Abstract
SEL1L is a putative tumor suppressor gene that is frequently down-regulated in pancreatic ductal adenocarcinoma (PDA). A single nucleotide polymorphism (SNP) rs12435998 in intron3 of SEL1L has previously been reported to be associated with susceptibility to Alzheimer’s disease. We hypothesized that this SNP may influence clinical outcomes of patients with PDA. We analyzed DNA samples from 497 Caucasian patients with pathologically confirmed primary PDA. Of these, 98 had been enrolled in a clinical trial of neoadjuvant chemo-radiotherapy and 77 of the 98 had subsequently undergone pancreaticoduodenectomy (PD). We performed Kaplan–Meier analysis to evaluate the correlation between different SNP genotypes and age at diagnosis, survival time after diagnosis, and survival time after PD. In nonsmokers, we found a significant difference in median age at diagnosis between variant genotypes (AG/GG) carriers and wild-type genotype (AA) carriers (58 versus 62 years; log-rank test, P=0.017). Patients with variant genotypes also showed an increased hazard ratio (HR) of 1.45 (95% confidence interval [CI], 1.07–1.97) relative to wild-type genotype. Among the patients in the clinical trial, the variant genotypes carriers had a median post-PD survival time that was 34.7 months shorter than wild-type genotype carriers (log-rank test, P=0.019; HR, 1.91; 95% CI, 1.09–3.34). Our results suggest that the rs12435998 SNP in SEL1L gene plays a role in modifying age at diagnosis of PDA in Caucasian nonsmokers. In addition, this SNP may serve as a prognostic marker in PDA patients who undergo the same or similar treatment as the clinical trials.
Keywords: pancreatic cancer, SNP, biologic marker, cigarette smoking, treatment
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-related deaths in both men and women in the United States [1]. The most common form of pancreatic cancer is pancreatic ductal adenocarcinoma (PDA), which accounts for >75% of all pancreatic tumors [2]. The prognosis for patients with pancreatic cancer is among the worst of all solid tumors, with a 5-year overall survival rate of 6% [3]. Pancreatic cancer has no reliable biomarkers for early detection and no disease-specific early symptoms, and it progresses rapidly. Therefore, the disease is usually diagnosed at a late stage after local or distant metastasis has already occurred [3, 4]. Thus, to prevent or improve clinical management of this dismal disease, it is critically important to identify factors that influence risk for PDA and clinical outcome of pancreatic cancer patients. Previous studies have shown that single nucleotide polymorphisms (SNPs) influence risk for PDA as well as clinical outcome [5–8]. Here, we analyzed the correlation between a SNP in the SEL1L (Sel-1-like) gene and age at diagnosis of PDA, as well as clinical outcome in a retrospective study of PDA patients.
In C. elegans, sel-1 has been identified as a negative regulator of the onco-proteins, lin-12 and glp-1, which are members of the lin-12/Notch family of receptors [9, 10]. The human homolog SEL1L is a component of the “unfolded protein response/endoplasmic reticulum associated protein degradation (UPR/ERAD) pathway” [11–13]. This pathway is activated in response to an accumulation of unfolded or misfolded proteins in the lumen of the endoplasmic reticulum (ER) and induces the activation of several additional signaling pathways to promote long-term stress adaptation or apoptotic cell death [14]. SEL1L is involved in recognizing misfolded proteins and transferring them into the cytosol for degradation. It has also been reported to interact and stabilize Hrd1p, a ubiquitin ligase [12, 13].
SEL1L messenger RNA is abundantly expressed in healthy pancreatic tissues from adult humans and is frequently downregulated in PDA tumors [15, 16]. SEL1L overexpression causes a decrease in the aggressive behavior of PDA tumor cells both in vitro and in vivo [17, 18]. This suggests that SEL1L plays a tumor-suppressive role in the pathogenesis of PDA. Therefore, we hypothesized that a SNP that is thought to be functional in the SEL1L gene may influence the clinical outcome of patients with PDA.
An adenine-to-guanine (A-to-G) SNP within SEL1L intron 3, rs12435998, has been described as an independent susceptibility factor for Alzheimer’s disease (AD), showing a higher frequency in cases than in controls [19]. Here, we analyzed this SNP to determine whether it is associated with age at diagnosis and clinical outcome of patients with PDA. In addition, we also analyzed this SNP in combination with smoking history, as cigarette smoking is a well recognized risk factor for pancreatic cancer [20–22]. Here we report for the first time that the SNP is associated with an earlier age at diagnosis of PDA in Caucasian nonsmokers, and also may serve as a promising prognostic marker for PDA.
MATERIALS AND METHODS
Study population
A total of 497 Caucasian patients with pathologically confirmed primary PDA of all stages were consecutively recruited at the University of Texas MD Anderson Cancer Center (UTMDACC) from January 1999 to May 2007. Of those patients, a subset of 98 had been enrolled in one of two phase II clinical trials (ID98-020 or ID01-341) of preoperative (neoadjuvant) chemoradiotherapy at the UTMDACC from February 1999 to August 2006. There were 41 (42%) patients enrolled in the ID98-020 trial and 57 (58%) patients enrolled in the ID01-341 trial. These 98 patients all had a diagnosis of potentially surgically resectable adenocarcinoma of the pancreatic head, and at the time of enrollment had not received any previous treatment for pancreatic cancer. Patients in the ID98-020 trial received gemcitabine-based chemoradiotherapy that consisted of weekly gemcitabine (400 mg/m2 by intravenous injection) for 4 weeks plus radiation (30 Gy in 10 fractions) over 2 weeks. Patients in the ID01-341 trial received induction therapy with gemcitabine (750 mg/m2/day) and cisplatin (30 mg/m2/day) every 2 weeks for 4 weeks plus radiation (30 Gy in 10 fractions) over 2 weeks. The same eligibility criteria for patient recruitment was applied for both protocols and no significant differences in any of the clinical features were observed between the two patient populations. Response was assessed by computerized tomography obtained before and 4–6 weeks after completion of the preoperative chemoradiotherapy. Patients who displayed response to the therapy underwent a potentially curative pancreaticoduodenectomy (PD).
Patient characteristics information was gathered from the questionnaire that the patients answered. Written informed consent was obtained from all patients enrolled in this study. The study was approved by the UTMDACC Institutional Review Board.
DNA extraction and genotyping
DNA was extracted from the blood samples with an Autopure LS automated DNA purification instrument (Gentra Systems, Minneapolis, MN). The genotypes were analyzed by pyrosequencing with a PSQ HS96A system (Biotage, Foxboro, MA) using the SNP mode according to the manufacturer’s instructions. The primer sequences for polymerase chain reaction (PCR) were: 5′-CCC AGT TAC AAA TCA GGC ATC AT-3′ (forward) and 5′-biotin-GGT CAA AGC TGG AAT GAC AAG AA-3′ (reverse). The PCR amplification was carried out in a final volume of 30 μL mixture, containing 10 ng of DNA, 0.4 mM of each primer, deoxynucleotide triphosphates (dATP, dCTP, dGTP, dTTP) each at 0.25 mM, KCl at 50 mM, MgCl2 at 1.5 mM, Tris-HCl at 10 mM (pH 8.3), and 1 unit of AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA). The reaction was started at 95°C for 5 min, followed by 35 cycles (95°C for 45 s, 61°C for 45 s, and 72°C for 45 s) and then an extension of 72°C for 7 min. The sequencing primer used was: 5′-GCA TCA TGA ATC TTT GCT-3′. For each assay, the genotypes were read independently by two people.
Statistical methods
The classical χ2 goodness-of-fit test was used to test for Hardy–Weinberg equilibrium. We used the Kaplan–Meier method to evaluate the correlation between the SNP genotypes and age at diagnosis, overall survival, and survival time after PD. Overall survival time was calculated from the date of diagnosis to the date of death or the date of the last follow-up. The log-rank test was used to evaluate the homogeneity of the Kaplan–Meier curves by genotype. Univariate and multivariate Cox proportional hazards regression analysis was used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs). Age, sex, diabetes history, smoking history, and family history of cancer were included in the multivariate model when appropriate. Stage and treatment were included for the clinical trials. The level of significance was set to P < 0.05 for all statistical analysis. All statistical analyses were two-sided and were performed using Stata 10.1 software (Stata, College Station, TX).
RESULTS
Patient characteristics
Our analysis revealed that for the 497 Caucasian patients in this study, the mean age at diagnosis of PDA was 61 years (yr) (range, 28–87 yr). The date of diagnosis was the date that a doctor first diagnosed the patient with pancreatic cancer. The mean survival time is 11.8 months (range, 0.4–84.3 months). As shown in Table 1, of the 497 patients, 318 (64%) were male and 179 (36%) were female. A smoker was defined as someone who had smoked at least 100 cigarettes during their life; our study population had 194 (39%) nonsmokers and 303 (61% smokers. Of the 98 patients enrolled in the clinical trials described above, 77 showed response to the treatment and then underwent PD surgery. The remaining 21 patients did not undergo resection because tumor progression was found at restaging after preoperative therapy.
Table 1.
Characteristics of the study population
| A. Characteristics of all patients
| |
|---|---|
| Variables | No. of patients (%) |
| Total | 497 (100) |
|
| |
| Age (years) | |
| ≤ 50 | 84 (17.5) |
| 51–60 | 147 (29.6) |
| 61–70 | 166 (33.4) |
| >70 | 97 (19.5) |
| Mean±SD | 60.9±10.6 |
| Sex | |
| Male | 318 (64.0) |
| Female | 179 (36.0) |
| Diabetes * | |
| No | 362 (74.0) |
| Yes | 127 (26.0) |
| Smoking | |
| Never | 194 (39.0) |
| Ever | 303 (61.0) |
| Family history of cancer | |
| No | 231 (46.4) |
| Yes | 266 (53.6) |
| B. Characteristics of patients in the trial
| |
|---|---|
| Variables | No. of patients (%) |
| Total | 98 (100) |
|
| |
| Age (years) | |
| ≤ 50 | 12 (12.2) |
| 51–60 | 28 (28.6) |
| 61–70 | 36 (36.7) |
| >70 | 22 (22.5) |
| Mean±SD | 62.1±9.6 |
| Sex | |
| Male | 60 (61.2) |
| Female | 38 (38.8) |
| Diabetes | |
| No | 74 (75.5) |
| Yes | 24 (24.5) |
| Smoking | |
| Never | 29 (29.6) |
| Ever | 69 (70.4) |
| Family history of cancer | |
| No | 48 (49.0) |
| Yes | 50 (51.0) |
| Tumor stage | |
| IA | 9 (9.2) |
| IB | 9 (9.2) |
| IIA | 34 (34.7) |
| IIB | 46 (46.9) |
| Protocol | |
| GEM/XRT† | 41 (41.8) |
| GEM/Cisp/XRT† | 57 (58.2) |
Data missing for 8 patients.
GEM, gemcitabine; XRT, radiotherapy; Cisp, cisplatin.
Genotype frequencies
Of the 497 patients, 64.0% had the SEL1L wild-type genotype (AA), 33.0% had the heterozygous variant genotype (AG), and 3.0% had the homozygous variant genotype (GG). These genotype frequencies were found to be consistent with Hardy–Weinberg equilibrium (χ2 = 1.260, P = 0.262). The allele frequencies were 80.5% for A and 19.5% for G. These frequencies were close to those in the NCBI SNP database from the CEU HapMap population: 81.9% for A and 18.1% for G.
Effect of genotype on age at diagnosis
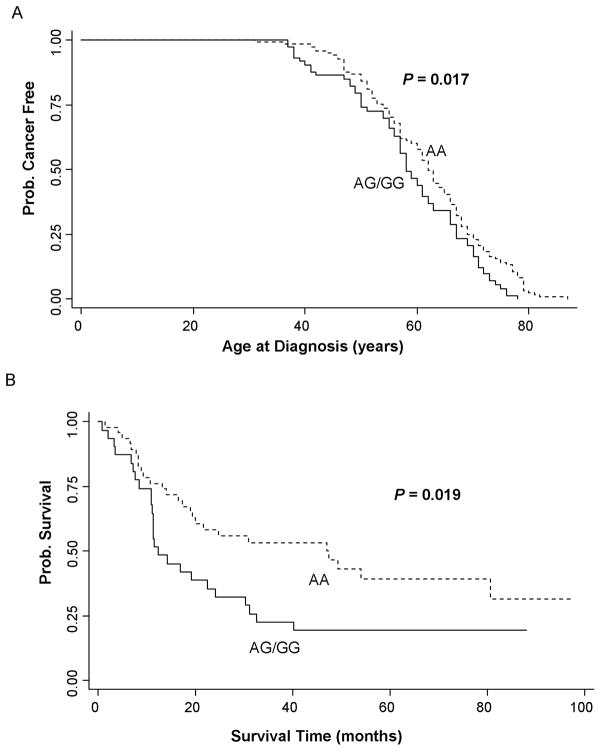
Because the number of patients with the GG genotype was too small to provide meaningful results, we combined the patients with the AG and GG genotypes for the analysis. We did not find a significant difference in median age at diagnosis between the AA and AG/GG genotypes for the 497 patients using Kaplan-Meier estimates (Log-rank test, P=0.140). The median age at diagnosis was 61 yr for both those with the AG/GG genotypes and those with the AA genotype. For the 303 smokers there was also no difference between the genotypes (Log-rank test, P=0.481). The median age at diagnosis was 62 yr for those with the AG/GG genotypes, and 61 yr for those with the AA genotype. However, as shown in Fig. 1A, for the 194 nonsmokers, Kaplan–Meier estimates showed a significant difference in the median age at diagnosis in patients with the AG/GG genotypes compared to the AA genotype, with the median age at diagnosis being 58 yr for those with the AG/GG genotypes, and 62 yr for those with the AA genotype (log-rank test, P=0.017; Table 2A). Using the nonsmoking patients with the AA genotype as a reference in the Cox proportional hazards regression model adjusting with sex, diabetes history, and family history of cancer, we found a HR of 1.45 (95% CI, 1.07–1.97; Table 2A). This result indicated that nonsmoking patients with the AG/GG genotypes had a significantly greater probability of being diagnosed with pancreatic cancer during any interval than those with the AA genotype.
Figure 1.
Kaplan–Meier curves for patients with the wild-type genotype (AA) and the variant genotypes (AG/GG): (A) Age at diagnosis of pancreatic cancer by genotype in nonsmokers (n = 194); (B) Survival time after pancreaticoduodenectomy by genotype (n = 77).
Table 2.
The correlation between genotypes and clinical outcomes of PDA patients
| A. Age at diagnosis in nonsmokers
| ||||
|---|---|---|---|---|
| Genotype | Patients No. (%) Total n = 194 |
MAD * (years) | Hazard ratio (95% CI) | P (Log-rank) |
| AA | 121 (62.4) | 62 | 1.00 | |
| AG/GG | 73 (37.6) | 58 | 1.45 (1.07–1.97) | 0.017 |
| B. Survival time after pancreaticoduodenectomy
| ||||
|---|---|---|---|---|
| Genotype | Patients No. (%) Total n = 77 |
MSP† (months) | Hazard ratio (95% CI) | P (Log-rank) |
| AA | 46 (59.7) | 47.0 | 1.00 | |
| AG/GG | 31 (40.3) | 12.3 | 1.91 (1.09–3.34) | 0.019 |
MAD, median age at diagnosis.
MSP, median survival time after pancreaticoduodenectomy.
Effect of genotype on survival time after PD
We did not find a significant correlation between the SEL1L genotype and overall survival time for the 497 patients, or for the 98 patients enrolled in the clinical trials (data not shown). Among the 77 patients who were treated in the clinical trials of preoperative chemoradiotherapy and subsequent PD surgery, we evaluated the effects of this SNP on survival time after PD. Kaplan–Meier estimates showed that the median survival time after PD for patients with the AG/GG genotypes was 34.7 months shorter than that for patients with the AA genotype, with the median survival being 12.3 months for those with the AG/GG genotypes, and 47.0 months for those with the AA genotype. Survival curves for these genotypes were significantly different (log-rank test, P=0.019; Fig. 1B). Using the patients with the AA genotype as a reference in the Cox proportional hazards regression model, we found a HR of 1.91 (95% CI, 1.09–3.34; Table 2B) for the AG/GG genotypes, after adjustment with age, sex, smoking, diabetes, family history of cancer, stage, and treatment.
DISCUSSION
In this case-only study of patients with PDA, we determined whether the SNP rs12435998, in the SEL1L gene was associated with age at diagnosis, and response to treatment of PDA. To our knowledge, ours is the first report to investigate the role of this SNP in cancer. We found that this SNP could serve as a predictive and prognostic marker in patients with PDA.
In the subgroup of nonsmokers, we found that the median age at diagnosis in patients with the variant genotypes was significantly younger than that of patients with the wild-type genotype for this SNP. The higher HR in the carriers of the variant genotypes suggests that patients with the variant genotypes had a significantly greater probability of being diagnosed with PDA during any time interval. These findings suggest that the variant genotypes may be useful in identifying those who are more likely to develop PDA among Caucasian nonsmokers. However, in smokers we did not find a significant difference between wild-type and variant genotypes in age at diagnosis. This may be because the influence of smoking on risk for PDA overwhelms the effect of the SNP. We used Mfold web server to predict the DNA folding with either A or G in the sequence [23]. The results indicate that the substitution of G for A results in a thermodynamically more stable DNA secondary structure. The structural change of the G configuration may interfere with the binding of trans-acting factors to the SEL1L DNA thereby leading to SEL1L downregulation. This is a possible mechanism by which SNP rs12435998 might influence the age at diagnosis of PDA in nonsmokers. Obesity/body mass index, diabetes mellitus, chronic pancreatitis, blood group, and potential dietary variables may be confounders for age at diagnosis of pancreatic cancer. Unfortunately, we did not have data on all of these possible confounders except for diabetes mellitus which was a limitation to our study.
We did not find a significant correlation between this SNP and overall survival time in the series of 497 patients that we studied. However, a limitation to our study was the lack of tumor size, stage and treatment information which could influence overall survival. In the 77 patients who were treated in the clinical trial of chemoradiotherapy and subsequent PD surgery, the variant G allele was associated with a reduced survival time after PD. This result suggests that the G allele for this SNP may serve as a negative prognostic marker in patients with PDA who undergoes the same or similar treatment as in the clinical trials.
There was no significant difference in age at diagnosis, sex, and smoking status between the patients participating in the clinical trials and those not in the trials. The patients participating in the trials all had a diagnosis of potentially surgically resectable adenocarcinoma of the pancreatic head. They were in either stage I or stage II. Regarding stage, we assume that most of the patients who were not part of the clinical trial were diagnosed at a more advanced stage. In general, most patients are usually diagnosed after the cancer has spread to distant sites. Cook et al. demonstrated that radiation induces increased expression of several genes associated with the UPR/ERAD pathway including ATF4 and Ero-1 like in murine squamous cell carcinoma cells. This finding suggested that radiation may cause accumulation of misfolded proteins and activation of UPR/ERAD pathway in cancer cells [24]. As radiotherapy was involved in our clinical trial, we presume that UPR/ERAD pathway was activated during the treatment. The SNP possibly influenced levels of SEL1L expression or function in misfolded protein degradation and had an impact on cancer cell fate, hence influencing the patients’ responses to the therapy.
In conclusion, our study provided evidence for the first time that the SEL1L SNP, rs12435998 is associated with age at diagnosis of PDA in nonsmokers, and with survival time after PD in the patients who received combined chemoradiotherapy and PD. Our findings suggest that SEL1L and thereby, the UPR/ERAD pathway plays an important role in the pathogenesis of PDA. This result has important clinical implications in the prediction and treatment of this deadly disease. A long-term goal in the prevention, early detection, and treatment of PDA is to identify a panel of risk markers combined with other characteristics (e.g. smoking, alcohol intake, diet) which will help to identify those who are at increased risk for developing pancreatic cancer, and also those who are more likely to develop it at an earlier age. The SEL1L SNP, rs12435998, is a likely candidate to be included in such a panel if validated in confirmatory studies.
Acknowledgments
Grant Support: This research was supported by National Cancer Institute U01 grant CA111302 (A. M. Killary, M. L. Frazier, and S. Sen), Cancer Center Support grant CA16672 (J. Mendelsohn); and National Institute of Health R25 educational grant CA56452 (R. M. Chamberlain, and S. Chang).
The authors would like to thank Domitila Patenia and Haidee Chancoco at UTMDACC for their technical support.
Abbreviations
- PDA
pancreatic ductal adenocarcinoma
- SNP
single nucleotide polymorphism
- PCR
polymerase chain reaction
- PD
pancreaticoduodenectomy
- HR
hazard ratio
- CI
confidence interval
- UPR/ERAD
unfolded protein response/endoplasmic reticulum associated protein degradation
- ER
endoplasmic reticulum
- A
adenine
- G
guanine
- AD
Alzheimer’s disease
- UTMDACC
University of Texas MD Anderson Cancer Center
- yr
years
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Types of Pancreas Tumors. Johns Hopkins University, The Sol Goldman Pancreatic Cancer Research Center; http://pathology.jhu.edu/pancreas/BasicTypes1.php. [Google Scholar]
- 3.American Cancer Society. Cancer Facts & Figures 2010. Atlanta: American Cancer Society; 2010. [Google Scholar]
- 4.Ghaneh P, Costello E, Neoptolemos JP. Biology and management of pancreatic cancer. Gut. 2007:1134–1152. doi: 10.1136/gut.2006.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Li D, Wei C, et al. Aurora-A and p16 polymorphisms contribute to an earlier age at diagnosis of pancreatic cancer in Caucasians. Clin Cancer Res. 2007;13:3100–3104. doi: 10.1158/1078-0432.CCR-06-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Li D, Killary A, et al. Polymorphisms of p16, p27, p73, and MDM2 Modulate Response and Survival of Pancreatic Cancer Patients Treated with Preoperative Chemoradiation. Ann Surg Oncol. 2009;16:431–439. doi: 10.1245/s10434-008-0220-8. [DOI] [PubMed] [Google Scholar]
- 7.Li D, Liu H, Jiao L, et al. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 2006;66:3323–3330. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McWilliams RR, Bamlet WR, et al. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res. 2008;68:4928–4935. doi: 10.1158/0008-5472.CAN-07-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant B, Greenwald I. The Caenorhabditis elegans sel-1 gene, a negative regulator of lin-12 and glp-1, encodes a predicted extracellular protein. Genetics. 1996;143:237–247. doi: 10.1093/genetics/143.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant B, Greenwald I. Structure, function, and expression of SEL-1, a negative regulator of LIN-12 and GLP-1 in C. elegans. Development. 1997;124:637–644. doi: 10.1242/dev.124.3.637. [DOI] [PubMed] [Google Scholar]
- 11.Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko M, Nomura Y. ER signaling in unfolded protein response. Life Sci. 2003;74:199– 205. doi: 10.1016/j.lfs.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 15.Biunno I, Appierto V, Cattaneo M, et al. Isolation of a pancreas-specific gene located on human chromosome 14q31: expression analysis in human pancreatic ductal carcinomas. Genomics. 1997;46:284–286. doi: 10.1006/geno.1997.5018. [DOI] [PubMed] [Google Scholar]
- 16.Harada Y, Ozaki K, Suzuki M, et al. Complete cDNA sequence and genomic organization of a human pancreas-specific gene homologous to Caenorhabditis elegans sel-1. J Hum Genet. 1999;44:330–336. doi: 10.1007/s100380050171. [DOI] [PubMed] [Google Scholar]
- 17.Cattaneo M, Orlandini S, Beghelli S, et al. SEL1L expression in pancreatic adenocarcinoma parallels SMAD4 expression and delays tumor growth in vitro and in vivo. Oncogene. 2003;22:6359–6368. doi: 10.1038/sj.onc.1206665. [DOI] [PubMed] [Google Scholar]
- 18.Cattaneo M, Fontanella E, Canton C, et al. SEL1L affects human pancreatic cancer cell cycle and invasiveness through modulation of PTEN and genes related to cell-matrix interactions. Neoplasia. 2005;7:1030–1038. doi: 10.1593/neo.05451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saltini G, Dominici R, Lovati C, et al. A Novel Polymorphism in SEL1L confers susceptibility to Alzheimer’s disease. Neurosci Lett. 2006;398:53–58. doi: 10.1016/j.neulet.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 20.Weiss W, Benarde MA. The temporal relation between cigarette smoking and pancreatic cancer. Am J Public Health. 1983;73:1403–1404. doi: 10.2105/ajph.73.12.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder I, Hoogenveen RT, van Genugten ML, et al. Smoking cessation would substantially reduce the future incidence of pancreatic cancer in the European Union. Eur J Gastroenterol Hepatol. 2002;14:1343–1353. doi: 10.1097/00042737-200212000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Wittel UA, Hopt UT, Batra SK. Cigarette smoke-induced pancreatic damage: experimental data. Langenbecks Arch Surg. 2008;393:581–588. doi: 10.1007/s00423-007-0273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook JA, Chuang EY, Tsai MH, et al. Radiation-induced changes in gene-expression profiles for the SCC VII tumor cells grown in vitro and in vivo. Antioxid Redox Signal. 2006;8:1263–1272. doi: 10.1089/ars.2006.8.1263. [DOI] [PubMed] [Google Scholar]