Abstract
Purpose
The aim of this study was to identify and validate novel predictive and/or prognostic serum proteomic biomarkers in patients with epithelial ovarian cancer (EOC) treated as part of the phase III international ICON7 clinical trial.
Experimental design
ICON7 was a phase III international trial in EOC which demonstrated a modest but statistically significant benefit in progression-free survival with the addition of bevacizumab to standard chemotherapy. Serum samples from 10 patients who received bevacizumab (5 responders, 5 non-responders) were analysed by mass spectrometry to identify candidate biomarkers. Initial validation and exploration by immunoassay was undertaken in an independent cohort of 92 patients, followed by a second independent cohort of 115 patients (taken from across both arms of the trial).
Results
Three candidate biomarkers were identified, mesothelin, fms-like tyrosine kinase-4 (FLT4) and α1-acid glycoprotein (AGP). Each showed evidence of independent prognostic potential when adjusting for high risk status in initial (p<0.02) and combined (p<0.01) validation cohorts. In cohort I individual biomarkers were not predictive of bevacizumab benefit; however, when combined with CA-125, a signature was developed that was predictive of bevacizumab response and discriminated benefit attributable to bevacizumab better than clinical characteristics. The signature showed weaker evidence of predictive ability in validation cohort II, but was still strongly predictive considering all samples (p=0.001), with an improvement in median PFS of 5.5 months in signature-positive patients in the experimental arm compared to standard arm.
Conclusions
This study demonstrates a discriminatory signature comprising mesothelin, FLT4, AGP and CA-125 as potentially identifying those patients with EOC more likely to benefit from bevacizumab. These results require validation in further patient cohorts.
Keywords: biomarkers, proteomics, bevacizumab, serum, predictive, ICON7
Introduction
Epithelial ovarian cancer (EOC) causes significant morbidity and mortality worldwide with an estimated 21,880 new cases and 13,850 attributable deaths in 2010 (1). Standard treatment for many years has involved debulking surgery combined with systemic platinum-based chemotherapy (2). However, despite EOC being very chemosensitive, most patients subsequently develop recurrent disease and die. Novel drug targets include vascular endothelial growth factor (VEGF), with progression of EOC commonly being VEGF-driven and increased VEGF expression is associated with more advanced disease, ascites and a worse overall prognosis (3, 4). Preclinical and early clinical data supported the further investigation of bevacizumab, a monoclonal antibody to VEGF, in the treatment of EOC.
ICON7 was a two-arm phase III international randomised open-label trial of 1528 women with high risk early-stage or advanced-stage EOC, comparing six cycles of standard chemotherapy (carboplatin and paclitaxel) to six cycles of chemotherapy plus the addition of concurrent and maintenance bevacizumab. Initial results demonstrated that bevacizumab improved progression-free survival (PFS), but although statistically significant, over the entire trial population the absolute PFS benefit was only 1.5 months (5). These data, along with the results of a similar trial, GOG218 (6), have led to bevacizumab being licensed in combination with carboplatin and paclitaxel in the first-line treatment of advanced EOC.
Subset analysis within ICON7 demonstrated increased benefit when the population was restricted to those patients at higher risk of disease progression (FIGO stage IV disease or FIGO stage III disease with >1cm residual disease after surgical debulking), with an increase in median PFS of 3.6 months and a trend towards increased overall survival (OS). Identifying novel biomarkers to select patients with EOC who will derive most benefit from bevacizumab is important, and is also required for patients with glioblastoma, colorectal, lung and renal cancer where bevacizumab is also licensed for use, as there are no predictive biomarkers in clinical use.
Patients participating in ICON7 were asked to donate tissue and longitudinal blood samples for use in future translational research projects. The ICON7 sample bank, with rigorous sample collection, processing and storage and associated high quality clinical data, provides an excellent opportunity to identify potential EOC prognostic and predictive biomarkers, and to perform initial validation. The aim of this study was to utilise serum samples from the ICON7 sample bank to identify, and subsequently validate, candidate biomarkers relating to bevacizumab use in EOC.
Methods
Patients and Samples
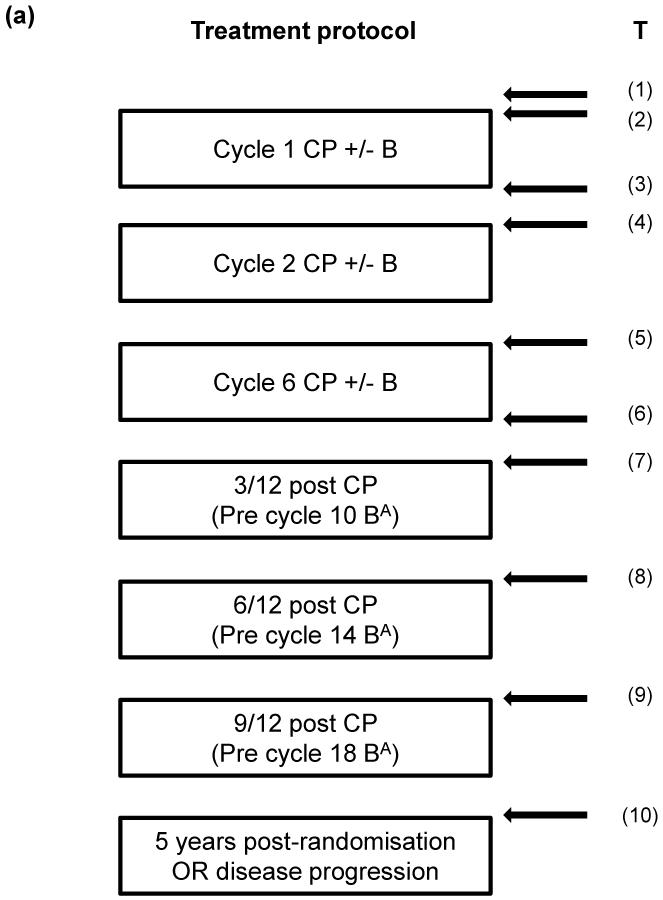
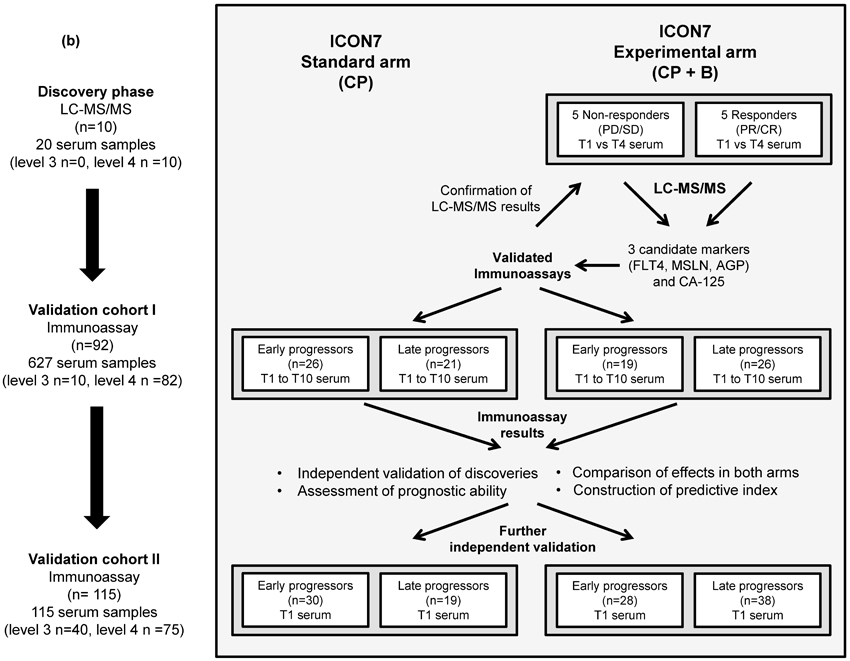
ICON7 patient recruitment, trial design and outcomes have been published elsewhere (5). Following ethical approval, blood samples were obtained from ICON7 patients following a schedule dependent on their level of consent (Figure 1a). Samples were collected into plain clot activator tubes, allowed to clot for 1 hour before centrifugation at 2000xg for 10 minutes at 20°C and serum aliquotted and stored at −80°C. This was performed according to standard operating procedures and samples transferred to the central ICON7 sample bank in Leeds. Following approval of this study by the ICON7 Trial Management Group, 762 serum samples from 217 patients were utilised (all patients with baseline samples from the total of 226 who donated serum; Figure 1a, b).
Figure 1. Timing of serum samples as part of the ICON7 trial and schematic diagram describing discovery and validation experiments.
(a). For patients consenting to level 4 sample collection, samples were taken pre-cycle 1 (T1 and T2), post-cycle 1 (T3), pre-cycles 2 (T4) and 6 (T5), post-cycle 6 (T6) and at 3, 6 and 9 months from completion of chemotherapy (T7-T9) and at either 5 years from randomisation or at progression (whichever occurred first) (T10). For patients consenting to level 3 sample collection, samples were taken pre-cycle 1 (T1), pre cycles 2 and 6 (T4 and T5) and at either 5 years from randomisation or at progression (whichever occurred first) (T10). A total of 226 patients consented to levels 3 or 4 with a further 536 patients consenting to the use of fresh-frozen paraffin embedded tissue and/or blood samples for genomic DNA analysis; (b). Discovery and validation experiments showing number of patients and serum samples and sample time points examined at each stage.
B = bevacizumab; C = carboplatin; P = paclitaxel; Aexperimental arm only. PR and CR = partial and complete response respectively, PD and SD = progressive and stable disease respectively.
Proteomic biomarker discovery by LC-MS/MS
Serum samples from 10 patients within the ICON7 experimental arm were selected for biomarker discovery, grouped as 5 responders (complete and partial response) and 5 non-responders (stable or progressive disease) defined by RECIST and/or CA-125 after 6 cycles of treatment. Selection on this basis was used as PFS data were not available at that time.
However, the median (range) PFS was 24.7 (12.0-25.8) months in the responder group and 12.8 (8.12-23.8) months in the non-responder group. All patients had grade 3 serous tumours and the two groups were matched as closely as possible by age, FIGO stage and surgical outcome (optimal or sub-optimal debulking) (Supplementary Table 1). Paired serum samples at time point 1 (baseline) and time point 4 (pre-cycle 2) from each of the selected patients were subjected to proteomic analysis by label-free MS and candidate biomarkers of response selected (Figure 1b).
For each sample, 200 μL of serum was filtered through 0.22 μm Spin-X filters (Corning) and 150 μL then depleted of the 14 most-abundant proteins using a Multiple Affinity Removal (MARS) human 14 column (7), leaving an average of 7% of the total protein in the samples. Samples were concentrated using 15 mL 10kDa MWCO filters (Millipore), desalted using 2 mL 7 kDa MWCO ZEBA spin-desalting columns (Thermo Scientific) and 150 μg protein was digested with trypsin using filter-aided sample preparation (FASP) (7, 8).
Peptides (triplicate injections each of 2 μg) were separated by online reversed-phase capillary liquid chromatography (LC) and analyzed by electrospray tandem mass spectrometry (MS) using a Thermo Orbitrap Velos (9). Data were searched against an International Protein Index (IPI 3.80) human protein sequence database with MaxQuant 1.1.1.36 software (10) and the Andromeda search engine (11). The initial maximal mass tolerance for MS scan was set to 10 ppm, the fragment mass tolerance for MS/MS was set to 0.5 Th. The maximum protein and peptide false discovery rates were set to 0.01. Label-free quantitation was performed with MaxQuant.
Results were subjected to initial exploratory data analysis using principal component analysis and hierarchical clustering considering the whole profile together to identify gross patterns and identify potential outliers. Each protein was then examined separately to identify differences in protein abundance between responders and non-responders at time points 1 and 4 (Mann-Whitney tests) and to identify differences between these time points (Wilcoxon signed-rank tests). False discovery rate was estimated by the q-value method.
Biomarker validation by immunoassay
The mass spectrometry results from the discovery analysis were confirmed by immunoassay. Validation and exploration of initial findings was undertaken using a cohort of 92 patients (627 longitudinal samples (Supplementary Table 2): validation cohort I), with further validation in an additional 115 patients (baseline samples only: validation cohort II). Samples in validation cohort I were selected to ensure similar numbers in each of the patient treatment and outcome groups described (limited by available assessable patients in the non-responder groups). Validation cohort II consisted of all remaining baseline samples from the biobank. Validation cohorts included patients from both arms of the trial (to enable markers differing specifically in response to bevacizumab and not just chemotherapy to be distinguished; Figure 1b), were independent of each other and the discovery set and were representative of the trial population (Table 1). Patients in the validation cohorts were separated around the median PFS (18 and 16.8 months in validation cohort I and II respectively) into early and late progressors (includes non-progressors). This differed from the RECIST-based criteria used for selection of patients for biomarker discovery as at the later time of validation sample analysis, the ICON7 trial PFS data, which is more clinically relevant than response or non-response, had become available.
Table 1.
Baseline characteristics of validation cohorts I and II at randomization and associated disease outcomes.
| Validation cohort I | Validation cohort II | |||
|---|---|---|---|---|
|
| ||||
| Characteristic | Standard arm (N=47) |
Experimental arm (N=45) |
Standard arm (N=49) |
Experimental arm (N=66) |
| Outcome | ||||
| Early progressors | 26 | 19 | 30 | 28 |
| Late progressors | 21 | 26 | 19 | 38 |
| Age – years | ||||
| Median (range) | 59 (38-75) | 53 (31-71) | 58 (38-79) | 56 (34-76) |
| Race – n (%) | ||||
| White | 44 (94) | 40 (89) | 48 (98) | 61 (92) |
| Asian/Black/Other | 3 (6) | 5 (11) | 1 (2) | 5 (8) |
| ECOG PS – n (%) | ||||
| 0 | 17 (36) | 20 (45) | 20 (41) | 29 (44) |
| 1 | 28 (60) | 23 (51) | 25 (51) | 35 (53) |
| 2 | 1 (2) | 2 (4) | 4 (8) | 2 (3) |
| Unknown | 1 (2) | 0 (0) | ||
| Origin of cancer – n (%) | ||||
| Ovary (epithelial) | 44 (94) | 36 (81) | 41 (84) | 60 (91) |
| Fallopian tube | 1 (2) | 2 (4) | 1 (2) | 1 (1) |
| Primary peritoneal | 1 (2) | 5 (11) | 5 (10) | 5 (8) |
| Multiple sites | 1 (2) | 2 (4) | 2 (4) | 0 (0) |
| Histology – n (%) | ||||
| Serous | 30 (64) | 30 (67) | 36 (74) | 43 (65) |
| Mucinous | 1 (2) | 0 (0) | 1 (2) | 0 (0) |
| Endometrioid | 5 (11) | 4 (9) | 4 (8) | 0 (0) |
| Clear cell | 6 (13) | 3 (7) | 5 (10) | 14 (21) |
| Mixed | 4 (8) | 7 (15) | 2 (4) | 9 (14) |
| Undefined | 1 (2) | 1 (2) | 1 (2) | 0 (0) |
| FIGO stage* – n (%) | ||||
| I/IIA | 4 (8) | 6 (13) | 5 (10) | 5 (8) |
| IIB/IIC | 5 (11) | 2 (4) | 7 (14) | 10 (15) |
| III | 34 (72) | 32 (72) | 33 (68) | 42 (64) |
| IV | 4 (9) | 5 (11) | 4 (8) | 9 (13) |
| Grade – n (%) | ||||
| Well differentiated | 1 (2) | 2 (4) | 2 (4) | 5 (8) |
| Moderately differentiated | 10 (21) | 5 (11) | 8 (16) | 9 (14) |
| Poorly differentiated | 36 (77) | 38 (85) | 36 (74) | 50 (76) |
| Unknown | 0 (0) | 0 (0) | 3 (6) | 2 (2) |
| Debulking surgery – n (%) | ||||
| No (inoperable) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Yes | 47 (100) | 45 (100) | 49 (100) | 66 (100) |
| >1cm residual disease | 23 (49) | 18 (40) | 20 (41) | 34 (52) |
| ≤1cm residual disease | 24 (51) | 27 (60) | 29 (59) | 32 (48) |
| High-risk of progression – n (%)* | ||||
| No | 25 (53) | 27 (60) | 30 (61) | 34 (52) |
| Yes | 22 (47) | 18 (40) | 19 (39) | 32 (48) |
| Progression free survival | ||||
| Median (range) | 17.3 (1.90-42.9) | 20.6 (6.30-33.30) | 12.1 (1.7-36.3) | 17.9 (3.7-41.8) |
| Number of events | 31 | 33 | 31 | 50 |
FIGO stage IV disease OR FIGO stage III disease AND >1.0 cm residual disease after debulking surgery
An ELISA for soluble FLT4 (VEGFR3) was developed (Supplementary Methods) using the human sVEGFR3 DuoSet (R&D Systems) but with a substituted standard due to validation issues. Soluble mesothelin-related peptides (sMRPs) were quantified using the FDA-approved MESOMARK® assay (Fujirebio Diagnostics) and AGP and CA-125 concentrations were measured using routine clinical assays (Behring Nephelometric Analyser II, Siemens and Siemens ADVIA Centaur CA-125 II assay respectively) at the Leeds General Infirmary.
Statistical analysis
Results were examined for predictive utility, either on the basis of baseline concentrations or patterns of longitudinal change of the proposed protein biomarkers. All analyses were performed according to REMARK criteria (12). Associations between the 4 biomarkers under study were visualised using a correlogram based on simple linear regression and Spearman’s rank correlation coefficient. Associations with demographic and clinical characteristics at baseline were investigated using Fisher’s exact test for categorical variables and Spearman’s rank correlation coefficient for continuous variables. Corrections for multiple testing were not applied due to the explorative nature of the study.
Estimates of intra- and inter-subject variation for each of the selected proteins were obtained using linear mixed effects models (13) considering biomarker concentrations from pre-treatment time points 1 and 2. Longitudinal line and box plots were used to assess trends in biomarker concentration over time to inform salient non-parametric significance testing of differences between time points. A linear mixed effect model was also used to investigate the rate of change of FLT4 concentration over time in patients in the experimental arm.
PFS was calculated from the date of randomization to the date of disease progression or death, whichever occurred first. Patients who were alive without disease progression were censored as of the date of their last assessment, with a cut-off date of November 30th 2010. Cox proportional hazards (PH) models were used to estimate the association between biomarker concentrations and PFS, the Kaplan-Meier (KM) method used to estimate survival functions and the logrank test used to compare survival functions. Markers were considered initially as continuous variates and then as binary factors using cut-points derived by maximising Harrell’s C-index (for visualisation of effects) in validation cohort I and subsequently by predictive index-defined cut points in the combined data. The predictive potential of markers was assessed using interaction terms for treatment arm and biomarker concentration in Cox PH models.
An additive marker index with scale 0-4 was calculated from the individual markers. Further details of marker index construction are provided in Supplementary Methods. Briefly, the optimum cut-point in the index was identified using Cox PH models (by examining the likelihood ratio test (LRT) on the interaction term for treatment arm and each level of the index), with indices below cut-points considered as signature-negative and those above as signature-positive. Internal validation of the optimal model was performed using R=1000 bootstrap resamples to estimate the optimism in model predictive ability. PH assumptions were tested for each model using tests based on Schoenfeld residuals (14). All analyses were initially carried out in validation cohort I and replicated in validation cohort II and the cohorts combined, with the exception of longitudinal analysis which was carried out only in validation cohort I. All statistical tests were two-sided and all analyses were undertaken in the R environment for statistical computing (15).
Results
Biomarker discovery phase
Serum profiling of the discovery cohort identified a total of 352 proteins with at least two significant and one unique peptide in the total data set (www.proteomics.leeds.ac.uk). From these, 4 proteins were selected on the basis of differences between responders and non-responders, either at time point 1 or longitudinally: mesothelin, FLT4/VEGFR3 and AGP1 and 2. In the discovery cohort, mesothelin did not differ at baseline between responders and non-responders, but decreased in both groups, and was not detected in responders at time point 4 (Supplementary Figure 1). FLT4 was only detected in two responders and at time point 1 only. Both these proteins were identified by only 2 peptides and therefore differences were potentially due to under-sampling during analysis, but given their biological relevance they were investigated further. Subsequent ELISA measurements of serum samples from the 10 patients in the discovery set confirmed the initial findings (Supplementary Figure 1 and Figure 2a). AGP1 and 2 were higher in three responders at time point 1 and decreased by time point 4. Although nominally removed by immunodepletion, their presence could reflect saturation of the column, with the residual amount detected being proportional to the starting concentration. The mass spectrometry results were confirmed using an immunoassay which detects both AGP1 and 2, which are referred to hereafter as AGP.
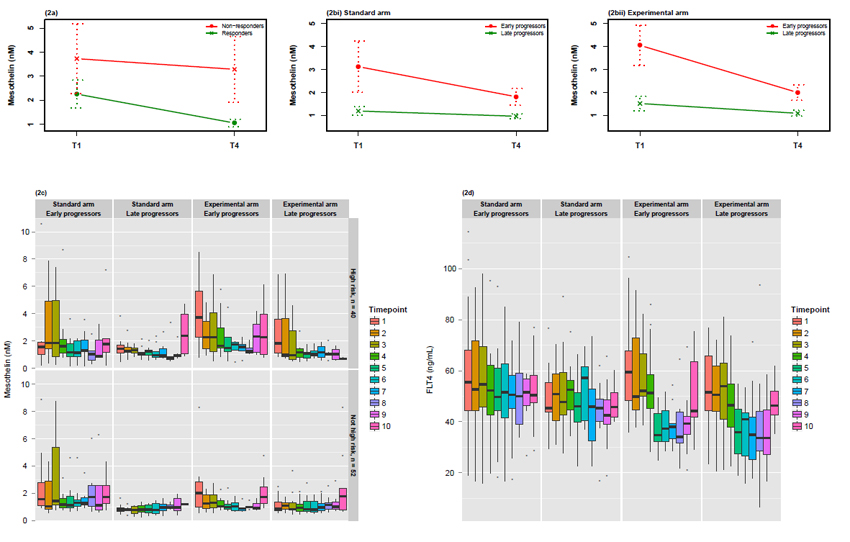
Figure 2. Relationship between mesothelin and response to treatment and the kinetics of change in FLT4 related to treatment in validation cohort I.
(a) Longitudinal changes in mesothelin between time points 1 and 4 in responders and non-responders in the discovery cohort as determined confirmed by ELISA. Longitudinal changes in mesothelin between time points 1 and 4 in early and late progressors in the validation cohort in the standard arm (b)[i] and experimental arm (b)[ii] measured by immunoassay. Data points are the mean with dotted lines representing ± standard error. (c) Relationship between mesothelin and response to treatment by risk of progression status in validation cohort. Longitudinal box plots* showing changes in mesothelin concentration across the sample time points in early and late progressors in the standard and experimental arms for patients at high risk of progression and patients not at high risk of progression (high risk of progression defined as FIGO stage IV disease or FIGO stage III disease AND >1.0cm residual disease after debulking surgery). (d) Longitudinal box plots* showing changes in FLT4 median concentration over time points in ICON7 sampling. Panels show early and late progressors in terms of PFS in standard and experimental arms of trial.
*Box plots show median, interquartile range (IQR) and 1.5×IQR. Dots represent outliers (defined as greater/less than 1.5×IQR).
Biomarker validation cohorts and clinical characteristics/associations
Biomarker validation cohort characteristics
The 3 candidate biomarkers together with CA-125 were examined in a further 207 patients (n=92 and 115 in validation cohort I and II respectively) from both arms of the trial in relation to disease progression. Baseline clinical and pathological characteristics and outcomes in the cohorts were similar to those in the overall trial population (Table 1) (5). Median PFS was longer in the experimental arm as in the trial, but was not significantly different between trial arms in either validation cohort (validation cohort I: p=0.860, Supplementary Figure 2a; validation cohort II: p=0.664, logrank test) due to the reduced power in these smaller cohorts. Treatment effect on PFS was also similar in both cohorts to that seen in the overall trial population (HR=0.956 validation cohort I; HR=0.923 validation cohort II cf. 0.81) with no evidence of being significantly different (validation cohort I: p=0.340; validation cohort II: p=0.664, Wald test). The lack of proportional hazards seen in the trial arms (5) was also apparent in the biomarker validation cohort, with comparable violation from proportionality observed (Supplementary Figure 2b).
Association of biomarkers with clinical characteristics
Examination of candidate biomarkers (Supplementary Figure 3) and clinical characteristics at baseline in validation cohort I (Table 2) showed an association between mesothelin and FLT4 and patients at high risk of progression (p=0.002 and p=0.043 respectively), with patients at higher risk of progression having higher concentrations. These associations were confirmed in validation cohort II (Supplementary Table 3) and the combined validation cohort (Supplementary Table 4) with all candidate markers and CA125 showing an association with high risk status (all p<0.02). Mesothelin and AGP were associated with FIGO stage in validation cohort I (p=0.008 and p=0.003 respectively); confirmed in validation cohort II (p<0.001 and p<0.001, Supplementary Table 3) and overall (p<0.001 and p<0.001, Supplementary Table 4). CA-125, as expected, was associated with FIGO stage (p<0.001) and histological subtype (p=0.052) with highest concentrations in more advanced stages and serous tumours, confirmed in validation cohort II and overall (Supplementary Tables 3 and 4). Baseline CA-125 concentrations were higher in patients in the experimental arm of validation cohort I (p=0.043) although ranges overlapped. This is likely an artefact of selection, supported by no such significant differences being seen in validation cohort II or overall. The markers showed no strong positive or negative associations (although correlations were significantly different from zero, all p<0.01 in validation cohort I, Supplementary Figure 4). Similar results were observed in validation cohort II and the combined validation cohort (data not shown).
Table 2.
Associations between the baseline concentrations (time point 1) of the serum biomarkers under investigation and the clinical characteristics in validation cohort I.
| Characteristic | N | Mesothelin (nM) median (range) |
p-value | FLT4 (ng/mL) median (range) |
p-value | AGP (g/L) median (range) |
p-value | CA-125 (ku/L) median (range) |
p-value |
|---|---|---|---|---|---|---|---|---|---|
|
All patients
Treatment |
88 | 1.33 (0.23, 23.28) | 52.72 (18.96, 114.37) | 1.29 (0.52, 2.74) | 76 (5,6333) | ||||
| Standard arm | 45 | 1.21 (0.23, 23.28) | 50.30 (18.96, 114.37) | 1.29 (0.52,2.74) | 57 (7,4996) | ||||
| Experimental arm | 43 | 1.44 (0.56, 14.43) | 0.433 | 56.08 (23.57, 104.46) | 0.295 | 1.29 (0.68, 2.73) | 0.970 | 109 (5,6333) | 0.043 |
| Age – years * | 88 | 0.16 (0.16, 0.16) | 0.145 | −0.04 (−0.04, −0.04) | 0.723 | 0.01 (0.01,0.01) | 0.929 | 0.05 (0.05,0.05) | 0.641 |
| Race - n (%) | |||||||||
| White | 81 | 1.35 (0.23, 23.28) | 52.74 (18.96, 114.37) | 1.29 (0.54,2.74) | 76 (7,6333) | ||||
| Asian/Black/Other | 7 | 1.24 (0.58, 5.21) | 0.951 | 50.02 (43.54, 59.46) | 0.441 | 1.29 (0.52,2.63) | 0.847 | 16 (5,970) | 0.280 |
| ECOG PS - n (%) | |||||||||
| 0 | 34 | 1.52 (0.45, 14.43) | 46.35 (18.96, 114.37) | 1.15 (0.52,2.32) | 76 (5,6333) | ||||
| 1 | 36 | 1.12 (0.23, 23.28) | 57.54 (32.19, 96.17) | 1.45 (0.68,2.63) | 101.5 (14,4996) | ||||
| 2 | 3 | 3.25 (1.54, 3.98) | 0.390 | 59.20 (52.48, 79.76) | 0.024 | 1.96 (0.94,2.73) | 0.088 | 284 (61,1168) | 0.328 |
| Origin of cancer - n (%) | |||||||||
| Ovary (epithelial) | 77 | 1.31 (0.23, 23.28) | 53.43 (18.96, 114.37) | 1.27 (0.52,2.74) | 67 (5,1593) | ||||
| Fallopian tube | 3 | 1.17 (0.83, 3.98) | 45.27 (35.85, 52.48) | 1.27 (0.97, 1.96) | 104 (57,284) | ||||
| Primary peritoneal | 6 | 2.43 (0.58, 8.85) | 56.73 (42.80, 76.60) | 1.60 (0.68,2.21) | 133.5 (14,4996) | ||||
| Multiple sites | 2 | 8.38 (2.33, 14.43) | 0.228 | 53.73 (45.02, 62.43) | 0.556 | 1.13 (0.82,1.44) | 0.627 | 3236 (139,6333) | 0.310 |
| Histology - n (%) | |||||||||
| Serous | 57 | 1.54 (0.23, 23.28) | 52.48 (18.96, 114.37) | 1.43 (0.68,2.73) | 90 (14,6333) | ||||
| Mucinous | 1 | 1.18 (1.18, 1.18) | 61.58 (61.58, 61.58) | 1.55 (1.55,1.55) | 42 (42,42) | ||||
| Endometrioid | 8 | 1.51 (0.96, 2.47) | 57.41 (29.45, 103.40) | 1.05 (0.54,2.74) | 46 (7,181) | ||||
| Clear cell | 9 | 0.96 (0.58, 2.22) | 48.82 (38.42, 59.95) | 1.22 (1.00,2.63) | 43 (5,970) | ||||
| Mixed | 11 | 1.21 (0.79, 5.21) | 59.46 (38.65, 89.01) | 0.99 (0.52,2.62) | 100 (13,518) | ||||
| Undefined | 2 | 1.08 (0.78, 1.38) | 0.594 | 39.89 (32.19, 47.58) | 0.642 | 1.88 (1.80,1.95) | 0.241 | 314 (204,424) | 0.052 |
| FIGO stage - n (%) | |||||||||
| l/IIA | 10 | 0.86 (0.45, 2.14) | 47.88 (29.31, 69.25) | 1.12 (0.81,1.6) | 56.60 (5,90) | ||||
| IIB/IIC | 7 | 1.12 (0.88, 1.58) | 44.30 (36.30, 58.82) | 1.02 (0.52,1.12) | 20 (7,112) | ||||
| III | 62 | 1.49 (0.46, 23.28) | 57.99 (23.57, 114.37) | 1.38 (0.68,2.74) | 89.5 (14,4996) | ||||
| IV | 9 | 3.25 (0.23, 14.43) | 0.008 | 47.40 (18.96, 79.76) | 0.030 | 1.56 (0.98,2.73) | 0.003 | 970 (43,6333) | <0.001 |
| Grade - n (%) | |||||||||
| Well differentiated | 3 | 1.58 (0.58, 8.85) | 58.82 (50.02, 76.60) | 1.63 (1.04,2.21) | 44 (14,4996) | ||||
| Moderately differentiated | 14 | 1.03 (0.23, 4.95) | 50.52 (32.35, 114.37) | 1.37 (0.83,2.29) | 109 (10,1352) | ||||
| Poorly differentiated | 71 | 1.38 (0.45, 23.28) | 0.719 | 52.74 (18.96, 104.46) | 0.631 | 1.27 (0.52,2.74) | 0.561 | 72 (5,6333) | 0.788 |
| Debulking surgery n (%) | |||||||||
| >1cm residual disease | 38 | 1.57 (0.23, 23.28) | 56.35 (18.96, 89.01) | 1.23 (0.72,2.63) | 64.5 (10,1352) | ||||
| ≤1cm residual disease | 50 | 1.15 (0.45, 14.43) | 0.160 | 48.80 (23.57, 114.37) | 0.171 | 1.40 (0.52,2.74) | 0.365 | 78 (5,6333) | 0.913 |
| High-risk of progression** n (%) | |||||||||
| No | 51 | 0.99 (0.45, 8.85) | 48.82 (23.57, 114.37) | 1.27 (0.52,2.74) | 66 (5,4996) | ||||
| Yes | 37 | 1.80 (0.23, 23.28) | 0.002 | 57.22 (18.96, 89.01) | 0.043 | 1.33 (0.72,2.73) | 0.657 | 100 (14,6333) | 0.243 |
Spearman’s rank correlation coefficient between analyte and age in years
FIGO stage IV disease or FIGO stage III disease AND >1.0 cm residual disease after debulking surgery
Biomarker validation - cohort I
Longitudinal changes in biomarkers during treatment
The inclusion in the validation group of 61 patients who had two baseline samples (ranging from 0 to 32 days apart) allowed for an estimate of “normal” intra-patient variation, important when considering biomarker changes in individuals during treatment. Intra-individual coefficients of variation (CV%) were 22.6% for mesothelin, 14.6% for FLT4, 12.0% for AGP and 16.3% for CA-125.
In validation cohort I, mesothelin differed in concentration between time points 1 and 4 in the early progressor subset of the experimental arm (p<0.001; Figure 2b), with slight but not statistically significant downward trends in the late progressor groups for each treatment arm. Comparing the difference in change between these time points in early and late progressors separately demonstrated evidence of a significantly different change in the experimental arm (p=0.003) not observed in the standard arm (p=0.945). The most striking decreases in mesothelin concentration were observed for patients at a high risk of progression, although they generally had higher baseline concentrations (Figure 2c).
Patients in the experimental (bevacizumab) arm of the trial had generally decreasing concentrations of FLT4 between time points 1 and 4, irrespective of the timing of disease progression (Figure 2d). This decrease was observed during treatment with plateauing during follow-up. At time point 10 (when 41 of the 45 patients with available samples have experienced disease progression) a rebound increase in concentration was apparent. Considering the longitudinal mean profile and using a simple trend variable for time point in a linear mixed effects model there was a significant decrease of 2.49 ng/mL per time point (time points 1-9, p<10−10). However, early progressors had higher concentrations overall as compared to late progressors even when taking this longitudinal effect into account (increased concentration of 9.99 ng/mL in early progressors, p=0.01) indicating a potential prognostic effect.
Concentrations of AGP also tended to reduce throughout the period of treatment, particularly between time points 1-6 (during systemic therapy) (Supplementary Figure 5a), with patients at higher risk of progression showing the greatest reductions (Supplementary Figure 5b).
Association between candidate biomarker concentrations at baseline and PFS
When using the 88 time point 1 samples for which analyte measurements were available, each of the candidate biomarkers and CA-125 showed evidence of prognostic potential upon univariate analysis in Cox PH models (Table 3: validation cohort I, column 1, all p<0.05), and independently of risk of progression status (Table 3, column 2) upon multivariable analysis.
Table 3.
Univariate and multivariable analysis of prognostic potential of biomarkers. Columns represent a univariate analysis in validation cohort followed by results when adjusting for risk in all patients and in each treatment arm separately. Univariate analysis of risk of progression in all patients and in each treatment arm separately is displayed in the fifth row of each cohort. HRs for biomarkers refer to increase in risk per unit increase in concentration.
| Univariate analysis | Multivariable analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| +adj for risk | +adjusted for risk (standard arm) |
+adjusted for risk (experimental arm) |
||||||||
| Level | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | ||
|
Validation cohort I
| ||||||||||
| Mesothelin (nM) | Risk | (-) | 1.08 (1.03,1.14) | 0.002 | 1.08 (1.02,1.14) | 0.005 | 1.04 (0.97,1.12) | 0.23 | 1.30 (1.13,1.50) | <0.001 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 1.05 (0.61,1.8) | 0.871 | 1.08 (0.51,2.28) | 0.84 | 0.82 (0.37,1.82) | 0.623 | |||
|
| ||||||||||
| FLT4 (ng/mL) | Risk | (-) | 1.02 (1.01,1.04) | 0.008 | 1.02 (1.00,1.04) | 0.011 | 1.02 (1.00,1.04) | 0.063 | 1.02 (1.00,1.05) | 0.102 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 1.14 (0.69,1.90) | 0.602 | 1.17 (0.58,2.38) | 0.663 | 1.14 (0.54,2.42) | 0.726 | |||
|
| ||||||||||
| AGP (g/L) | Risk | (-) | 2.29 (1.40,3.75) | 0.001 | 2.25 (1.37,3.69) | 0.001 | 2.06 (1.04,4.08) | 0.038 | 2.65 (1.21,5.82) | 0.015 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 1.17 (0.71,1.95) | 0.538 | 1.27 (0.62,2.59) | 0.514 | 1.05 (0.48,2.27) | 0.906 | |||
|
| ||||||||||
| CA-125 (ku/L) | Risk | (-) | 1.05 (1.03,1.08) | <0.001 | 1.05 (1.03,1.07) | <0.001 | 1.04 (1.01,1.08) | 0.024 | 1.07 (1.03,1.11) | 0.001 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 1.40 (0.85,2.31) | 0.189 | 1.37 (0.67,2.82) | 0.384 | 1.48 (0.70,3.13) | 0.302 | |||
|
| ||||||||||
| Risk | (-) | (-) | (-) | (-) | (-) | |||||
| Not high | 1 | (-) | 1 | 1 | ||||||
| High | 1.51 (0.92.2.48) | 0.103 | (-) | 1.28 (0.64,2.61) | 0.482 | 1.86 (0.92,3.76) | 0.0842 | |||
|
| ||||||||||
|
Validation cohort II
| ||||||||||
| Mesothelin | Risk | (-) | 1.10 (1.07,1.14) | <0.001 | 1.06 (1.02,1.1) | 0.002 | 1.01 (0.96,1.06) | 0.663 | 1.06 (0.96,1.17) | 0.216 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 3.30 (2.00,5.47) | <0.001 | 6.80 (2.69,17.22) | <0.001 | 2.69 (1.43,5.03) | 0.002 | |||
|
| ||||||||||
| FLT4 | Risk | (-) | 1.01 (1.00,1.02) | 0.050 | 1.01 (0.99,1.02) | 0.286 | 1.01 (0.99,1.03) | 0.32 | 1.01 (0.99,1.03) | 0.423 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 3.96 (2.47,6.33) | <0.001 | 7.51 (3.41,16.54) | <0.001 | 2.86 (1.57,5.22) | 0.001 | |||
|
| ||||||||||
| AGP | Risk | (-) | 2.03 (1.44,2.86) | <0.001 | 1.44 (0.95,2.16) | 0.083 | 1.67 (0.96,2.93) | 0.071 | 1.33 (0.74,2.37) | 0.341 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 3.48 (2.11,5.74) | <0.001 | 6.75 (3.04,14.99) | <0.001 | 2.61 (1.34,5.05) | 0.005 | |||
|
| ||||||||||
| CA-125 | Risk | (-) | 1.03 (1.01,1.04) | <0.001 | 1.02 (1.00,1.03) | 0.028 | 1.00 (0.98,1.02) | 0.792 | 1.06 (1.02,1.10) | 0.001 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 3.81 (2.37,6.15) | <0.001 | 7.83 (3.46,17.68) | <0.001 | 2.54 (1.37,4.68) | 0.003 | |||
|
| ||||||||||
| Risk | (-) | (-) | (-) | (-) | (-) | |||||
| Not high | 1 | (-) | 1 | 1 | ||||||
| High | 4.11 (2.58,6.54) | <0.001 | (-) | 7.56 (3.47,16.5) | <0.001 | 3.03 (1.69,5.43) | <0.001 | |||
|
| ||||||||||
|
Validation cohorts I
and II combined | ||||||||||
| Mesothelin | Risk | (-) | 1.10 (1.07,1.13) | <0.001 | 1.07 (1.04,1.11) | <0.001 | 1.05 (1.02,1.09) | 0.004 | 1.11 (1.03,1.21) | 0.01 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 2.04 (1.42,2.91) | <0.001 | 2.10 (1.2,3.68) | 0.009 | 2.00 (1.22,3.28) | 0.006 | |||
|
| ||||||||||
| FLT4 | Risk | (-) | 1.01 (1.00,1.02) | 0.003 | 1.01 (1.00,1.02) | 0.009 | 1.01 (1.00,1.03) | 0.014 | 1.01 (1.00,1.02) | 0.183 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 2.46 (1.77,3.44) | <0.001 | 2.98 (1.78,5) | <0.001 | 2.33 (1.48,3.68) | <0.001 | |||
|
| ||||||||||
| AGP | Risk | (-) | 2.16 (1.63,2.86) | <0.001 | 1.86 (1.38,2.51) | <0.001 | 2.12 (1.41,3.18) | <0.001 | 1.69 (1.08,2.65) | 0.021 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 2.14 (1.52,3.02) | <0.001 | 2.68 (1.62,4.46) | <0.001 | 2.00 (1.23,3.26) | 0.005 | |||
|
| ||||||||||
| CA-125 | Risk | (-) | 1.03 (1.02,1.04) | <0.001 | 1.00 (1.00,1.00) | <0.001 | 1.01 (1.00,1.03) | 0.045 | 1.05 (1.03,1.08) | <0.001 |
| Not high | (-) | 1 | 1 | 1 | ||||||
| High | (-) | 2.32 (1.65,3.26) | <0.001 | 2.53 (1.52,4.24) | <0.001 | 2.13 (1.34,3.37) | 0.001 | |||
|
| ||||||||||
| Risk | (-) | (-) | (-) | (-) | (-) | |||||
| Not high | 1 | (-) | 1 | 1 | ||||||
| High | 2.53 (1.81,3.52) | <0.001 | (-) | 2.69 (1.62,4.46) | <0.001 | 2.51 (1.61,3.91) | <0.001 | |||
Similar results were observed when stratifying analysis by treatment arm (Table 3, column 3-4) and adjusting for high risk of progression status with the exception of mesothelin in the standard arm. This evidence of independent prognostic potential for FLT4 and AGP warrants further investigation in a larger cohort and was also evident in KM survival functions for optimal cut-points in biomarker concentration (Supplementary Figure 6). None of these models showed evidence of violating the proportional hazards assumption.
Predictive potential of individual biomarkers
All three candidate biomarkers and CA-125 showed evidence of prognostic potential (Table 3). However, to determine if a marker has predictive potential a contrasting effect in treatment arms must be identified (usually through a significant treatment/biomarker interaction term in a Cox PH model). Cox PH models with terms for marker concentration, treatment and the relevant interaction term gave non-significant Wald tests on the interaction term, indicating that individually the markers showed no evidence of predictive potential (Supplementary Figure 6, column d). However, examination of plots for treatment effects separated by optimal cut-points (derived by maximising Harrell’s C-index) for biomarker concentrations (Supplementary Figure 6, column b, c) showed some evidence of non-additive effects, suggesting further investigation to combine the effect of biomarkers in a predictive score.
A predictive biomarker index to inform treatment decisions
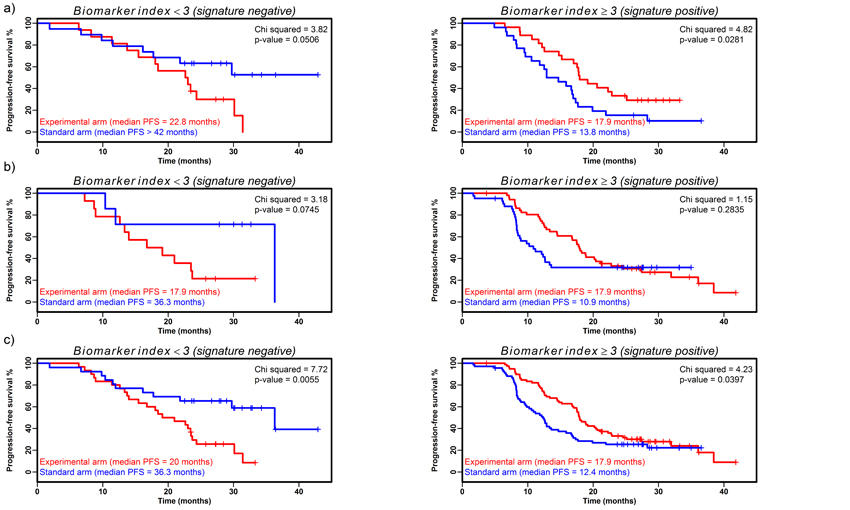
A biomarker index was constructed as described and when Cox PH models with index, experimental arm and interaction terms were estimated for the dichotomized index at each potential cut-point (no patients had index=0, implying dichotomization around 1, 2 and 3) the optimum model had a significant interaction term (p=0.006, LRT; full model shown in Supplementary Table 5) indicating potential predictive ability. When patients were separated at this optimum cut-point (index<3; index≥3), Cox’s PH models for each treatment arm yielded significant results with contrasting beneficial treatments (signature-negative (n=35): HR=2.38; 95%CI 0.97-5.81; p=0.058 and signature-positive (n=53): HR=0.51; 95%CI 0.28-0.94; p=0.031, Supplementary Table 5). KM estimates of survival functions highlighted the difference between treatment arms in each signature group (Figure 3a). In the signature-negative group, patients responded better to the standard therapy (median PFS standard arm not attained, but >42 months; experimental arm 22.8 months; p=0.051, logrank test), whereas in the signature-positive group the median PFS for patients on the experimental arm was 4.1 months longer (median PFS standard arm 13.8 months; experimental arm 17.9 months; p=0.028, logrank test). Constructing the index similarly excluding CA-125 resulted in a significant interaction in the Cox PH model (p=0.035), but an index with lesser predictive ability.
Figure 3. Predictive potential of biomarker index in signature-negative and signature-positive patient groups.
Kaplan-Meier estimates of survival functions for biomarker index in signature-negative and positive groups separated by treatment arm in a) validation cohort I, b) validation cohort II and c) validation cohorts I and II combined. Index constructed by calculating cut-points for each marker (mesothelin = 0.96 nM; FLT4 = 45.45 ng/ml; AGP = 1.29 g/L; CA-125 = 295 kU/L; all p<0.05) converting to binary and summing for each patient before dichotomising around the optimal index cut-point to give signature-negative and signature-positive patient groups (index construction described fully in Supplementary Methods).
The biomarker index showed weak evidence of prognostic potential overall (p=0.099; logrank test; Supplementary Figure 7a) representing the combined and conflicting effects in each arm (Supplementary Figure 7b). Considering age, FIGO stage, histology, ECOG performance status, surgical outcome and those patients classified clinically to be at higher risk of progression, there was no significant difference in the composition of the groups which could explain the better prognosis in the signature-negative, standard arm (age p>0.05, Kruskal-Wallis test, all others p>0.05, Fisher’s exact test).
The final Cox PH model for the biomarker index was internally validated using bootstrap resamples to estimate the optimism in the model (16). The final model had an estimated Harrell’s concordance index (C) of 0.62 and the optimism (O) was estimated to be 0.05 from 1000 resamples. As the estimated C-O was >0.5, this implies that the final model has predictive ability. The second baseline sample at time point 2 also allowed the evaluation of the model in a smaller subset of patients (N=58 with 38 PFS events). The results were similar with coefficients of the same sign and similar magnitude, but with increased Wald test p-values (all p<0.150) most likely caused by the lack of power given the smaller cohort.
Biomarker validation - cohort II
Association between candidate biomarker concentrations at baseline and PFS
The univariate associations seen between candidate biomarkers and PFS observed in validation cohort I were confirmed in validation cohort II (Table 3: validation cohort II, p≤0.05). Multivariable associations overall and in each trial arm also showed similar effects, but were not statistically significant (p>0.05).
Validating the biomarker index
The biomarker index was re-calculated using the 115 time point 1 samples in validation cohort II with the same methods and cut-points as outlined above. The subsequent model showed further evidence for a predictive effect for the biomarker index (interaction term from the LRT, p=0.060, full model shown in Supplementary Table 5). Separating at the optimal cut-point, there was further evidence from the Cox’s PH models of contrasting benefit in each treatment arm (signature-negative (n=21): HR=3.64; 95%CI 0.80-16.57; p=0.095 and signature-positive (n=94): HR=0.76; 95%CI 0.47-1.25; p=0.283). KM estimates of median survival indicated differences between treatment arms in each signature group (Figure 3b) with patients in the signature-negative standard arm responding better than patients in the experimental arm of the same signature group (median PFS in the standard arm 36.3 months, experimental arm 17.9 months) with the converse result seen in the signature-positive group (median PFS in the standard arm 10.9 months, experimental arm 17.9 months).
Biomarker validation - combined cohorts I and II
Association between candidate biomarker concentrations at baseline and PFS
Using the data from 203 time point 1 samples from both validation cohorts allowed a more powerful assessment of prognostic potential of the proteins. Significant univariate associations were again seen (Table 3: combined data, p≤0.003). Upon multivariate analysis the candidate markers and CA-125 were independently prognostic overall and in each arm separately (p≤0.045), the exception being FLT4 in the experimental arm (p=0.183).
Further validation of biomarker index
The biomarker index was recalculated using time point 1 samples from the 207 patients from validation cohorts I and II combined, using the same methods and cut-points as described above. The resultant model had a significant interaction term from the LRT (p=0.001, full model shown in Supplementary Table 5) and KM survival estimates show differences between treatment arm for both signature groups (Figure 3c), again with the standard arm responding better in the signature negative group (median PFS in the standard arm 36.3 months, experimental arm 20 months; p=0.005, logrank test) and the experimental arm responding better in the signature positive group (median PFS in the standard arm 12.4 months, experimental arm 17.9 months; p=0.040, logrank test).
Further analysis of the candidate markers in the combined validation cohorts (with increased statistical power) suggested that mesothelin and AGP combined may perform equally as well as the biomarker index (Supplementary Tables 6-8, Supplementary Figure 8).
Discussion
Bevacizumab has demonstrated efficacy in a number of different solid tumours (17-20) but identification of patients who would derive most benefit would allow more selective and appropriate usage, improving efficacy and cost-effectiveness. In this exploratory translational study focussed on proteomic identification of relevant serum biomarkers in ICON7 patients, we identified 3 candidate biomarkers (FLT4, AGP and mesothelin) which we then sought to validate on larger cohorts. Although baseline values of all candidate markers appeared to have prognostic value, unfortunately none were individually predictive of benefit from the addition of bevacizumab. However, combining these with CA-125 in a biomarker index, a clear predictive value was seen in validation cohort I with signature-positive patients shown to benefit from the addition of bevacizumab.
We performed additional validation of the biomarker index in a further independent cohort of ICON7 patients. Using an approximation based on the LRT for interaction (21) with the hazard ratios observed in validation cohort I for the signature negative (HR1=2.38) and signature positive (HR2=0.51) groups, a sample size of 15 PFS events in each group (signature positive or negative crossed with trial arm) would be required at a significance level of 5% and 80% power. However, post-hoc power analysis based on the true number of events observed in each group and the reduced interaction effect (HR1=2.58 and HR2=0.75) meant that the sample had a power of only 43%. In some part this explains the disparate significance levels for the interaction effect in validation cohorts I and II as a larger and more matched cohort would have been more appropriate.
The identification of a biomarker index with potential clinical utility demonstrates the potential of proteomics. However, limitations need to be acknowledged. Firstly, with practical limitations influencing sample number in the discovery phase, only patients on the experimental arm were included. Potential markers were then examined for predictive utility by analysing samples from patients in both arms in the subsequent validation phase. Ideally comparing both arms at the discovery phase would have allowed earlier highlighting of bevacizumab-specific response biomarkers. Secondly, although the initial immunoassay results on the discovery samples did reproduce the MS findings, not all findings replicated independently within the larger validation cohorts, presumably due to biological variation and the small size of the discovery set. For example, changes seen between time points 1 and 4 for the various proteins were reproduced in the validation cohort. However, the finding of higher baseline mesothelin being associated with poorer outcome/response in the validation set was not apparent in the MS data.
Selection of candidate biomarkers for further validation included consideration of biological relevance. Mesothelin is over-expressed in EOC, involved in tumour progression (22, 23) and associated with chemoresistance and a poorer overall prognosis (24). It has also been proposed as a potential biomarker for EOC identification (25, 26), and in mesothelioma as a putative biomarker to monitor response to treatment (27). Our association between baseline mesothelin and risk of EOC progression may relate to increased disease burden. Mesothelin is synthesised as a precursor and cleaved to form soluble megakaryocyte potentiating factor and membrane-bound mesothelin. The two peptides identified in the MS study were both in the mesothelin part of the molecule and are present in variants including the soluble forms. The immunoassay used detects forms including variant 1, most frequently found in ascitic fluid due to shedding from EOC cells (28), and variant 3.
FLT4 (VEGFR3) is a receptor for VEGF-C and VEGF-D, and plays a key role in angiogenesis, being implicated in breast cancer for example (29). FLT4 signalling leads to increased VEGF-C, and VEGF-A. The reduction of soluble FLT4 here only in patients treated with bevacizumab is presumably due to the effect of bevacizumab on the VEGF-related angiogenic pathway. However, as it is not independently predictive of response, non-responders may have alternative angiogenic pathways predominating. Similar changes are seen in serum VEGFR2 in patients with renal cancer treated with sorafenib, an anti-angiogenic tyrosine kinase inhibitor (30).
AGP is an acute phase protein which is elevated in several cancers (31, 32), and although not previously linked with EOC, it has been reported to have pro-angiogenic properties and to support the pro-angiogenic effect of VEGF-A (33).
The predictive ability of the biomarker index is promising and a strength is the derivation from a major RCT, allowing a comparison between the standard and experimental arms, high quality clinical data and stringent sample processing (34). The signature requires further validation including eventual prospective validation within a clinical trial, for example randomising to bevacizumab based on biomarker index, and possible investigation of relevance in other solid tumours treated with bevacizumab, should be considered. Circulating protein and cellular biomarkers, single nucleotide polymorphisms (SNPs), expression arrays, and clinical correlates such as hypertension have been examined for potential association with bevacizumab effectiveness (reviewed in (35)) but no definitive biomarkers have yet been found. Higher baseline VEGF-A is most promising as potentially predictive of bevacizumab benefit in breast cancer (AVADO) (36), gastric cancer (AVAGAST) (37) and pancreatic cancer (AViTA) (38). However, in some studies it has appeared only prognostic rather than predictive, for example in colorectal cancer (AVF2107g) (39) and renal cancer (AVOREN) (38), possibly reflecting study design, specific disease type or assay. Other promising markers include delta-like ligand-4, VEGF-C, neuropilin (40) and a VEGFR2 SNP (41) in breast cancer, high circulating intercellular adhesion molecule (ICAM) in non-small cell lung cancer (although as there was still benefit in the low ICAM group this appeared to be prognostic rather than predictive) (42) and day 4 circulating endothelial progenitor cells and proportion of baseline CXCR4-positive circulating endothelial cells in colorectal cancer (43). Hypertension has also been proposed (44, 45), although again not borne out in all studies (46).
Predictive biomarkers relating to bevacizumab in EOC are exploratory and limited to smaller phase II studies (reviewed in (47)). High tumour VEGF levels have been reported to be associated with early progression in patients treated with bevacizumab and erlotinib (48), whereas in patients treated with gemcitabine/oxaliplatin and bevacizumab (n=19), increases in plasma placental growth factor and VEGFR2-expressing monocytes correlated with EOC outcomes (49). Examination of biomarkers relating to alternative angiogenic pathways (for example platelet-derived growth factor and fibroblast growth factor) found none to be of predictive value in a study of 106 patients with chemotherapy-resistant EOC treated with single agent bevacizumab (50).
Based on the results from ICON7 and GOG218, bevacizumab is the first targeted therapy to be licensed for use in the first line treatment of EOC, in combination with carboplatin and paclitaxel. Current clinical practice is variable, with some clinicians using it in all patients with advanced EOC, and others using it only in those clinically identified at high risk of progression. There is clearly still a need to identify biomarkers predictive of response to bevacizumab, to enable avoidance of potential toxicities in those least likely to benefit and also to maximise the cost effectiveness of this drug. Our study has identified a biomarker index which appears predictive of benefit from bevacizumab but further validation of clinical utility in a larger population, including assessment of the predictive ability of all possible biomarker combinations, is needed.
Supplementary Material
Translational relevance.
The benefit seen from the addition of bevacizumab to standard chemotherapy in the ICON7 trial was relatively modest in the overall population studied in terms of progression-free survival (1.5 months). When additional clinical factors were used to identify the population of patients at high risk of progression, the benefit specifically to this group increased to 3.6 months. Several biomarkers predictive of bevacizumab response have been reported in a number of different tumours, with promising preliminary results. This translational research study aligned to the ICON7 trial identified a biomarker signature which appears to assist in identifying patients with ovarian cancer most likely to benefit from bevacizumab treatment.
Acknowledgments
We would like to thank all patients and staff at the various sites who participated in this research, the MRC CTU and Dr. Helene Hoegsbro Thygesen and Prof. Jenny Barrett for statistical discussions.
Financial Support: This research study was funded by Cancer Research UK [C2075/A7966]. The ICON7 trial was sponsored by the Medical Research Council and managed internationally by the MRC Clinical Trials Unit in London with the clinical trial and sample collection funded by Hoffmann-La Roche. DAC is supported by a UK Medical Research Council Career Development Fellowship [G0802416].
Footnotes
Conflicts of Interest: GJ and JL have sat on Advisory Boards for Roche and JL has lectured at a Roche-sponsored educational meeting, with any payments relating to these activities being made to the Institutions. GJ has received research funds from Roche. None of the other authors have anything to declare.
References
- [1].Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- [2].Stuart GC, Kitchener H, Bacon M, duBois A, Friedlander M, Ledermann J, et al. 2010 Gynecologic Cancer InterGroup (GCIG) consensus statement on clinical trials in ovarian cancer: report from the Fourth Ovarian Cancer Consensus Conference. Int J Gynecol Cancer. 2011;21:750–5. doi: 10.1097/IGC.0b013e31821b2568. [DOI] [PubMed] [Google Scholar]
- [3].Shen GH, Ghazizadeh M, Kawanami O, Shimizu H, Jin E, Araki T, et al. Prognostic significance of vascular endothelial growth factor expression in human ovarian carcinoma. Br J Cancer. 2000;83:196–203. doi: 10.1054/bjoc.2000.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Han ES, Burger RA, Darcy KM, Sill MW, Randall LM, Chase D, et al. Predictive and prognostic angiogenic markers in a gynecologic oncology group phase II trial of bevacizumab in recurrent and persistent ovarian or peritoneal cancer. Gynecol Oncol. 2010;119:484–90. doi: 10.1016/j.ygyno.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484–96. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- [6].Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–83. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- [7].Smith MP, Wood SL, Zougman A, Ho JT, Peng J, Jackson D, et al. A systematic analysis of the effects of increasing degrees of serum immunodepletion in terms of depth of coverage and other key aspects in top-down and bottom-up proteomic analyses. Proteomics. 2011;11:2222–35. doi: 10.1002/pmic.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- [9].Craven RA, Cairns DA, Zougman A, Harnden P, Selby PJ, Banks RE. Proteomic analysis of formalin-fixed paraffin-embedded (FFPE) renal tissue samples by label-free mass spectrometry: assessment of overall technical variability and the impact of block age. Proteomics Clin Appl. 2012 doi: 10.1002/prca.201200065. [DOI] [PubMed] [Google Scholar]
- [10].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- [11].Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- [12].McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Urol. 2005;2:416–22. [PubMed] [Google Scholar]
- [13].Cairns DA, Perkins DN, Stanley AJ, Thompson D, Barrett JH, Selby PJ, et al. Integrated multi-level quality control for proteomic profiling studies using mass spectrometry. BMC Bioinformatics. 2008;9:519–519. doi: 10.1186/1471-2105-9-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–26. [Google Scholar]
- [15].R Development Core Team . Vienna, Austria: 2011. R: A language and environment for statistical computing. [Google Scholar]
- [16].Harrell FE, Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [17].Miller KD, Chap LI, Holmes FA, Cobleigh MA, Marcom PK, Fehrenbacher L, et al. Randomized phase III trial of capecitabine compared with bevacizumab plus capecitabine in patients with previously treated metastatic breast cancer. J Clin Oncol. 2005;23:792–9. doi: 10.1200/JCO.2005.05.098. [DOI] [PubMed] [Google Scholar]
- [18].Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27:4733–40. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- [19].Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–42. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- [20].Rini BI, Halabi S, Rosenberg JE, Stadler WM, Vaena DA, Ou SS, et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol. 2008;20:5422–8. doi: 10.1200/JCO.2008.16.9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Peterson B, George SL. Sample size requirements and length of study for testing interaction in a 2 × k factorial design when time-to-failure is the outcome. Control Clin Trials. 1993;14:511–22. doi: 10.1016/0197-2456(93)90031-8. [DOI] [PubMed] [Google Scholar]
- [22].Tang Z, Qian M, Ho M. The role of mesothelin in tumor progression and targeted therapy. Anticancer Agents Med Chem. 2012 doi: 10.2174/1871520611313020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chang MC, Chen CA, Chen PJ, Chiang YC, Chen YL, Mao TL, et al. Mesothelin enhances invasion of ovarian cancer by inducing MMP-7 through MAPK/ERK and JNK pathways. Biochem J. 2012;442:293–302. doi: 10.1042/BJ20110282. [DOI] [PubMed] [Google Scholar]
- [24].Cheng WF, Huang CY, Chang MC, Hu YH, Chiang YC, Chen YL, et al. High mesothelin correlates with chemoresistance and poor survival in epithelial ovarian carcinoma. Br J Cancer. 2009;100:1144–53. doi: 10.1038/sj.bjc.6604964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fritz-Rdzanek A, Grzybowski W, Beta J, Durczynski A, Jakimiuk A. HE4 protein and SMRP: Potential novel biomarkers in ovarian cancer detection. Oncol Lett. 2012;4:385–9. doi: 10.3892/ol.2012.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst. 2010;102:26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Franko A, Dolzan V, Kovac V, Arneric N, Dodic-Fikfak M. Soluble mesothelin-related peptides levels in patients with malignant mesothelioma. Dis Markers. 2012;32:123–31. doi: 10.3233/DMA-2011-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hellstrom I, Raycraft J, Kanan S, Sardesai NY, Verch T, Yang Y, et al. Mesothelin variant 1 is released from tumor cells as a diagnostic marker. Cancer Epidemiol Biomarkers Prev. 2006;15:1014–20. doi: 10.1158/1055-9965.EPI-05-0334. [DOI] [PubMed] [Google Scholar]
- [29].Valtola R, Salven P, Heikkila P, Taipale J, Joensuu H, Rehn M, et al. VEGFR-3 and its ligand VEGF-C are associated with angiogenesis in breast cancer. Am J Pathol. 1999;154:1381–90. doi: 10.1016/S0002-9440(10)65392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zurita AJ, Jonasch E, Wang X, Khajavi M, Yan S, Du DZ, et al. A cytokine and angiogenic factor (CAF) analysis in plasma for selection of sorafenib therapy in patients with metastatic renal cell carcinoma. Ann Oncol. 2012;23:46–52. doi: 10.1093/annonc/mdr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].El-Najjar N, Ketola RA, Nissila T, Mauriala T, Antopolsky M, Janis J, et al. Impact of protein binding on the analytical detectability and anticancer activity of thymoquinone. J Chem Biol. 2011;4:97–107. doi: 10.1007/s12154-010-0052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ongay S, Martin-Alvarez PJ, Neususs C, de FM. Statistical evaluation of CZE-UV and CZE-ESI-MS data of intact alpha-1-acid glycoprotein isoforms for their use as potential biomarkers in bladder cancer. Electrophoresis. 2010;31:3314–25. doi: 10.1002/elps.201000244. [DOI] [PubMed] [Google Scholar]
- [33].Irmak S, Oliveira-Ferrer L, Singer BB, Ergun S, Tilki D. Pro-angiogenic properties of orosomucoid (ORM) Exp Cell Res. 2009;315:3201–9. doi: 10.1016/j.yexcr.2009.07.024. [DOI] [PubMed] [Google Scholar]
- [34].Jackson DH, Banks RE. Banking of clinical samples for proteomic biomarker studies: a consideration of logistical issues with a focus on pre-analytical variation. Proteomics Clin Appl. 2010;4:250–70. doi: 10.1002/prca.200900220. [DOI] [PubMed] [Google Scholar]
- [35].Murukesh N, Dive C, Jayson GC. Biomarkers of angiogenesis and their role in the development of VEGF inhibitors. Br J Cancer. 2010;102:8–18. doi: 10.1038/sj.bjc.6605483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jayson GC, Hicklin DJ, Ellis LM. Antiangiogenic therapy--evolving view based on clinical trial results. Nat Rev Clin Oncol. 2012;9:297–303. doi: 10.1038/nrclinonc.2012.8. [DOI] [PubMed] [Google Scholar]
- [37].van Cutsem E, de HS, Kang YK, Ohtsu A, Tebbutt NC, Ming XJ, et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a biomarker evaluation from the AVAGAST randomized phase III trial. J Clin Oncol. 2012;30:2119–27. doi: 10.1200/JCO.2011.39.9824. [DOI] [PubMed] [Google Scholar]
- [38].Lambrechts D, Claes B, Delmar P, Reumers J, Mazzone M, Yesilyurt BT, et al. VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol. 2012;13:724–33. doi: 10.1016/S1470-2045(12)70231-0. [DOI] [PubMed] [Google Scholar]
- [39].Holden SN, Ryan E, Kearns A, Holmgren E. Hurwitz. Benefit from bevacizumab (BV) is independent of pretreatment plasma vascular endothelial growth factor-A (pl-VEGF) in patients (pts) with metastatic colorectal cancer (mCRC) J Clin Oncol. 2005;23:16S. Abstract 3555. [Google Scholar]
- [40].Jubb AM, Miller KD, Rugo HS, Harris AL, Chen D, Reimann JD, et al. Impact of exploratory biomarkers on the treatment effect of bevacizumab in metastatic breast cancer. Clin Cancer Res. 2011;17:372–81. doi: 10.1158/1078-0432.CCR-10-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26:4672–8. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dowlati A, Gray R, Sandler AB, Schiller JH, Johnson DH. Cell adhesion molecules, vascular endothelial growth factor, and basic fibroblast growth factor in patients with non-small cell lung cancer treated with chemotherapy with or without bevacizumab--an Eastern Cooperative Oncology Group Study. Clin Cancer Res. 2008;14:1407–12. doi: 10.1158/1078-0432.CCR-07-1154. [DOI] [PubMed] [Google Scholar]
- [43].Matsusaka S, Mishima Y, Suenaga M, Terui Y, Kuniyoshi R, Mizunuma N, et al. Circulating endothelial progenitors and CXCR4-positive circulating endothelial cells are predictive markers for bevacizumab. Cancer. 2011;117:4026–32. doi: 10.1002/cncr.25977. [DOI] [PubMed] [Google Scholar]
- [44].de Stefano A, Carlomagno C, Pepe S, Bianco R, De PS. Bevacizumab-related arterial hypertension as a predictive marker in metastatic colorectal cancer patients. Cancer Chemother Pharmacol. 2011;68:1207–13. doi: 10.1007/s00280-011-1604-1. [DOI] [PubMed] [Google Scholar]
- [45].Osterlund P, Soveri LM, Isoniemi H, Poussa T, Alanko T, Bono P. Hypertension and overall survival in metastatic colorectal cancer patients treated with bevacizumab-containing chemotherapy. Br J Cancer. 2011;104:599–604. doi: 10.1038/bjc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dewdney A, Cunningham D, Barbachano Y, Chau I. Correlation of bevacizumab-induced hypertension and outcome in the BOXER study, a phase II study of capecitabine, oxaliplatin (CAPOX) plus bevacizumab as peri-operative treatment in 45 patients with poor-risk colorectal liver-only metastases unsuitable for upfront resection. Br J Cancer. 2012;106:1718–21. doi: 10.1038/bjc.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Raja FA, Hook JM, Ledermann JA. Biomarkers in the development of anti-angiogenic therapies for ovarian cancer. Cancer Treat Rev. 2012;38:662–72. doi: 10.1016/j.ctrv.2011.11.009. [DOI] [PubMed] [Google Scholar]
- [48].Chambers SK, Clouser MC, Baker AF, Roe DJ, Cui H, Brewer MA, et al. Overexpression of tumor vascular endothelial growth factor A may portend an increased likelihood of progression in a phase II trial of bevacizumab and erlotinib in resistant ovarian cancer. Clin Cancer Res. 2010;16:5320–8. doi: 10.1158/1078-0432.CCR-10-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Horowitz NS, Penson RT, Duda DG, di TE, Boucher Y, Ancukiewicz M, et al. Safety, Efficacy, and Biomarker Exploration in a Phase II Study of Bevacizumab, Oxaliplatin, and Gemcitabine in Recurrent Mullerian Carcinoma. Clin Ovarian Cancer Other Gynecol Malig. 2011;4:26–33. doi: 10.1016/j.cloc.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Madsen CV, Steffensen KD, Olsen DA, Waldstrom M, Adimi P, Smerdel M, et al. Serial measurements of serum PDGF-AA, PDGF-BB, FGF2, and VEGF in multiresistant ovarian cancer patients treated with bevacizumab. J Ovarian Res. 2012 doi: 10.1186/1757-2215-5-23. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.