Abstract
Although nonnative protein conformations, including intermediates along the folding pathway and kinetically trapped misfolded species that disfavor the native state, are rarely isolated in the solution phase, they are often stable in the gas phase, where macromolecular ions from electrospray ionization can exist in varying charge states. Differences in the structures of nonnative conformations in the gas phase are often large enough to allow different shapes and charge states to be separated because of differences in their mobilities through a gas. Moreover, gentle collisional activation can be used to induce structural transformations. These new structures often have different mobilities. Thus, there is the possibility of developing a multidimensional separation that takes advantage of structural differences of multiple stable states. This review discusses how nonnative states differ in the gas phase compared with solution and presents an overview of early attempts to utilize and manipulate structures in order to develop ion mobility spectrometry as a rapid and sensitive technique for separating complex mixtures of biomolecules prior to mass spectrometry.
Keywords: mass spectrometry, electrospray ionization, protein conformation, multidimensional analysis
1. INTRODUCTION
1.1. Scope of This Review
Ion mobility spectrometry (IMS), sometimes called plasma (1, 2) or ion chromatography (3, 4), has been utilized for many analytical applications, ranging from the detection of chemical warfare agents (5, 6) to particle sizing (7, 8). In the 1990s, several important advances made it possible to use IMS for analyzing biomolecules. Specifically, researchers coupled soft macromolecular sources (9–11) with IMS (12–16), developed theoretical methods for elucidating ion structure from comparisons of mobility measurements with calculated mobilities for computer-generated structures1 (17–21), and implemented a range of mass spectrometry (MS) techniques (22–24). In the past five years, combinations of IMS with MS and liquid chromatography (LC) have emerged as powerful, hyphenated platforms for examining complex biomolecular mixtures (25–30), and mobility-based MS instruments have recently become commercially available [e.g., the Sionex Corporation microDMx differential mobility sensor (31), the Ionalytics Selectra FAIMS system (32), and the Synapt HDMS system (33)]. Applications ranging from the high-throughput detection of tens of thousands of peptide ions (30) to the obtaining of structural insight about large protein complexes (34, 35) have stimulated significant excitement.
Whereas many important applications of IMS to biomolecular analysis are now becoming routine, the early development of this field was driven by curiosity about the structures, stabilities, and reactivities of protein ions in the absence of solvent (36–47). In the gas phase, proteins display many nonnative conformations that are stable during the millisecond timescales of ion mobility experiments. Resolving different conformations is readily achieved and requires that some structures are not in equilibrium with one another. This behavior is different from the solution-phase equilibrium of states that is responsible for the cooperative transitions normally observed in solution (48, 49). The ability to examine stable populations raises an opportunity to select specific ion shapes for activation and then examine the new conformations that are formed in nondissociative collisions. The vacuum environment not only allows one to examine intramolecular interactions in the absence of solvent effects, it arguably may be the only place where it is possible to select many different types of precursors and intermediates in a way that allows the step-by-step motions of folding and unfolding transitions to be delineated. These curiosity-driven studies are intimately tied to the development of the next generation of multidimensional [IMS-MS, IMS-IMS and IMS-IMS-IMS (50–55)] techniques that are likely to lead to new forms of biomolecule analysis.
This review begins with a short background discussion of protein structure intended as a framework for understanding IMS separations of biomolecules. We present general theoretical considerations associated with IMS and describe a modern instrumental design as an operational example. We have tried to provide references that are representative of the many advances within the past decade that have led to the flurry of activity in this field. Instrumental operation is illustrated with several example data sets, including applications involving what is perhaps one of the most complex biomolecular mixtures: proteins from plasma. Anderson & Anderson (56) have argued that because plasma is in contact with all cells, it may contain proteins from the entire human proteome. It presents an extraordinary challenge for analysis, and although IMS analyses are at an early stage, including such work in a review is an important milestone. Although we mention these experiments, the focus of this review is intended to illuminate the next steps of IMS evolution that will allow new generations of multidimensional IMS experiments to be conducted. The analysis of biomolecules by IMS is an area that builds on many diverse fields and important studies by others. We refer the interested reader to several other valuable reviews about ion thermochemistry (41, 57, 58), macromolecular conformations (59, 60), MS instrumentation (61, 62), condensed-phase separations (63, 64), and proteomics (65).
1.2. General Features of Macromolecular Structures in Solution and in the Gas Phase
The structures of large biological molecules such as proteins are often described as native or denatured. The native state implies a conformation that is capable of biological function; investigators have now obtained thousands of detailed geometries (66, 67) of what are usually assumed to be native structures from nuclear magnetic resonance (68–70) and crystallographic techniques (71, and references therein; 72–74). Far less is conveyed about structure from the term denatured. Lumry & Eyring (75) began their 1954 paper “Conformation changes of proteins” with “[t]he term protein denaturation even in its original meaning included all those reactions destroying the solubility of native proteins and has since acquired so many other meanings as to become virtually useless.” In the half-century since this statement, little has changed. The nature of macromolecules in solution is such that transitions between denatured and native states occur rapidly and presumably through a plurality of intermediates; however, intermediates along folding pathways are rarely stabilized to a degree that allows them to be isolated in the quantities and lifetimes necessary for structural characterization by conventional methods. Rather, macromolecular systems appear to rapidly establish an equilibrium in distributions of structures to optimize appropriate interactions with the environment (76–78).
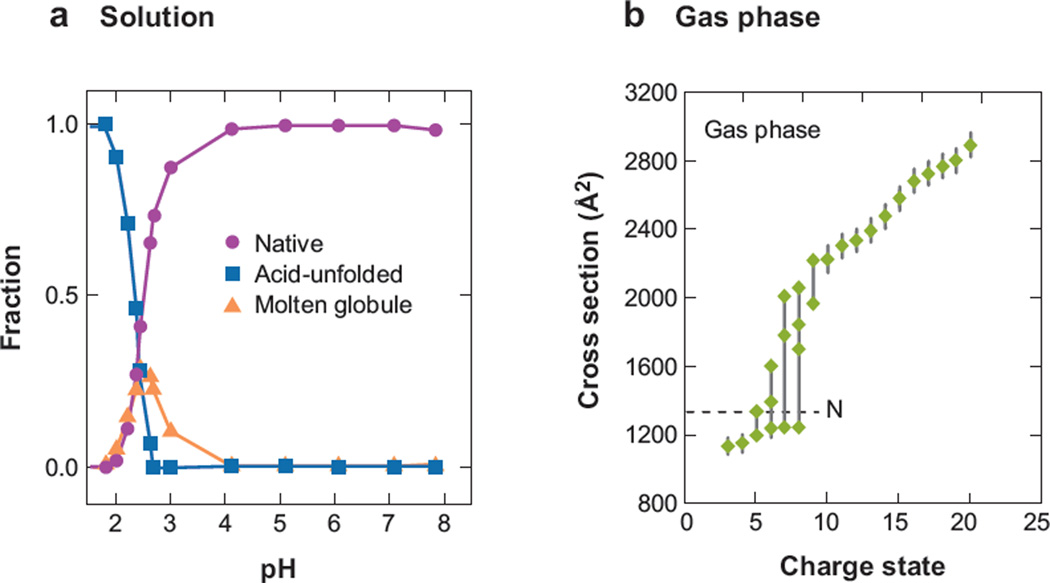
As an example of such behavior, let us consider the acid denaturation curve for cytochrome c in Figure 1 that is measurable by a number of techniques, in this case Soret absorption (79). As the pH of the solution is lowered to a value of ~4, the fraction of the native state begins to decrease; this continues as pH is dropped to ~2 until essentially no signal associated with the native conformation is detectable. The sigmoidal shape associated with this transition is characteristic of many macromolecules, regardless of the approach used to induce denaturation (e.g., temperature or solvent denaturation) (80–83), and is considered a signature of a cooperative transition (48, 49). In this case, the transition involves two other types of states: (a) a molten globule state (48, 80, 84) observed from pH ~2 to 4 and believed to correspond to a set of relatively compact, although not native, distributions and (b) another corresponding to a distribution of unfolded states, which dominates the pH scale below a pH of 2.
Figure 1.
(a) The relative abundances of three ensembles of structures (native, acid-unfolded, and molten globule) observed by Soret absorption analysis of the acid denaturation of cytochrome c in water. (b) The collision cross sections for the most intense features observed as a function of charge state for protonated cytochrome c in the gas phase. The dotted line denotes the cross section of the native state at 1334 Å2. Figure 1a reprinted from Reference 79. Copyright American Chemical Society 1993. The value for the dotted line in Figure 1b comes from Reference 88.
Such transitions for macromolecules in solution are well known, and usually only few states coexist. In the absence of solvent, large molecules display many properties that resemble those of their solution-phase counterparts, but also many that are different. As an example, we consider the gas-phase collision cross sections of cytochrome c as a function of the protonation state produced by electrospray ionization (ESI) (42, 60). The overall appearance of these data shows a roughly sigmoidal shape, as observed for decreasing pH in solution. In the absence of solvent, low-charge states of cytochrome c (e.g., the [M + 3H]3+ to [M + 7H]7+ species) show features in the ion mobility distributions corresponding to ions with cross sections ranging from ~1000 to 1200 Å2, values that are near the cross section expected for compact states that are similar in conformation to the native solution structure. As the number of protons added during electrospray increases, the ions adopt geometries with larger cross sections. For example, cross sections for highly charged ions [M + 12H]12+ indicate that extended states (with cross sections that are more than twice the value anticipated for the native conformations) are favored. The [M + 6H]6+ to [M + 9H]9+ species exist as structures that range in cross sections from ~1200 to 2000 Å2.
The transition from compact to extended states observed with an increasing protonation state has been explained by considering the forces involved in stabilizing conformations. The folding free energy of cytochrome c in solution is −37.1 kJ mol−1 (85), whereas in vacuo values range from −2182 to −3497 kJ mol−1 (85–87). In the gas phase, the energetics of these structural differences are not mitigated by solvation effects. The structure of a gas-phase protein having a net charge of zero is established only by intramolecular interactions, such as zwitterion formation, hydrogen bonding, and van der Waals contacts (88). Excess protons presumably disrupt solution-phase structure; that is, a protonated basic site that would normally be solvated in solution must be accommodated by intramolecular interactions, primarily involving polar side chains and backbone N-H or C-O groups. This internal solvation of charged residues in the gas phase causes the conformations of low-charge-state ions in the gas phase to contract and become more compact than the native solution structures (85, 89, 90). As the number of excess protons increases, the structure becomes sensitive to differences in the dielectric of the surrounding media (~80 for water and 1.0 for a vacuum). In the low dielectric of the vacuum, high-charge states adopt highly extended conformations to minimize repulsive Coulombic interactions that are induced upon desolvation, giving rise to the sigmoidal shaped curve in Figure 1.
Having pointed out the similarities in the shapes of these curves associated with changes in structure upon acid denaturation in solution and the increased state of protonation in the gas phase, we need to stress a key difference. Although the sigmoidal shape associated with the solution-phase denaturation implies an equilibrium and cooperativity, the similar shape of the curve as a function of protonation state for ions in the gas phase does not. Ion shapes in the gas phase are often stable for extended time periods (substantially longer than the millisecond time periods necessary for analysis). This difference makes it possible to utilize the gas-phase conformations for a number of different applications.
2. EXPERIMENTAL CONSIDERATIONS
2.1. Mobility Measurements
When a packet of ions in a buffer gas is exposed to a weak electric field (E), it drifts with a velocity vD = K·E, where K corresponds to the mobility constant of a specific ion in the buffer gas. Because K is specific to interactions between the ion and the gas, individual components within packets that contain a mixture of species may be separated owing to differences in the mobilities of the components. One determines the value of K by measuring the time (tD) required for ions to drift through a specified distance. Measurements between laboratories can be compared by normalizing values to standard conditions that produce the reduced mobility (K0) using the relation
| (1) |
where the variables L and V correspond to the length of the drift region and the voltage applied across it, respectively; and P and T correspond to buffer gas pressure and temperature, respectively (91).
It is also possible to report values as experimental cross sections by the relation
| (2) |
where ze corresponds to the charge on the ion; kB is Boltzmann’s constant; N is the number density of the buffer gas; and mI and mB correspond to the mass of the ion and buffer gas, respectively (91). One can easily rearrange this equation to solve for tD. For a macromolecular ion drifting in He buffer gas, the expression associated with the reduced mass shows that this separation is dominated by the cross section (rather than the ion mass).We point this out because when coupling IMS separations with MS techniques, the strong correlation of the cross section and mass arises because the mass and size increase are intrinsically coupled (92). It is often useful to convert the time axis of an IMS directly to a cross-section axis via Equation 2. We note that this can be done only for ions having the same net charge.
2.2. Weak and Strong Fields
An important definition is associated with the drift field. The applied field is considered to be weak if the buffer gas density is high enough to collisionally dampen ions such that the internal ion temperature is that of the bulk buffer gas (60, 91, 93). Under these conditions, vD is small compared with the thermal velocity of the gas, and ions are not expected to align with the field in the drift region. In this case, the cross-section measurement corresponds to the average of all orientations of the ion as it passes through the drift region and can be used to infer information about the average shape of the ion.
Although a wide range of applied fields satisfies the low-field definition, many interesting phenomena occur as the applied field reaches and then exceeds the low-field limit and ions move in the high-field regime. For example, the field associated with the transition region is different for each ion and depends on the ion charge state, as well as the stability and dynamics of different structures. Under high-field conditions, reduced mobilities may increase or decrease relative to those measured at low fields owing to processes that are not completely understood and are currently under investigation (94–98). It is possible to induce structural transitions as well as dissociation in this region. Differences in mobilities at low and high fields are analytically valuable and have led to the development of the field asymmetric (FA)IMS technique (31, 32, 94–96), as well as hybrid FAIMS-IMS approaches (24).
2.3. Peak Shape and Resolving Power Considerations
As a complement to MS methods, we are especially interested in utilizing IMS for separating isobaric ions. To this end, highly folded (compact) conformations have smaller cross sections (or larger mobilities) than unfolded (extended) states. Often ions with similar structures exist over several charge states. In this case, more highly charged ions have higher mobilities because they experience a different drift force (qeV).
For a single isomer, the theoretical shape of a packet of ions exiting the drift region is determined by the flux
| (3) |
where r0 is the radius of the drift tube entrance aperture; P(tp)dtp is the time-dependent shape of the packet as it enters the drift region; C is a constant; and D is the diffusion constant, given by K0kbT/ze (99). If more than one structure is present, the experimental peak shape may be broader than that calculated for a single structure. Although good agreement with the experimental and calculated peak shapes is suggestive of a single structure, this is not required because two geometries could have identical mobilities; additionally, if multiple structures interconvert on timescales that are much shorter than the millisecond timescale associated with the experiment, then they appear as a single sharp peak.
Revercomb&Mason (93) showed that the theoretical resolving power (R = t/Δt, where Δ t corresponds to the full width at half-maximum of a peak) of a drift tube can be approximated from
| (4) |
which shows that increasing the drift field or length, or decreasing the temperature, leads to an increase in resolution. It is unfortunate that this increase scales as the square root of the experimental parameters. However, it provides an understanding of the impetus to build longer, high-field drift regions.
Although not shown in Equation 4, a parameter that ultimately plays a key role in defining instrument design and resolving power is buffer gas pressure. At high fields the gas must remain stable, so it is necessary to operate at a pressure where the gas does not discharge. Above a few torr, the breakdown potential of most buffer gasses increases with increasing pressure. Thus, it is possible to obtain very high resolving powers (~80 to 300) by utilizing relatively high drift voltages (10 to 30 KV) over ~1-m drift regions at high buffer gas pressures (~100 to 760 torr). One drawback of the high-pressure approach is that it is difficult to store ions. Thus, for a continuous ion source, the introduction of a short pulse of ions upon initiation of experiments often limits duty cycles. Researchers have used a number of approaches (including utilizing multiple injections, as well as Fourier and Hadamard transform approaches) to improve duty cycles (100–102). Alternatively, at low pressures, it is possible to store ions for extended times to accumulate continuous signals into concentrated packets. However, at low pressures, drift fields are typically limited to ~10 to 30 V cm−1.
2.4. Utilizing Nonuniform Fields
In the past decade, a number of separation devices that utilize nonuniform fields to separate ions based on differences in mobilities have emerged. These include FAIMS (31, 32, 94–96), as well as an approach in which ions are exposed to a sequence of accelerating voltages and dampening collisions, referred to as a traveling-wave IMS (33, 103). These instruments often yield results for biomolecules that are similar in appearance to those that utilize uniform fields. Some theoretical treatments of the separation mechanisms have been presented. However, in general, this treatment is at a relatively early stage, and calibrations to uniform field instruments are often employed in data interpretation.
2.5. Combining Ion Mobility Spectrometry with Mass Spectrometry Technologies
The analysis of biomolecules by IMS has been accelerated by advances in MS-based technologies. Specifically, macromolecular ions are created by gentle ionization sources, such as ESI [and more recently desorption (D)ESI (104)] and matrix-assisted laser desorption/ionization. Interestingly, Dole and colleagues’ (105) early work to develop an ESI source for biomolecules investigated lysozyme and utilized IMS rather than MS detection. This pioneering analysis did not resolve charge-state distributions; a later interpretation suggests that although protein ions were observed, the assignment of peaks as low-charge-state protein clusters is probably incorrect (92). Hill and coworkers (12) were the first to resolve ESI charge states with IMS. Clemmer and Jarrold, et al. (13) were the first to resolve different protein ion conformations. Bowers’ (14, 15) and Russell’s (16) groups developed early matrix-assisted laser desorption/ionization sources with IMS. Recently, Clemmer’s (106) group coupled desorption ESI with IMS. Once the ion source is separated from the tube, it is possible to couple essentially any ionization source or MS technique with IMS. To this end, IMS has been coupled with Fourier transform ion cyclotron resonance (107), linear quadrupoles (108), and trapping devices (109–111), as well as time-of-flight (TOF) (112) mass spectrometers.
2.6. Methods for Determining Ion Structure
Arguably one of the most important advances achieved with IMS technologies is associated with the ability to determine information about ion shape. The first description of such a comparison was given in 1925 by Mack (17), who projected the shadows of models at different orientations onto a screen to determine an orientationally averaged cross section, which could then be used to back out a calculated diffusion constant. In the mid-1990s, Jarrold’s (19) and Bowers’ (20) groups developed computer algorithms for calculating cross sections. These calculations were initially used to investigate the structures of a number of atomic clusters and attracted considerable attention with the elucidation of a series of structures associated with carbon clusters as a function of cluster size (including a family of fullerenes). At about this time, advances in biological ion sources and computations of molecular structure that made it possible to rapidly generate molecular coordinates of biomolecules became available. The combination of technologies led to rapid advances in understanding biomolecular ion structure. Most of what is now understood about the shapes of macromolecules in the absence of solvent was determined by comparing calculated cross sections for trial geometries with experimental values. In many cases, it is only possible to estimate a general conformation type; however, in favorable cases, the comparison helps to guide theory in such a way that a low-energy structure that fits the experiment can be obtained. There is now substantial evidence for sequences that form compact globular, helical, and helical-coil conformations. Recently, Robinson and coworkers (34) used a similar approach to determine that the overall geometry of the trp RNA binding protein complex favored a ring-like structure.
2.7. Combining Ion Mobility Spectrometry with Liquid Chromatography Separations
Because of the complexity of biological systems, essentially all analyses involve some form of chromatography. A number of studies have utilized combinations of LC with IMS-MS. As discussed in more detail below, a primary advantage of these techniques for the analysis of complex samples is that multiple dimensions provide enhanced analytical peak capacity. With more analytical space available for peaks upon inclusion of IMS, there are advantages of reduced spectral congestion and new information content. Recently, attempts to modulate conditions between precursor and fragment ion formation using two-dimensional LC, combined with an additional IMS separation, have resulted in a highly parallel approach for identifying large mixtures of peptides. Early examples include analyses of the complex mixtures of proteins from human plasma (as tryptic peptides) (29, 30), as well as attempts to characterize the proteome of the model Drosophila organism (26).Two-dimensional LC (strong cation exchange coupled offline with reversed-phase LC) has been combined with IMS-MS to produce a comprehensive list of peptides detected from human plasma. The list includes more than 9000 entries, 2928 of which are believed to be high-confidence assignments (30).
3. INSTRUMENTATION, EXAMPLES OF ION MOBILITY SPECTROMETRY DATA, AND APPLICATIONS
3.1. Description of an Ion Mobility Spectrometry–Mass Spectrometry Instrument
The constraints associated with buffer gas discharge upon the application of high fields have led to two general instrumental designs: high-pressure, high-field instruments that are capable of generating high resolving powers (113, 114) and low-pressure, low-field configurations with lower resolving powers (115). There are advantages to each type of instrument; many of the desirable features as well as limitations have been discussed previously (60). A significant attraction to low-pressure instruments is that it is straightforward to couple them with existing MS ion sources, trapping devices, and analyzers.
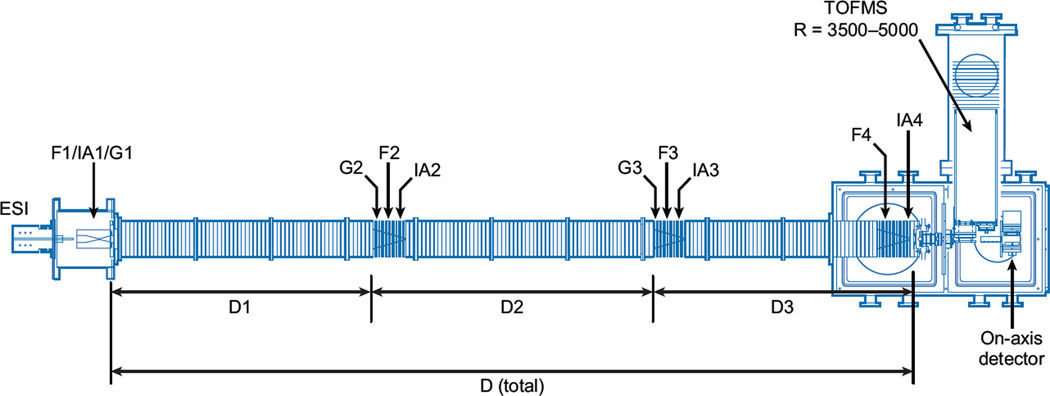
Figure 2 shows an example of an instrument we constructed at Indiana University in 2005 (50, 51), which builds on a design that incorporated an ion funnel at the exit of the drift tube (23). This instrument utilizes a 3-m drift tube with several ion funnels and activation regions along the drift axis. The drift tube can be operated as a single long instrument or broken into three independent drift regions. We begin by considering operation as a single IMS separation in combination with the TOFMS analysis. This is intended to capture the advances in IMS-TOFMS techniques developed from ~1997 to 2005. The utilization of independent drift regions is described below.
Figure 2.
Schematic of the IMS-IMS-IMS-TOF instrument built at Indiana University. Ions are accumulated in the ion funnel F1 and pulsed into the drift tube for experiments by G1. Ions can be mobility selected in two regions (G2, G3); the mobility-selected structures can then be collisionally activated (IA2, IA3). Fragmentation of ions can be performed at IA4, prior to mass analysis in the time-of-flight (TOF) mass analyzer.
Because flight times in the evacuated flight tube of the mass spectrometer (on the order of microseconds) are substantially shorter than drift times through the He-filled drift tube (milliseconds), it is possible to record mass spectra within individual drift windows across the IMS spectrum. This is referred to as a nested measurement because m/z information is nested within individual time windows of the IMS distribution (112). The collection of nested IMS-MS data commences when ions are gated into the entrance of the drift region (using a standard electrostatic gating strategy). The pulse that initiates this gate is synchronized with a pulse in the source region of the TOF mass spectrometer. The repetition rate of the MS pulser is fixed to allow the mass range of interest to be analyzed. This range (usually 5 to 50 µs) becomes the width of the individual time elements that make up the IMS distribution.
3.2. Nested Ion Mobility Spectrometry–Mass Spectrometry Data Set Recorded for a Mixture of Tryptic Peptides
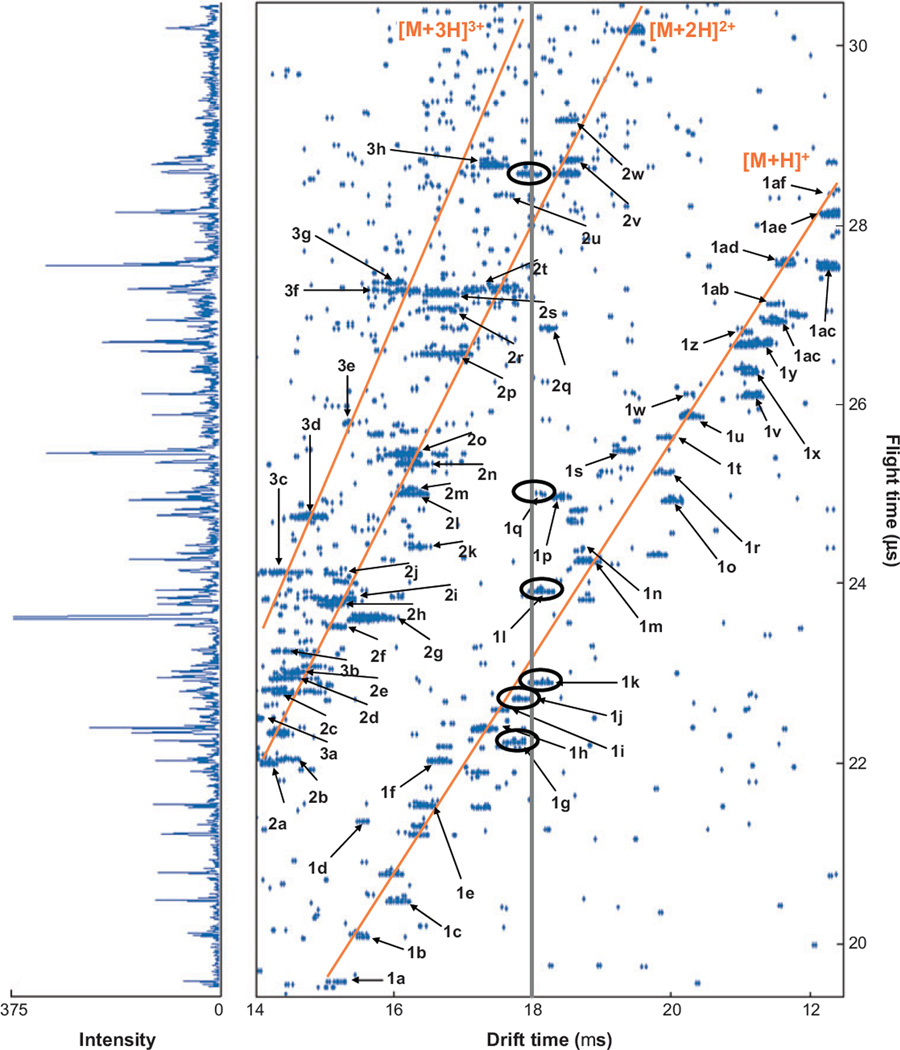
A nested IMS-MS measurement is best illustrated by an example. Figure 3 shows a nested IMS-MS data set for a mixture of tryptic peptides (in this case obtained upon digestion of a standard mixture of commercially available proteins). The drift times of ions (associated with the IMS separation) range from 14 to 22.2 ms, whereas flight times in the mass spectrometer range from 19.5 to 30.5 µs. As individual components associated with the mixture of ions exit the drift region, they enter the source region of the MS instrument where they are orthogonally accelerated into the TOF instrument and the flight times are recorded. For example, ions that exit the drift tube at a single drift time (e.g., 18 ms as in Figure 3) split into multiple peaks over the ~10.5-µs range of flight times shown.
Figure 3.
Nested td (tf) distribution for the direct infusion of a mixture of ions produced by electrospray of a tryptic peptide mixture from the digestion of dog and pig hemoglobin, bovine and pig albumin, and horse cytochrome c. The solid lines in the two-dimensional plot indicate the positions of the [M + H]+, [M + 2H]2+, and [M + 3H]3+ charge-state families. The vertical line at 18 ms highlights the overlap of different m/z species within the mobility distribution; several of the ions having drift times similar to this value are circled. The numbers and letters label peaks and correspond to the assignments given in table 1 of Reference 25. On the left is a mass spectrum obtained by summing the intensities at a given flight time across all drift time windows. These data were acquired using a drift field of 171.7 V cm−1 with a 300-K helium pressure of 150.3 torr. The data were acquired in ~100 min. Figure adapted from Reference 25.
There are a number of apparent advantages associated with this approach. The nested measurement allows mobilities and m/z values for all the ions present in the mixture to be recorded in a single experiment. This has led to large databases of cross sections for different peptide sequences and charge states (116). The availability of cross sections for many sequences makes it possible to extract information about how the amino acid composition, as well as the position of a specific amino acid in a sequence, influences cross section (117). The ability to predict mobilities from sequences is an exciting advance as it provides a constraint for assignment that is not available from MS or MS/MS studies.
Additionally, the IMS separation reduces spectral congestion. Also plotted vertically in Figure 3 is the mass spectrum that would be obtained if no IMS separation was used. Although many peaks in the mass spectrum are apparent, the ability to pull the distribution of peaks apart prior to MS analysis allows many features (especially small peaks) that would otherwise overlap to be resolved (118). In total, 98 resolved peaks were observed in the two-dimensional, corresponding to 60 of the 187 peptides expected upon complete digestion of all five proteins.
Finally, in many cases, ions appear to fall into families. Figure 3 illustrates families of ions having the same charge states (in this case either [M + H]+, [M + 2H]2+, or [M + 3H]3+ produced during ESI). In other cases, it is possible to resolve families of molecules that have different chemical properties (119). For example, mixtures of lipids (120), glycans (121), peptides with specific post-translational modifications (122), and other types of polymers often fall into families. Thus, there is significant new information associated with the combined measurement.
3.3. Instruments That Incorporate Additional Dimensionality
The relatively long times associated with IMS compared with MS offer other advantages for resolving complex mixtures. As long as dissociation techniques are carried out rapidly (on submillisecond timescales and with no disruption of the time resolution after the fragments are formed), it is possible to gain a parallel advantage of generating fragmentation spectra. In this case, fragment ions appear along the vertical dimension of IMS-MS data sets. By modulating voltages at the exit of the drift tube, it is possible to use conditions that favor precursors and those that favor collision-induced dissociation (CID). The coincidence of drift times is used to unite peaks formed during dissociation with their antecedent precursors (from back-to-back, modulated condition experiments). This is called a parallel fragmentation approach (123, 124) to emphasize that all ions are examined as precursors and fragments after ionization (unlike MS selection of precursor ions used for traditional MS/MS studies). However, the approach is really a very fast (submillisecond timescale) serial approach for those ions having different mobilities. Similar to MS/MS, the fidelity of the information is limited by the ability to resolve components prior to CID. There is clearly a need to improve IMS resolving power as mixture complexity increases.
A range of dissociation approaches has been demonstrated, including traditional collisions in an octopole collision cell (123, 124), surface-induced dissociation (125, 126), and fragmentation induced by an orifice skimmer cone mounted at the exit of a drift tube (127), as well as fragmentation induced in the high-pressure exit region of the drift tube (128, 129). It should be straightforward to include photodissociation approaches (130) and, to the extent that time resolution is not lost, new techniques such as electron capture (131) or transfer (132) dissociation as well. Parallel dissociation is now obtainable using the traveling-wave IMS approach that is available commercially, in which it is referred to as time-aligned-parallel fragmentation (133).
Although 1 to 100 ms is a long time to manipulate ions in a mass spectrometer, the combined IMS-MS timescale allows it to still function as a very-high-speed detector for condensed-phase analyses. Thus, one can utilize the nested advantage for higher-dimensionality experiments by coupling IMS with slower techniques. We discuss the data set in Figure 3 above in terms of an IMS-MS analysis; however, it was actually the first IMS-MS data set recorded as a single frame of a higher-dimensionality separation involving LC. In this case, as peptides eluted from the LC column, they were ionized by ESI and analyzed by IMS-MS. The combined LC-IMS-MS approach can be modulated to include CID between the IMS and MS instruments [an LC-IMS(CID)MS analysis]. Thus, although no MS is used for precursor ion selection, the combined resolution of the LC and IMS dimensions often provides an opportunity to correlate precursor and fragment ion spectra for ion identification. The approach has been automated to an extent that database-searching methods (134, 135) can now be employed.
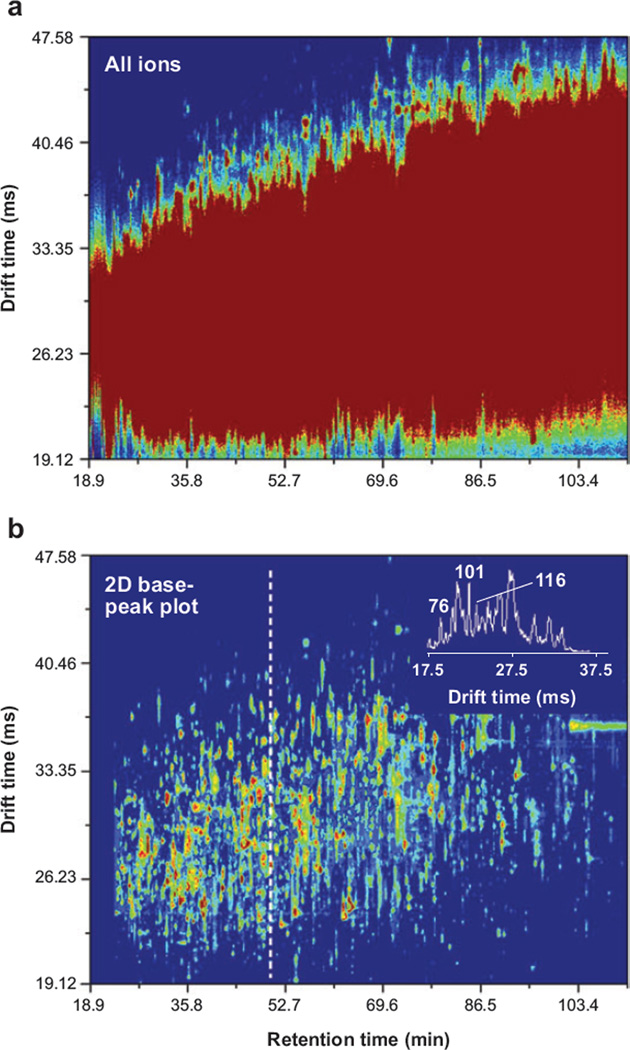
The combination of IMS with LC, parallel CID, and MS is emerging as an especially powerful approach for examining complex mixtures of proteins from biological sources. One of the most formidable challenges in proteomics experiments is to develop reproducible, information-rich, tremendously high-peak-capacity analytical platforms that operate quickly. Figure 4a shows a plot of the LC and IMS dimensions for an LC-IMS(CID)MS analysis of peptides obtained upon tryptic digestion of proteins isolated from plasma. In this case, we show only the precursor data set (the modulated data associated with the CID spectra appear similar when plotted in these dimensions). We have used a high-resolution LC separation that over several hours shows a peak capacity of >300. Clearly this sample is complex, and the number of components far exceeds this capacity. Even with the additional high-resolution IMS separation (having a resolving power in excess of 100 for most peaks and providing a two-dimensional peak capacity of >15,000), the main portion of the spectrum appears as a large unresolved feature. We note that a few species on the leading and trailing edges of the IMS distribution are resolved. A feeling for the shapes of peaks associated with individual components within the data set can be obtained by extracting and plotting the most intense features (a two-dimensional base peak plot) shown in Figure 4b.
Figure 4.
Two-dimensional retention time (drift time) contour plots of the high-resolution ion mobility spectrometry–mass spectrometry analysis of a digest of proteins from plasma. (a) The summed intensity of all mass spectral features at each retention time and drift time value. (b) A two-dimensional base-peak plot in which only the most intense mass spectral feature is plotted at each retention time and drift time value. An intensity threshold of four was used to construct both plots. The inset in panel b shows the drift time distribution for species observed at a retention time of 48 min, along with the resolution for several species. Figure reprinted from S.J. Valentine, S.L. Koeniger & D.E. Clemmer, unpublished data.
A primary consideration in developing this analysis is that no additional time is required for the experiment. This is not the case for the interpretation of results. It is possible to pick large features (precursors and CID data sets) quickly and carry out database-search analyses for assignments relatively quickly (and, in doing so, several hundred proteins can be confidently identified); however, a more detailed analysis is time intensive owing to the large size of the data sets.
3.4. New Developments Involving IMS-IMS and IMS-IMS-IMS Instrumentation
In analogy with condensed-phase chromatography, in which it is possible to change the mobile phase during a separation (or use different stationary phases to influence the ability to separate different components), a number of studies have attempted to change the IMS separation of ions in the gas phase by changing the buffer gas (136). There are many interesting factors that arise as the buffer gas is varied, and it is possible to substantially shift the mobilities of ions by using different gasses. Asbury & Hill (136) have shown that it is possible to change the order of the drift times of chloroaniline and iodoaniline by varying the buffer gas from helium to carbon dioxide. However, in general, mobility separations are dominated by momentum transfer processes during the ion-neutral collisions; because this depends largely on the shapes of ions, the ability to substantially change the resolution of two ions that have similar components is limited (137).
As described above, an important difference between the behavior of macromolecules in the gas phase compared with solution is that in the absence of solvent, many different conformations appear to be stable. Early ESI-IMS work on proteins showed that upon high-energy injection of ions into low-pressure drift tubes, it was possible to induce unfolding and refolding transitions (115, 138). Other studies of ions in Paul-type and ion-cyclotron-resonance traps also indicated that different types of conformations were stable over long time periods (139–142).
This curious property of biomolecules in the absence of solvent provided the rationale to develop a multidimensional IMS-IMS instrument that would utilize differences in ion cross sections (before and after activation) for separation. The schematic shown for the 3-m long drift tube in Figure 2 is designed specifically for this type of measurement. Although the instrument can be operated as a single separation device to produce IMS-MS data, it comprises three separate drift regions (D1–D3) and ion gates (G1–G3), as well as four ion activation regions (IA1–IA4). These regions are designed so that following an initial IMS separation (in the D1 region), ions of a specified mobility can be selected at G2, activated at IA2, and separated again in D2 and D3. If desired, one can carry out an additional selection and activation process in the G3/IA3 region and follow this with a final separation in D3. Finally, after the final IMS separation, it is possible to induce fragmentation at IA4 (and carry this out in a modulated fashion to generate precursor and CID spectra).
A drift tube of this length and capability is possible because of the development of the ion funnel by Smith’s group (22, 23). The instrument shown in Figure 2 uses four of these funnels, F1 to F4. A combination of radio frequency and direct current fields in the funnel allows ions to be focused in the off-axis dimensions so that ion transmission is high. Tang et al. (23) demonstrated that inclusion of a funnel inside a drift region does not significantly change the resolving power of the measurements.
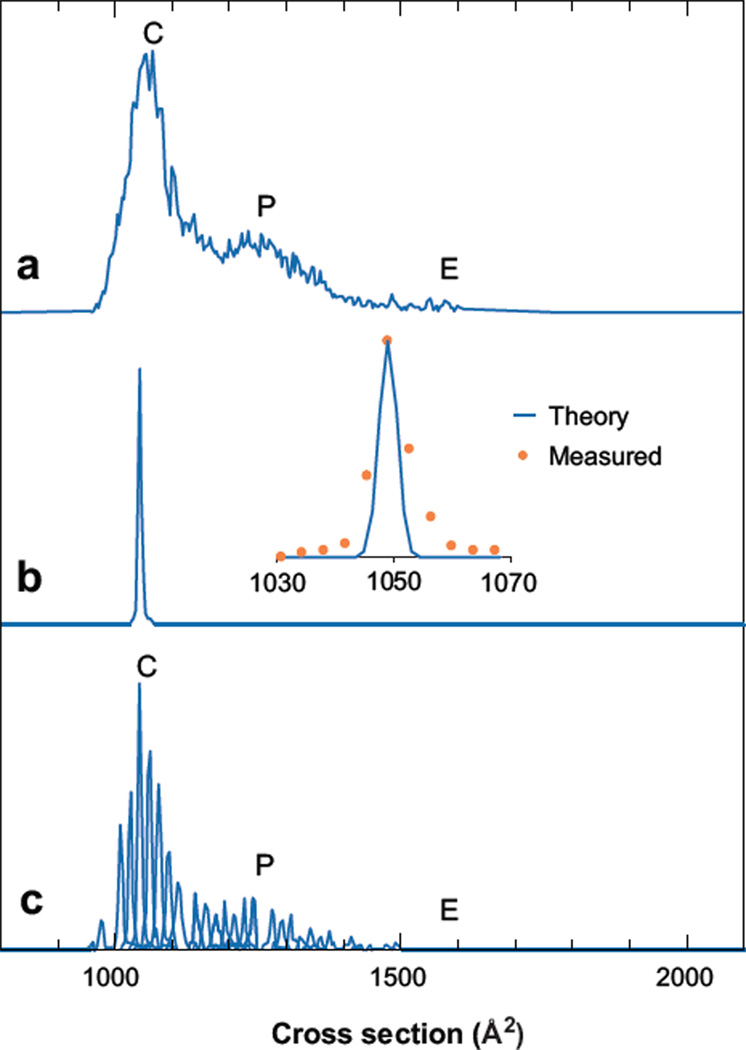
The ability to select and activate ions inside of the drift tube has led to a number of interesting findings. Koeniger et al. (52) examined the broad IMS peaks observed for specific charge states of ubiquitin (a small 8.6-kDa protein that has been studied extensively). Figure 5 shows that the IMS spectrum for [M + 7H]7+ (obtained by transmitting ions through the entire 3-m drift region) is essentially identical to [M + 7H]7+ distributions recorded using earlier low-pressure, low-resolution as well as a high-pressure, high-resolution instruments. Interestingly in the prior work, although the instrumental resolving powers differed by an order of magnitude, the spectra looked similar in that the [M + 7H]7+ ion exhibited at most three fairly broad features. Comparisons of the cross sections with values calculated for the native solution coordinates (as well as other structures generated by molecular modeling) suggested that these peaks corresponded to compact (tightly folded) conformers and partially folded states; the position of a peak corresponding to an elongated state (approximately 1600 to 1700 Å2) is also indicated. This state can be observed by injecting ions into a low-pressure drift tube at high energies or heating the capillary ion source inlet, but it is not observed in significant abundance in the IMS distribution shown.
Figure 5.
Drift time distributions of the [M + 7H]7+ of ubiquitin. (a) The total mobility distribution shows that the +7 charge state exists mostly as compact structures (C), some partially folded structures (P), and minimal elongated structures (E). (b) A single peak is observed when a narrow distribution (50 µs) of the compact structure is isolated with mobility selection at 7.8 ms. The inset compares the theoretical to the experimentally measured peak shape. (c) The total mobility distribution for the +7 charge state is reconstructed with 28 mobility-selected distributions acquired every ~0.125 ms. This demonstrates that this distribution arises from many overlapping structures that are stable over the course of the two-dimensional acquisition. Figure reprinted from Reference 52.
The question that emerged from these and early data was, what limits the widths of these features? Are the peaks broad because multiple conformations interconvert during the experiment on timescales that are similar to the separation? Or are the peaks broad because there are many unresolved conformations within the broader conformation type? Selection of a narrow ion pulse at G2 reveals that the peak remains sharp (Figure 5b) as selected ions drift the remaining distances through the drift tube. Figure 5c shows that this is the case at essentially any point across the distribution, requiring that many unresolved structures within the broader conformation type are stable (thus peaks are broad because these structures are not resolved).
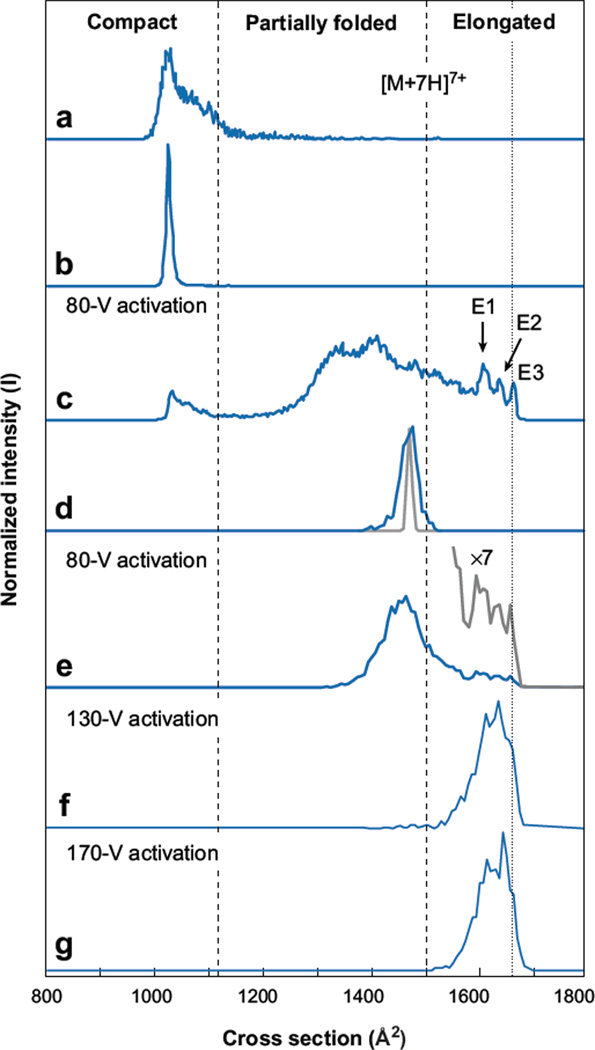
Once it was known that it is possible to select subsets of conformations, it is possible to investigate how activation influences IMS distributions. Figure 6 shows a study demonstrating the ability to select and vary the conformation of [M + 7H]7+ across a broad range of conformations (53). As in Figure 5, it is possible to select any region of ions (here a subset of compact states); upon activation (in this case an 80-V bias applied across a short 0.3-cm activation region), the compact states produce broad peaks associated with a range of partially folded and elongated states. Which states are produced depend on what part of the distribution is selected, as well as the solution conditions used to produce the ions. From the distribution shown in Figure 6c, it is possible to further isolate ions (Figure 6d) by selection at G3. For example, activation of the partially folded conformers can produce a range of other structures for separation. Under low-energy activation conditions, partially folded intermediates (formed from compact precursors) primarily lead to products that are partially folded. At higher energies along this pathway, elongated states can be favored (53).
Figure 6.
Ion mobility distributions of ubiquitin [M + 7H]7+ ions obtained in IMS-IMS-IMS/MS experiments. The distribution in panel a is the initial distribution consisting of primarily compact conformers. Upon selection of a narrow distribution of compact ions at G2 (100 µs), the distribution in panel b is obtained. Activation of the selected compact ions at IA2 produces the distribution in panel c. A second selection of an intermediate within the partially folded structures performed at G3 (150 µs) is shown in panel d, with the diffusion-limited peak width of the selected ion (gray line). Upon activation of the partially folded structures at IA3, the distribution in panel e is produced, consisting of a broader distribution of partially folded structures and a smaller distribution of elongated states (gray line). The distributions in panels f and g are obtained upon higher-energy activation of the partially folded structures shown in panel d. Figure reprinted from Reference 53.
These pathways are clearly complex, and we are at an early stage in understanding these data. However, it is apparent that IMS-IMS and IMS-IMS-IMS studies can offer information about how ions with specified cross sections change through specified intermediates to reach product states. Such step-by-step structural transitions are rare, and this type of study is impossible in solution because of difficulties in trapping the intermediate states. One intriguing finding of the early work is that the distribution of elongated states appears to retain information relevant to the compact precursor ions that produced it. Presumably, this memory exists as retained elements of secondary structure (53, 54).
3.5. Extension of IMS-IMS Techniques for Complex Mixture Analysis
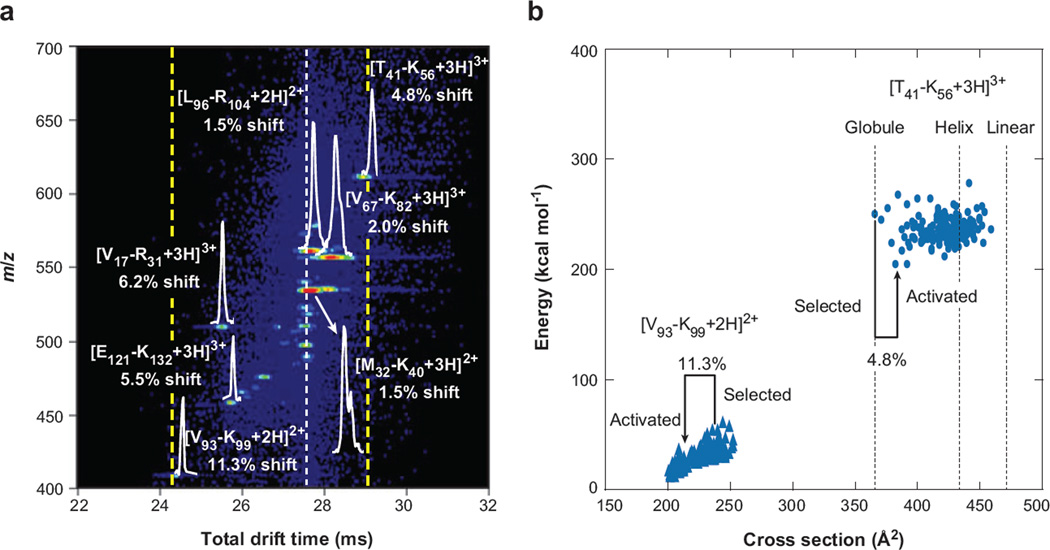
The ability to separate, select, and activate different conformations may also have merit for the analysis of complex mixtures. Figure 7 shows an example of such an experiment for a mixture of tryptic peptides from human hemoglobin. Here, we represent the final IMS-IMS separation for a subset of separated ions that were selected and activated at the G2/IA2 region. With no activation, this set of ions would arrive along the line at 27.5 ms. With activation, some ions shift to slightly shorter or longer times. These shifts require that the conformations have changed such that ions have higher or lower mobilities through the remaining D2 and D3 regions of the instrument.
Figure 7.
(a) An expanded view of an activated selection of human hemoglobin tryptic peptides. The white dotted line denotes the time at which mobility-selected ions with no activation are observed, whereas the dashed yellow lines show the new effective separation space of the second ion mobility spectrometry experiment. Drift distributions for several peptides are shown, along with shifts from original (inactivated) drift times for all ions. (b) Energy versus conformer cross section of the 150 lowest-energy structures post–simulated annealing for two peptides from panel a ([V93 − K99 + 2H]2+ and [T41 − K56 + 3H]3+) as well as cross sections of the energy-minimized structures of the [T41 − K56 + 3H]3+ ion modeled as a helix and linear structure, denoting the range of structures available to each sequence. Cross sections of the selected and activated structures for each sequence are highlighted, along with percent shifts from the mobility-selected structure. Note that the nomenclature used refers to the position of the peptide by providing the location (with respect to the intact protein sequence) and single letter abbreviation of the N- and C-terminal residues, respectively. Figure reprinted from Reference 55.
In the distribution shown in Figure 7, peaks have been assigned based on their masses and drift times, and are consistent with previous work in our laboratory. The origin shifts for peaks associated with two peptides, [V93 − K99 + 2H]2+ and [T41 − K56 + 3H]3+, have been studied using molecular modeling and collision cross-section calculations (and the results are also shown in Figure 7). The molecular modeling protocol is intended to start with a range of states and simulate an annealing process. In the case of [V93 −K99 + 2H]2+, an array of relatively high-energy structures generated by molecular modeling anneals into lower-energy structures upon application of the heating/cooling protocol used in the calculation. The data are consistent with the experimental findings. It appears that upon exiting the ion source, [V93 − K99 + 2H]2+ favors a relatively open conformation having a cross section of 240 Å2; as these ions are accelerated through the IA2 region (at 80 V × 0.3 cm−1), they are rapidly heated, and as they exit the region, they cool down. This process allows the ions to anneal to a lower-energy state. In this case, [V93 − K99 + 2H]2+ adopts a conformation that has a cross section that is 11.3% smaller than the state favored from the source. In contrast, the [T41 − K56 + 3H]3+ species shifts to lower mobilities because it can form more extended states upon activation.
3.6. Peak Capacity Estimates for IMS-IMS Separations
For multidimensional separations, it is relevant to describe the analytical peak capacity. Specifically, it is important to understand what can be gained by the two-dimensional IMS-IMS compared with the single-dimension of IMS separation. Giddings and others (25, 143, 144) have discussed these ideas for multidimensional chromatography. Figure 7 shows insets and shifts associated with the peaks for specific peptides after initial IMS separation, selection, and activation, followed by additional IMS separation. A careful analysis of the peak shapes and ranges over which peaks are observed (before and after activation) indicates that two-dimensional IMS-IMS peak capacities can be surprisingly high. This increase occurs because the ion’s mobility is influenced by the change in structure. In general, separations are carried out under equilibrium conditions such that adding a second identical separation would lead to little, if any, improvement. Values of ~480 to 1360 have been reported for mixtures of tryptic peptides. We note that this is for IMS-IMS alone. The inclusion of MS and LC will greatly increase these values.
3.7. Developing Shift Reagents
In the case of peptide ions, structural differences between states are much smaller than for larger protein ions. As we look to the future of these techniques, it would be advantageous to develop approaches that maximize changes in structures that can be used for multidimensional IMS separations. Another approach to modifying the ion structure is to produce noncovalent molecular adducts and then remove them to specific locations within the IMS instrument. The nature of the ion-neutral interactions is relatively long range; thus, the choice of system allows a means to tune dissociation energies and may also make it possible to access different predissociative states having different structures. A number of studies have investigated peptideion interactions with neutrals that may yield binding to specific structural motifs (145–147). One set of molecules to emerge as candidates for shift reagents for IMS-IMS (and higher-order IMS) separations is the crown ethers.
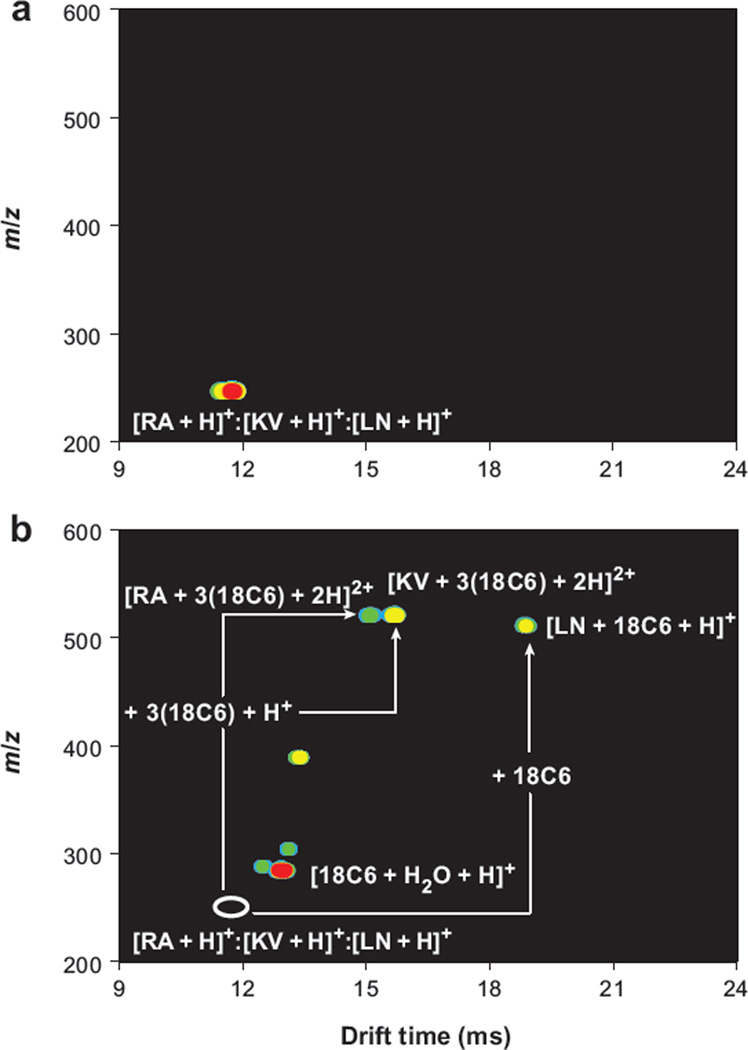
Figure 8 shows the first study specifically designed to incorporate molecular adducts as shift reagents for IMS. Hildebrand et al. (148) examined three dipeptides ions: [RA + H]+, [KV + H]+, and [LN + H]+. These ions are isobaric and also have the same mobilities; thus, even with a two-dimensional high-resolution IMS-MS analysis, only a single peak is observed. Upon the addition of 18-crown-6 ether (18C6) to the mixture of analytes, we find substantially different properties in the ESI-generated species. Specifically RA and KV accommodate three crowns to produce [M + 3(18C6) + 2H]2+, whereas LN incorporates only a single 18C6. The result is that all three peptides can be resolved as adducts. In the same paper, Hilderbrand et al. also showed that it was possible to remove adducts (by low-energy CID) from tripeptides designed by combinatorial synthesis to selectively bind different numbers of crowns. By incorporating CID following the IMS separation (but prior to MS analysis), the mobilities of the peptide-adduct ions are shifted relative to the peptides (because of adduct formation), but the measured m/z values are those of peptides without adducts.
Figure 8.
(a) Nested plot showing the protonated forms of the dipeptides RA, KV, and LN, which are irresolvable in the drift dimension. (b) The addition of 18-crown-6 ether (18C6) to the electrospray solution creates noncovalent complexes that shift the mobilities of the three peptides from that of the bare species (open white oval) enough that they are now mobility resolved under the same instrumental conditions. Figure reprinted from Reference 148.
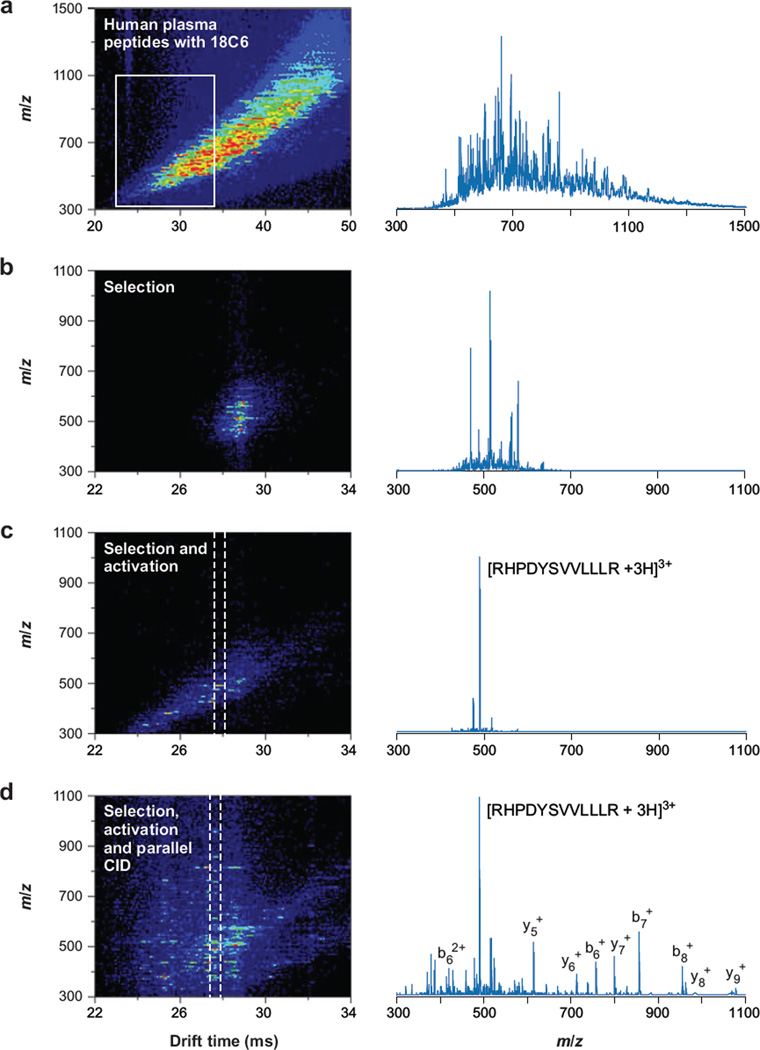
Bohrer and coworkers (B.C. Bohrer, S.J. Valentine, S. Naylor & D.E. Clemmer, unpublished data) have extended the use of crown ethers as shift regents for the analysis of peptides from human plasma. Figure 9 shows one result for an analysis of peptides obtained by digesting a mixture of proteins from human plasma (following depletion of six abundant proteins) that was carried out using the instrument in Figure 2. IMS-MS analysis of ions produced by direct-infusion ESI of the peptide-crown mixture leads to a complex pattern of peaks (Figure 9a). Examination of the mass spectrum shows that the overlap of many features leads to a large baseline signal. This baseline is reduced substantially when the G2 gate is used to introduce a small fraction of ions into the remaining portion of the drift tube (Figure 9b). Selection at G2 and activation at IA2 lead to large changes in mobilities (associated with losses of different numbers of 18C6 units from the adducts). The simplification of the spectrum is remarkable. It is possible to extract regions that have only a few abundant ions (Figure 9c). When the selected and collisionally activated ions are exposed to the parallel CID analysis (at the exit of the drift tube), there is clear evidence for many different peptides; however, some ranges still show fragmentation data that can be interpreted. For example, a database search of the peaks found in the extracted regions of Figure 9c,d allows us to assign the [RHPDYSVVLLLR + 3H]3+ ion a sequence from albumin. The potential to selectively move across this spectrum with a controlled approach holds promise for developing analyses aimed at determining the presence of a component in a complex sample.
Figure 9.
Ion mobility spectrometry–mass spectrometry nested plot of human plasma peptides, electrosprayed from solution containing 18-crown-6 ether (a) with integrated mass spectrum (right column). The white box indicates the region focused on in the other panels of this figure. Mobility-selected complex ions gated through G2 (b) are activated with 160 V at IA2 to dissociate the complex ions into bare peptide ions, which resolve further in mobility (c). Integration of the data between the dashed lines shows that [RHPDYSVVLLLR + 3H]3+ is practically isolated in the drift dimension (c, right column). Parallel dissociation at the end of the drift tube renders mobility-labeled fragments (d). Integration about the dashed lines now provides fragmentation spectrum for [RHPDYSVVLLLR + 3H]3+ (d, right column). Abbreviation: CID, collision-induced dissociation. Figure taken from B.C. Bohrer, S.J. Valentine, S. Naylor & D.E. Clemmer, unpublished manuscript.
4. CONCLUSIONS
The above discussion investigates the analytical utility of exploring the curious nature of macromolecules in the gas phase. Primarily, the importance of these studies arose because the shapes of large ions in the absence of solvent were relatively unknown and the new information obtained appears to complement MS analyses. That the ion shapes do not appear to readily reach equilibrium is also interesting as this makes the IMS separation highly complementary to condensed-phase-separation approaches. Another interesting parameter that has been stressed is the ability to place the IMS separation between an LC separation and MS detection. In general IMS separations require longer acquisition times than the flight times required for MS analysis, but are still much faster than those required for LC. This results from the differences in number density for the environment of each analysis. Moreover, the experimental variables associated with the IMS measurement can be controlled in a manner that makes measurements highly reproducible. Alternatively, the coupling of FAIMS to IMS, as well as the ability to change the structures of mobility-selected ions between IMS drift regions, introduces the concept of high-speed, high-peak capacity multidimensional analyses solely in the gas phase.
One imagines that the availability of commercial instruments will lead to rapid advances and many new applications for IMS as it pertains to analyzing biomolecules. This also leads to opportunities to take advantage of the coupling of IMS to other orthogonal approaches, including ion-molecule reactions (41, 59) and fluorescence (149). This type of analysis provides additional dimensionality for biomolecules with several stable structures to the extent that these structures react differently. An early example of this combination of experiments involved hydrogen-deuterium exchange reactions on different conformations of protein ions (109). Another area gaining interest is the examination of large proteins and macromolecular complexes with IMS techniques, as demonstrated by work in the Robinson (34) and Loo (35) groups.
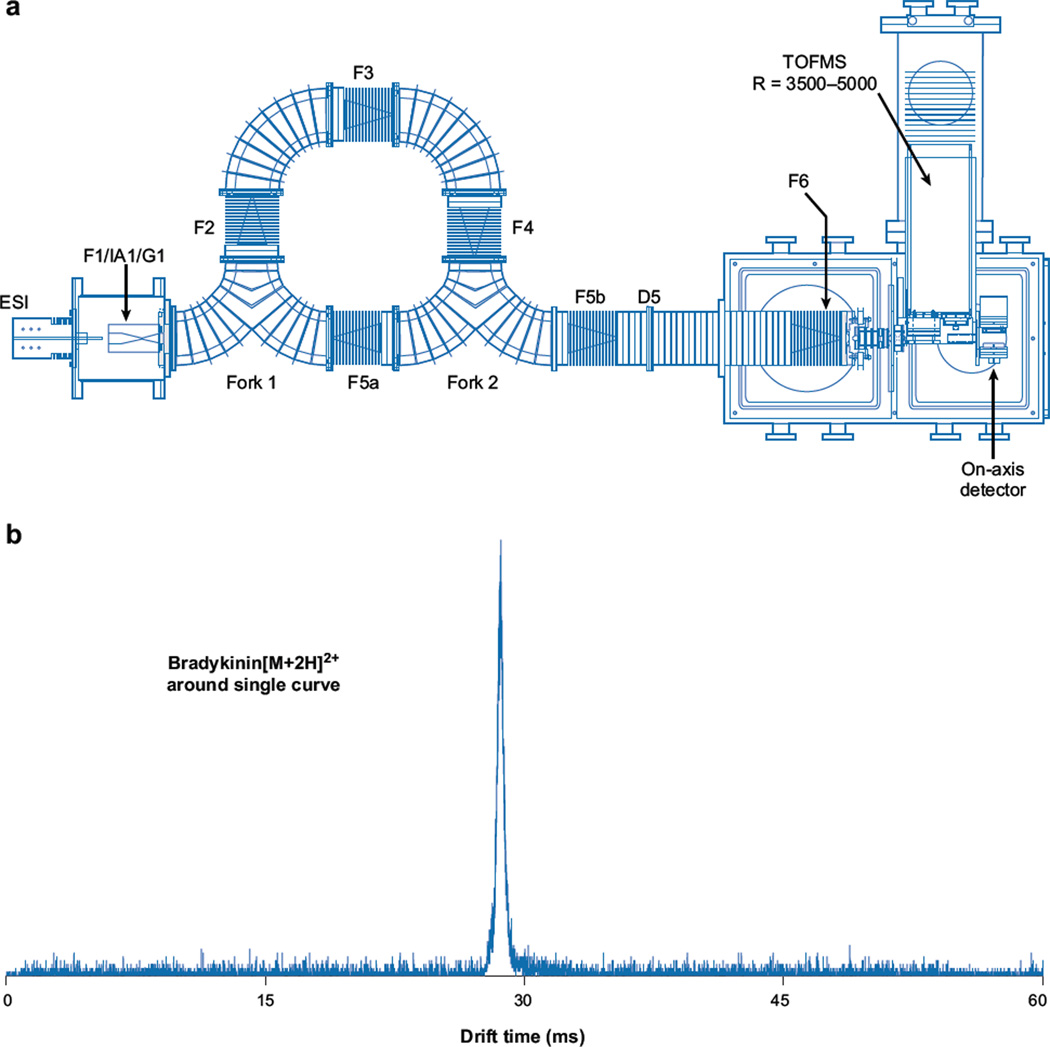
With growing interest in higher-dimensionality IMS, we (S.I. Merenbloom & D.E. Clemmer, unpublished results) have recently constructed the circular instrument shown in Figure 10. The idea behind this design is to bring ions in from the source and allow them to separate around the circle multiple times before changing the focusing such that ions exit to the mass spectrometer. Results from a previous instrument configuration featuring a single 90° curve show little loss in drift resolution (Figure 10b).We also have demonstrated that it is possible to get ions around multiple turns with little loss of signal. We anticipate that once functional, this instrument will be of great importance because it allows ions to be introduced and analyzed for extended periods. Initial studies of the curved regions suggest that it will be possible to move ions through curves and, by doing so, take advantage of longer drift lengths to enhance IMS resolution. One can see that the limit of resolution of ions moving in this system occurs when the leading edge of the diffusing ion cloud catches up with its own trailing edge. When this occurs, one can eject the ion packet into the remaining regions of the instrument for subsequent mass analysis. The ability to cycle ions should allow higher-order IMS experiments to be carried out. It should also allow ions to be stored at well-defined temperatures in the presence of reactant gasses for much longer times.
Figure 10.
(a) Schematic of a circular ion mobility spectrometry time-of-flight instrument. (b) The mobility distribution for mobility-selected bradykinin [M + 2H]2+ ions that have traversed a drift tube containing a single 90° turn. Figure reprinted from S.I. Merenbloom & D.E. Clemmer, unpublished data.
SUMMARY POINTS.
Large macromolecules in the gas phase exhibit a myriad of structures that can be monitored by IMS and are related to solution-phase structures, including nonnative states that may be intermediates along the folding pathway or trapped species.
IMS coupled to MS provides a two-dimensional separation space in which ions can be resolved on the basis of size, charge state, and chemical properties.
Mobility selection of a macromolecular ion between drift regions that results in a sharp peak is indicative of a stable structure, collisional activation of which can probe folding or unfolding pathways.
The coupling of IMS with additional separations such as LC or additional IMS leads to very high peak capacities without significant increases in acquisition time. Incorporating shift reagents improves mobility-based separation performance.
FUTURE ISSUES.
IMS may be coupled to other orthogonal techniques including ion-molecule reactions, such as electron-transfer dissociation and hydrogen-deuterium exchange, and fluorescence analyses.
Increasingly higher IMS resolutions and dimensionalities will drive the further development of emerging technologies and instrumentation.
Analysis of large protein complexes by IMS will aid in purifying and understanding structures.
ACKNOWLEDGMENTS
The authors are grateful for many valuable contributions of our colleagues and reviewers of our papers, specifically, the early work involving protein conformations and instrumentation carried out by Stephen J.Valentine, Anne E. Counterman, Cherokee S. Hoaglund-Hyzer, Catherine Srebalus-Barnes, John Taraszka, and Sunnie Myung. We also thank Emily Vincent for her meticulous editing and proofreading work. New instrumentation has been supported by grants from the NIH (P41 RR018942) and the Indiana 21st Century fund.
Glossary
- IMS
ion mobility spectrometry
- Plasma/ion chromatography
gas-phase separation of ions based on size and charge; synonyms for ion mobility spectrometry
- MS
mass spectrometry
- LC
liquid chromatography
- Native state
a conformation in its natural form capable of biological function; nonnative indicates any other form
- Molten globule
compact form of a protein that does not retain its biological function
- ESI
electrospray ionization
- TOF
time of flight
- Nested measurement
recording a longer timescale experiment as increments of a shorter timescale experiment
- CID
collision-induced dissociation
Footnotes
A number of software packages are available for calculating molecular structure, including the molecular mechanics package available through the Insight II suite of programs [Insight II 2000 (Accelerys Software, San Diego, California, 2001)] or the AMBER suite of programs [AMBER 7 (University of California, San Francisco, California, 2002)] and quantum chemical calculations from Gaussian [GAUSSIAN 03 (Gaussian, Inc., Wallingford, Connecticut, 2004)] or Jaguar [Jaguar 5.5 (Schrödinger, LLC, Portland, Oregon, 1991–2003)].
DISCLOSURE STATEMENT
D.E.C. is a scientific cofounder of BG Medicine and founder of Predictive Physiology and Medicine, two early-stage biotechnology companies. He is also currently a consultant for Waters Corporation. The authors have filed a number of patents related to ion mobility.
Contributor Information
Brian C. Bohrer, Email: bbohrer@indiana.edu.
Samuel I. Merenbloom, Email: smerenbl@indiana.edu.
Stormy L. Koeniger, Email: stormy.koeniger@abbott.com.
Amy E. Hilderbrand, Email: ahilderb@email.arizona.edu.
David E. Clemmer, Email: clemmer@indiana.edu.
LITERATURE CITED
- 1.Cohen MJ, Karasek FW. Plasma chromatography: a new dimension for gas chromatography and mass spectrometry. J. Chromatogr. Sci. 1970;8:330–337. [Google Scholar]
- 2.Karasek FW. Plasma chromatography. Anal. Chem. 1974;46:A710–A720. doi: 10.1021/ac60345a031. [DOI] [PubMed] [Google Scholar]
- 3.Kemper PR, Bowers MT. Electronic state chromatography: application to 1st-row transition-metal ions. J. Phys. Chem. 1991;95:5134–5146. [Google Scholar]
- 4.Bowers MT, Kemper PR, von Helden G, van Koppen PAM. Gas-phase ion chromatography: transition metal state selection and carbon cluster formation. Science. 1993;260:1446–1451. doi: 10.1126/science.260.5113.1446. [DOI] [PubMed] [Google Scholar]
- 5.Hill HH, Siems WF, St. Louis RH, McMinn DG. Ion mobility spectrometry. Anal. Chem. 1990;62:A1201–A1209. doi: 10.1021/ac00222a001. [DOI] [PubMed] [Google Scholar]
- 6.Chen YH, Hill HH, Wittmer DP. Thermal effects on electrospray ionization ion mobility spectrometry. Int. J. Mass Spectrom. Ion Process. 1996;154:1–2. [Google Scholar]
- 7.Sanders TM, Forrest SR. Small particle-size distributions from mobility measurements. J. Appl. Phys. 1989;66:3317–3323. [Google Scholar]
- 8.Whitby KT, Clark WE. Electronic aerosol particle counting and size distribution measuring system for 0.015 to 1 mu size range. Tellus. 1966;18:573. [Google Scholar]
- 9.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T. Protein and polymer analyses up to m/z 100000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 11.Whitehouse CM, Dreyer RN, Yamashuta M, Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal. Chem. 1985;57:675–679. doi: 10.1021/ac00280a023. [DOI] [PubMed] [Google Scholar]
- 12.Wittmer D, Luckenbill BK, Hill HH, Chen YH. Electrospray-ionization ion mobility spectrometry. Anal. Chem. 1994;66:2348–2355. [Google Scholar]
- 13.Clemmer DE, Hudgins RR, Jarrold MF. Naked protein conformations: cytochrome c in the gas phase. J. Am. Chem. Soc. 1995;117:10141–10142. [Google Scholar]
- 14.von Helden G, Wyttenbach T, Bowers MT. Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science. 1995;267:1483–1485. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 15.von Helden G, Wyttenbach T, Bowers MT. Inclusion of a MALDI ion-source in the ion chromatography technique: conformational information on polymer and biomolecular ions. Int. J. Mass Spectrom. Ion Process. 1995;146:349–364. [Google Scholar]
- 16.Gillig KJ, Ruotolo B, Stone EG, Russell DH, Fuhrer K, et al. Coupling high-pressure MALDI with ion mobility/orthogonal time-of flight mass spectrometry. Anal. Chem. 2000;72:3965–3971. doi: 10.1021/ac0005619. [DOI] [PubMed] [Google Scholar]
- 17.Mack E. Average cross-sectional areas of molecules by gaseous diffusion methods. J. Am. Chem. Soc. 1925;47:2468–2482. [Google Scholar]
- 18.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem. Phys. Lett. 1996;261:86–91. [Google Scholar]
- 19.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem. 1996;100:16082–16086. [Google Scholar]
- 20.Wyttenbach T, von Helden G, Batka JJ, Carlat D, Bowers MT. Effect of the long-range potential on ion mobility measurements. J. Am. Chem. Soc. 1997;8:275–282. [Google Scholar]
- 21.Shvartsburg AA, Hudgins RR, Dugourd P, Jarrold MF. Sturctural information from ion mobility measurements: applications to semiconductor clusters. Chem. Soc. Rev. 2001;30:26–35. [Google Scholar]
- 22.Shaffer SA, Tang KQ, Anderson GA, Prior DC, Udseth HR, Smith RD. A novel ion funnel for focusing ions at elevated pressure using electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1997;11:1813–1817. [Google Scholar]
- 23.Tang K, Shvartsburg AA, Lee HN, Prior DC, Buschbach MA, Li FM, et al. High-sensitivity ion mobility spectrometry/mass spectrometry using electrodynamic ion funnel interfaces. Anal. Chem. 2005;77:3330–3339. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang KQ, Li FM, Shvartsburg AA, Strittmatter EF, Smith RD. Two-dimensional gas-phase separations coupled to mass spectrometry for analysis of complex mixtures. Anal. Chem. 2005;77:6381–6388. doi: 10.1021/ac050871x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentine SJ, Kulchania M, Barnes CA, Clemmer DE. Multidimensional separations of complex peptide mixtures: a combined high-performance liquid chromatography/ion mobility/time-of-flight mass spectrometry approach. Int. J. Mass Spectrom. 2001;212:97–109. [Google Scholar]
- 26.Taraszka JA, Kurulagama R, Sowell RA, Valentine SJ, Koeniger SL, et al. Mapping the proteome of Drosphila melanogaster: analysis of embryos and adult heads by LC-ion mobility-MS methods. J. Proteome Res. 2005;4:1223–1237. doi: 10.1021/pr050038g. [DOI] [PubMed] [Google Scholar]
- 27.Valentine SJ, Liu XY, Plasencia MD, Hilderbrand AE, Kurulugama RT, et al. Developing liquid chromatography ion mobility mass spectrometry techniques. Expert Rev. Proteomics. 2005;2:553–565. doi: 10.1586/14789450.2.4.553. [DOI] [PubMed] [Google Scholar]
- 28.Venne K, Bonneil E, Eng K, Thibault P. Improvement in peptide detection for proteomics analyses using nanoLC-MS and high-field asymmetry waveform ion mobility mass spectrometry. Anal. Chem. 2005;77:2176–2186. doi: 10.1021/ac048410j. [DOI] [PubMed] [Google Scholar]
- 29.Valentine SJ, Plasencia MD, Liu X, Krishnan M, Naylor S, et al. Toward plasma proteome profiling with ion mobility-mass spectrometry. J. Proteome Res. 2006;5:2977–2984. doi: 10.1021/pr060232i. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Valentine SJ, Plasencia MD, Trimpin S, Naylor S, Clemmer DE. Mapping the human plasma proteome by SCX-LC-IMS-MS. J. Am. Soc. Mass Spectrom. 2007;18:1249–1264. doi: 10.1016/j.jasms.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller RA, Eiceman GA, Nazarov EG, King AT. A novel micromachined high-field asymmetric waveform-ion mobility spectrometer. Sens. Actuators B. 2000;67:300–306. [Google Scholar]
- 32.Guevremont R, Barnett DA, Purves RW, Vandermey J. Analysis of a tryptic digest of pig hemoglobin using ESI-FAIMS-MS. Anal. Chem. 2000;72:4577–4584. doi: 10.1021/ac0000271. [DOI] [PubMed] [Google Scholar]
- 33.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, et al. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int. J. Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 34.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 35.Loo JA, Berhane B, Kaddis CS, Wooding KM, Xie YM, et al. Electrospray ionization mass spectrometry and ion mobility analysis of the 20S proteasome complex. J. Am. Soc. Mass Spectrom. 2005;16:998–1008. doi: 10.1016/j.jasms.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Chowdhury SK, Katta V, Chait BT. Probing conformational changes in proteins by mass spectrometry. J. Am. Chem. Soc. 1990;112:9012–9013. [Google Scholar]
- 37.Suckau D, Shi Y, Beu SC, Senko MW, Quinn JP, et al. Coexisting stable conformations of gaseous protein ions. Proc. Natl. Acad. Sci. USA. 1993;90:790–793. doi: 10.1073/pnas.90.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loo RRO, Smith RD. Investigation of the gas-phase structure of electrosprayed proteins using ion-molecule reactions. J. Am. Soc. Mass Spectrom. 1994;5:207–220. doi: 10.1016/1044-0305(94)85011-9. [DOI] [PubMed] [Google Scholar]
- 39.Wolynes PG. Biomolecular folding in vacuo. Proc. Natl. Acad. Sci. USA. 1995;92:2426–2427. doi: 10.1073/pnas.92.7.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross DS, Schnier PD, Rodriguez-Cruz SE, Fagerquist CK, Williams ER. Conformation and folding of lysozyme ions in vacuo. Proc. Natl. Acad. Sci. USA. 1996;93:3143–3148. doi: 10.1073/pnas.93.7.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams ER. Proton transfer reactivity of large multiply charged ions. J. Mass Spectrom. 1996;31:831–842. doi: 10.1002/(SICI)1096-9888(199608)31:8<831::AID-JMS392>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shelimov KB, Clemmer DE, Hudgins RR, Jarrold MF. Protein structure in vacuo: gas-phase conformations of BPTI and cytochrome c. J. Am. Chem. Soc. 1997;119:2240–2248. [Google Scholar]
- 43.Hudgins RR, Woenckhaus J, Jarrold MF. High resolution ion mobility measurements for gas phase proteins: correlation between solution phase and gas phase conformations. Int. J. Mass Spectrom. Ion Process. 1997;165:497–507. [Google Scholar]
- 44.Gross DS, Zhao YX, Williams ER. Dissociation of heme-globin complexes by blackbody infrared radiative dissociation: molecular specificity in the gas phase? J. Am. Soc. Mass Spectrom. 1997;8:519–524. doi: 10.1016/S1044-0305(97)00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirza UA, Chait BT. Do proteins denature during droplet evolution in electrospray ionization? Int. J. Mass Spectrom. Ion Process. 1997;162:173–181. [Google Scholar]
- 46.Hoaglund-Hyzer CS, Counterman AE, Clemmer DE. Anhydrous protein ions. Chem. Rev. 1999;99:3037–3079. doi: 10.1021/cr980139g. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Taraszka JA, Counterman AE, Clemmer DE. Influence of solvent composition and capillary temperature on the conformations of electrosprayed ions: unfolding of compact ubiquitin conformers from pseudonative and denatured solutions. Int. J. Mass Spectrom. Ion Process. 1999;185–187:37–47. [Google Scholar]
- 48.Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular protein structure. Proteins. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 49.Dill KA, Fiebig KM, Chan HS. Cooperativity in protein folding kinetics. Proc. Natl. Acad. Sci. USA. 1993;90:1942–1946. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth H, et al. An IMS-IMS analogue of MS-MS. Anal. Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 51.Merenbloom SI, Koeniger SL, Valentine SJ, Plasencia MD, Clemmer DE. IMS-IMS and IMS-IMS-IMS/MS for separating peptide and protein fragment ions. Anal. Chem. 2006;78:2802–2809. doi: 10.1021/ac052208e. [DOI] [PubMed] [Google Scholar]
- 52.Koeniger SL, Merenbloom SI, Clemmer DE. Evidence for many resolvable structures within conformation types of electrosprayed ubiquitin ions. J. Phys. Chem. B. 2006;110:7017–7021. doi: 10.1021/jp056165h. [DOI] [PubMed] [Google Scholar]
- 53.Koeniger SL, Merenbloom SI, Sevugarajan S, Clemmer DE. Transfer of structural elements from compact to extended states in unsolvated ubiquitin. J. Am. Chem. Soc. 2006;128:11713–11719. doi: 10.1021/ja062137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koeniger SL, Clemmer DE. Resolution and structural transitions of elongated states of ubiquitin. J. Am. Soc. Mass Spectrom. 2007;18:322–331. doi: 10.1016/j.jasms.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 55.Merenbloom SI, Bohrer BC, Koeniger SL, Clemmer DE. Assessing the peak capacity of IMS-IMS separations of tryptic peptide ions in 300 K He. Anal. Chem. 2007;79:515–522. doi: 10.1021/ac061567m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 57.Harrison AG. The gas-phase basicities and proton affinities of amino acids and peptides. Mass Spectrom. Rev. 1997;16:201–217. [Google Scholar]
- 58.McLuckey SA, Goeringer DE. Slow heating methods in tandem mass spectrometry. J. Mass Spectrom. 1997;32:461–474. [Google Scholar]
- 59.Green MK, Lebrilla CB. Ion-molecule reactions as probes of gas-phase structures of peptides and proteins. Mass Spectrom. Rev. 1997;16:53–71. doi: 10.1002/(SICI)1098-2787(1997)16:2<53::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 60.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 61.Marshall AG, Grosshans PB. Fourier transform ion cyclotron resonance mass spectrometry: the teenage years. Anal. Chem. 1991;63:A215–A259. [Google Scholar]
- 62.March RE. An introduction to quadrupole ion trap mass spectrometry. J. Mass Spectrom. 1997;32:351–369. [Google Scholar]
- 63.Gorg A, Obermaier C, Boguth G, Harder A, Scheibe B, et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 64.Wang H, Hanash S. Multi-dimensional liquid phase based separations in proteomics. J. Chromatogr. B. 2003;787:11–18. doi: 10.1016/s1570-0232(02)00335-5. [DOI] [PubMed] [Google Scholar]
- 65.Aebersold R, Mann M. Mass spectrometry–based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 66.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:535–542. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 68.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 69.Bax A. Two-dimensional NMR and protein structure. Annu. Rev. Biochem. 1989;58:223–256. doi: 10.1146/annurev.bi.58.070189.001255. [DOI] [PubMed] [Google Scholar]
- 70.Wüthrich K. The development of nuclear magnetic resonance spectroscopy as a technique for protein structure determination. Acc. Chem. Res. 1989;22:36–44. [Google Scholar]
- 71.Lipscomb WN. Experimental crystallography. Annu. Rev. Phys. Chem. 1953;4:253–266. [Google Scholar]
- 72.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North ACT. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5 Å resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 73.Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, et al. Structure of myoglobin: a three-dimensional Fourier synthesis at 2 Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- 74.Blundell TL, Johnson LN. Protein Crystallography. New York: Academic; 1976. p. 56. [Google Scholar]
- 75.Lumry R, Eyring H. Conformation changes of proteins. J. Phys. Chem. 1954;58:110–120. [Google Scholar]
- 76.Levinthal C. Are there pathways for protein folding? J. Chim. Phys. 1968;65:44–45. [Google Scholar]
- 77.Anfinsen CB. Principles that govern folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 78.Roder H, Elöve GA, Englander SW. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988;335:700–704. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goto Y, Hagihara Y, Hamada D, Hoshino M, Nishii I. Acid-induced unfolding and refolding transitions of cytochrome c: a three-state mechanism in H2O and D2O. Biochemistry. 1993;32:11878–11885. doi: 10.1021/bi00095a017. [DOI] [PubMed] [Google Scholar]
- 80.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas AF, Gilmanshin RI. Study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 81.Mirza UA, Cohen SL, Chait BT. Heat-induced conformational changes in proteins studied by electrospray ionization mass spectrometry. Anal. Chem. 1993;65:1–6. doi: 10.1021/ac00049a003. [DOI] [PubMed] [Google Scholar]
- 82.Bai YW, Milne JS, Mayne L, Englander SW. Protein stability parameter measured by hydrogen exchange. Proteins. 1994;20:4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- 83.Dobson CM, Sali A, Karplus M. Protein folding: a perspective from theory and experiment. Angew. Chem. Int. Ed. Engl. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 84.Ohgushi M, Wada A. ‘Molten-globule’ state: a compact form of globular proteins with mobile side-chains. FEBS Lett. 1983;164:21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- 85.Makhatadze GI, Privalov PL. Energetics of protein structure. Adv. Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 86.Lazaridis T, Archontis G, Karplus M. Enthalpic contribution to protein stability: insights from atom-based calculations and statistical mechanics. Adv. Protein Chem. 1995;47:231–306. doi: 10.1016/s0065-3233(08)60547-1. [DOI] [PubMed] [Google Scholar]
- 87.Woenckhaus J, Mao Y, Jarrold MF. Hydration of gas phase proteins: folded +5 and unfolded +7 charge states of cytochrome c. J. Phys. Chem. B. 1997;101:847–851. [Google Scholar]
- 88.Mao Y, Woenkhaus J, Kolafa J, Ratner MA, Jarrold MF. Thermal unfolding of unsolvated cytochrome c: experiment and molecular dynamics simulations. J. Am. Chem. Soc. 1999;121:2712–2721. [Google Scholar]
- 89.van Gunsteren WF, Karplus M. Protein dynamics in solution and in a crystalline environment: a molecular dynamics study. Biochemistry. 1982;21:2259–2274. doi: 10.1021/bi00539a001. [DOI] [PubMed] [Google Scholar]
- 90.Levitt M, Sharon R. Accurate simulation of protein dynamics in solution. Proc. Natl. Acad. Sci. USA. 1988;85:7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. New York: Wiley & Sons; 1988. p. 560. [Google Scholar]
- 92.Counterman AE, Hilderbrand AE, Srebalus Barnes CA, Clemmer DE. Formation of peptide aggregates during ESI: size, charge, composition, and contributions to noise. J. Am. Soc. Mass Spectrom. 2001;12:1020–1035. [Google Scholar]
- 93.Revercomb HW, Mason EA. Theory of plasma chromatography/gaseous electrophoresis: a review. Anal. Chem. 1975;47:970–983. [Google Scholar]
- 94.Buryakov IA, Krylov EV, Nazarov EG, Rasulev UK. A new method of separation of multi-atomic ions by mobility at atmospheric pressure using a high-frequency amplitude-asymmetric strong electric field. Int. J. Mass Spectrom. Ion Process. 1993;128:143–148. [Google Scholar]
- 95.Purves RW, Guevremont R, Day S, Pipich CW, Matyjaszczyk MS. Mass spectrometric characterization of a high-field asymmetric waveform ion mobility spectrometer. Rev. Sci. Instrum. 1998;69:4094–4105. [Google Scholar]
- 96.Guevremont R, Purves RW. Atmospheric pressure ion focusing in a high-field asymmetric waveform ion mobility spectrometer. Rev. Sci. Instrum. 1999;70:1370–1383. [Google Scholar]
- 97.Shvartsburg AA, Mashkevich SV, Smith RD. Feasibility of higher-order differential ion mobility separations using new asymmetric waveforms. J. Phys. Chem. A. 2006;110:2663–2673. doi: 10.1021/jp055349t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shvartsburg AA, Bryskiewicz T, Purves RW, Tang KQ, Guevremont R, Smith RD. Field asymmetric waveform ion mobility spectrometry studies of proteins: dipole alignment in ion mobility spectrometry? J. Phys. Chem. B. 2006;110:21966–21980. doi: 10.1021/jp062573p. [DOI] [PubMed] [Google Scholar]
- 99.Moseley JT, Gatland IR, Martin DW, McDaniel EW. Measurement of transport properties of ions in gases: results for K+ ions in N2. Phys. Rev. 1969;178:234–239. [Google Scholar]
- 100.Knorr FJ, Eatherton RL, Siems WF, Hill HH. Fourier transform ion mobility spectrometry. Anal. Chem. 1985;57:402–406. doi: 10.1021/ac50001a018. [DOI] [PubMed] [Google Scholar]
- 101.Szulmas AW, Ray SJ, Hieftje GM. Hadamard transform ion mobility spectrometry. Anal. Chem. 2006;78:4474–4481. doi: 10.1021/ac051743b. [DOI] [PubMed] [Google Scholar]
- 102.Belov ME, Buschbach MA, Prior DC, Tang KQ, Smith RD. Multiplexed ion mobility spectrometry-orthogonal time-of-flight mass spectrometry. Anal. Chem. 2007;79:2451–2462. doi: 10.1021/ac0617316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a traveling wave-based radio frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 104.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Ambient mass spectrometry. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 105.Gieniec J, Mack LL, Nakamae K, Gupta C, Kumar V, Dole M. Electrospray mass spectroscopy of macromolecules: application of an ion-drift spectrometer. Biomed. Mass Spectrom. 1984;11:259–268. [Google Scholar]
- 106.Myung S, Wiseman JM, Valentine SJ, Zoltán T, Cooks RG, Clemmer DE. Coupling desorption electrospray ionization (DESI) with ion mobility/mass spectrometry for analysis of protein structure: evidence for desorption of folded and denatured states. J. Phys. Chem. B. 2006;110:5045–5051. doi: 10.1021/jp052663e. [DOI] [PubMed] [Google Scholar]
- 107.Bluhm BK, Gillig KJ, Russell DH. Development of a Fourier-transform ion cyclotron mass spectrometer-ion mobility spectrometer. Rev. Sci. Instrum. 2000;71:4078–4086. [Google Scholar]
- 108.Lawrence AH, Barbour RJ, Sutcliffe R. Identification of wood species by ion mobility spectrometry. Anal. Chem. 1991;63:1217–1221. [Google Scholar]
- 109.Valentine SJ, Clemmer DE. H/D exchange levels of shape-resolved cytochrome c conformers in the gas phase. J. Am. Chem. Soc. 1997;119:3558–3566. [Google Scholar]
- 110.Creaser CS, Benyezzar M, Griffiths JR, Stygall JW. A tandem ion trap/ion mobility spectrometer. Anal. Chem. 2000;72:2724–2729. doi: 10.1021/ac991409d. [DOI] [PubMed] [Google Scholar]
- 111.Clowers BH, Hill HH. Mass analysis of mobility-selected ion populations using dual gate, ion mobility, quadrupole ion trap mass spectrometry. Anal. Chem. 2005;77:5877–5885. doi: 10.1021/ac050700s. [DOI] [PubMed] [Google Scholar]
- 112.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-dimensional ion mobility TOFMS analysis of electrosprayed biomolecules. Anal. Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 113.Howorka F, Fehsenfeld FC, Albritton DL. H+ and D+ ions in He: observations of a runaway mobility. J. Phys. B. 1979;12:4189–4197. [Google Scholar]
- 114.Dugourd Ph, Hudgins RR, Clemmer DE, Jarrold MF. High-resolution ion mobility measurements. Rev. Sci. Instrum. 1997;68:1122–1129. [Google Scholar]
- 115.Valentine SJ, Clemmer DE. H/D exchange levels of shape-resolved cytochrome c conformers in the gas phase. J. Am. Chem. Soc. 1997;119:3558–3566. [Google Scholar]
- 116.Valentine SJ, Counterman AE, Clemmer DE. A database of 660 peptide ion cross sections: use of intrinsic size parameters for bona fide predictions of cross sections. J. Am. Soc. Mass Spectrom. 1999;10:1188–1211. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 117.Counterman AE, Clemmer DE. Volumes of individual amino acid residues in gas-phase peptide ions. J. Am. Chem. Soc. 1999;121:4031–4039. [Google Scholar]
- 118.Valentine SJ, Counterman AE, Hoaglund CS, Reilly JP, Clemmer DE. Gas-phase separations of protease digests. J. Am. Soc. Mass Spectrom. 1998;9:1213–1216. doi: 10.1016/S1044-0305(98)00101-9. [DOI] [PubMed] [Google Scholar]
- 119.Woods AS, Ugarov M, Egan T, Koomen J, Gillig KJ, et al. Lipid/peptide/nucleotide separation with MALDI-ion mobility-TOF MS. Anal. Chem. 2004;76:2187–2195. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- 120.Jackson SN, Wang HYJ, Woods AS. Direct tissue analysis of phospholipids in rat brain using MALDI-TOFMS and MALDI-ion mobility-TOFMS. J. Am. Soc. Mass Spectrom. 2005;16:133–138. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 121.Dwivedi P, Bendiak B, Clowers BH, Hill HH. Rapid resolution of carbohydrate isomers by electrospray ionization ambient pressure ion mobility spectrometry-time-of-flight mass spectrometry (ESI-APIMS-TOF) J. Am. Soc. Mass Spectrom. 2007;18:1163–1175. doi: 10.1016/j.jasms.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 122.Ruotolo BT, Verbeck GF, Thomson LM, Woods AS, Gillig KJ, Russell DH. Distinguishing between phosphorylated and nonphosphorylated peptides with ion mobility mass spectrometry. J. Proteome Res. 2002;1:303–306. doi: 10.1021/pr025516r. [DOI] [PubMed] [Google Scholar]
- 123.Hoaglund-Hyzer CS, Li JW, Clemmer DE. Mobility labeling for parallel CID of ion mixtures. Anal. Chem. 2000;72:2737–2740. doi: 10.1021/ac0000170. [DOI] [PubMed] [Google Scholar]
- 124.Hoaglund-Hyzer CS, Clemmer DE. Ion trap/ion mobility/quadrupole/time of flight mass spectrometry for peptide mixture analysis. Anal. Chem. 2001;73:177–184. doi: 10.1021/ac0007783. [DOI] [PubMed] [Google Scholar]
- 125.Stone E, Gillig KJ, Ruotolo B, Fuhrer K, Gonin M, et al. Surface-induced dissociation on a MALDI-ion mobility-orthogonal time-of-flight mass spectrometer: sequencing peptides from an “in-solution” protein digest. Anal. Chem. 2001;73:2233–2238. doi: 10.1021/ac001430a. [DOI] [PubMed] [Google Scholar]
- 126.Sun WJ, May JC, Russell DH. A novel surface-induced dissociation instrument for ion mobility-time-of-flight mass spectrometry. Int. J. Mass Spectrom. 2007;259:79–86. [Google Scholar]
- 127.Lee YJ, Hoaglund-Hyzer CS, Taraszka JA, Zientara GA, Counterman AE, Clemmer DE. Collision-induced dissociation of mobility-separated ions using an orifice-skimmer cone at the back of a drift tube. Anal. Chem. 2001;73:3549–3555. doi: 10.1021/ac010295e. [DOI] [PubMed] [Google Scholar]
- 128.Steiner WE, Clowers BH, Fuhrer K, Gonin M, Matz LM, et al. Electrospray ionization with ambient pressure ion mobility separation and mass analysis by orthogonal time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2001;15:2221–2226. doi: 10.1002/rcm.495. [DOI] [PubMed] [Google Scholar]
- 129.Valentine SJ, Koeniger SL, Clemmer DE. A split-field drift tube for separation and efficient fragmentation of biomolecular ions. Anal. Chem. 2003;75:6202–6208. doi: 10.1021/ac030111r. [DOI] [PubMed] [Google Scholar]
- 130.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 131.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations: a nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 132.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen W, Olivova P, Doneanu CE, Gebler JC. A novel approach for identification and characterization of glycoproteins using a quadrupole ion-mobility time-of-flight mass spectrometer; Presented at Annu. Meet. Am. Soc. Mass Spectrom.55th; Indianapolis. 2007. [Google Scholar]
- 134.Eng JK, McCormack AL, Yates JRI. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 135.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 136.Asbury GR, Hill HH. Using different drift gases to change separation factors (α) in ion mobility spectrometry. Anal. Chem. 2000;72:580–584. doi: 10.1021/ac9908952. [DOI] [PubMed] [Google Scholar]
- 137.Ruotolo BT, McLean JA, Gillig KJ, Russell DH. Peak capacity of ion mobility mass spectrometry: the utility of varying drift gas polarizability for the separation of tryptic peptides. J. Mass Spectrom. 2004;39:361–367. doi: 10.1002/jms.592. [DOI] [PubMed] [Google Scholar]
- 138.Shelimov KB, Jarrold MF. Conformations, unfolding, and refolding of apomyoglobin in vacuum: an activation barrier for gas-phase protein folding. J. Am. Chem. Soc. 1997;119:2987–2994. [Google Scholar]
- 139.Freitas MA, Hendrickson CL, Emmett MR, Marshall AG. Gas-phase bovine ubiquitin cation conformations resolved by gas-phase hydrogen/deuterium exchange rate and extent. Int J Mass Spectrom. 1999;187:565–575. [Google Scholar]
- 140.Badman ER, Hoaglund-Hyzer CS, Clemmer DE. Monitoring structural changes of proteins in an ion trap over ~10–200 ms: unfolding transitions in cytochrome c ions. Anal. Chem. 2001;73:6000–6007. doi: 10.1021/ac010744a. [DOI] [PubMed] [Google Scholar]
- 141.Myung S, Badman ER, Young JL, Clemmer DE. Structural transitions of electrosprayed ubiquitin ions stored in an ion trap over ~10 ms to 30 s. J. Phys. Chem. A. 2002;106:9976–9982. [Google Scholar]
- 142.Badman E, Myung S, Clemmer DE. Evidence for unfolding and refolding of gas phase cytochrome c ions in a Paul trap. J. Am. Soc. Mass Spectrom. 2005;16:1493–1497. doi: 10.1016/j.jasms.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 143.Giddings JC. Concepts and comparisons in multidimensional separation. J. High Resolut. Chromatogr. Chromatogr. Commun. 1987;10:319–323. [Google Scholar]
- 144.Giddings JC. Unified Separation Science. New York: Wiley; 1991. Steady-state, two-dimensional, and overlapping zones; p. 112.p. 140.p. 352. [Google Scholar]
- 145.Julian RR, Beauchamp JL. Site specific sequestering and stabilization of charge in peptides by supramolecular adduct formation with 18-crown-ether by way of electrospray ionization. Int J Mass Spectrom. 2001;210:613–623. [Google Scholar]
- 146.Colgrave ML, Bramwell CJ, Creaser CS. Nanoelectrospray ion mobility spectrometry and ion trap mass spectrometry studies of the noncovalent complexes of amino acids and peptides with polyethers. Int J Mass Spectrom. 2003;229:209–216. [Google Scholar]
- 147.Ly T, Julian RR. Using ESI-MS to probe protein structure by site-specific noncovalent attachment of 18-crown-6. J. Am. Soc. Mass Spectrom. 2006;17:1209–1215. doi: 10.1016/j.jasms.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 148.Hilderbrand AE, Myung S, Clemmer DE. Exploring crown ethers as shift reagents for ion mobility spectrometry. Anal. Chem. 2006;78:6792–6800. doi: 10.1021/ac060439v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iavarone AT, Patriksson A, van der Spoel D, Parks JH. Fluorescence probe of Trp-cage protein conformation in solution and in gas phase. J. Am. Chem. Soc. 2007;129:6726–6735. doi: 10.1021/ja065092s. [DOI] [PubMed] [Google Scholar]