Abstract
Objective
Estradiol (E2) regulates gene transcription by activating estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ). Many of the genes regulated by E2 via ERs are repressed, yet the molecular mechanisms that mediate E2-induced gene repression are currently unknown. We hypothesized that E2, acting through ERs, regulates expression of microRNAs (miRs) leading to repression of expression of specific target genes.
Methods and Results
Here, we report that E2 significantly up-regulates the expression of 26 miRs and down-regulates the expression of 6 miRs in mouse aorta. E2 mediated up-regulation of one of these miRs, miR-203, was chosen for further study. In cultured vascular smooth muscle cells (VSMC), E2-mediated up-regulation of miR-203 is mediated by ERα (but not ERβ) via transcriptional up-regulation of the primary miR. We demonstrate that the transcription factors Zeb-1 and AP-1 play critical roles in mediating E2-induced up-regulation of miR-203 transcription. We show further that miR-203 mediates E2-induced repression of Abl1 and p63 protein abundance in VSMC. Finally, knocking-down miR-203 abolishes E2-mediated inhibition of VSMC proliferation, and over-expression of miR-203 inhibits cultured VSMC proliferation, but not vascular endothelial cell proliferation.
Conclusions
Our findings demonstrate that E2 regulates expression of miRs in the vasculature, and support that ER-dependent induction of miRs is a mechanism for E2-mediated gene repression. Furthermore, our findings demonstrate that miR-203 contributes to E2-induced inhibition of VSMC proliferation and highlight the potential of miR-203 as a therapeutic agent in the treatment of proliferative cardiovascular diseases.
Keywords: estrogen; gene regulation; microRNA; muscle, smooth; proliferation
Estrogen regulates diverse biological processes involving the reproductive, skeletal, neurologic, and cardiovascular systems via activation of the two known estrogen receptors, estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ).1, 2Estrogen has important effects on vascular physiology and pathophysiology, with potential therapeutic implications. ERα and ERβ are both expressed in vascular smooth muscle cells (VSMC) and vascular endothelial cells (VEC), identifying these cells as direct targets of E2 action.3-9 Estrogen inhibits VSMC proliferation and migration, in vivo and in vitro.8, 10-13 In contrast, estrogen increases VEC proliferation and migration, and induces nitric oxide production.14, 15 ERs are ligand-activated transcription factors, and as such, they regulate gene expression in response to estrogen. The molecular mechanisms by which ERs up-regulate gene expression are now understood in some detail.9, 16, 17 However, we recently demonstrated that many ER-regulated genes in the mouse aorta are down-regulated rather than up-regulated.18, 19 Similar findings have been demonstrated in other, non-vascular cells as well.20 The molecular mechanisms by which ERs repress gene expression are currently unknown.
Micro-RNAs (miRs) are small non-coding RNAs that repress gene expression at the post-transcriptional level.21, 22 Recent studies have shown that miRs are involved in the regulation of a variety of biologic processes including development, signal transduction, apoptosis, cell proliferation, and tumorigenesis. Though miRs are postulated to participate in estrogen regulation of gene expression in other systems, this has not been examined in vascular cells or tissues. Further, though estrogen has recently been shown to alter expression of specific miRs in non-vascular cells, the molecular mechanisms by which estrogen regulates these miRs have not been explored, nor have any physiologic consequences of these changes been shown.23-28
In this report, we demonstrate that ex vivo estrogen treatment of mouse aorta regulates miR expression. Among the estrogen up-regulated miRs, miR-203 has been reported to inhibit cell proliferation in a variety of malignant tumor cells.29-32 We then focused our study on estrogen regulation of miR-203 in cultured VSMC and elucidate some of the molecular mechanisms by which E2 regulates miR-203. We demonstrate further that miR-203 contributes to estrogen-mediated inhibition of VSMC proliferation, and that miR-203 can, by itself, specifically inhibit VSMC proliferation.
Methods
Mice and Tissue collection
Aortas harvested from 10-week old wild-type C57BL/6 mice 7 days after ovariectomy, were treated ex vivo with 10 nM 17β-estradiol (E2) or vehicle for 4 hr as previously described.19 3 aortas were pooled for each assay. For each treatment group, miR arrays and real-time PCR were performed in duplicate and quadruplicate, respectively, as described below. The Animal Care and Use Committee at Tufts Medical Center approved all animal procedures.
Plasmid constructions
Plasmids for promoter activity assays were cloned into pGL3-basic vector (Promega). The region 1.1 kb upstream to the mmu-miR-203 gene was generated by PCR using primers 5′-GCTAGCGCTGTGGAACCTCTGAGAAG-3′(NheI), 5′-AAGCTTGCCTGCACACAGAAGTTCTG-3′ (HindIII), with template of genomicl DNA. The PCR fragments were ligated into Zero Blunt TOPO vector (Invitrogen), and subcloned into pGL3-basic (NheI/HindIII sites), creating construct pGl3-miR-203-pro2. To generate pGl3-miR-pro1, a 1.9kb upstream region of mmu-miR-203 gene was amplified by PCR using primers 5′- GCTAGCCCCATGAGAATTTACACCCT-3′ (NheI), and 5′-GAGGACCAGAAGGCTGTCAT-3′, with template of genomic DNA. The PCR fragments were ligated into Zero Blunt vector and sub-cloned into pGL3-miR-203-pro2 (NheI/NsiI sites). Constructs for deletion analysis were generated by PCR reaction with appropriate primers. Plasmids bearing specific mutations were generated from construct miR-203pro1 using Quick Change Site-Directed mutagenesis approach (Stratagene).
Mouse aortic smooth muscle cell (MAoSMC) harvest and culture
MAoSMC were obtained using the explant procedure as previously published. 3 Briefly, several mouse aortas or carotid arteries were obtained sterilely and placed into a 100mm dish containing media. The adventia was cleaned off and the aorta was cut horizontally into 10-15 pieces. Each piece was placed into a 6 or 12 well collagen Biocoat plate (Fisher). Explants were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing antibiotics and 10% bovine growth serum (BGS) for 3-7 days. When the well was 50-75% confluent, the explants were removed, and the SMCs were cultured in low glucose phenol red-free DMEM containing antibiotics and 10% fetal bovine serum (FBS). Before experimental use, SMCs were grown in low glucose DMEM supplemented with 5% dextran-coated charcoal-treated fetal bovine serum.
Human aortic smooth muscle cell (HAoSMC) harvest, culture and immortalization
HAoSMC were derived by the explant method from discarded aortic tissue from Tufts Medical Center operating room and immortalized at passage 2 with retrovirus E6/E7 ELXN and selected with G418 (300ug/ml). The cells were cultured in low glucose phenol red-free DMEM containing antibiotics and 10% FBS.
Cell transfection
Plasmid transfection was performed with Lipofectamine 2000 (Life Technologies) following the manufacturer’s protocol. Transfection with anti-miR (Life Technologies) and miR mimics (Life Technologies) was performed using RNAiMAX (Invitrogen) according to the supplier’s instructions.
Antibodies
The following antibodies were used: ERα (Santa Cruz Biotechnology, sc-7207), ERβ (Affinity BioReagents, PA1-310B), alpha smooth muscle actin (SMA) (Sigma-Aldrich, A2547), Flag M2 (Sigma-Aldrich, F1804), c-Abl (Calbiochem, OP20), p63 (Thermo Scientific, MS-1084), Zeb-1 (Santa Cruz Biotechnology, sc-10573), and β-actin (Santa Cruz Biotechnology, sc-1616) for westernblot; ERα (Santa Cruz Biotechnology, sc-542) for chromatin immunoprecipitation; Zeb-1 (Santa Cruz Biotechnology, sc-25388) for westernblot, immunoprecipitation and chromatin immunoprecipitation.
Array-based miR profiling
Aortas were treated ex vivo with 10 nM E2 or vehicle for 4 hr. 3 aortas were pooled and total RNA was isolated using the miRNeasy kit (Qiagen). MiRs expression profiling was performed utilizing the TaqMan Rodent MicroRNA Array Set (Applied Biosystems). This array contains 517 mouse miRs as well as endogenous controls. Normalization was performed with snoRNA202. For each treatment group, miR arrays were performed in duplicate.
Quantitative Real-time PCR
Quantitative real-time PCR confirmation of miR array results was performed in quadruplicate by using RNA isolated from independently obtained aortas treated as described above. MAoSMC were infected with adenovirus expressing human ERα (Ad-ERα), ERβ (Ad-ERβ) or green fluorescent protein (Ad-GFP) and treated with 10nM E2 for 8 hr or 24 hr. Total RNA was extracted from aorta or MAoSMCs using the miRNeasy kit (Qiagen). Mature miR and pri-miR-203 expression were measured using Taqman® MicroRNA Assay and TaqMan® Pri-miRNA Assay, respectively (Applied Biosystems). Quantitative real-time PCR analysis was performed using the Mastercycler ep realplex (Eppendorf).
Luciferase assays
Firefly luciferase assays were carried out as follows: 20ul cell lysate was added to 100ul of firefly luciferase assay buffer (Promega). The samples were placed in a luminometer (Luminoscan Ascent, Labsystems) and light output was determined over a 10 second interval. β-galactosidase activity was measured with a Tropix Galacto-Light Plus kit (Applied biosystems) following the manufacturer’s instructions. Firefly luciferase activity was normalized to β-galactosidaseactivity.
Cell proliferation assay
MAoSMC, HAoSMC, and human umbilical vein endothelial cell (HUVEC) were transiently transfected with miR-203 precursor molecule (pre-miR-203, Ambion), or Pre-miR precursor’s negative control no.1 (negative control, Ambion) using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s instructions. The final concentration of pre-miR was 50nM. Cells were seeded in 96-well plates. Cell proliferation was measured at 0, 2, 3 and 4 days post seeding using Cell TiterGlo Luminescent Cell Viability assay (Promega) following the manufacturer’s protocol. Each assay was performed in tetracate and repeated at least 3 times independently. To study the effects of E2 on MAoSMC in which miR-203 is depleted, the Ad-ERα infected MAoSMC were transfected with anti-miR-203 or anti-miR control (Ambion) and seeded into 96-well plates. E2 treatment was initiated 6 hr after transfection. Cell proliferation was measured at 0, 2, 3 and 4 days post seeding.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the protocol for ChIP Assay Kit (Millipore). Immunoprecipitates were dissolved into 50μL of distilled water. 2μL of the DNA was used for a 20μL PCR reaction using the QuantiTect SYBR Green PCR Kits (Qiagen). Primers sequence for amplification of miR-203 promoter region are ERBS-For:5′-TGATTCCAATCGCTCTCCTTTTCC-3′, ERBS-Rev:5′-GCGCCCTGCCCTCTTAGTCACC -3′; E-Box1-For:5′-TTCCAATCGCTCTCCTTTTCC-3′; E-Box1-Rev:5′-GTCCTTGTGCCATGTACCAG-3′; E-Box3-For:5′-CCCTTGGCGAGCATTGCAAAG-3′; E-Box3-Rev:5′-CGCGCTGGCCTGACCGGTTT-3′. Amplification of an intergenic region of ~170kb upstream of the Progesterone Receptor gene was used as a negative control.
Co-immunoprecipitation
MAoSMCs were infected with Ad-ERα and treated with E2. At 24h post-treatment, cells were washed once with cold PBS and lysed with cold RIPA buffer at 4°C for 30 min. The lysates were incubated with anti-zeb1 antibody (sc-25388, Santa Cruz) overnight at 4°C, followed by incubation with protein G-coated agarose beads (Millipore) for another 1 h at 4°C. After samples were washed five times with ice-cold RIPA buffer and supernatants were removed by centrifugation at 2,000×g for 1min. The proteins were then separated from beads using immunoblot loading buffer for 5 min at 95°C. The supernatants were collected for immunoblotting.
Apoptosis assay
Apoptotic cells were measured by TUNEL assay (In Situ Cell Death Detection Kit, Roche) according to the manufacturer’s instructions.
Bioinformatic analysis
ER binding elements (ERE) analysis
ERE analysis was carried out using the CREAD suite of programs (STORM and MULTISTORM) that allows for the prediction of transcription factor binding sites and their conservation among various species. Mouse Genome Assembly mm9 (July 2007, NCBI Build 37) was used for all the calculations.
Transcription factor binding site (TFBS) analysis
TFBS analysis was carried out using the program STORM in the CREAD suite of programs and TRANSFAC matrices for TFBS in the 91 bp sequence of interest. Only those matches that had a functional depth of at least 0.9 were considered as significant. The functional depth is defined as F=(S - Smin)/ (Smax -Smin), where S is the score of occurrence for a motif in a sequence and Smin and Smax are the minimum and maximum that a motif match can score. A functional depth of 1.0 signifies the best match of a motif to the sequence.
Statistical Analysis
Between group differences were tested for using the Student’s t-test. All data are shown as mean ± standard deviations. A P value <0.05 was considered statistically significant. Significantly differentially expressed miR from TaqMan miR array results was defined as a mean fold change ≥1.4.
Results
E2 regulates miRs expression profiles in mouse aorta
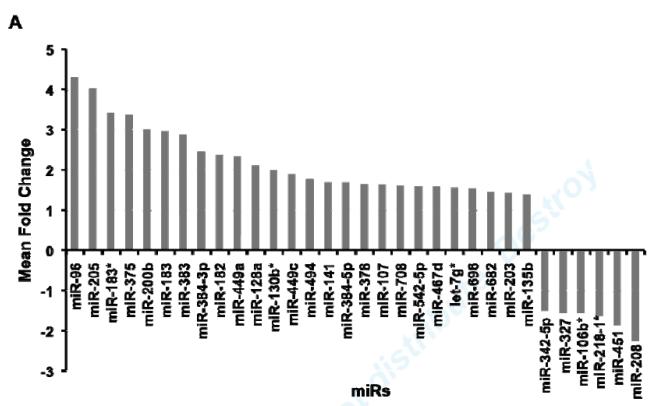
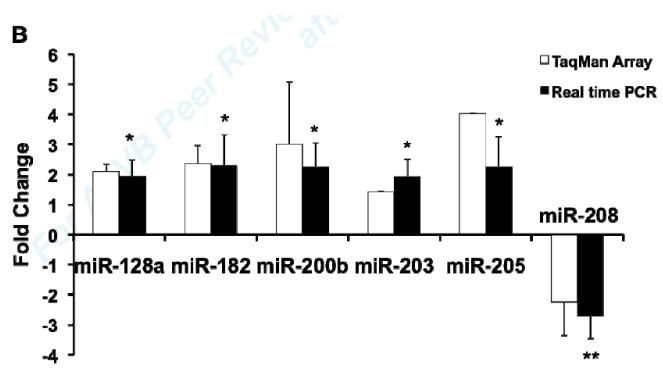
MiR expression profile analysis revealed that 318 out of 517 miRs were detected at a significant level in vehicle treated mouse aorta. 32 miRs were significantly regulated by 4 hours of ex vivo E2 treatment, including 26 up-regulated and 6 down-regulated miRs (Fig. 1A). Using independently obtained RNA samples, real-time RT-PCR confirmed E2-mediated regulation of 6 of the 10 miRs chosen for testing, including E2-mediated up-regulation of 5 miRs (miR-128a, miR-182, miR-200b, miR-203, miR-205) and down-regulation of 1 miR (miR-208) compared with vehicle treated aorta (Fig. 1B).
Fig1. MiR expression profilling analysis in mouse aorta.
(A) Significantly differentially expressed miRs in E2-treated mouse aortas. Relative quantification of miR expression was calculated by the 2−ΔΔCt method. Data normalization was done using snoRNA202 as the endogenous control and ath-miR159a as the negative control. Results were shown as mean fold change (MFC) of two independent experiments. (B) Validation of E2-regulated miRs in mouse aorta. 10 miRs were selected from microarray data for qRT-PCR validation. Data were normalized with the vehicle-treated controls. Results were shown as mean ± S.D. of four independent experiments. *P<0.05, **P<0.01, refer to a comparison between E2- and vehicle-treated samples for qRT-PCR.
ERα mediates E2 regulation of miR-203 expression in VSMC at the transcriptional level
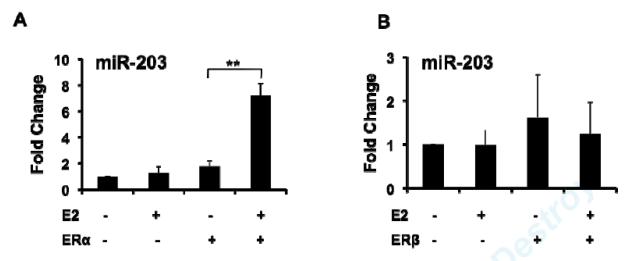
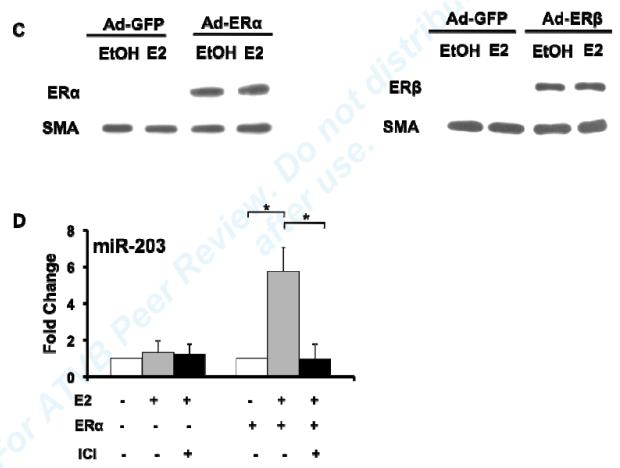
Given prior reports demonstrating that miR-203 inhibits proliferation in a variety of malignant tumor cells, we hypothesized that E2-mediated up-regulation of miR-203 contributes to E2’s inhibitory effects on VSMC proliferation, and therefore focused the remainder of our studies on E2-mediated regulation of miR-203 in VSMC. To investigate whether E2 can regulate miR-203 expression in cultured vascular cells, we performed real-time RT-PCR for miR-203 in MAoSMC. To allow us to determine which ER subtype mediates specific effects of E2, we used passaged MAoSMC that express low levels of ERs (Fig. 2C), and infected them with adenovirus expressing ERα (Ad-ERα) or ERβ (Ad-ERβ) as specified below. After 8h of treatment, E2 significantly up-regulated miR-203 expression in MAoSMC infected with Ad-ERα (Fig. 2A), but not Ad-ERβ (Fig. 2B), despite similar levels of over-expression of each receptor (Fig. 2C), and ERα-mediated up-regulation was blocked by the ER anatagonist ICI 182780 (Fig. 2D). Similar results were observed after 24h and 48h E2 treatment (data not shown).
Fig2. E2 up-regulation of miR-203 expression in MAoSMCs is mediated by ERα.
(A) QRT-PCR showing E2 significantly up-regulates miR-203 expression at 8h treatment in MAoSMC infected with Ad-ERα. (B) QRT-PCR showing E2 has no effect on miR-203 expression in MAoSMC infected with Ad-ERβ. (C) Immunoblot showing ERα and ERβ protein abundance were increased in Ad-ERα and Ad-ERβ infected MAoSMC, respectively. (D) Pre-treatment of ICI for 30 min completely blocks E2 induction of miR-203 expression in Ad-ERα infected MAoSMC. All quantitative values show mean ± S.D. of three independent experiments. *P<0.05, **P<0.01.
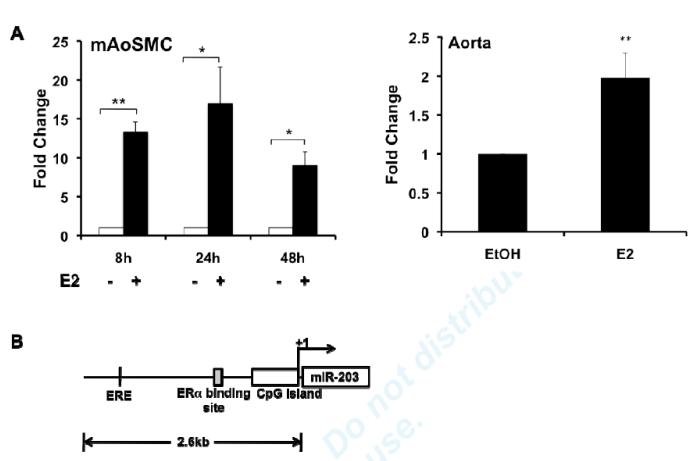
To begin to study the molecular mechanisms by which E2 regulates miR-203 biogenesis, we examined E2’s effects on primary miR-203 (pri-miR-203), the precursor to mature miR-203. E2 significantly increased the abundance of pri-miR-203 in MAoSMC (Fig. 3A left panel) and in aorta (Fig. 3A right panel), suggesting that E2 regulation of miR-203 expression occurs at the transcriptional level.
Fig3. E2 up-regulates miR-203 expression at the transcriptional level.
(A) Left panel: E2 increases pri-miR203 expression at 8h, 24h, and 48h treatment in Ad-ERα infected MAoSMCs. Right Panel: 4h ex vivo E2 treatment increases pri-miR-203 expression in aortas. (B) Schematic representation of mouse miR-203 gene, predicted ERE, and the known ERα binding site. (C) Transcription activation effects of E2 on the two putative promoter regions of the mouse miR-203 gene (pro1 and pro2). E2 was added to the media at the same time as transfection of luciferase constructs. Firefly luciferase activity was measured 24 h after E2 treatment. (D) Mapping of transcriptional regulation region of E2 by promoter deletion analysis and luciferase assay. Portions of the miR-203 promoter region were deleted according to the location of the known ERα binding sites. Values shown are mean±SD of at least three independent experiments. *P<0.05, ** P<0.01, N.S., non-significant.
Identification of the promoter region for miR-203
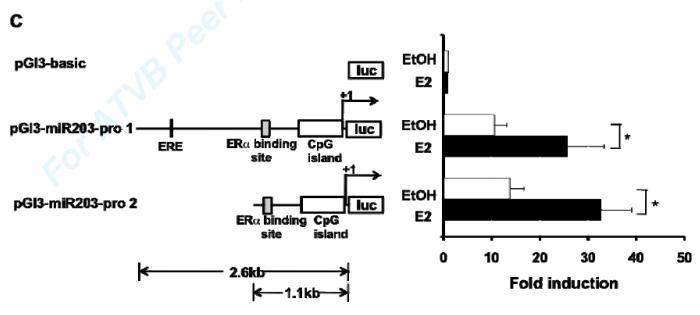
In order to confirm transcriptional regulation of miR-203 by E2, and to identify important transcriptional regulatory control elements for miR-203, we analyzed the 5kb upstream region of mouse miR-203 gene using the CpG Island track, conservation track, and ORegAnno track (open regulatory annotation database) in the UCSC genome browser and identified several potentially interesting regulatory elements including: 1. a highly conserved CpG island (−474~+154), with 52% identical sequence between human and mouse, 2. an Estrogen Response Element (ERE) (−1,725~-1,711), and 3. a previously published ERα binding site in liver33 in the region (−936~−716bp) close to 5′ end of the CpG island. The schematic representation of the predicted ERE, potential regulatory elements and CpG island in the upstream region of mouse miR-203 gene are shown in Fig. 3B.
Characterization of the miR-203 gene promoter
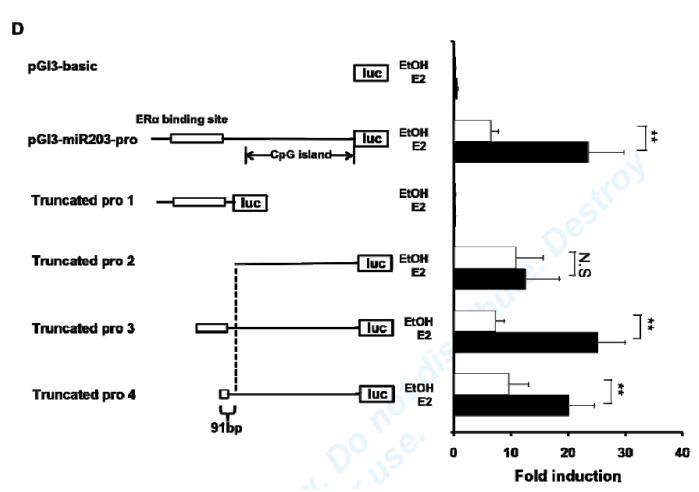
In order to characterize the miR-203 gene promoter, two DNA regions, which we named miR-203-pro-1 (a 2.6 kb upstream fragment) and miR-203-pro-2 (a 1.1 kb upstream fragment), were fused to a firefly luciferase reporter gene and transfected into E2- or vehicle-treated MAoSMC (Fig. 3C). The reporter activity of both promoter constructs was increased similarly, ~3-fold by E2 treatment, in Ad-ERα infected cells, supporting that the predicted ERE is not required for E2-mediated induction of miR-203.
To determine whether E2 regulation of miR-203 transcription is mediated by the known ERα binding site in the promoter region, deletion analyses were performed (Fig. 3D). Several truncated reporter constructs, named Truncated pro 1-4, were transfected into E2- or vehicle-treated mouse carotid artery smooth muscle cells (MCASMCs). The ERα-binding site alone was insufficient to mediate E2’s induction of miR-203 expression (Truncated pro 1), and removal of the ERα-binding site resulted in loss of E2 induction (Truncated pro 2). These findings suggest that the known ERα binding site has no promoter activity by itself, but is required for E2’s effect on miR-203 expression. With Truncated pro3 and 4, E2 still significantly increased reporter activity. Comparison of results with Truncated pro2 and 4 localized the minimal region required for E2-mediated regulation to a 91bp DNA sequence located at −752~−662 in the miR-203 promoter.
AP-1 is required for E2 regulation of miR-203 expression
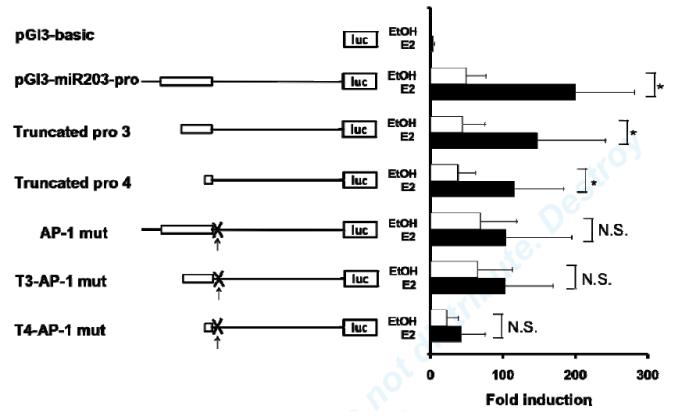
In order to identify other co-factors for E2 regulation of miR-203 transcription, we performed a transcription factor binding site (TFBS) analysis of the 91bp DNA sequence required for E2 induction of miR-203 promoter. Computational analysis revealed potential binding sites for AP-1, a previously identified ERα binding protein.34 We examined E2’s effect on miR-203 promoter reporters bearing a mutated AP-1 binding site. Mutation of the AP-1 binding site suppressed E2 induction of reporter activity (Fig. 4). These findings suggest that the AP-1 binding site is required for E2 regulation of miR-203 expression.
Fig4. AP-1 binding site is required for E2 induction of miR-203 promoter.
E2-mediated transcriptional activation of different miR-203 promoter constructs with and without mutated AP-1 binding site. Values shown are mean±SD of five independent transfection experiments. *P<0.05, N.S., non-significant.
Zeb-1 is involved in E2 regulation of miR-203 expression
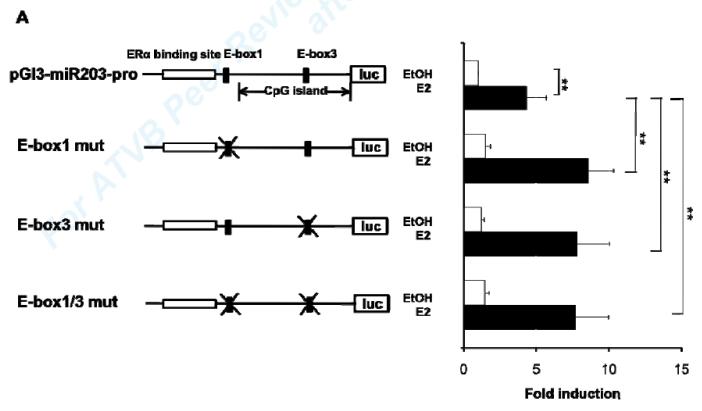
A previous study showed that the transcription factor Zeb-1 inhibits transcription of the human miR-203 gene by binding to conserved E-Boxes 1 and 3 in the promoter region of miR-203. 35 To investigate whether Zeb-1 is involved in E2 regulation of miR-203 in VSMC, we mutated the 2 conserved E-Boxes 1 and 3 in the mouse miR-203 promoter in a luciferase-driven reporter plasmid and transfected it into vehicle- or E2-treated MCASMCs. Mutations in either E-Box or in both E-Boxes significantly increased E2 activation of the miR-203 promoter reporter construct (Fig. 5A). These results support that Zeb-1 represses E2 up-regulation of miR-203 expression by binding to the two conserved E-Boxes.
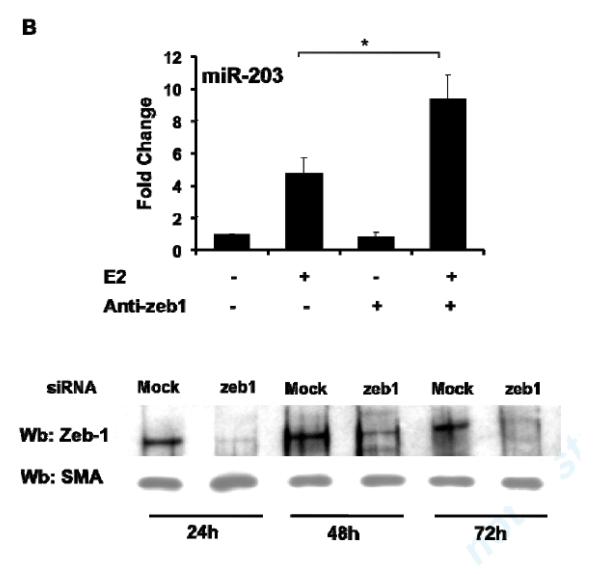
Fig5. Zeb-1 is involved in E2 regulation of miR-203 expression in MAoSMCs.
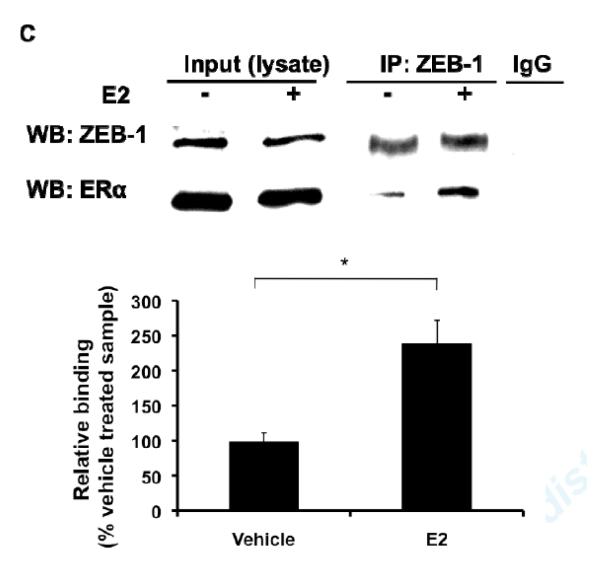
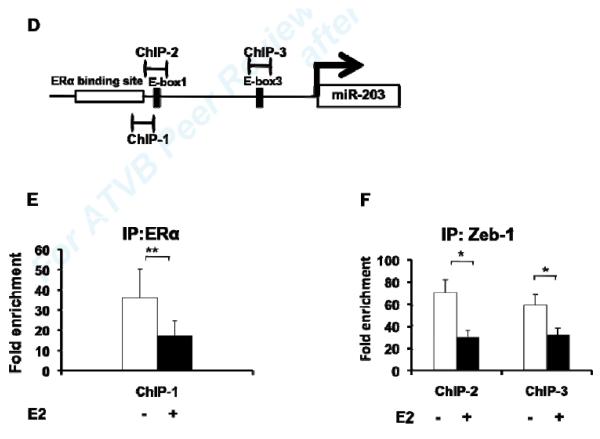
(A) Mutations of single or double E-Boxes 1 and 3 resulted in significantly increased E2 induction of transcriptional activation of the miR-203 promoter. Values shown are mean ± SD of seven independent experiments. (B) Inhibition of Zeb-1 expression by introducing siRNA in MAoSMCs enhanced E2 induction of miR-203 gene expression. Values shown are mean ± SD of three independent experiments. Immunoblots in the lower panel show Zeb-1 expression was significantly reduced 24h, 48h, and 72h after transfection of siRNA. (C) Co-immunoprecipitation of endogenous Zeb-1 and ectopically expressed flag-tagged ERα in MAoSMCs. MAoSMCs were infected with Ad-ERα and treated with E2 for 24h. Immunoprecipitation was performed with Zeb-1 and western blotting was performed with a monoclonal anti-Flag antibody to detect the co-immunoprecipitated ERα protein. One-twentieth of the input cell lysate were loaded in the first two panels and blotted for the presence of Zeb-1 and ERα. Representative data of three independent experiments are shown. Densitometric analysis of gel lanes was performed using AlphaImager Gel Imaging System (Alpha Innotech). Data were corrected both for the ERα input and Zeb-1 pull down. (D) The locations of three primer pairs used for the ChIP analysis (ChIP-1, ChIP-2, and ChIP-3) are depicted. Arrow represents transcription start site of miR-203 gene. (E,F) ChIP analysis of the miR-203 locus in MAoSMCs. Cells were infected with Ad-ERα and treated with E2 for 24 h. Cross-linked chromatin was immunoprecipitaed with antibodies specific for the mouse ERα or Zeb-1. Purified DNA was amplified with the three ChIP primer pairs shown in Fig 5D. Results are shown as fold enrichment over an intergenic region serving as negative control. Values shown are mean ± SD of at least four independent experiments. *P<0.05, **P<0.01.
To confirm that Zeb-1 inhibits E2-mediated induction of miR-203 expression, we knocked down Zeb-1 in Ad-ERα infected MAoSMC and quantified the effect of E2 treatment on miR-203 expression. The real-time RT-PCR results showed that knockdown of Zeb-1 enhances E2 induction of miR-203 expression (Fig 5B), supporting that Zeb-1 is a repressor of E2 regulation of miR-203 expression in VSMC.
To further study if Zeb-1 exerts its function on E2 regulation of miR-203 by interacting with ERα, we performed co-immunoprecipitation experiments in MAoSMC. Zeb-1 co-immunoprecipitated with ERα and E2 treatment enhanced the co-immunoprecipitation (Fig. 5C). These data suggested that Zeb-1 forms a complex with ERα in VSMC and that E2 promotes this interaction.
E2 reduces ERα and Zeb-1 binding to the miR-203 promoter region
Having demonstrated that ERα and Zeb-1 are involved in regulating miR-203 promoter activity, and that unliganded ERα at its binding site may inhibit miR-203 promoter activity, we performed ChIP assay with antibodies against ERα and Zeb-1 in vehicle- or E2-treated MAoSMC. The locations of primers used for ChIP assays are shown in Fig. 5D. Enrichment of ERα and Zeb-1 associated promoter fragments were confirmed by real-time PCR. Interestingly, we found less enrichment of both promoter fragments in E2 treated samples (Fig. 5E,F). These results indicate that both ERα and Zeb-1 bind to the miR-203 promoter, and E2 treatment, while increasing ERα-Zeb-1 complex formation, reduces their binding to the miR-203 promoter.
E2 down-regulation of Abl1 and p63 in MAoSMC are mediated by miR-203
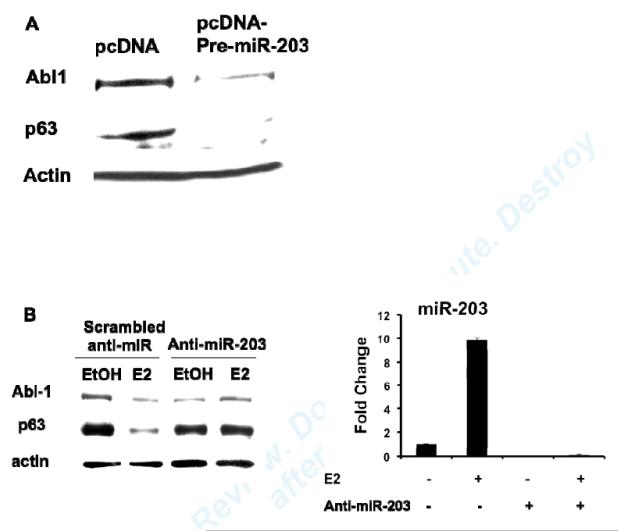
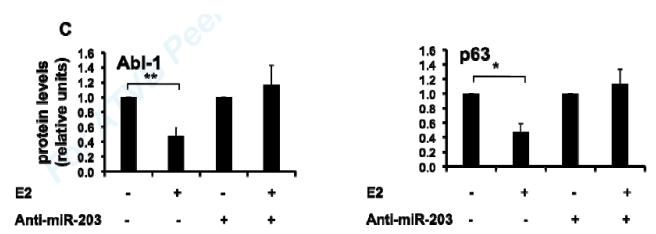
In order to explore whether E2 up-regulation of miR-203 leads to repression of target gene expression, we chose two previously identified miR-203 targets, Abl1 and p63, 29, 32 and studied the relationship between their protein abundance and E2 treatment in VSMC. Over-expression of miR-203 by transfecting cells with a plasmid expressing precursor miR-203 (pre-miR-203) inhibited Abl1 and p63 protein abundance in MAoSMC (Fig. 6A), which confirmed that both genes are miR-203 targets in VSMC. E2 treatment decreased both Abl1 and p63 protein abundance in Ad-ERα infected MAoSMC (Fig. 6B, left panel, scrambled anti-miR). E2-mediated repression of Abl1 and p63 gene expression was abolished in miR-203 depleted MAoSMC (Fig. 6B, left panel, anti-miR-203), supporting that E2 down-regulation of Abl1 and p63 in VSMC are mediated by miR-203. MiR-203 expression was significantly reduced as quantified by qRT-PCR in anti-miR-203 treated cells (Fig. 6B, right panel). The densitometry results of Abl1 and p63 blots are shown in Fig. 6C.
Fig6. E2 down-regulation of Abl1 and p63 are mediated by miR-203.
(A) Immunoblots showing over-expression of miR-203 by transfection of a pre-miR-203 containing plasmid decreased Abl1 and p63 protein abundance in MAoSMCs. (B) Left panel: MAoSMC over-expressing ERα was transfected with scrambled anti-miR or anti-miR-203 and treated with E2 or vehicle for 48 hours. Abl1 and p63 expression was monitored at the protein level as detected by immunoblotting. Actin was used as a loading control. Right panel: qRT-PCR results of miR-203 after anti-miR transfection. (C) Densitometry results of B, left panel. Diagrams show the results of three independent experiments ± SD. *P<0.05, **P<0.01.
E2 inhibition of VSMC proliferation is diminished by miR-203 knockdown
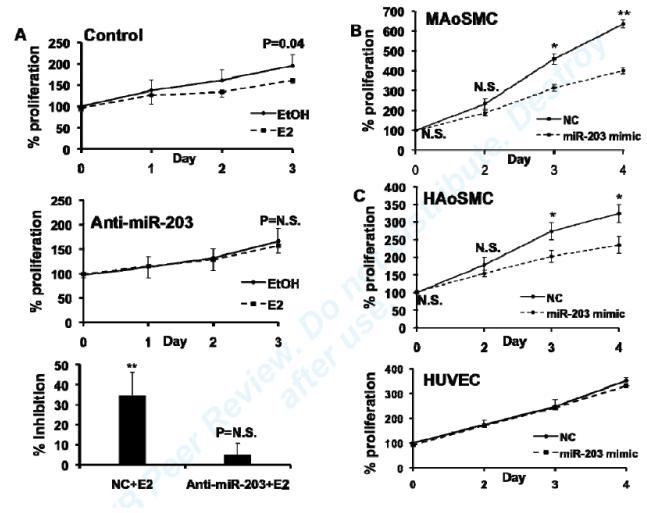
MiR-203 has been shown to inhibit proliferation in a variety of malignant tumor cells.29-32 Given that E2 has previously been shown to inhibit VSMC proliferation, both in vitro and in vivo, 8, 10-13 we hypothesized that miR-203 contributes to the inhibitory effect of E2 on VSMC proliferation. To test this hypothesis, we studied the effects of E2 on proliferation in ERα-overexpressing MAoSMC in which miR-203 was knocked down. In control cells, E2 significantly inhibited MAoSMC proliferation on day 3, and this effect was abolished by depletion of miR-203 (Fig. 7A). These results indicated that E2 inhibition of VSMC proliferation is partially mediated by miR-203.
Fig7. MiR-203 specifically inhibits VSMC proliferation and is involved in E2 inhibition of VSMC proliferation.
(A) Effects of miR-203 knockdown on E2 inhibition of MAoSMCs proliferation. Top and middle figures show E2 effects on cell proliferation in MAoSMCs infected with Ad-ERα and transfected with a negative control or anti-miR-203. Bottom figure shows percent inhibition of MAoSMCs proliferation by E2 in anti-miR-203 or negative control transfected cells on day 3. Values are presented as mean ± SD of four independent experiments. P values refer to a comparison between E2-treated and vehicle-treated cells. N.S., non-significant. (B) Proliferation of MAoSMCs after over-expression of miR-203 by introducing pre-miR-203 into the cells. Cell proliferation was measured at 0, 2, 3, and 4 days using Cell TiterGlo and read for luminescence. Data are presented as percent proliferation compared to the proliferation of control pre-miR transfected cells on day 0. (C) Effects of over-expressing miR-203 on HAoSMC and HUVEC proliferation. Values are presented as mean ± SD of three independent experiments. N.S., nonsignificant, *P<0.05, **P<0.01, refer to a comparison between control pre-miR and pre-miR-203-treated cells.
Over-expression of miR-203 specifically inhibits VSMC proliferation
To examine whether miR-203 by itself has an anti-proliferative effects on VSMC, we introduced pre-miR-203 into MAoSMC and did proliferation assays. Over-expression of miR-203 significantly reduced proliferation up to 4 days after transfection (Fig.7B). We performed TUNEL assays to determine whether inducing apoptosis might be a mechanism of miR-203’s anti-proliferative effect. The mean percentage of TUNEL-positive cells in scrambled miR-treated and miR-203 treated cells was 0.54±0.62% and 0.24±0.40%, which suggested that over-expression of miR-203 did not result in an induction of apoptosis. To determine whether the anti-proliferative effect of miR-203 is specific to VSMC, we studied the effect of over-expressing miR-203 in HAoSMC and HUVEC. MiR-203 significantly inhibited HAoSMC proliferation, but caused no change in HUVEC cell growth (Fig. 7C), despite similar expression levels of miR-203 (data not shown). These results indicate that the anti-proliferative effect of miR-203 is specific for VSMC.
Discussion
We have previously shown that many vascular genes are down regulated by E2 19, 20, but the molecular mechanisms by which E2 elicits target gene repression are poorly understood. So far, several mechanisms have been proposed for E2-mediated gene repression, including physiological interference of cofactors, direct action of co-repressors, and participation of elements of the basal transcriptional machinery.36 Given the known role of miRs in other cells and tissues, we hypothesized that E2-mediated regulation of miR expression might be a mechanism for gene repression in the vasculature and thereby be involved in suppressing VSMC proliferation. Therefore, we studied the role of E2 in regulating miRs expression in the vasculature. Our screen of E2 treated mouse aortas revealed that 26 of the 32 E2 responsive miRs were up-regulated, while only 6 were down-regulated. Several previous studies of E2 regulation of miRs expression in breast cancer cells26 and mouse uterus28 also showed that the majority of the E2-regulated miRs are up-regulated. Though several of the E2-regulated miRs were of potential interest, we chose to focus on miR-203 as it had previously been identified as regulating cell proliferation in tumor cells, and E2 is known to inhibit VSMC proliferation. In miR-203 target gene studies, we found that E2 represses previously identified miR-203 targets, p63 and Abl gene expression, and miR-203 knockdown fully prevented the E2-mediated repression of their gene repression, providing molecular evidence for a new mechanism of E2-mediated gene repression in vascular cells. The addition of miR regulation to the canonical pathway of E2 and ERα signaling may allow greater insight into the mechanism of E2’s tissue specific effects.
It has been demonstrated that ERs interact with the transcription factor AP-1 and regulate target gene expression through a tethering mechanism in which AP-1 directly binds to DNA. AP-1 binding sites have been shown to be significantly enriched at ER binding in sites that lack recognizable EREs.37 In our study, an AP-1 binding site in the miR-203 promoter region was required for E2 activation of miR-203 expression. Since no functional ERE was identified in the predicted miR-203 promoter region, ERα binding to the miR-203 promoter is likely mediated by AP-1. A recent study showed that the AP-1 proteins c-Jun and JunB oppositely regulate miR-203 expression in keratinocytes by binding to the AP-1 site in miR-203 promoter.38 Future study is required to elucidate whether AP-1 acts as more than a DNA binding moiety in E2 regulation of miR-203 expression in VSMC.
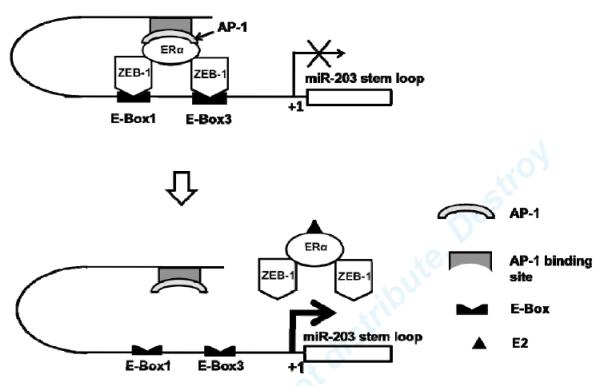
Zeb-1 has been demonstrated to inhibit miR-203 expression in pancreatic carcinoma cells.35 Although we didn’t see direct regulation of miR-203 by Zeb-1 in our study, we identified Zeb-1 as a repressor in E2 regulation of miR-203 expression through interacting with ERα at miR-203 promoter. The proposed molecular mechanism by which E2 regulates miR-203 transcription is shown in Fig. 8. In the absence of E2, an ERα and Zeb-1 complex binds to the miR-203 promoter and inhibits gene transcription. ERα-mediated regulation requires other co-factors, such as AP-1. In the presence of E2, the liganded ERα and Zeb-1 complex disassociates from the miR-203 promoter, presumably removing this repression and increasing gene transcription. Whether ERα/Zeb-1 is involved in regulation of other miRs transcription remains to be elucidated.
Fig8. Proposed mechanism by which E2 regulates miR-203 expression in VSMC.
In the absence of E2, ERα binds to miR-203 promoter region through other cofactors, such as Ap-1, and interacts with Zeb-1, which binds to the E-Boxes. This complex inhibits miR-203 gene transcription. In the presence of E2, E2 promotes dissociation of the ERα-Zeb1 complex from the miR-203 promoter, and this then activates miR-203 gene transcription.
A role for E2 transcriptional regulation of miR-203 in multiple biological systems is emerging. A recently published study showed that miR-203 was identified as the most significantly up-regulated miR in ERα-positive human mammary cells when compared with ERα-negative cell lines.39 Another previous study also identified miR-203 as an E2-inducible miR in MCF-7 cells.26 The current study confirms a role for E2 and ERα in transcriptional regulation of miR-203 in vascular cells and tissues.
VSMC proliferation contributes significantly to the pathobiology of vascular proliferative disorders such as atherosclerosis, post-angioplasty restenosis, and transplant vasculopathy.40 Estrogen has previously been shown to inhibit VSMC proliferation, both in vitro and in vivo. 8, 10-13 In the current study, we demonstrate that knock-down of miR-203 abolished E2-mediated inhibition of VSMC proliferation in vitro (without increasing apoptosis), identifying miR-203 as contributing to this anti-proliferative effect of E2.
Drug-eluting stents (DES) have been increasingly used to reduce VSMC proliferation after percutaneous coronary intervention (PCI). Recent studies have demonstrated that these DES may increase the risk of late thrombosis, which partially results from inhibition of VEC proliferation, thereby impairing re-endotheliazation.41-43 Identification of anti-proliferative agents selective for VSMC will represent a significant advance in the treatment of restenosis and other VSMC proliferative diseases. In the present study, we found miR-203 specifically inhibits proliferation in VSMC but not VEC in vitro. A limitation of the current study includes that we have only partially characterized the role of miR-203 on VEC proliferation. Further studies are required to determine whether miR-203 may serve as valuable therapeutic agent in the treatment of proliferative cardiovascular disease.
In conclusion, our study provides the first evidence that E2 selectively regulates miRs that in turn contribute to repression of target gene expression in vascular cells and tissues, and identify an important role for miR-203 in E2-mediated inhibition of VSMC proliferation. The specific inhibitory effect of miR-203 on VSMC proliferation supports its potential as a therapeutic agent in proliferative cardiovascular diseases.
Acknowledgments
We are grateful to Dr. Akiko Hata and Dr. Brandi Davis for their invaluable suggestions and generous help.
Funding Sources This work is supported by NIH grants RO1 HL61298 and T32 HL069770.
Footnotes
Disclosures None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 2.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol. 2007;25:5815–5824. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 3.Karas RH, Patterson BL, Mendelsohn ME. Human vascular smooth muscle cells contain functional estrogen receptor. Circulation. 1994;89:1943–1950. doi: 10.1161/01.cir.89.5.1943. [DOI] [PubMed] [Google Scholar]
- 4.Losordo DW, Kearney M, Kim EA, Jekanowski J, Isner JM. Variable expression of the estrogen receptor in normal and atherosclerotic coronary arteries of premenopausal women. Circulation. 1994;89:1501–1510. doi: 10.1161/01.cir.89.4.1501. [DOI] [PubMed] [Google Scholar]
- 5.Venkov CD, Rankin AB, Vaughan DE. Identification of authentic estrogen receptor in cultured endothelial cells. A potential mechanism for steroid hormone regulation of endothelial function. Circulation. 1996;94:727–733. doi: 10.1161/01.cir.94.4.727. [DOI] [PubMed] [Google Scholar]
- 6.Kim-Schulze S, McGowan KA, Hubchak SC, Cid MC, Martin MB, Kleinman HK, Greene GL, Schnaper HW. Expression of an estrogen receptor by human coronary artery and umbilical vein endothelial cells. Circulation. 1996;94:1402–1407. doi: 10.1161/01.cir.94.6.1402. [DOI] [PubMed] [Google Scholar]
- 7.Lindner V, Kim SK, Karas RH, Kuiper GG, Gustafsson JA, Mendelsohn ME. Increased expression of estrogen receptor-beta mRNA in male blood vessels after vascular injury. Circ Res. 1998;83:224–229. doi: 10.1161/01.res.83.2.224. [DOI] [PubMed] [Google Scholar]
- 8.Karas RH, Hodgin JB, Kwoun M, Krege JH, Aronovitz M, Mackey W, Gustafsson JA, Korach KS, Smithies O, Mendelsohn ME. Estrogen inhibits the vascular injury response in estrogen receptor beta-deficient female mice. Proc Natl Acad Sci U S A. 1999;96:15133–15136. doi: 10.1073/pnas.96.26.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendelsohn ME, Karas RH. Molecular and cellular basis of cardiovascular gender differences. Science. 2005;308:1583–1587. doi: 10.1126/science.1112062. [DOI] [PubMed] [Google Scholar]
- 10.Pare G, Krust A, Karas RH, Dupont S, Aronovitz M, Chambon P, Mendelsohn ME. Estrogen receptor-alpha mediates the protective effects of estrogen against vascular injury. Circ Res. 2002;90:1087–1092. doi: 10.1161/01.res.0000021114.92282.fa. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan TR, Jr., Karas RH, Aronovitz M, Faller GT, Ziar JP, Smith JJ, O’Donnell TF, Jr., Mendelsohn ME. Estrogen inhibits the response-to-injury in a mouse carotid artery model. J Clin Invest. 1995;96:2482–2488. doi: 10.1172/JCI118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhalla RC, Toth KF, Bhatty RA, Thompson LP, Sharma RV. Estrogen reduces proliferation and agonist-induced calcium increase in coronary artery smooth muscle cells. Am J Physiol. 1997;272:H1996–2003. doi: 10.1152/ajpheart.1997.272.4.H1996. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura Y, Suzuki T, Miki Y, Tazawa C, Senzaki K, Moriya T, Saito H, Ishibashi T, Takahashi S, Yamada S, Sasano H. Estrogen receptors in atherosclerotic human aorta: inhibition of human vascular smooth muscle cell proliferation by estrogens. Mol Cell Endocrinol. 2004;219:17–26. doi: 10.1016/j.mce.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Morales DE, McGowan KA, Grant DS, Maheshwari S, Bhartiya D, Cid MC, Kleinman HK, Schnaper HW. Estrogen promotes angiogenic activity in human umbilical vein endothelial cells in vitro and in a murine model. Circulation. 1995;91:755–763. doi: 10.1161/01.cir.91.3.755. [DOI] [PubMed] [Google Scholar]
- 15.Krasinski K, Spyridopoulos I, Asahara T, van der Zee R, Isner JM, Losordo DW. Estradiol accelerates functional endothelial recovery after arterial injury. Circulation. 1997;95:1768–1772. doi: 10.1161/01.cir.95.7.1768. [DOI] [PubMed] [Google Scholar]
- 16.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 17.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 18.O’Lone R, Knorr K, Jaffe IZ, Schaffer ME, Martini PG, Karas RH, Bienkowska J, Mendelsohn ME, Hansen U. Estrogen receptors alpha and beta mediate distinct pathways of vascular gene expression, including genes involved in mitochondrial electron transport and generation of reactive oxygen species. Mol Endocrinol. 2007;21:1281–1296. doi: 10.1210/me.2006-0497. [DOI] [PubMed] [Google Scholar]
- 19.Schnoes KK, Jaffe IZ, Iyer L, Dabreo A, Aronovitz M, Newfell B, Hansen U, Rosano G, Mendelsohn ME. Rapid recruitment of temporally distinct vascular gene sets by estrogen. Mol Endocrinol. 2008;22:2544–2556. doi: 10.1210/me.2008-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 23.Cohen A, Shmoish M, Levi L, Cheruti U, Levavi-Sivan B, Lubzens E. Alterations in micro-ribonucleic acid expression profiles reveal a novel pathway for estrogen regulation. Endocrinology. 2008;149:1687–1696. doi: 10.1210/en.2007-0969. [DOI] [PubMed] [Google Scholar]
- 24.Kovalchuk O, Tryndyak VP, Montgomery B, Boyko A, Kutanzi K, Zemp F, Warbritton AR, Latendresse JR, Kovalchuk I, Beland FA, Pogribny IP. Estrogen-induced rat breast carcinogenesis is characterized by alterations in DNA methylation, histone modifications and aberrant microRNA expression. Cell Cycle. 2007;6:2010–2018. doi: 10.4161/cc.6.16.4549. [DOI] [PubMed] [Google Scholar]
- 25.Dai R, Phillips RA, Zhang Y, Khan D, Crasta O, Ahmed SA. Suppression of LPS-induced Interferon-gamma and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: a novel mechanism of immune modulation. Blood. 2008;112:4591–4597. doi: 10.1182/blood-2008-04-152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, Nakshatri H. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2008;12:227–240. doi: 10.1111/j.1582-4934.2007.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Nothnick WB, Healy C. Estrogen induces distinct patterns of microRNA expression within the mouse uterus. Reprod Sci. 2010;17:987–994. doi: 10.1177/1933719110377472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernandez-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 31.Viticchie G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli LG, Knight RA, Candi E, Melino G. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- 32.Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J, Cheng HZ, Huang SD. MicroRNA-203 inhibits cell proliferation by repressing DeltaNp63 expression in human esophageal squamous cell carcinoma. BMC Cancer. 2011;11:57. doi: 10.1186/1471-2407-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao H, Falt S, Sandelin A, Gustafsson JA, Dahlman-Wright K. Genome-wide identification of estrogen receptor alpha-binding sites in mouse liver. Mol Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 35.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 36.Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol Cell Biol. 2009;29:1749–1759. doi: 10.1128/MCB.01476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin CY, Vega VB, Thomsen JS, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Torma H, Stahle M, Pivarcsi A. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol. 2010;130:124–134. doi: 10.1038/jid.2009.294. [DOI] [PubMed] [Google Scholar]
- 39.de Souza Rocha Simonini P, Breiling A, Gupta N, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 40.Dzau VJ, Braun-Dullaeus RC, Sedding DG. Vascular proliferation and atherosclerosis: new perspectives and therapeutic strategies. Nat Med. 2002;8:1249–1256. doi: 10.1038/nm1102-1249. [DOI] [PubMed] [Google Scholar]
- 41.McFadden EP, Stabile E, Regar E, Cheneau E, Ong AT, Kinnaird T, Suddath WO, Weissman NJ, Torguson R, Kent KM, Pichard AD, Satler LF, Waksman R, Serruys PW. Late thrombosis in drug-eluting coronary stents after discontinuation of antiplatelet therapy. Lancet. 2004;364:1519–1521. doi: 10.1016/S0140-6736(04)17275-9. [DOI] [PubMed] [Google Scholar]
- 42.Pfisterer M, Brunner-La Rocca HP, Buser PT, Rickenbacher P, Hunziker P, Mueller C, Jeger R, Bader F, Osswald S, Kaiser C. Late clinical events after clopidogrel discontinuation may limit the benefit of drug-eluting stents: an observational study of drug-eluting versus bare-metal stents. J Am Coll Cardiol. 2006;48:2584–2591. doi: 10.1016/j.jacc.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 43.Serruys PW, Kutryk MJ, Ong AT. Coronary-artery stents. N Engl J Med. 2006;354:483–495. doi: 10.1056/NEJMra051091. [DOI] [PubMed] [Google Scholar]