Abstract
Three-dimensional in vitro extracellular matrix models provide a physiological alternative to regular two-dimensional cell culture, though they lack the full diversity of molecular composition and physical properties of whole-animal systems. Cell-derived matrices are extracellular matrices that are the product of matrix secretion and assembly by cells cultured at high density in vitro. After the removal of the cells that produced the matrix, an assembled matrix scaffold is left that closely mimics native stromal fiber organization and molecular content. Cell-derived matrices have been shown to impart in vivo-like responses to cells cultured in these matrices. In this review, we focus on mechanisms through which the distinct molecular and topographical composition of cell-derived matrices directs cellular behavior, specifically through regulation of cell-matrix adhesions and subsequent contributions to the process of cell migration.
Keywords: cell-derived matrix (CDM), extracellular matrix, 3D cell migration, matrix adhesions, fibrillar topography, matrix elasticity
Introduction
The use of 3D in vitro models to bridge the gap between cell culture and in vivo systems permits greater understanding of the physical and chemical interactions between cells and the 3D extracellular matrix (ECM). Cell-derived matrices (CDMs) provide a more complex, physiological alternative to purified 3D matrix protein scaffolds, such as polymerized type I collagen or fibrin gels; CDMs consist of a heterogeneous mixture of proteins, proteoglycans and growth factors similar to the native stromal environment [1]. CDMs are naturally produced by the continued secretion and organization of ECM components into a 3D matrix when certain cell types are cultured at high density in vitro. Within several (3–10) days, a thick 3D matrix is generated that can be denuded of cells to yield a 3D ECM substrate [2]. Fibroblasts are responsible for in vivo stromal ECM deposition and maintenance and are commonly used for CDM generation, though 3D CDMs have been produced from endothelial, epithelial, stem cell, and cancer cell cultures [3–6].
Biochemical and biophysical characterization of CDMs
Fibroblast-generated CDM is a heterogeneous fibrous matrix, consisting primarily of a meshwork of linear fibronectin fibrils that can be oriented in parallel or more random in organization (Fig. 1, magenta fibronectin fibers). Additional matrix proteins such as collagen I and IV, perlecan, tenascin-C, hyaluronic acid, and heparin sulfate proteoglycans are present in lower abundance, as well as sequestered growth factors [3, 7]. This diversity and spatial heterogeneity of CDM components mimic what is found in in vivo matrix, providing physiological properties not commonly found in traditional polymerized or synthetic 3D in vitro scaffolds. Because CDM is generated by the secretion and assembly of matrix fibers from layers of confluent cells in vitro, its topography consists of arrays of fibronectin fibers that are stacked to an approximate depth of 5–20 microns [5, 8–10]. These fibers have an average inter-fiber spacing of approximately 1–2 microns [9], resulting in larger internal spaces in comparison to other 3D ECM models. This distinct matrix architecture provides an advantage for microscopy by minimizing light scatter through the matrix and additionally ensures relatively constant nutrient delivery and physical characteristics throughout the CDM model.
Figure 1. Fibroblast migration in 3D cell-derived matrix.
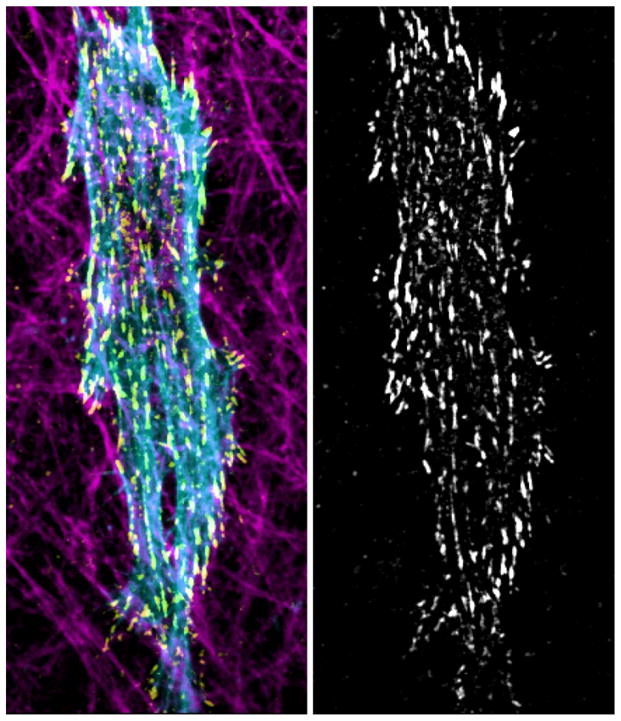
Left: Maximum projection confocal image of a human foreskin fibroblast (HFF) embedded within a matrix containing oriented CDM fibronectin fibrils (magenta, anti-fibronectin immunostaining). Cytoskeletal architecture visualized by staining for actin (blue, phalloidin) and paxillin-containing focal adhesions (yellow, anti-paxillin). Right: Maximum projection image of focal adhesion profile of paxillin-containing adhesions during migration in CDM.
One concern raised about most 3D in vitro matrix models, whether comprised of a purified protein or CDM, is the physiological relevance of its matrix stiffness, particularly regions in close proximity to a rigid underlying glass or plastic surface (>1 GPa). This issue of local matrix properties is relevant biologically, since ECM stiffness is sensed by cells through cell-matrix adhesions and can alter intracellular signaling pathways, such as by Rho GTPases, leading to changes in migration, differentiation and proliferation [11–13]. Atomic force microscopy measurements of CDM have reported a Young’s modulus in the range of 200–600 Pa, which is comparable to reconstituted basement membrane (175 Pa), 3D collagen gels (concentration dependent, 15–1000 Pa), and dermal tissue explants (> 300 Pa) [8, 9, 13, 14]. Despite their relatively shallow depth and proximity to an artificial, rigid surface, CDMs are a physiologically-compliant matrix, allowing for flexibility and malleability similar to in vivo ECM. An additional physical parameter that has recently been identified as a regulator of cell migration and signaling is the elastic behavior of the matrix, i.e., non-linear or linear elasticity. In contrast to non-linearly elastic 3D collagen matrices, CDMs are linearly elastic and do not undergo strain stiffening under increasing force, perhaps due to its distinct composition and less crosslinking between fibers compared to meshwork collagen matrices. Using CDM, Petrie et al. demonstrated that fibroblasts have the ability to discern the elastic behavior of the ECM and accordingly switch modes of migration [14].
Besides possessing biochemical and physical characteristics that mimic in vivo matrices, CDM is by nature an in vitro system and thus more accessible to experimental manipulation. The composition of the medium used for culturing cells during CDM production can be adjusted to alter matrix composition, thickness, and topography. For example, the addition of ascorbic acid to culture media will increase the collagen content of the CDM and addition of epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) will increase overall ECM production and matrix thickness [8, 9]. Genetic ablation and RNA interference analyses of the cultured fibroblasts during CDM production can permit direct examination of the pathways involved in ECM synthesis, deposition, and organization. Recently, Goetz el al. discovered a role for caveolin-1 in the topographical assembly and remodeling of 3D microenvironments by examining the organization and compliance of CDM fibronectin fibers derived from wild-type and caveolin-1 knockout fibroblasts [15]. Additionally, post-synthesis chemical manipulation of CDM by crosslinking the matrix or by treating with trypsin can modulate the stiffness and elastic behavior of the CDM [14]. Taken together, CDM provides an in vitro 3D model that closely mimics the chemical and physical heterogeneity found in vivo, but which also allows for experimental manipulation in composition and physical properties to test hypotheses in ways that are not possible in vivo.
Cell-derived matrices regulate adhesion composition and stability
It is well known that mesenchymally derived fibroblasts require attachment or adhesion to the surrounding extracellular matrix for cell migration as well as maintenance of cell homeostasis and survival. Other cell types, such as many cancer cell lines and hematopoietic cells can not only migrate, but can survive in the absence cell-matrix attachment [16]. In this section, we will focus solely on cell-CDM adhesions made by fibroblasts and other anchorage-dependent cell types.
Cell-matrix interactions at cell adhesions not only provide physical support for cell migration, but can also act as outside-in signaling hubs that initiate or modulate a wide range of intracellular signaling cascades. Linkage to the surrounding matrix is often initiated by integrins, a group of heterodimeric transmembrane proteins consisting of thirteen α and seven β subunits, whose different combinations dictate their ECM specificity [17]. While multiple pairs of α and β integrin subunits can mediate adhesion to the same type of ECM molecule, the strength of their association can vary greatly. For example, both αvβ3 and α5β1 can bind to fibronectin, the primary component of 3D CDMs, yet the adhesion strength of α5β1-containing adhesions can be six-fold higher [18]. Within 3D CDMs, the high quantity of fibronectin tends to favor α5β1 associated attachments, yet Chinese hamster ovary (CHO) cells that lack the α5 subunit can still attach and become imbedded in the matrix through αvβ3-matrix interactions [19]. A key connection between integrins and the surrounding ECM is through the β subunit, though the α subunits are also essential for function. A number of integrins can bind to short peptide sequences in matrix proteins, such as the Arginine-Glycine-Aspartic acid (RGD) sequence found on fibronectin, vitronectin, and other ECM proteins [20].
The cytoplasmic tails of integrins interact with numerous intracellular proteins in large multimolecular complexes at cell adhesions. These proteins vary widely in function, from mechanosensitive proteins such as vinculin, to signaling molecules like Src, to cytoskeletal adaptor proteins such as paxillin (Fig. 1, yellow adhesions). These proteins initially localize or accumulate at clustered integrins to form small foci that can become directly linked to the actin cytoskeleton. It is at these cellular adhesions that actomyosin contractility can be propagated to the external environment. Hence, cellular adhesions are the sites of mechanotransduction between the cell and its surroundings. We use the term cell adhesions as a loose general classification that include nascent adhesions, focal complexes, focal adhesions, fibrillar adhesions, and 3D matrix adhesions each can be considered a distinct subset of adhesions with different molecular components and associations with the actin cytoskeleton [21]. The majority of adhesion studies have been performed on 2D rigid glass surfaces, often coated with an adsorbed globular ECM molecule, such as fibronectin. In general, 2D cell-ECM adhesions proceed through a maturation process; that is they grow in size, increase the number of proteins that associate within them, and progress in order from focal contacts through fibrillar adhesions [22]. While both nascent adhesions and focal complexes have been observed and well-characterized on 2D substrates, they have not been identified within 3D CDM to date. This is likely due to their small size, relatively short longevity (under 90 seconds), and absence of requirement for actin association [23]. However, the absence of nascent adhesions in 3D CDMs suggests that nearly all develop into actin-linked, contractility-dependent adhesions [24]. 3D matrix adhesions (3D-MA) formed by cells with 3D matrix fibrils have characteristics of both focal (FA) and fibrillar (FX) adhesions [10]. More than a dozen years ago, Cukierman et al. [10] determined several key protein components of 3D-MA in CDM including: activated β1 integrin, α5 integrin, α-actinin, actin, talin, tensin, vinculin, focal adhesion kinase (FAK) and paxillin. Interestingly, FAK in 3D-MA is not phosphorylated at tyrosine 397, similar to fibrillar adhesions in 2D, while 2D FAs have highly phosphorylated FAK. Phosphorylation of FAK at this site has been correlated with rigidity sensing on 2D rigid surfaces, and is down-regulated on soft 2D substrates or in 3D collagen gels [25, 26]. Additionally, even though β3 integrin was plentiful in 2D FAs, it is absent from 3D-MA structures in CDM [21].
3D CDMs have been utilized to elucidate mechanisms of cancer cell invasion. Deakin and Turner have shown that the adaptor proteins hic-5 and paxillin are crucial for cancer cell attachment or for mediating 3D-MA adhesion dynamics in 3D CDMs, respectively [27]. Recently, proteomic analysis has identified greater than 700 different proteins and/or macromolecular complexes that reside within myosin II-dependent FAs formed on 2D rigid surfaces with 283 known to be a part of the integrin adhesome [28]. While this feat has yet to be accomplished for cells embedded within 3D CDMs, it seems likely that many FA-associated proteins will be found in 3D-MAs.
The coupling that occurs within cellular adhesions links integrins and the actin cytoskeleton. This integration site has been dubbed the molecular clutch, where relatively immobile integrins are indirectly coupled to rapidly flowing actin through binding interactions involving the numerous cell adhesion-associated proteins discussed above. It is now known that cellular adhesions are highly dynamic structures that assemble, grow or mature, become stable, and finally disassemble [24] with the latter three stages being highly dependent on actin-based contractility. Hence, nascent adhesions that are contractility-independent have a short lifespan or stability compared to contractility dependent FAs. Recent evidence suggests that the coupling between 3D-MAs to CDM is relatively high, showing a three-fold increase in length of time that these adhesions exist compared to 2D FAs [23]. One possible explanation is the high concentration of fibronectin within 3D CDMs: increasing ligand density on 2D surfaces leads to a decrease in FA turnover [29]. In addition, the fibrillar structure of fibronectin may also be a regulating factor. When fibronectin is physically stretched during the process of fibrillogenesis, interactions occur between multiple cryptic cystine residues within several fibronectin-type III (FN III) repeats between molecules, which has been shown to increase the ability of fibronectin to withstand forces compared to single molecules alone [30]. These ideas suggest that adhesion stability within 3D CDMs may be intrinsically related to the concentration of ECM proteins (specifically fibronectin) and their fibrillar nature.
One very important feature of 3D-MAs that is not normally found in 2D FAs is the abundance of α5β1 integrin localizing to adhesion sites. This localization is likely due to the high concentrations fibronectin in 3D CDMs. Hence, research investigating the interactions between integrins and fibronectin, regardless of ECM dimensionality, can contribute to our understanding of cellular adhesion to 3D CDMs. For example, increasing the concentration of fibronectin (from ~ 10 to 50 μg/ml) on 2D surfaces results in the appearance of α5 integrin in 2D FAs (A.D.D., unpublished results), which could explain the high amounts of this integrin in 3D-MAs interacting with thick fibrils of fibronectin. In contrast, αVβ3 is normally present in 2D FAs at low fibronectin concentrations of 1–10 μg/ml. Chemical crosslinking of 2D FN-coated dishes also produces a similar migration of α5 into FAs [31]. The amino terminal domain of the α5 subunit is known to bind directly to fibronectin at the FN III9 repeat, also known as the synergy site [32]. Although the RGD-binding site on β1 is the most crucial for binding ligand, the synergy site considerably promotes binding to fibronectin [33], especially when a cell is under tension. Friedland et al. [34] demonstrated that when bound to fibronectin, elevating cytoskeletal or ECM tension results in a switch of the α5β1 integrin from a relaxed to a tension state. This switch is associated with engagement of α5 with the synergy site. Mutating the synergy site resulted in weakening of the integrin-fibronectin linkage, suggesting that it is required for increasing ligand-receptor bond strength and likely the stabilization of cellular adhesions.
In summary, the adhesion components that constitute 3D-MAs show similarities with 2D FAs and FXs [23], but how these components are utilized for intracellular signaling remains to be determined. The major differences in molecular composition between these subtypes of cell adhesions are likely due to the ECM itself, where high concentrations of organized fibrillar fibronectin promote α5β1 integrin association, increasing cell adhesiveness and likely altering outside-in signaling events.
Fibrillar topography dictates cell migration in cell-derived matrix
Cell migration studies in 3D have achieved prominence due to the stark differences in morphology, motility, adhesion, and signaling observed in the same cell type by simply altering matrix dimensionality. These distinct phenotypes result from differences between 2D and 3D migratory environments, including presence of dorsal and ventral matrix adhesions, fibrillar topography of the matrix proteins, and steric inhibition of spreading and migration by matrix fibers and porosity [1, 35]. As discussed below, cell migration in CDM is regulated not only by chemical information (molecular composition), but particularly the topographical cues provided by the physical organization of the ECM ligands to which the cells adhere.
In oriented 3D CDMs, such as those generated by primary human fibroblasts that grow in strongly parallel patterns, cells adhere to the linear arrays of fibronectin fibrils, rapidly align and polarize along the fiber, and migrate in a high directional manner. This elongated, uniaxial migratory phenotype mimics the behavior of fibroblasts in vivo migrating along stromal fibrils [10, 36]. Unlike migration in 3D fibrous matrices such as collagen or fibrin, fibroblasts do not significantly remodel these highly organized CDM fibers. This finding is potentially explained by the inherent linear topography of the fibronectin fibrils generated during CDM production by parallel arrays of cells and the large interfiber spacing (≥2 um) [9] of CDM in comparison to the dense lattices of fibrous collagen and fibrin gels. Interestingly, this elongated, spindle-like morphology of cells in 3D CDM contrasts strikingly with the shapes of fibroblasts migrating on 2D fibronectin or on the 2D upper surface of the CDM, both of which are characterized by broad lamellipodia, lower migration rates, and decreased directional persistence in comparison to 3D CDM [10, 36]. Recent studies have tested the hypothesis that the fibrillar topography of 3D CDM is the driving mechanism behind this phenotypic switch in migration by mimicking 3D matrix fibrils with 1D fibronectin-coated micropatterned lines. By generating 1.5 micron wide fibronectin-coated lines, either as single lines or multiple two micron-spaced arrays, the ligand-independent, rapid uniaxial migratory phenotype observed in 3D CDM was recapitulated on the 1D lines. Examining fibroblasts in 1D and 3D CDM revealed that in comparison to 2D migration, the processes of protrusion, cell translocation, and tail retraction are highly efficiently coordinated in 1D and 3D. Additionally, oriented 1D and 3D matrices provided cues to promote linear actomyosin and microtubule networks, leading to the uniaxial phenotype as well decreased levels of Rac1, which had been previously demonstrated to control directional persistence [36, 37]. Taken together, these studies showed 3D CDM migration is highly dependent on physical cues from the linear, fibrillar topography to regulate cell behavior.
Another significant observation made in fibroblasts migrating in 3D CDM is the dependence on myosin II contractility for efficient migration in comparison to 2D. Inhibition of myosin II ATPase activity by the drug blebbistatin produced a 60% decrease in cell velocity during migration in 3D CDM, but in contrast led to a slight increase in velocity and protrusive activity in 2D, indicating a requirement for contractile forces in 3D CDM [23, 37]. Leading edge analysis showed a dependence on the spatial distribution of cell adhesions: during fibrillar migration, adhesions formed within one micron of the leading edge and their assembly rates matched the rate of leading edge protrusion. Mechanistically, cell adhesions in 3D CDM are stabilized for prolonged periods as indicated by low turnover rates, and actin retrograde flow is significantly decreased, further demonstrating enhanced coupling between the molecular clutch guiding protrusion and adhesion. Genetic ablation of myosin IIA or IIB isoforms revealed that myosin IIA is critical for maintaining the contractile requirements for efficient migration in CDM. Loss of myosin IIA results in both a loss of the force that is applied to the fibrillar environment and loss of adhesion stability observed during CDM migration [23, 37]. Therefore, the physically restricting cues of 3D CDM fibrils require cellular contraction through myosin IIA to allow for increased mechanical coupling and efficient, rapid migration.
Recently, a novel mode of fibroblast motility was identified in both in vivo-like dermal explant and CDM models. Originally observed as a distinct population of fibroblasts migrating in CDM, this lobopodial form of migration is defined by the absence of the characteristic lamellipodia at the leading edge of the cell observed during mesenchymal cell migration. Instead, the leading edge of the cell is characterized by a blunt, cylindrical protrusion that is devoid of lamellipodial actin and cortactin; in contrast, blebs appear at the sides of the cell. Interestingly, unlike canonical lamellipodia-based migration, Rac1 and Cdc42 activity are no longer polarized toward the leading edge of the cell. Instead, lobopodial migration is driven by RhoA signaling to downstream effectors Rho-associated protein kinases (ROCK) and myosin II. Through experimental modulation of CDM, lobopodia-based migration was found to be dictated by the linear elasticity of CDM, while lamellipodial-based migration was associated with nonlinear elastic matrix environments, such as polymerized 3D collagen or trypsin-treated CDM [14]. These findings demonstrate that similar to in vivo situations, 3D CDM is able to trigger migratory plasticity within the same cell type. Further insight into the molecular mechanisms guiding these migratory switches will be crucial for understanding the processes driving cellular migration through heterogeneous, native stromal tissue.
Applications of cell-derived matrix
CDMs have been shown to impart in vivo-like responses to cells cultured inside the matrix. Recent studies have also shown that the CDM produced by cells effectively recapitulates the distinct in vivo stromal characteristics of the parental environment of the cells generating that matrix. In vivo comparisons of normal and tumor-associated stroma have shown different stromal fiber organization and content, fibroblast morphology, and gene expression. CDM generated from fibroblasts derived from tumor-associated stroma matched the architecture and content observed in vivo [6, 7, 10]. Additionally, tumor-associated CDM was necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts as well as to trigger invasive behavior of breast cancer cells [7, 38], indicating that these matrices contain the necessary physical and molecular cues to induce transformation events observed in vivo during stromatogenesis. These matrix-encoded signals are also found using in vitro models for understanding ECM roles during developmental processes. Studies using CDM generated from young mesenchymal stem cells proved they were able to restore the replication and osteogenic functions of aged mesenchymal stem cells [4]. Fibroblast CDM has also been shown to promote morphogenesis and capillary-like tubule formation of endothelial cells in vitro [3, 39]. These results with different types of CDMs provide intriguing new approaches to study distinct in vivo ECM effects on a variety of cellular processes in a controlled in vitro setting.
Conclusion
The many discoveries from in vivo and 3D in vitro analyses of cellular processes have generated a need for specific, physiologically relevant in vitro ECM models. Different types of CDMs can provide the complex composition, stiffness, and topographical architecture found in native stromal ECM. Cells cultured in CDMs adopt the morphological and migratory phenotypes commonly observed in vivo. Compared to traditional 2D migration, cells in 3D CDM can utilize distinct adhesive components and engage the contractile machinery more efficiently, driving persistent, rapid migration. This migratory behavior is directly related to the restrictive fibrillar topography of CDM. These approaches provide insight into the mechanisms driving fibroblast and other mesenchymal cell migration in native stroma. With increasing evidence that CDMs can retain the molecular cues of native stromal tissue, CDMs provide robust in vitro models capable of analyzing complex physiological processes in an ex vivo setting. The same transfection, RNAi, genetic, and immunological tools used so effectively for understanding cellular functions in regular cell culture can be applied directly to cells growing in CDMs. These approaches should permit more rapid dissection of molecular mechanisms that are difficult to study in vivo, and provide new insights into the regulation of cellular functions by the ECM.
Acknowledgments
We thank Ryan Petrie and Will Daley for critical reading of the manuscript. Supported by the Intramural Research Program of the NIDCR, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soucy PA, Romer LH. Endothelial cell adhesion, signaling, and morphogenesis in fibroblast-derived matrix. Matrix Biol. 2009;28:273–283. doi: 10.1016/j.matbio.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Li W, Lu Z, Chen R, Ling J, Ran Q, Jilka RL, Chen XD. Rescuing replication and osteogenesis of aged mesenchymal stem cells by exposure to a young extracellular matrix. Faseb J. 2011;25:1474–1485. doi: 10.1096/fj.10-161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwolinski CM, Ellison KS, Depaola N, Thompson DM. Generation of cell-derived three dimensional extracellular matrix substrates from two dimensional endothelial cell cultures. Tissue Eng Part C Methods. 2011;17:589–595. doi: 10.1089/ten.TEC.2010.0619. [DOI] [PubMed] [Google Scholar]
- 6.Quiros RM, Valianou M, Kwon Y, Brown KM, Godwin AK, Cukierman E. Ovarian normal and tumor-associated fibroblasts retain in vivo stromal characteristics in a 3-D matrix-dependent manner. Gynecol Oncol. 2008;110:99–109. doi: 10.1016/j.ygyno.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amatangelo MD, Bassi DE, Klein-Szanto AJ, Cukierman E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol. 2005;167:475–488. doi: 10.1016/S0002-9440(10)62991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlfors JE, Billiar KL. Biomechanical and biochemical characteristics of a human fibroblast-produced and remodeled matrix. Biomaterials. 2007;28:2183–2191. doi: 10.1016/j.biomaterials.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 9.Soucy PA, Werbin J, Heinz W, Hoh JH, Romer LH. Microelastic properties of lung cell-derived extracellular matrix. Acta biomaterialia. 2011;7:96–105. doi: 10.1016/j.actbio.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 11.Heck JN, Ponik SM, Garcia-Mendoza MG, Pehlke CA, Inman DR, Eliceiri KW, Keely PJ. Microtubules regulate GEF-H1 in response to extracellular matrix stiffness. Molecular biology of the cell. 2012;23:2583–2592. doi: 10.1091/mbc.E11-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessey EC, Guilluy C, Burridge K. From Mechanical Force to RhoA Activation. Biochemistry. 2012;51:7420–7432. doi: 10.1021/bi300758e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cukierman E, Bassi DE. Physico-mechanical aspects of extracellular matrix influences on tumorigenic behaviors. Seminars in cancer biology. 2010;20:139–145. doi: 10.1016/j.semcancer.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. The Journal of cell biology. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibanez T, Pellinen T, Echarri A, Cerezo A, Klein-Szanto AJ, Garcia R, Keely PJ, Sanchez-Mateos P, Cukierman E, Del Pozo MA. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 17.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell and tissue research. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao Y, Schwarzbauer JE. Accessibility to the fibronectin synergy site in a 3D matrix regulates engagement of alpha5beta1 versus alphavbeta3 integrin receptors. Cell communication & adhesion. 2006;13:267–277. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- 20.Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. Journal of cell science. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Current opinion in cell biology. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 22.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration--myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. Journal of cell science. 2012;125:2244–2256. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nature cell biology. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Deakin NO, Ballestrem C, Turner CE. Paxillin and Hic-5 interaction with vinculin is differentially regulated by Rac1 and RhoA. PLoS One. 2012;7:e37990. doi: 10.1371/journal.pone.0037990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo JC, Han X, Hsiao CT, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nature cell biology. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 30.Peleg O, Savin T, Kolmakov GV, Salib IG, Balazs AC, Kroger M, Vogel V. Fibers with integrated mechanochemical switches: minimalistic design principles derived from fibronectin. Biophysical journal. 2012;103:1909–1918. doi: 10.1016/j.bpj.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz BZ, Zamir E, Bershadsky A, Kam Z, Yamada KM, Geiger B. Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Molecular biology of the cell. 2000;11:1047–1060. doi: 10.1091/mbc.11.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mould AP, Askari JA, Aota S, Yamada KM, Irie A, Takada Y, Mardon HJ, Humphries MJ. Defining the topology of integrin alpha5beta1-fibronectin interactions using inhibitory anti-alpha5 and anti-beta1 monoclonal antibodies. Evidence that the synergy sequence of fibronectin is recognized by the amino-terminal repeats of the alpha5 subunit. The Journal of biological chemistry. 1997;272:17283–17292. doi: 10.1074/jbc.272.28.17283. [DOI] [PubMed] [Google Scholar]
- 33.Aota S, Nomizu M, Yamada KM. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. The Journal of biological chemistry. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 34.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 35.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. The Journal of cell biology. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. The Journal of cell biology. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. The Journal of cell biology. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castello-Cros R, Khan DR, Simons J, Valianou M, Cukierman E. Staged stromal extracellular 3D matrices differentially regulate breast cancer cell responses through PI3K and beta1-integrins. BMC Cancer. 2009;9:94. doi: 10.1186/1471-2407-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthod F, Germain L, Tremblay N, Auger FA. Extracellular matrix deposition by fibroblasts is necessary to promote capillary-like tube formation in vitro. J Cell Physiol. 2006;207:491–498. doi: 10.1002/jcp.20584. [DOI] [PubMed] [Google Scholar]


