin pathway can play both inductive and repressive roles in cardiomyogenesis, depending on the temporal onset and duration of transcriptional activity [28,42,43]. Early activation of β-catenin within differentiating ESCs has been linked to increased mesoderm differentiation and proliferation of Mesp-1+, Isl-1+, and Mef-2c+ cardiomyocyte progenitor cells [10,30,42–45]. However, persistent β-catenin signaling (i.e., beyond day 4 of differentiation) inhibits subsequent cardiomyocyte maturation [42,43]; conversely, inhibition of β-catenin signaling through natural (e.g., Dkk-1) or small-molecule Wnt inhibitors at later stages of differentiation (day 4+) increases mature cardiomyocyte development within EBs [28,46–48]. A similar biphasic regulation of β-catenin activity and inhibitor expression was observed in this study in response to accelerated cellular adhesion kinetics. Nuclear total β-catenin expression was modulated by ESC aggregation kinetics (Fig. 4B), with increased expression of signaling active β-catenin within 25 rpm rotary conditions at early time points of differentiation (days 2 and 4, Fig. 3), significantly increased TCF/LEF activation at day 3 of differentiation (Fig. 4C), and increased Mesp-1 and Mef-2c expression (day 4) compared with other conditions (Fig. 5). Subsequently, expression of β-catenin signaling inhibitors Dkk-1 and Nkx2.5 [49] was also increased within the same rotary orbital condition, concomitant with decreased Wnt expression levels (days 4–6). The increase in Dkk-1 expression may be related to increased Mesp-1 expression within 25 rpm rotary conditions, as Mesp-1 is a transcription factor that is capable of directly increasing Dkk-1 expression [40]. Thus, the expression of Mesp-1, Dkk-1, and Nkx2.5 at later stages of differentiation supports the maturation of cardiomyocyte progenitors, as evidenced by increased expression of MLC-2v in 25 rpm rotary conditions (Fig. 5I). Consistent with our previous results [36,37], rotary orbital culture promoted cardiomyogenic gene transcription in a speed-dependent manner, and the results from this study suggest that β-catenin signaling may be implicated in the regulation of mesoderm differentiation and maturation of cardiomyocyte progenitors (Fig. 6) in response to changes in early ESC aggregation kinetics.
Until recently, the membrane-associated and nuclear signaling roles of β-catenin have been largely studied independently; however, increasing evidence suggests that the two processes may be intimately coordinated through competition for the total pool of available β-catenin in the cell [31,33]. Studies using recombinant forms of β-catenin indicate that the binding domains of β-catenin which mediate interactions with cadherins, TCF/LEF, and APC are mutually exclusive [50]. The loss of E-cadherin expression is correlated with invasive phenotypes during cancer progression and is exhibited concomitant with up-regulation of the Wnt/β-catenin signaling, suggesting the possible interplay between E-cadherin expression and β-catenin transcriptional activation [12,51–55]. In the undifferentiated state, ESCs largely express β-catenin at the plasma membrane, presumably bound to E-cadherin, with little nuclear localization and signaling (Fig. 1) [39]. Moreover, the culture of ESCs in a three-dimensional microwell format increased E-cadherin expression and subsequently increased Wnt signaling on EB formation, suggesting a link between intercellular adhesions, Wnt signaling, and cardiogenesis in ESCs [56]. Similarly, in this study, E-cadherin was dynamically regulated, including significant correlations between E-cadherin (Fig. 2) and β-catenin (Fig. 3) expression over time (Table 1), concurrent with increased TCF/LEF transcriptional activity (Fig. 4C). The regulation of intercellular adhesions in EBs is consistent with changes during embryonic development, whereby E-cadherin is down-regulated as cells traverse the primitive streak and β-catenin signaling is increased to promote axis formation and mesoderm differentiation [57–62]. It is likely that similar interrelated signaling between cadherins and β-catenin may enable the pool of cadherin-bound β-catenin to become available for nuclear translocation and signaling on remodeling of cadherins after EB formation. Although the direct relationship between cadherin and TCF/LEF-mediated regulation of the β-catenin pool remains unclear [33,63,64], evidence from this study suggests a correlation between E-cadherin and β-catenin signaling during EB formation and ESC differentiation.
Rotary orbital suspension culture has previously demonstrated the modulation of EB formation kinetics across a range of mixing conditions, with slower speeds resulting in increased EB formation kinetics [34,36]. The changes in β-catenin transcriptional activity as a function of rotary speed, in conjunction with changes in E-cadherin expression, indicate that cell association kinetics during EB formation can impact the subsequent pathway activation and ESC differentiation. β-catenin transcriptional activity was significantly increased at early stages of differentiation in EBs that exhibit faster cell association kinetics during EB formation at slower rotary speeds. Studies indicate that mixed culture conditions also modulate EB size, which may be partially responsible for changes in ESC differentiation [36]. Mesoderm induction in different sized EBs, however, is thought to result from paracrine actions of endoderm cells at the exterior of EBs [65], which do not arise until later stages of differentiation. In the context of this study, the initial cell specification resulting from changes in β-catenin signaling at early time points (days 2–4) is likely independent of signaling from divergent cell populations. The observed changes in cell aggregation kinetics due to rotary orbital suspension culture conditions may also be modulated across a range of suspension culture formats [66]; β-catenin transcriptional activity may be implicated in the modulation of cardiogenic differentiation as a function of bioreactor configuration, mixing speed, and cell density [2,67]. Hanging drop cultures exhibit an increased endogenous propensity to differentiate toward mesoderm lineages and develop functional cardiomyocytes compared with other formats [37,68]. In contrast to suspension cultures, which rely on the random collision of cells in larger-volume suspensions to form EBs, hanging-drop cultures force ESC aggregation in small volumes to more precisely control EB size, which likely results in faster association kinetics compared with suspension cultures. The increased cardiac differentiation yielded by hanging drops, therefore, is consistent with the observed increase in cardiomyogenic gene transcription in response to slower rotary speed and faster aggregation kinetics. Many recently developed technologies for spheroid formation may also alter the formation kinetics, by controlling aggregation within microwells or on micropatterned surfaces [69–71]; EBs formed via forced aggregation exhibited increased homogeneity and endogenous differentiation capacities across a similar range of rotary conditions examined in this study [35]. Thus, techniques to control intercellular adhesion dynamics may impact the differentiation capacity of stem and progenitor cells to either aid or hinder directed differentiation protocols, depending on the application. The results from this study, coupled with the developing understanding of the complex relationship between E-cadherin and β-catenin, highlight the important roles of β-catenin signaling in ESCs, which may be modulated by increasing cell association kinetics during EB formation to enhance cardiomyogenic differentiation.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH R01 EB010061) and by the National Science Foundation (NSF CBET 0939511). M.A.K. is currently supported by an American Heart Association (AHA) Pre-Doctoral Fellowship and earlier by an NSF Graduate Research Fellowship. C.Y.S. was also previously supported by an NSF Graduate Research Fellowship and an AHA Pre-Doctoral Fellowship.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Larue L. Antos C. Butz S. Huber O. Delmas V. Dominis M. Kemler R. A role for cadherins in tissue formation. Development. 1996;122:3185–3194. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- 2.Itskovitz-Eldor J. Schuldiner M. Karsenti D. Eden A. Yanuka O. Amit M. Soreq H. Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 3.Doetschman TC. Eistetter H. Katz M. Schmidt W. Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 4.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 5.Yost C. Torres M. Miller JR. Huang E. Kimelman D. Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed OA. Clarke HJ. Dufort D. Beta-catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Dev Dyn. 2004;231:416–424. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- 7.Haegel H. Larue L. Ohsugi M. Fedorov L. Herrenknecht K. Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 8.Sato N. Meijer L. Skaltsounis L. Greengard P. Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 9.Kelly KF. Ng DY. Jayakumaran G. Wood GA. Koide H. Doble BW. B-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell. 2011;8:214–227. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindsley RC. Gill JG. Kyba M. Murphy TL. Murphy KM. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 11.ten Berge D. Koole W. Fuerer C. Fish M. Eroglu E. Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottardi CJ. Gumbiner BM. Distinct molecular forms of beta-catenin are targeted to adhesive or transcriptional complexes. J Cell Biol. 2004;167:339–349. doi: 10.1083/jcb.200402153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 14.Aberle H. Schwartz H. Kemler R. Cadherin‐catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 15.Yamada S. Pokutta S. Drees F. Weis WI. Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat Cell Biol. 2000;2:653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- 17.Behrens J. von Kries JP. Kühl M. Bruhn L. Wedlich D. Grosschedl R. Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 18.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 19.van Noort M. Meeldijk J. van der Zee R. Destree O. Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem. 2002;277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- 20.Hart MJ. de los Santos R. Albert IN. Rubinfeld B. Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 21.Orford K. Crockett C. Jensen JP. Weissman AM. Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 22.Cong F. Schweizer L. Varmus H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, frizzled and LRP. Development. 2004;131:5103–5115. doi: 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- 23.Mao J. Wang J. Liu B. Pan W. Farr GH. Flynn C. Yuan H. Takada S. Kimelman D. Li L. Wu D. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 24.Munemitsu S. Albert I. Souza B. Rubinfeld B. Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci (USA) 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korinek V. Barker N. Morin PJ. van Wichen D. de Weger R. Kinzler KW. Vogelstein B. Clevers H. Constitutive transcriptional activation by a beta-catenin-tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 26.Hurlstone AFL. Haramis A-PG. Wienholds E. Begthel H. Korving J. Van Eeden F. Cuppen E. Zivkovic D. Plasterk RHA. Clevers H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 27.Lin L. Cui L. Zhou W. Dufort D. Zhang X. Cai C-L. Bu L. Yang L. Martin J, et al. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci (USA) 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian X. Hsiao C. Wilson G. Zhu K. Hazeltine LB. Azarin SM. Raval KK. Zhang J. Kamp TJ. Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci (USA) 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T. Sano M. Songyang Z. Schneider MD. A Wnt- and beta -catenin-dependent pathway for mammalian cardiac myogenesis. Proc Natl Acad Sci (USA) 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paige SL. Osugi T. Afanasiev OK. Pabon L. Reinecke H. Murry CE. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson WJ. Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maretzky T. Reiss K. Ludwig A. Buchholz J. Scholz F. Proksch E. de Strooper B. Hartmann D. Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci (USA) 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kam Y. Quaranta V. Cadherin-bound beta-catenin feeds into the Wnt pathway upon adherens junctions dissociation: evidence for an intersection between beta-catenin pools. PLoS ONE. 2009;4:e4580. doi: 10.1371/journal.pone.0004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenedo RL. Sargent CY. McDevitt TC. Rotary suspension culture enhances the efficiency, yield, and homogeneity of embryoid body differentiation. Stem Cells. 2007;25:2224–2234. doi: 10.1634/stemcells.2006-0523. [DOI] [PubMed] [Google Scholar]
- 35.Kinney MA. Saeed R. McDevitt TC. Systematic analysis of embryonic stem cell differentiation in hydrodynamic environments with controlled embryoid body size. Int Biol. 2012;4:641–650. doi: 10.1039/c2ib00165a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargent CY. Berguig GY. Kinney MA. Hiatt LA. Carpenedo RL. Berson RE. McDevitt TC. Hydrodynamic modulation of embryonic stem cell differentiation by rotary orbital suspension culture. Biotechnol Bioeng. 2010;105:611–626. doi: 10.1002/bit.22578. [DOI] [PubMed] [Google Scholar]
- 37.Sargent CY. Berguig GY. McDevitt TC. Cardiomyogenic differentiation of embryoid bodies is promoted by rotary orbital suspension culture. Tissue Eng Part A. 2009;15:331–342. doi: 10.1089/ten.tea.2008.0145. [DOI] [PubMed] [Google Scholar]
- 38.Carpenter AE. Jones TR. Lamprecht MR. Clarke C. Kang IH. Friman O. Guertin DA. Chang JH. Lindquist RA, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anton R. Kestler HA. Kühl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 40.David R. Brenner C. Stieber J. Schwarz F. Brunner S. Vollmer M. Mentele E. Müller-Höcker J. Kitajima S, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 41.Kwon C. Arnold J. Hsiao EC. Taketo MM. Conklin BR. Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci (USA) 2007;104:10894–10899. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naito AT. Shiojima I. Akazawa H. Hidaka K. Morisaki T. Kikuchi A. Komuro I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci (USA) 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueno S. Weidinger G. Osugi T. Kohn AD. Golob JL. Pabon L. Reinecke H. Moon RT. Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci (USA) 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon C. Qian L. Cheng P. Nigam V. Arnold J. Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qyang Y. Martin-Puig S. Chiravuri M. Chen S. Xu H. Bu L. Jiang X. Lin L. Granger A, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 46.Ren Y. Lee MY. Schliffke S. Paavola J. Amos PJ. Ge X. Ye M. Zhu S. Senyei G, et al. Small molecule Wnt inhibitors enhance the efficiency of BMP-4-directed cardiac differentiation of human pluripotent stem cells. J Mol Cell Cardiol. 2011;51:280–287. doi: 10.1016/j.yjmcc.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H. Hao J. Hong CC. Cardiac induction of embryonic stem cells by a small molecule inhibitor of Wnt/β-catenin signaling. ACS Chem Biol. 2011;6:192–197. doi: 10.1021/cb100323z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willems E. Spiering S. Davidovics H. Lanier M. Xia Z. Dawson M. Cashman J. Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riazi AM. Takeuchi JK. Hornberger LK. Zaidi SH. Amini F. Coles J. Bruneau BG. Van Arsdell GS. NKX2-5 regulates the expression of beta-catenin and GATA4 in ventricular myocytes. PLoS One. 2009;4:e5698. doi: 10.1371/journal.pone.0005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orsulic S. Huber O. Aberle H. Arnold S. Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 51.Gavard J. Mège R-M. Once upon a time there was beta-catenin in cadherin-mediated signalling. Biol Cell. 2005;97:921–926. doi: 10.1042/BC20040531. [DOI] [PubMed] [Google Scholar]
- 52.Pálmer HG. González-Sancho JM. Espada J. Berciano MT. Puig I. Baulida J. Quintanilla M. Cano A. de Herreros AG. Lafarga M. Muñoz A. Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. J Cell Biol. 2001;154:369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamora C. Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–8. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- 54.Nakanishi Y. Ochiai A. Akimoto S. Kato H. Watanabe H. Tachimori Y. Yamamoto S. Hirohashi S. Expression of E-cadherin, alpha-catenin, beta-catenin and plakoglobin in esophageal carcinomas and its prognostic significance: immunohistochemical analysis of 96 lesions. Oncology. 1997;54:158–165. doi: 10.1159/000227681. [DOI] [PubMed] [Google Scholar]
- 55.Gottardi CJ. Wong E. Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azarin SM. Lian X. Larson EA. Popelka HM. De Pablo JJ. Palecek SP. Modulation of Wnt/β-catenin signaling in human embryonic stem cells using a 3-D microwell array. Biomaterials. 2012;33:2041–2049. doi: 10.1016/j.biomaterials.2011.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burdsal CA. Damsky CH. Pedersen RA. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829–844. doi: 10.1242/dev.118.3.829. [DOI] [PubMed] [Google Scholar]
- 58.Cadigan KM. Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 59.Ciruna B. Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 60.Hecht A. Litterst CM. Huber O. Kemler R. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- 61.Hecht A. Vleminckx K. Stemmler MP. van Roy F. Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barker N. Hurlstone A. Musisi H. Miles A. Bienz M. Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonvini P. An WG. Rosolen A. Nguyen P. Trepel J. Garcia de Herreros A. Dunach M. Neckers LM. Geldanamycin abrogates ErbB2 association with proteasome-resistant beta-catenin in melanoma cells, increases beta-catenin-E-cadherin association, and decreases beta-catenin-sensitive transcription. Cancer Res. 2001;61:1671–1677. [PubMed] [Google Scholar]
- 64.Marambaud P. Shioi J. Serban G. Georgakopoulos A. Sarner S. Nagy V. Baki L. Wen P. Efthimiopoulos S, et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bauwens CL. Song H. Thavandiran N. Ungrin M. Masse S. Nanthakumar K. Seguin C. Zandstra PW. Geometric control of cardiomyogenic induction in human pluripotent stem cells. Tissue Eng Part A. 2011;17:1901–1909. doi: 10.1089/ten.TEA.2010.0563. [DOI] [PubMed] [Google Scholar]
- 66.Kinney MA. Sargent CY. McDevitt TC. The multiparametric effects of hydrodynamic environments on stem cell culture. Tissue Eng Part B Rev. 2011;17:249–262. doi: 10.1089/ten.teb.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shafa M. Krawetz R. Zhang Y. Rattner JB. Godollei A. Duff HJ. Rancourt DE. Impact of stirred suspension bioreactor culture on the differentiation of murine embryonic stem cells into cardiomyocytes. BMC Cell Biol. 2011;12:53. doi: 10.1186/1471-2121-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon BS. Yoo SJ. Lee JE. You S. Lee HT. Yoon HS. Enhanced differentiation of human embryonic stem cells into cardiomyocytes by combining hanging drop culture and 5-azacytidine treatment. Differentiation. 2006;74:149–159. doi: 10.1111/j.1432-0436.2006.00063.x. [DOI] [PubMed] [Google Scholar]
- 69.Mohr JC. De Pablo JJ. Palecek SP. 3-D microwell culture of human embryonic stem cells. Biomaterials. 2006;27:6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 70.Ungrin MD. Joshi C. Nica A. Bauwens C. Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park J. Cho CH. Parashurama N. Li Y. Berthiaume F. Toner M. Tilles AW. Yarmush ML. Microfabrication-based modulation of embryonic stem cell differentiation. Lab Chip. 2007;7:1018–1028. doi: 10.1039/b704739h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.