Abstract
Interactions between hosts and pathogens are complex, so understanding the events that govern these interactions requires the analysis of molecular mechanisms operating in both organisms. Many pathogens use multiple strategies to target a single event in the disease process, confounding the identification of the important determinants of virulence. We developed a genetic screening strategy called insertional Mutagenesis And Depletion (iMAD) that combines bacterial mutagenesis and RNA interference, to systematically dissect the interplay between a pathogen and its host. We used this technique to resolve the network of proteins secreted by the bacterium Legionella pneumophila to promote intracellular growth, a critical determinant of pathogenicity of this organism. This strategy is broadly applicable, allowing the dissection of any interface between two organisms involving numerous interactions.
The outcome of most parasitic relationships is decided by a series of molecular interactions involving hundreds of proteins. Through specialized secretion systems, intracellular pathogens deploy an arsenal of proteins that modulate numerous host cell processes to establish growth (1). For many pathogens, redundancy complicates the analysis of these secreted virulence proteins (2), and no systematic strategy exists that allows specific roles to be ascribed to proteins that, based on mutant analysis, appear to be dispensable for growth within the host.
Legionella pneumophila is a parasite of a broad range of free-living amoebae (3). Human disease occurs after inhalation of contaminated water aerosols followed by replication of the bacteria in alveolar macrophages (4) which results in pneumonia. Within the host cell, L. pneumophila establishes replication within a membrane-bound compartment by preventing the delivery of this vacuole to the lysosome (5), while recruiting host material from the endoplasmic reticulum (ER) (6). This latter event is accomplished by manipulating host proteins involved in the early secretory pathway (7), including the small GTPases Rab1, Arf1 and Sar1 as well as Sec22 and Bet5, proteins involved in tethering and fusion of ER-derived vesicles to target membranes (7–9).
Intracellular replication of L. pneumophila depends on the Dot/Icm Type IVb secretion system (10, 11). To date, over 270 L. pneumophila proteins have been shown to be substrates of Dot/Icm (12), several of which directly modulate the activity of GTPases that control host vesicle trafficking. Although the biochemical activity of more than a dozen Dot/Icm translocated substrates (TS) and their host targets are known, their roles in disease are unclear, because deletion of their encoding genes rarely causes a detectable defect in intracellular growth (13). The most likely explanation for this is redundancy, which extends beyond multiple paralogs that can perform the same function (13, 14) to the use of seemingly unrelated proteins to manipulate complementary host cell pathways (9). Although this has impeded understanding how these proteins contribute to pathogenesis, there has been no attempt to systematically resolve this problem.
Studies using RNA interference (RNAi) in cultured Drosophila cells demonstrate that wild-type L. pneumophila grow efficiently in cells depleted of one of the membrane trafficking proteins Arf1, Sec22 or Bet5 (9); however, simultaneous depletion of pairs of these proteins impairs intracellular replication of the bacterium (9). These data are consistent with several different pathways promoting intracellular replication, with bacterial growth inhibition requiring disruption of at least two pathways. We reasoned that mutation of a bacterial gene encoding a Dot/Icm TS that targets a host pathway distinct from one targeted by RNAi should effectively abolish the contribution of both pathways (Fig. S1).
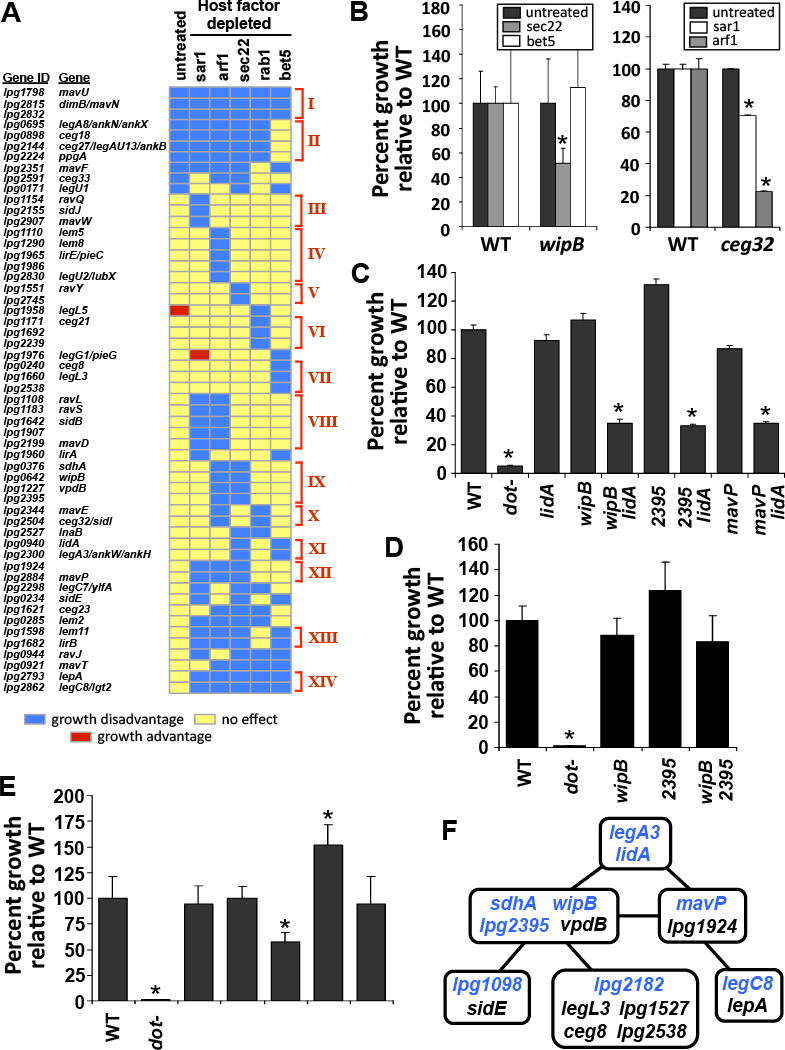
Using transposon site hybridization (TraSH) (15), a library of L. pneumophila transposon mutants (16) was screened for mutations that render the bacterium defective for growth in Drosophila cells depleted of various component proteins of the early secretory system (Fig. S1). A total of 678 genes were identified as being important for intracellular growth in Drosophila cells in the presence of dsRNA treatment (Table S1). Many of these encode proteins that are not predicted to function in the host cytoplasm. For example, several genes encode proteins involved in nutrient acquisition and metabolism (Table S1), consistent with disruptions in vacuole remodeling limiting the availability of metabolites required for growth. To address the inability to connect phenotypes with mutations in Dot/Icm TS genes, one of the central problems in the field, we decided to focus on the 55 Dot/Icm TS-encoding genes identified (Fig. 1A), 44 of which had not previously been shown to have a role in promoting intracellular growth of L. pneumophila.
Figure 1. iMAD identifies redundant relationships between Dot/Icm translocated substrates.
(A) Defective intracellular growth of L. pneumophila resulting from mutations in Dot/Icm TS genes combined with depletion of host cell proteins associated with the early secretory pathway. Cultured Drosophila cells treated with dsRNA were challenged with a library of L. pneumophila transposon mutants and the relative intracellular growth under each depletion condition was determined by TraSH. A growth disadvantage is defined as a 3.7 ± 0.2-fold growth defect, equivalent to an average TraSH ratio > 1.75 ± 0.1 standard deviations from the mean, which varies depending on the dsRNA treatment. Individual genes are clustered into distinct functional groups (red brackets, I-XIV) based on common behavioral patterns across all host conditions examined. (B) Host-condition specific growth defects for L. pneumophila null mutants. Growth of wipB (lpg0642) (Group IX) (left panel) and ceg32/sidI (lpg2504) (Group X) (right panel) null mutants in cultured Drosophila cells depleted of the indicated proteins by RNAi was compared to the wild-type (WT) strain. (C) The combined deletion of bacterial genes from separate functional groups impaired intracellular replication of L. pneumophila in untreated Drosophila cells. (D) Deletion of genes belonging to the same functional group did not adversely affect bacterial growth in untreated Drosophila cells. (E) Genetic interactions define distinct relationships between different functional groups. (F) Summary of redundant relationships between individual functional groups defined by genetic interaction mapping. A solid black line indicates aggravating genetic interactions on intracellular growth of the bacterium in untreated Drosophila cells when two L. pneumophila genes, one from each of the corresponding functional groups (indicated by blue lettering), are deleted in combination. (B–E) Bacterial growth was determined by colony forming units (cfus) on solid media from lysed host cells 48 hours post infection relative to the number of cfus recovered 1 hour post infection. Data are means from at least 2 independent experiments ± standard deviation of 3 replicates. *p <0.05 relative to the wild-type (WT) strain.
Data from the iMAD screen could be recapitulated for selected genes when Drosophila cells treated with the appropriate dsRNA were challenged with L. pneumophila deletion mutants (Fig. 1B). For example, a L. pneumophila ΔwipB (lpg0642) mutant was defective for growth in cells depleted of Sec22 but not in untreated Drosophila cells or in cells depleted of Bet5, as predicted from our screen. Similarly, growth of Δceg32/sidI (lpg2504) was more defective in cells depleted of Arf1 than in untreated cells or cells depleted of Sar1 (Fig. 1B). Thus, we have mapped a series of aggravating genetic interactions (17) that identify specific host factors required for intracellular replication of bacterial mutants. These results defined host conditions under which individual bacterial proteins were important for growth and provided the basis for identifying Dot/Icm TS that function in complementary host pathways.
We predicted that if a set of Dot/Icm TS targets a particular host pathway, mutations in individual members of that set should result in similar phenotypes. Our iMAD screen examined growth of L. pneumophila mutants in Drosophila cells under five different conditions of dsRNA treatment. Using these data, we performed cluster analysis to group bacterial mutations based on common behavior patterns. This identified 14 distinct functional groups of Dot/Icm TS with more than one member (Fig. 1A; Groups I–XIV). For example, mutations in the L. pneumophila gene lidA impaired intracellular growth if either Bet5 or Sec22 was depleted, and mutations in legA3 (lpg2300) behaved similarly, placing legA3 and lidA in the same functional group (Fig. 1A; Group XI). Conversely, mutation of wipB caused defective intracellular growth in cells depleted of either Arf1 or Sec22, placing it in a separate functional group (Fig. 1A; Group IX). If two functional groups consist of bacterial proteins that modulate redundant host pathways, we predicted that the combined deletion of one bacterial gene from each functional group should result in defective intracellular replication of L. pneumophila in untreated Drosophila cells. LidA and WipB clustered to separate functional groups predicting that they act on potentially redundant host cell pathways, so growth of a ΔwipBΔlidA double mutant was assessed in untreated Drosophila cells. Consistent with the interpretation that these two proteins target redundant pathways, the double mutant showed reduced intracellular replication in untreated Drosophila cells compared to either wild-type L. pneumophila or strains bearing mutations in only one of these genes (Fig. 1C). Mutations in a newly identified Dot/Icm TS-encoding gene lpg2395 (Fig. S2), clustered to the same functional group as wipB mutations (Fig. 1A: Group IX), and as predicted by this strategy, deletion of lpg2395 impaired growth of L. pneumophila when combined with ΔlidA (Fig. 1C). In contrast, a strain containing lesions in two genes assigned to the same functional group, ΔwipBΔlpg2395, grew as proficiently as the wild-type strain in untreated Drosophila cells, consistent with the model that growth defects are only observed when targeting of multiple host pathways by Dot/Icm TS is disrupted (Fig. 1D). This demonstrated that cluster analysis can predict pairs of seemingly unrelated Dot/Icm TS that perform compensatory functions.
Defective intracellular growth was not limited to strains that combine mutations from functional groups IX and XI. Mutations in mavP (lpg2884) clustered in a third functional group, Group XII (Fig. 1A), and the combined deletion of mavP with lidA similarly impaired bacterial growth in untreated Drosophila cells (Fig. 1C). We also observed specificity between functional groups. For example, reduced growth of L. pneumophila was observed when legC8 (lgt2/lpg2862) (Group XIV) was deleted in combination with mavP (Fig. 1E). However, neither ΔlegC8ΔwipB nor ΔlegC8Δlpg2395 double mutants were attenuated despite loss of gene pairs from separate functional groups (Fig. 1E). Instead, we observed a positive interaction between mutations in wipB and legC8 in which intracellular growth was enhanced by combining the two mutations (Fig. 1E). Thus, cluster analysis of mutations allows functional relationships between bacterial proteins to be revealed.
By analyzing genetic interactions between members of several functional groups, we were able to identify a network of functional relationships between L. pneumophila proteins, in which pairs of mutations in Dot/Icm TS genes were predicted to have aggravating effects on intracellular growth (Fig. 1F, lines represent predicted aggravating genetic interactions). Many of these predicted interactions were verified by constructing defined double deletions (Fig. 1F, blue font). The cluster analysis also assigned genes encoding proteins of unknown function, such as lpg2182 and lpg1098 (Table S1), to specific functional groups (Fig. 1F) allowing these proteins to be assigned to particular steps in intracellular growth, defined by the members of their respective functional groups.
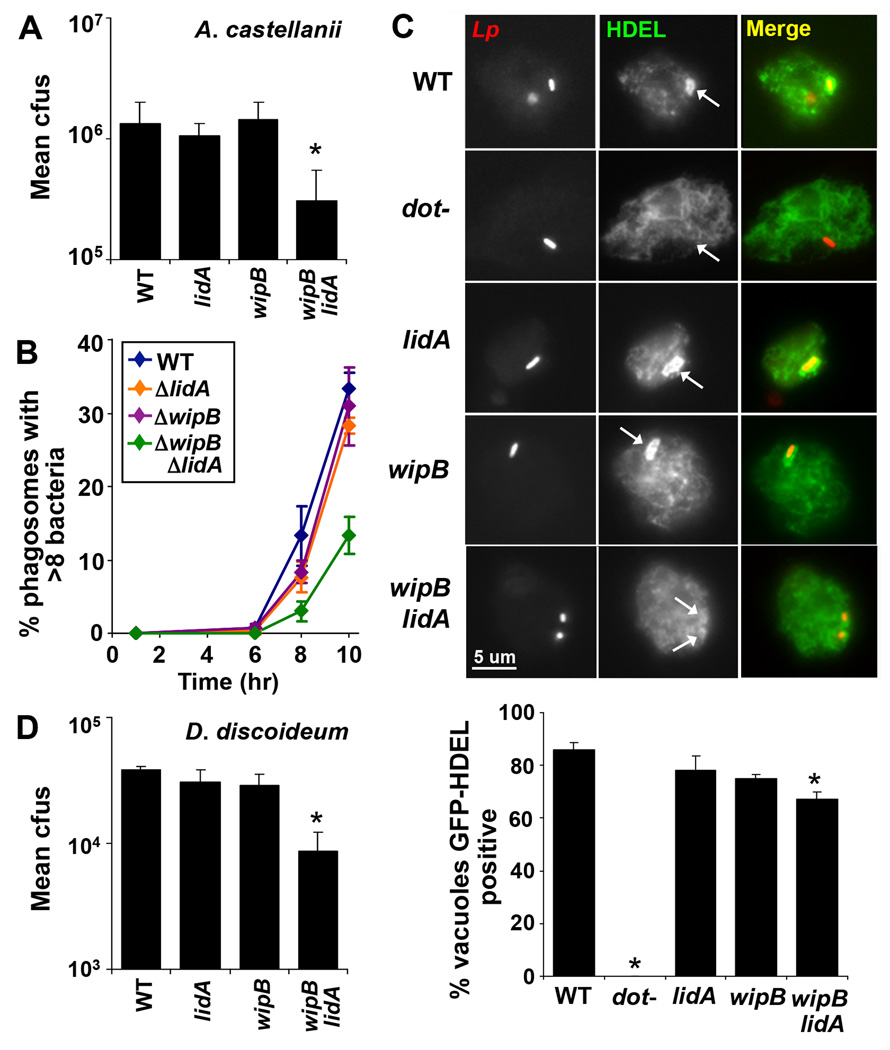
To show that the functional relationships between Dot/Icm TS identified using iMAD were similarly important in a natural host of L. pneumophila, the growth of double mutants in Acanthamoebae castellanii was examined. Each of the double mutants shown to be defective for replication in Drosophila cells exhibited impaired intracellular replication in the amoebal host (Fig. 2A and Fig. S3). Growth of all of the double mutants could be rescued by reintroducing either of the deleted genes individually, and no growth defects were observed for any of the double mutants when grown in bacteriological medium (Fig. S3). A more detailed examination through a single round of infection in A. castellanii revealed that all three double mutants generated vacuoles that contained fewer bacteria than the wild-type strain (Fig. 2B and Fig. S3). This phenotype correlated with inefficient recruitment of ER-derived material, as determined in Dictyostelium discoideum by measuring accumulation of the GFP-HDEL protein about the Legionella-containing vacuole (LCV). GFP-HDEL is a fusion protein of the green fluorescent protein (GFP) and the ER localization signal HDEL, which is used as a proxy for the presence of ER-resident proteins in amoebae (5, 18, 19). In D. discoideum harboring GFP-HDEL, fewer ΔwipBΔlidA-containing vacuoles stained positively for this marker compared to amoebae challenged with either wild-type or bacteria containing a single mutation (Fig. 2C) supporting the model that the activities of WipB and LidA can compensate for each other’s absence. As expected, the ΔwipBΔlidA mutant grew less robustly than either the wild-type or single deletion strains in this host (Fig. 2D). These results are consistent with at least two routes for generating a replication compartment that are targeted by L. pneumophila proteins in all the host cells examined. The partial defect in GFP-HDEL recruitment and intracellular growth of the ΔwipBΔlidA mutant likely reflects the existence of additional pathways that can partially compensate for loss of WipB and LidA.
Figure 2. Impaired growth of L. pneumophila mutants in natural hosts.
(A) The ΔwipBΔlidA mutant shows a growth defect in A. castellanii relative to the wild-type (WT) and corresponding single deletion strains. (B) The ΔwipBΔlidA mutant is impaired in the accumulation of vacuoles containing large numbers of bacteria. A. castellanii were infected with L. pneumophila strains expressing GFP, fixed 1, 6, 8 or 10 hours post infection and the number of bacteria per phagosome was scored using fluorescence microscopy. (C) The ΔwipBΔlidA mutant is defective for recruitment of the ER-targeted fusion protein GFP-HDEL in D. discoideum. D. discoideum expressing GFP-HDEL were infected with L. pneumophila strains expressing the red fluorescent protein tdTomato for 4 hours, fixed and visualized by fluorescence microscopy (upper panel). The number of GFP-HDEL positive Legionella vacuoles was scored, counting 100 vacuoles per replicate (lower panel). (D) Defective intracellular growth of the ΔwipBΔlidA mutant in D. discoideum. (A, D) A. castellanii and D. discoideum were infected with L. pneumophila and bacterial growth was monitored as described in Fig. 1 over 3 days, equivalent to 3 consecutive rounds of replication and plotted as in Fig. 1. Data are representative of at least 2 independent experiments ± standard deviation of 3 replicates. *p <0.05 relative to the wild-type strain.
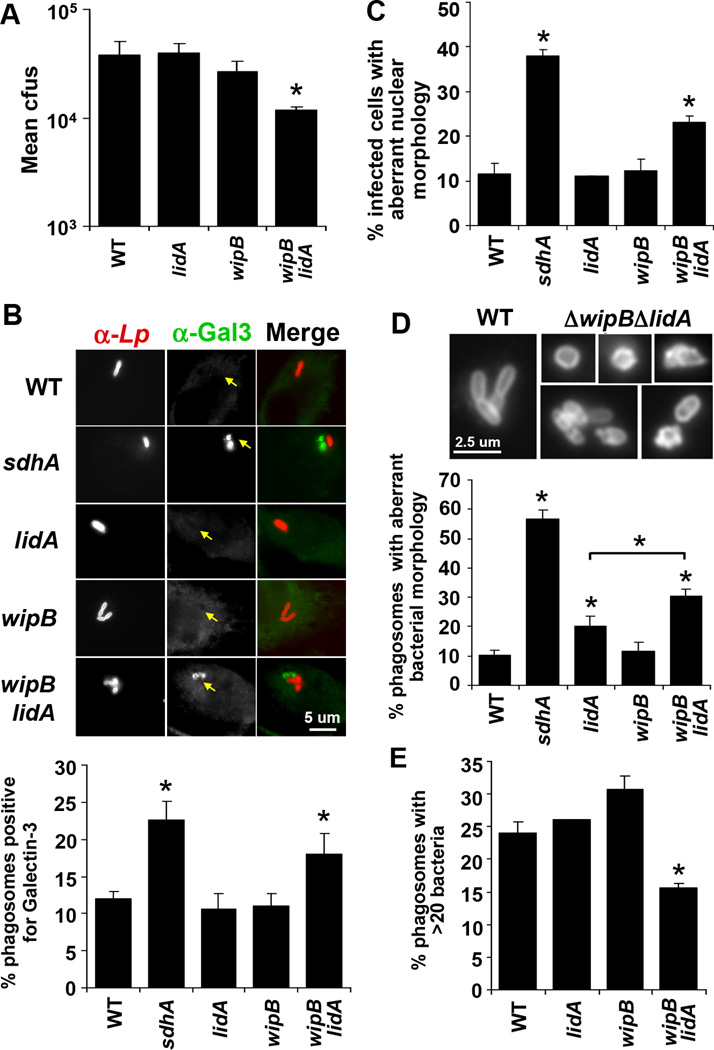
Mutations placed in the same group by hierarchical cluster analysis identify proteins that participate in the same process in the cell or comprise a single protein complex (20). We applied the same reasoning to the cluster analysis data obtained from our iMAD screen in Drosophila cells, which indicated that mutations in wipB and lpg2395 belonged to the same functional group as sdhA mutations (Fig. 1A), consistent with all three proteins contributing to a common step in LCV formation. SdhA plays a crucial role in maintaining LCV integrity in bone marrow-derived murine macrophages (21). Vacuoles harboring ΔsdhA mutant bacteria are more permeable than those harboring wild-type bacteria (21), and as a consequence co-localize with host proteins associated with partially degraded phagosomes (21, 22). This vacuolar disruption leads to host cell death, bacterial degradation and poor bacterial growth in macrophages challenged with ΔsdhA mutants (21, 23). Therefore, we tested whether mutations that cluster with sdhA lesions could identify other L. pneumophila proteins that promote vacuole integrity. Consistent with other cell types, the ΔwipBΔlidA double mutant exhibited reduced replication in bone marrow-derived murine macrophages relative to either the wild-type strain or strains having single mutations in each of these genes (Fig. 3A). Since a L. pneumophila mutant lacking 31% of Dot/Icm TS grows robustly in macrophages (16) this finding confirmed the power of the cluster analysis to identify functionally related Dot/Icm TS. The growth defect was associated with a loss of membrane integrity of the LCV based on colocalization with Galectin-3, a marker for vacuole lysis (22). Macrophages challenged with the ΔwipBΔlidA mutant showed an increase relative to wild-type bacteria in the number of vacuoles staining positively for Galectin-3 (Fig. 3B). Localization of this marker was comparable to challenge with the ΔsdhA mutant, while the ΔwipB and ΔlidA single deletion mutants resembled the wild-type strain in this assay (Fig. 3B).
Figure 3. Hierarchial cluster analysis predicts double mutants defective in maintaining vacuole integrity.
(A) Growth of the ΔwipBΔlidA mutant was reduced in bone marrow-derived murine macrophages compared to the wild-type (WT) and single deletion mutant strains. Bacterial growth was determined as described in Fig. 2. (B) ΔwipBΔlidA mutant-containing vacuoles showed enhanced recruitment of Galectin-3. Macrophages were infected with L. pneumophila for 6 hours, fixed and stained for Legionella and Galectin-3 then visualized by fluorescence microscopy (upper panel). The number of Legionella vacuoles staining positive for Galectin-3 were scored (lower panel). (C) Cells infected with the ΔwipBΔlidA mutant exhibit increased host cell death based on aberrant nuclear morphology characteristic of apoptosis. Macrophages were infected with L. pneumophila for 8 hours, fixed and stained for Legionella then treated with Hoechst stain. (D) ΔwipBΔlidA mutant bacteria exhibited aberrant morphology after challenge of macrophages. Bacteria visualized by fluorescence microscopy as in (B) showed both swelling and blebbing in vacuoles containing either single or multiple bacteria relative to the smooth rod shaped morphology of wild-type bacteria (upper panel). The number of vacuoles in which at least one bacterium exhibited aberrant morphology was scored (bottom panel). (E) The ΔwipBΔlidA mutant showed a defect in the number of vacuoles containing large numbers of bacteria in a macrophage host. Macrophages were infected with L. pneumophila for 10 hours, fixed and stained for Legionella and the number of bacteria per vacuole was scored. (A–E) Data are representative of at least 2 independent experiments ± standard deviation of 3 replicates, scoring 100 vacuoles (B, D, E) or infected cells (C) per replicate. *p <0.05 relative to the wild-type strain unless indicated otherwise.
Analysis of morphological markers was consistent with the ΔwipBΔlidA strain causing a breakdown in vacuole integrity (21). We observed an increase in the number of infected host cells undergoing cell death relative to cells harboring wild-type bacteria (Fig. 3C), indicated by aberrant nuclear morphology characteristic of apoptosis, while the number of bacteria appearing degraded, based on aberrant bacterial morphology (Fig. 3D), was similarly increased. The amount of bacterial degradation and host cell death was less extensive than that observed for the ΔsdhA mutant, consistent with the relative severity of the growth defects of these two strains. For the ΔsdhA mutant, the vast majority of vacuoles contained a single bacterium, while vacuoles harboring the ΔwipBΔlidA mutant often had more than one bacterium, many of which appeared degraded (Fig. 3D). This is consistent with failure of the double mutant to maintain vacuole integrity, even though there was sufficient vacuolar remodeling to support initial replication events (Fig. 3D and 3E). Enhanced recruitment of Galectin-3 and host cell death relative to the wild-type strain was similarly observed in macrophages challenged with the ΔmavPΔlidA mutant, accompanying defective growth of this mutant in macrophages (Fig. S4). Assigning genes to specific functional groups based on the behavior of their corresponding mutants, therefore, identified microbial proteins that were required for similar steps during replication vacuole biogenesis.
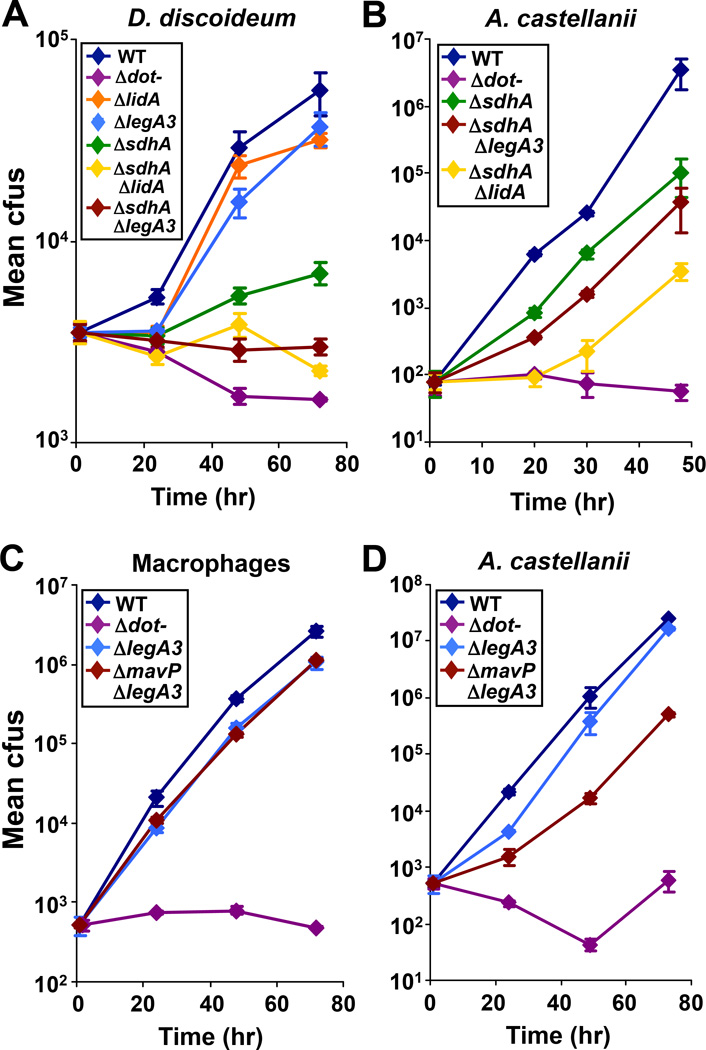
Despite its severe growth defect in macrophages, an L. pneumophila ΔsdhA mutant is only partially attenuated for growth in D. discoideum, presumably because the host cell death pathways that inhibit growth of this mutant in macrophages are not present in amoebae (23). The cluster analysis predicted that this partial defect should be potentiated by the addition of a second mutation known to aggravate mutations that belong to the same functional group as sdhA (Group IX) (Fig. 1A). As mutations in Group IX (wipB and lpg2395) were aggravated by mutations in Group XI (lidA and legA3) (Fig. 2B and 2D), we predicted that a Group XI mutation, such as deletion of lidA, should exacerbate the growth defect of the ΔsdhA mutant in D. discoideum. Indeed, the ΔsdhAΔlidA double mutant exhibited reduced growth relative to the ΔsdhA mutant alone (Fig. 4A). Moreover, a mutant lacking sdhA and legA3, another member of Group XI, was also attenuated for growth in this host (Fig. 4A). Thus, the presence of Group XI proteins could compensate for loss of SdhA in this amoebal host.
Figure 4. Host-specific aggravating genetic interactions between mutations in genes encoding Dot/Icm translocated substrates.
(A) Deletion of lidA or legA3 (Group XI) attenuated the growth defect of the ΔsdhA mutant (Group IX) in D. discoideum. (B) The ΔsdhAΔlidA mutant was more defective for growth than the ΔsdhAΔlegA3 mutant in A. castellanii. (C) A ΔmavPΔlegA3 mutant grew as well as a ΔlegA3 single mutant in bone marrow-derived murine macrophages. (D) The ΔmavPΔlegA3 mutant showed reduced growth relative to the ΔlegA3 mutant in A. castellanii. (A–D) Bacterial growth was monitored as described in Fig. 2. WT: wild-type. Data are representative of at least 2 independent experiments ± standard deviation of 3 replicates.
Phenotypic analysis of L. pneumophila double mutants showed that Group XI in its entirety was required to bypass the loss of single members of Group IX, however the extent to which individual members were able to compensate varied. For example, in A. castellanii, the ΔsdhAΔlidA mutant was more attenuated for growth than the ΔsdhAΔlegA3 mutant (Fig. 4B), arguing for a more significant role for LidA than LegA3 in promoting growth in the absence of SdhA in this species. This phenomenon was dependent on the host examined, as we did not observe this hierarchial relationship in D. discoideum (Fig. 4A). Similarly, in bone marrow-derived macrophages, loss of mavP had no effect on the growth of a ΔlegA3 mutant (Fig. 4C) whereas the ΔmavPΔlegA3 mutant showed reduced growth in A. castellanii (Fig. 4D). Thus, optimal growth in a particular host depends on a specific set of Dot/Icm TS. Similar to the partial defect in GFP-HDEL recruitment observed for the ΔwipBΔlidA mutant (Fig. 2C), the residual growth of specific double mutants in A. castellanii or macrophages (Fig. 4C and 4D) is consistent with existence of additional compensatory pathways promoted by other functional groups.
Maintaining an extensive repertoire of Dot/Icm TS enables L. pneumophila to grow in a broad assortment of amoebal hosts (16). As it is common for many TS to be present in only a subset of isolates (24), much of the selective pressure for preserving Dot/Icm TS appears to be centered on maintaining sets of functions, rather than individual proteins. For example, activation of Rab1 appears to be a central feature of L. pneumophila pathogenesis. A key player in this event in the Philadelphia 1 strain is SidM, which acts at two levels to activate and recruit Rab1 to the LCV (25, 26), but is only encoded by a subset of clinical strains (24). Presumably, the presence of TS such as AnkX, which activates Rab1 via a phosphocholine transfer mechanism (27), allows conservation of function. Therefore, activation of Rab1 is conserved without conserving specific proteins, or even the enzymatic activities they perform to activate this modulator of vesicle trafficking.
The accumulation of a large repertoire of Dot/Icm TS to cope with host variation allows different combinations of proteins to promote growth, arming the bacterium with multiple mechanisms for accomplishing a single task as long as the appropriate host targets are present. This flexibility is limited however, as it was possible to impair intracellular growth by disrupting more than one pathway (Fig. 1C). The host cell also restricts the degree of flexibility the bacterium can exert, as the specific combinations of Dot/Icm TS that were required for optimal growth varied between hosts (Fig. 4).
We have developed a strategy to elucidate the network of functional interactions central to Legionella’s manipulation of host cells. This approach defines sets of proteins that participate in common pathways, groups of proteins that modulate redundant virulence strategies and predicts cell biological defects based on hierarchical clustering. For pathogens that employ redundant virulence mechanisms, similar to L. pneumophila, simple strategies such as the construction of strains with random sets of genetic lesions, have failed to identify critical bacterial determinants of pathogenicity and have given no information on the interrelationships between members of a large group of proteins known to interact with the host cell. Our integrated approach can be applied to other bacterial pathogens, as well as other systems that have convergent biochemical pathways, and shows that it is now possible to define roles for individual proteins and their relative contributions to pathogenesis in different hosts. The utility of this strategy can be extended to any interface between two organisms involving a large number of interactions.
Supplementary Material
Acknowledgements
We thank E. Rubin and C. Sassetti for helpful advice regarding the TraSH protocol, plasmids and equipment use; J. Vogel for plasmids; K. Bodi, TUCF genomics core facility and A. Ensminger for scientific advice with genome sequencing protocols and analyses; A. Camilli for equipment use. We thank D. Daigle, K. Davis, E. Haenssler, A. Hempstead, D. DeJesus, S. Mohammadi and E. Creasey for reviewing the manuscript. This work was supported by the Howard Hughes Medical Institute (T.J.O. and M.S.D.), by a Natalie V. Zucker Fellowship (T.J.O.) and a grant from the National Institutes of Health (#GMO41883) (D.B.). R. Isberg is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Materials
Materials and Methods
Figs. S1 to S4
Tables S1 to S2
References (28–38)
References and Notes
- 1.Galàn JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kvitko BH, et al. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 1980;33:1179. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen TM, et al. A community-wide outbreak of Legionnaires disease linked to industrial cooling towers-how far can contaminated aerosols spread? J. Infect. Dis. 2006;193:102. doi: 10.1086/498575. [DOI] [PubMed] [Google Scholar]
- 5.Swanson MS, Isberg RR. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect. Immun. 1996;64:2585. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell. Sci. 2001;114:4637. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- 7.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 2002;4:945. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 8.Kagan JC, Stein MP, Pypaert M, Roy CR. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 2004;199:1201. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e4. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand BC, Sadosky AB, Shuman HA. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 1994;14:797. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 11.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 12.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 2005;56:90. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 15.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor TJ, Adepoju Y, Boyd D, Isberg RR. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. USA. 2011;108:14733. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;134:507. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Robinson CG, Roy CR. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol. 2006;8:793. doi: 10.1111/j.1462-5822.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- 19.Monnat J, et al. Identification of a novel saturable endoplasmic reticulum localization mechanism mediated by the C-terminus of a Dictyostelium protein disulfide isomerase. Mol. Biol. Cell. 2000;11:3469. doi: 10.1091/mbc.11.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 21.Creasey L, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci USA. 2012;109:3481. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paz I, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12:530. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 23.Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA. 2006;103:18745. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2006;36:1165. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 25.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell. 2006;11:47. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 26.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee S, et al. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477:103. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz MA, Silverstein SC. Intracellular multiplication of Legionnaires' disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J. Clin. Invest. 1983;71:15. doi: 10.1172/JCI110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feeley JC, et al. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 1979;10:437. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 1997;65:2497. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolter R, Inuzuka M, Helinski DR. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 32.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 1993;7:7. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 33.Solomon JM, Rupper A, Cardelli JA, Isberg RR. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun. 2000;68:2939. doi: 10.1128/iai.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Solomon JM, Isberg RR. Dictyostelium discoideum strains lacking the RtoA protein are defective for maturation of the Legionella pneumophila replication vacuole. Cell Microbiol. 2005;7:431. doi: 10.1111/j.1462-5822.2004.00472.x. [DOI] [PubMed] [Google Scholar]
- 35.Moffat JF, Tompkins LS. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 1992;60:296. doi: 10.1128/iai.60.1.296-301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaner, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 37.Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 2009;11:230. doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.