Abstract
Killer Ig-like receptors (KIRs) are innate immune receptors expressed by NK and T cells classically associated with the detection of missing self through loss of their respective MHC ligand. Some KIR specificities for allelic classical class I MHC (MHC-I) have been described, whereas other KIR receptor–ligand relationships, including those associated with nonclassical MHC-I, have yet to be clearly defined. We report in this article that KIR3DL2 and KIR2DS4 and the nonclassical Ag HLA-F, expressed as a free form devoid of peptide, physically and functionally interact. These interactions extend to include classical MHC-I open conformers as ligands, defining new relationships between KIR receptors and MHC-I. The data collectively suggest a broader, previously unrecognized interaction between MHC-I open conformers—including prototypical HLA-F—and KIR receptors, acting in an immunoregulatory capacity centered on the inflammatory response.
Introduction
Natural killer cells are an important component of the innate immunity that allows the immune system to respond to changes in class I MHC (MHC-I) expression, which can occur with tumor transformation or viral infection (1). Changes in MHC-I expression are sensed by polymorphic receptors on NK and T cells that are specific for allelic determinants on the MHC molecule itself (2, 3). Among these, the killer cell Ig-like receptors (KIRs) show the most developed ability to differentiate between different MHC allotypes (4) and are therefore of considerable interest for their involvement in transplantation (5, 6), pregnancy (7), and infectious disease (8), in which MHC allogenicity is a critical determinant. The genetic variation of KIR is extensive with respect to both gene content and allelic variation to a degree similar in magnitude to that of MHC class I and II (4, 9). The classically defined allospecific pairs are KIR2DL1 and HLA-C group 2 (C2), and KIR2DL2/3 and HLA-C group 1 (C1), defined by asparagine or lysine at position 80 of HLA-C (10). KIR3DL1-mediated recognition of the Bw4 epitope present in a subset of HLA-A and -B alleles contributes a second well-characterized interaction (11, 12). Specific interactions of these KIRs with their respective HLA ligand inhibit NK activity, which forms the basis for recognition of missing self.
NK cells also interact with nonclassical MHC-I, including HLA-E and -G. Unlike classical MHC-I, HLA-G is expressed on placental trophoblast cells and apparently does not participate in classical Ag presentation but rather may function as an important tolerogenic immunoregulator during pregnancy through interactions with ILT2 and ILT4 and KIR2DL4 (13–15). HLA-E, which has been shown to present Ags to T cells in certain circumstances (16), functions as a ligand for the CD94/NKG2 lectin receptors to regulate the activity of NK cells and subsets of NKT cells (17, 18). The third human nonclassical class I HLA-F is expressed on proliferating lymphoid and monocyte cells as a molecule devoid of bound peptide with or without β2-microglobulin (β2m) (19, 20). HLA-F associates with other MHC-I proteins as open conformers (OCs) without peptide but not with peptide-bound complexes (21). Recent data suggest HLA-F and MHC-I OCs operate together on activated lymphocytes and monocytes in a novel pathway for Ag cross-presentation (22). Together, the restriction of HLA-F expression to activated cells and the ability of HLA-F to bind free forms of MHC-I suggest that HLA-F and possibly heterodimers of HLA-F and MHC-I may represent another class of ligands for novel receptor–MHC interactions.
In consideration of the known receptor–ligand interactions between classical KIR and MHC-I complex, potential interactions between KIR and HLA-F were explored through tetramer and recombinant receptor binding, followed by specific receptor–ligand blocking to identify functional interactions. A tumor cell line expressing KIR3DL2 and NK clones expressing combinations of KIR3DL2 and other KIRs were used to demonstrate specific interactions between KIR3DL2 and MHC-I OC and HLA-F. Recombinant forms of both KIR3DL2 and KIR2DS4 were tested for interactions with HLA-F and other MHC-I as OCs. These findings were extended with NK and T cell lines in which functional responses correlated with specific interference of receptor–ligand interaction occurring between KIR3DL2 and both HLA-F and free forms of MHC-I that resembled, but did not precisely mirror, the function of inhibitory KIR. The data collectively support a model for a broader interaction between MHC-I OCs—for which HLA-F may serve as the prototypical example—and KIR receptors that was not previously recognized and thus may contribute to a more precise understanding of the functional interactions between MHC-I and KIR.
Materials and Methods
Cells and cell lines
B-LCL cell lines were previously collected and analyzed by the International Histocompatibility Workshops and Conference and obtained directly from the International Histocompatibility Working Group in Seattle, WA (www.ihwg.org). Cell lines were grown in RPMI 1640 supplemented with 15% FBS, 100 U/ml penicillin, and 100 U/ml streptomycin. The B-LCL/T cell hybrid cell line T2 (hemizygous chromosome 6 derived from 0.174, MHC class II deficient) was the generous gift of Thomas Spies (Fred Hutchinson Cancer Research Center, Seattle, WA). The KIR3DL2+ T cell acute lymphoblastic leukemia line TALL104 was obtained from American Type Culture Collection and cultured in complete medium (American Type Culture Collection–formulated IMDM with 20% FCS; 2.5 μg/ml human albumin; 0.5 μg/ml d-mannitol; 100 U rIL-2) at a minimum cell density of 5.0 × 105 cells per milliliter.
Pall filters of random healthy blood donors were provided by the Puget Sound Blood Center (Seattle, WA) with institutional review board approval. PBMCs were separated from whole blood using density gradient centrifugation with Lymphocyte Separation Medium (Cellgro, Manassas, VA) and ACCUSPIN columns (Sigma-Aldrich, St. Louis, MO). NK and T cells were negatively selected using either the NK Isolation Kit II (Miltenyi Biotec, Cambridge, MA) from enriched PBMCs or the RosetteSep Human NK cell, CD4 T cell, or CD8 T cell Enrichment mixture (STEMCELL Technologies, Vancouver, BC, Canada) from whole blood. Purified NK cells were incubated with 100 U rIL-2 for 5–7 d prior to use.
NK cell cloning was performed as previously described (23) after prior enrichment for KIR3DL2+ NK cells. Negatively enriched NK cells were labeled with mAb Q66 and separated using Anti-Mouse IgM Microbeads (Miltenyi Biotec, Cambridge, MA). Enriched KIR3DL2+ cells were seeded at a concentration of 1 cell per well with 1 × 105 γ-irradiated PBMCs from at least two donors and 1 × 104 γ-irradiated T2 cells as feeders in complete media supplemented with 2 μg/ml PHA and 500 U rIL-2. Cells from wells with an enlarged pellet were expanded, confirmed as NK cells (CD3−CD56+), and screened for expression of KIR.
Functional assays
Cytotoxicity.
B-LCL cell lines were prelabeled with 50 μCi [51Cr] for 1 h at 37°C, incubated with blocking mAb or recombinant protein, and plated at 5 × 103 with effector cells at the indicated E:T ratio. After 16 h, 30 μl supernatant was collected and applied to lumaplates, dried, and counted using a TopCount scintillation counter (PerkinElmer, San Jose, CA). Background release of chromium was measured from target cells without effectors, and full release was taken from targets lysed with 5% Triton X-100. Specific lysis was calculated using the formula [(sample − background)/(full release − background)] * 100.
CD107A and intracellular IFN-γ.
Assays were performed with purified NK cells at day +7 stimulation with 100 U rIL-2. Target T2 cells were incubated with blocking mAb at saturating concentration, as indicated. NK cells were washed and stained with mouse anti-human CD107A-PCy5 (H4A3; BD Pharmingen, San Jose, CA) and added to target cells at an E:T ratio of 1:2 for 1 h at 37°C, and then incubated with monensin (GolgiStop, BD) for 5 h. The cells were then stained and analyzed as described below.
To measure intracellular IFN-γ, targets and effectors were incubated together for 6 h at 37°C and then with 10 μg/ml brefeldin A for 6 h at 37°C. Cells were stained for surface phenotype as described below, followed by intracellular staining with mouse anti-human IFN-γ PE-Cy7 (4SB3; BD Pharmingen) using FACS lysing and FACS Permeabilizing solutions (BD). CD4+ and CD8+ T cells were incubated with plate-bound mouse anti-human CD3 (OKT-3) and 40 U rIL-2 for 7 d prior to assay. IFN-γ assay was performed as for the NK cell assay minus target cells and incubation of the T cells directly with blocking Abs.
Statistical testing.
Functional assays involving random healthy donors were analyzed using the paired t test for statistical significance. Data sets showing significant change (p < 0.05) in functional response under different conditions are indicated on the graph. All statistical analysis was performed using GraphPad Prism (San Diego, CA).
Cellular staining and phenotyping
Phenotyping of purified whole NK cells for functional analysis was performed with direct conjugates of anti-CD56pacific blue (HCD56), anti-KIR2DL1/S1FITC (HP-MA4), anti-KIR2DL2/3FITC (DX27), anti-KIR3DL1A700 (DX9) (BioLegend, San Diego, CA), and anti-NKG2AAPC (Z199; Beckman Coulter, Brea, CA). For CD4 and CD8 T cell assays, mouse anti-human CD4 PE-Cy5 or mouse anti-human CD8 PE-Cy5 was included. Cells were analyzed using a BD LSR II Flow Cytometer and FlowJo (TreeStar, Ashland, OR).
For each tetramer staining reaction, 1 μg BirA biotinylated monomer was incubated with 1.5 μg streptavidin-PE (SAPE; Invitrogen, Carlsbad, CA) for 30 min at room temperature. Tetramers were incubated with target cells for 25 min at 4°C. Tetramer binding was blocked by incubating target cells with blocking mAb or dilution factor (hybridoma supernatant) for 25 min at 4°C prior to staining with tetramer.
Staining with recombinant KIR was performed by first incubating target cells with blocking mAb or isotype control for 25 min at 4°C, and then with recombinant KIR for 25 min at 4°C. KIR binding was detected using streptavidin-PE, and stained cells were analyzed using a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) and FlowJo (TreeStar).
Two-step Ab staining was performed with saturating concentrations of biotinylated mAb (typically, 10 μg/ml) followed by washing and labeling with streptavidin-PE. The mAbs 3D11 and 6A4 specific for HLA-F were generated as previously described (19). HCA2 was a gift from Thomas Spies. mAb 5.133 (KIR3DL1, KIR3DL2, and KIR2DS4 specific) and mAb DX31 (KIR3DL2 specific) were kind gifts from Kerry Campbell and Kalle Malmberg, respectively. Q66 hybridoma supernatant was provided by Daniela Pende, Istituto Nazionale per la Ricerca sul Cancro, Genoa, Italy.
The HC10 Fab fragment was generated by immobilized papain at 37°C for 5 h using the Fab Micro Preparation Kit (Pierce, Rockford, IL) according to the manufacturer’s suggestions. The Mouse IgG1 Fab Preparation Kit (Pierce) was used for 3D11 and 6A4 Fab fragmentation. Briefly, Abs were digested by immobilized ficin in the presence of 25 mM cysteine at 37°C for 4.5 h. Sample preparation, digestion, and purification were performed according to the manufacturer’s instructions.
Protein analysis
Refolding and purification.
KIR3DL2 cDNA encoding residues 96–319 (D1D2 domains and stem region) was synthesized (Blue Heron, Bothell, WA) and cloned with a C terminus HIS-tag in pET-22b vector. KIR2DS4 and KIR2DL2 cDNA, both encoding residues 1–224 (D1D2 domains and stem region), were constructed without tags for use in surface plasmon resonance (SPR) experiments. The KIR2DS4 construct was also tagged with an N-terminal HIS-tag for pull-down experiments. BL21 (DE3) pLysS cells carrying the plasmid were grown to logarithmic phase, induced with 1.0 mM isopropyl β-d-thiogalactoside, and lysed by freezing and thawing. Inclusion bodies were washed extensively to remove contaminating proteins, dissolved in 6 M guanidine hydrochloride, and refolded by dilution into 100 ml refolding buffer (100 mM Tris⋅HCl, pH 8.2; 500 mM L-arginine HCl; 2 mM EDTA; 6.4 mM cysteamine; 3.6 mM cystamine⋅2 HCl; and 0.1 mM PMSF) to a final concentration of 4 μM and incubated with stirring at 4°C for 72 h. Refolded protein was dialyzed at 4°C against 100 mM urea and then against 10 mM Tris⋅HCl, 10 mM MES, and 100 mM urea before being concentrated using a 0.22-μm filter. KIR recombinant protein was purified by ion metal affinity chromatography using Ni-NTA resin (QIAGEN, Valencia, CA) and then on a Superdex 200 10/300 GL (GE Healthcare) liquid chromatography gel filtration column.
Biotinylation.
The following were added stepwise to 3.3 ml KIR3DL2-D1D2stem-bio: 10 mM MgOAc, 10 mM ATP, 50 μM d-biotin, 9 μg BirA, 0.1 mM PMSF, 50 μg leupeptin, and 5 μg pepstalin. After incubation at 21°C for 17 h, protein was buffer exchanged into 3.5 ml 20 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 2 mM EDTA, using a PD10 column and then concentrated to 250 μL.
3DL2 pull-down and Western blotting.
Whole cells were incubated with purified KIR3DL2-D1-D2-His or His-KIR2DS4-stem for 1 h at 4°C, were lysed in Dulbecco’s PBS containing 1% Nonidet P-40, and the proteins of interest were captured by Ni-NTA agarose (QIAGEN). After washing with lysis buffer and then lysis buffer with 10 mM imidazole, protein complexes were eluted with Dulbecco’s PBS with 300 mM imidazole. Proteins were separated on 10% Bis-Tris gels (Invitrogen, Grand Island, NY) and analyzed by Western blot using Abs 3D11 and HCA2.
SPR analysis
Interaction between KIRs and HLA-F/MHCs was analyzed by SPR using a Biacore 3000 system at 25°C in HBS-EP buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4). SPR ligands—HLA-F, MHC-I, and KIR3DL2—were biotinylated through an engineered C-terminal BirA biotinylation site. Biotinylated MHCs were purified at the National Institutes of Health Tetramer Core Facility or the Immune Monitoring Laboratory at Fred Hutchinson Cancer Research Center (Seattle, WA). Ligands were capture immobilized (at 10 μl/min) on an SA sensor chip (∼1000 RU) immediately following repurification by size exclusion chromatography to remove any aggregation products. Reference flow cells were left blank. SPR analytes—KIR3DL2-D1D2stem, HLA-F, KIR2DS4-D1D2stem, KIR2DL2-D1D2stem, ILT2-D1D2, and ILT4-D1D2—were repurified by size exclusion chromatography in HBS-EP buffer within 48 h of use.
Experiments were performed in duplicate and run at 20 μl/min. After running analytes over the captured MHC surface, a mild acid treatment consisting of two consecutive injections of 10 mM glycine-HCl at pH 2.0 (flow rate of 100 μl/min for 5 s) was performed, followed by a 15-min HBS-EP buffer stabilization, and then all analytes were injected again. Analytes and buffer blank injections were randomized before and after acid treatment and between the two sets. Abs known to bind to the surfaces were injected at the end of each injection set. Sensorgrams obtained from SPR measurements were analyzed using the double-subtraction method described by Myszka (24). The specific immobilization conditions, analyte concentrations, and interaction parameters are summarized in Supplemental Table I. DNA encoding the first two extracellular domains of ILT2 and ILT4 were a gift from Dr. Katsumi Maenaka (Laboratory of Biomolecular Science, Hokkaido University, Sapporo, Japan). ILT2-D1D2 and ILT4-D1D2 were purified as described (13).
Results
HLA-F tetramer binds specifically to KIR3DL2-expressing cells
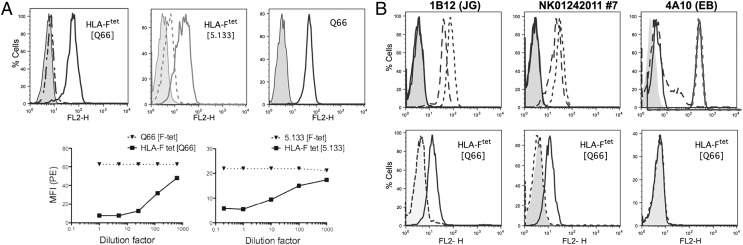
The well-studied receptor–ligand relationship between MHC-I and KIR, combined with putative structural similarities between HLA-F and MHC-I OCs, suggested a potential relationship between HLA-F and KIR. On the basis of prior data implicating interactions between the 3-domain KIR and HLA-B*27 homodimers, which may resemble MHC-I OCs (25) and the well-characterized relationships between 2-domain KIR and MHC-I complex, we suspected that the 3-domain KIRs were more likely to play a role in recognition of HLA-F and possibly free forms of MHC-I. To test this, HLA-F tetramer was used to query cell lines that expressed KIRs but that did not express MHC-I OC, as measured by binding to mAb HCA2 or HC10. Binding of HLA-F tetramer to cells that express MHC-I OC owing to a natural physical interaction between these molecules had been previously established (21). Specific staining with HLA-F tetramer was observed with human leukemic T cell line TALL-104, known to express KIR3DL2 and found to be negative for MHC-I OC (HCA2 and HC10). Further, HLA-F tetramer binding to TALL-104 was blocked using anti-3DL2 mAbs (3DL2-specific Q66 or 3DL1/2-specific 5.133). Blocking titrated with decreasing mAb concentration, whereas the prior application of HLA-F tetramer did not block respective mAb binding (Fig. 1A).
FIGURE 1.
Physical interactions between HLA-F and KIR3DL2. (A) HLA-F tetramer binding to TALL-104 is blocked by mAbs Q66 and 5.133. FACS profiles are shown of HLA-F tetramer binding to TALL-104 expressing KIR3DL2 without (solid lines) and after addition (dotted lines) of KIR3DL2-specific mAb Q66 or KIR3DL1/2-specific 5.133 (indicated within brackets). Control HLA-G tetramer staining is shown in gray. The rightmost profile shows TALL-104 stained with Q66 (solid line), with mouse IgM as control in gray. Immediately below the profiles are titrations of HLA-F tetramer and KIR3DL2 mAbs. Blocking conditions are indicated in brackets in the legend above each graph. (B) NK and T cell clones expressing KIR3DL2 specifically bind HLA-F tetramer. The upper row of panels shows NK clones stained with the indicated mAb specific for KIR3DL1 (DX9, dotted line), KIR3DL2 (Q66, dashed line), or KIR3DL1/2 (5.133, solid line). Controls were isotype-matched irrelevant mAb (gray fill). The lower row of panels shows profiles of cells stained with HLA-F tetramer before (solid line) and after (dashed line) incubation with mAb, as indicated within brackets. Control HLA-G tetramer staining is in gray.
Next, NK clones were generated and screened for expression of KIR3DL2 or KIR3DL1. Three appropriate clones were examined, two KIR3DL2+, as measured by reactivity with both Q66 and 5.133, and one KIR3DL1+, as measured by reactivity with only 5.133. Clones were tested in the resting state, in which they lacked expression of HLA-F or MHC-I OC. HLA-F tetramer stained both KIR3DL2+ clones, whereas no staining was detected on the clone expressing KIR3DL1. HLA-F tetramer binding to the KIR3DL2+ clones was reversed by prior addition of anti-3DL2 mAb (Fig. 1B).
Recombinant KIR3DL2 binds to HLA-F and MHC-I OC
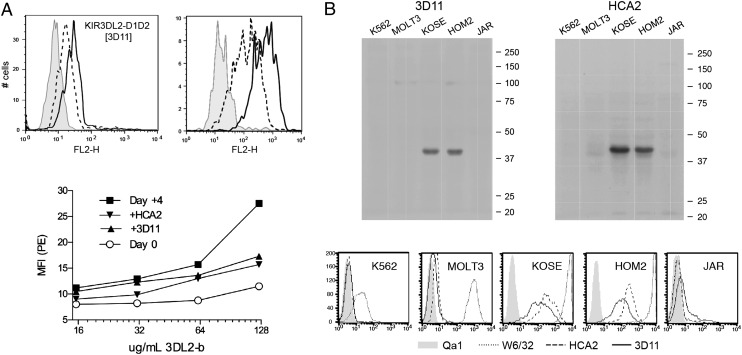
To confirm HLA-F binding to KIR3DL2, we used a recombinant form of a truncated KIR3DL2 protein in binding assays. Through testing of various recombinant forms, we found that the KIR3DL2 sequence containing the D1 and D2 domains and stem region, with or without a BirA tail, could be stably refolded, whereas we were unable to refold other forms, including those with the D0 domain. Although the D0 domain does enhance the affinity of ligand binding, it may have less involvement in the specificity of ligand recognition because binding of the D0 domain to HLA ligands appears to be independent of sequence variation in the HLA allotypes (26, 27). The KIR3DL2-D1D2stem-BirA recombinant protein bound to an activated T cell clone expressing both HLA-F and MHC-I OC, but not to the same cell in the resting state (Fig. 2A). Binding of recombinant KIR3DL2 could be blocked with either anti–HLA-F mAb 3D11 or anti–MHC-I HC mAb HCA2 (Fig. 2A), suggesting an interaction with both HLA-F and MHC-I OC either individually or possibly as a heterodimer. We next tested the same recombinant KIR3DL2-D1D2stem protein for the ability to bind and precipitate ligand from cell lines with and without HLA-F and MHC-I OC expression.
FIGURE 2.
Cell surface binding of recombinant KIR3DL2-D1D2stem to HLA-F and MHC-I. (A) Soluble KIR3DL2-D1D2stem-bio binding to activated T cell clone 7D9 is blocked by anti–HLA-F and anti–MHC-I mAbs. FACS profile of refolded KIR3DL2-D1D2 staining activated 7D9 without (solid lines) and following addition (dashed lines) of anti–HLA-F mAb 3D11 (indicated within brackets). Control is clone 7D9 before activation stained with KIR3DL2-D1D1stem-bio construct (gray). To the right is a FACS showing the same clone stained with 3D11 (dashed lines) and HCA2 (solid lines), with control 16G1 in gray shading. T cells were stimulated with IL-2 and anti-CD3 for 4 d before staining. The graph immediately below shows a titration of KIR3DL2-D1D2-stem-bio staining of clone 7D9 before and after activation and with blocking by anti–MHC-I mAb HCA2 or anti–HLA-F mAb 3D11 (conditions indicated in the legend). (B) Western analysis with the indicated mAbs of gel fractionated after pull-down with KIR3DL2-D1D2stem-His. Five cell lines were incubated with KIR3DL2-D1D2stem-His and pull-downs performed followed by Western blot analysis with HCA2 and 3D11. In the panels below, cell lines were examined for surface expression of MHC-I HC (HCA2, dashed line), HLA-F (3D11, solid line), and MHC-I complex (W6/32, dotted line). Control mAb (QA-1) is in gray fill.
The KIR3DL2-D1D2stem-BirA pull-down precipitate from this panel of cells was fractionated and probed for the presence of MHC-I and HLA-F, using Western blot analysis. Detectable levels of both MHC-I and HLA-F were precipitated only from cells that expressed both HLA-F and MHC-I OC (Fig. 2B), consistent with surface binding of KIR3DL2-D1D2stem and the respective specific mAb blocking. Although these experiments confirmed binding between MHC-I OC/HLA-F and KIR3DL2, they do not distinguish between direct binding to each individually or an interaction dependent upon dimerization between HLA-F and MHC-I, a possible structure suggested by the physical binding observed between these molecules.
SPR confirms interactions of MHC-I and HLA-F with KIR3DL2
To further test the interactions between HLA-F and KIR3DL2, we performed SPR measurements using either recombinant KIR3DL2-D1D2stem as analyte over immobilized HLA-F surfaces or refolded HLA-F as analyte over biotinylated KIR3DL2-D1D2stem surface. SPR measurements were performed with multiple concentrations of KIR3DL2-D1D2stem as analyte and two surface densities of HLA-F and with multiple concentrations of HLA-F as analyte and a single surface density of KIR3DL2-D1D2stem (Fig. 3A). The results of these experiments were consistent with concentration-dependent binding of both HLA-F and KIR3DL2-D1D2stem. The findings are also consistent with the ability of HLA-F ligand to bind KIR3DL2 receptor directly without dependence on other MHC-I.
FIGURE 3.
MHC-I and KIR3DL2 interactions measured by SPR. (A) Left, Sensorgrams of KIR3DL2-D1D2stem binding to HLA-F immobilized at two different densities, as indicated in the legend. Serial 2-fold dilutions of analyte were used, starting at 2.6 μM. Right, Sensorgrams of HLA-F analyte series binding to immobilized KIR3DL2. Serial 2-fold dilutions of HLA-F were used, starting at 3.0 μM. (B) Sensorgrams of KIR3DL2 analyte binding to immobilized HLA refolded with conditional ligand. KIR3DL2-D1D2stem was injected over one surface immobilized with complex of HLA H chain, β2m, and conditional ligand before and after acid treatment and another surface immobilized with refolded conditional ligand complex after UV treatment (indicated in legend). HLA molecules used in this experiment were HLA-A*03 and -B*07, and the conditional ligand sequences are described in Supplemental Table I. (C) KIR3DL2 differential binding to MHC-I OCs. Shown are sensorgrams of KIR3DL2 analyte binding to immobilized refolded HLA H chain, β2m, and respective peptide for three HLA molecules before (blue) and after (red) acid treatment. KIR3DL2-D1D2stem concentrations are indicated by light and dark colors, respectively. Controls for the integrity of the complex and of the free H chain/OC generated by acid treatment and UV treatment are presented in Supplemental Fig. 1.
To examine MHC-I binding to KIR3DL2, we tested two different methods to generate MHC-I OC, given our inability to refold stable OCs. Previous work had shown that OC could be formed from complex refolded with conditional ligand after UV treatment and used as analyte in SPR experiments (21), suggesting that similarly treated MHC-I could be immobilized directly on surfaces. Acid treatment was used as an alternative means of generating OCs on surfaces to include MHC-I alleles for which allele-specific conditional ligands have not been designed. Biotinylated HLA-A*03 and HLA-B*07 proteins refolded with conditional ligand peptides before and after UV treatment were immobilized on different surfaces. KIR3DL2-D1D2stem binding was examined before and after acid treatment. KIR3DL2-D1D2stem bound specifically to both HLA-A*03 and HLA-B*07 after either acid treatment or UV exposure but had reduced or absent binding to complex (Fig. 3B). In parallel with the observed KIR3DL2 binding, examination of surfaces with control proteins and mAbs that recognized complex (ILT2, ILT4, W6/32, and anti-β2m BB2M) and OC (ILT4, HCA2, and HC10) confirmed the structures and showed that acid treatment was quantitatively more effective at producing OC (Supplemental Fig. 1A).
To examine additional MHC-I alleles, including alleles that had previously been implicated as ligands for KIR3DL2, we immobilized different surfaces with recombinant HLA-A*03, A*11, and A*74, each refolded with an allele-specific high-affinity peptide (see Supplemental Table I). Again, analyte KIR3DL2-D1D2stem bound only HLA-A*03 acid-treated surface and not complex, consistent with the results found using conditional ligand peptide (Fig. 3C). Similar results were obtained with HLA-A*11 surfaces; however, no binding was apparent on HLA-A*74 surfaces either as complex or after acid treatment as OC. Control experiments confirmed that complex and OC surfaces exhibited essentially similar binding parameters for all three alleles (Supplemental Fig. 1B, Supplemental Table I). Allele-specific binding of MHC-I to KIRs has been clearly defined in the KIR2DL1/2/3 and HLA-C1, C2 receptor–ligand pairs and differential binding of the HLA-Bw4, Bw6 ligands with KIR3DL1.
Recombinant KIR2DS4 binds to HLA-F and MHC-I OC
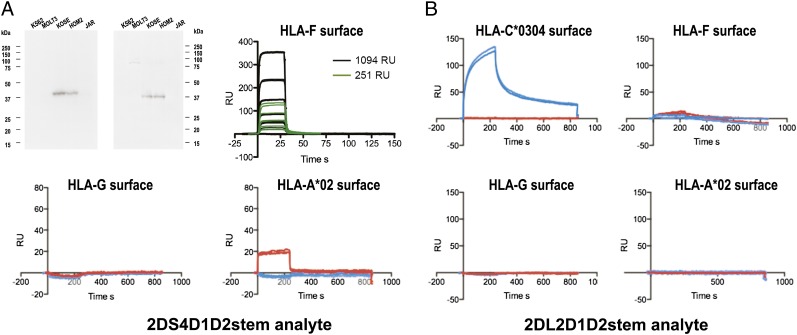
As mentioned above, a limitation of the biophysical measurements of ligand interactions with KIR3DL2-D1D2stem was the use of a truncated protein. Thus, although the biophysical interactions observed confirmed the biochemical and functional interactions we observed with intact KIR3DL2 expressed on cells, we further examined other KIRs that could be refolded as intact proteins, as an indirect means to confirm the biophysical measurements and to extend our knowledge of HLA-F and KIR interactions. We examined the KIR2DS4-stem recombinant protein based on the structural similarity between KIR2DS4 and KIR3DL2 in the putative ligand binding domains and functional binding with common allelic MHC-I, which suggested the possibility of overlapping ligands (28). It was possible to refold recombinant KIR2DS4-stem, including the complete extracellular domains and stem structure, which was tested in biophysical measurements essentially similar to those using KIR3DL2-D1D2stem. Pull-down experiments using KIR2DS4-stem-His and the HLA-F and MHC-I OC+ and MHC-I OC− cell lines used in the experiments described in Fig. 2B were performed as a first measurement of the interaction of KIR2DS4-stem with native HLA-F and MHC-I OC. As with KIR3DL2-D1D2stem, only cells expressing HLA-F and MHC-I OC yielded detectable MHC-I and HLA-F protein upon pull-down with KIR2DS4-stem (Fig. 4A).
FIGURE 4.
KIR2DS4 receptor binds to MHC-I OCs. (A) Western blot analysis of gel fractionated protein after pull-down with His-KIR2DS4-stem. Five cell lines were incubated with His-KIR2DS4-stem and pull-downs performed, followed by gel fractionation and Western blot analysis with mAbs HCA2 and 3D11. Sensorgrams of KIR2DS4-stem binding to HLA-F, HLA-G, and HLA-A*02, as indicated above each graph. Serial 2-fold dilutions of KIR2DS4-stem, starting at 24 μM, were used for binding to two RU surface densities of HLA-F, as indicated in the legend. (B) Sensorgrams of KIR2DL2-stem binding to natural ligand HLA-C*0304 and to HLA-F, HLA-G, and HLA-A*02, as indicated above each graph. All sensorgrams were performed in duplicate and where indicated before (blue) and after (red) acid treatment. Controls for the integrity of the complex and of the free H chain/OC generated by acid treatment are presented in Supplemental Fig. 2.
We next performed SPR measurements using recombinant KIR2DS4-stem as analyte in 2-fold serial dilutions over two different densities of immobilized HLA-F surface, and a single analyte concentration over different MHC-I surfaces with and without acid treatment to generate OC forms of MHC-I. Overall results paralleled those for KIR3DL2-D1D2stem in showing binding of KIR2DS4-stem to HLA-F in its native OC form, whereas binding to HLA-A*02 was detected only after acid treatment generating the OC form and not as complex (Fig. 4A). In addition, HLA-G did not bind KIR2DS4-stem either before or after acid treatment, consistent with the possibility that KIR2DS4 has specificity for a subset of MHC-I OCs. OC and complex forms for all surfaces were confirmed as shown in Supplemental Fig. 2.
These SPR binding data combined with the pull-down precipitations support our qualitative interpretations of HLA-F and MHC-I OC binding to KIR2DS4. The SPR experiments were limited by the highest concentration of KIR2DS4-stem available for testing, which precipitated at > 30 μM. This concentration was insufficient to achieve saturation of the surface; thus we could not determine the KD using equilibrium analysis. In addition, because of the fast on-and-off rates of the interaction, it was not possible to fit the binding curves to a simple binding model, precluding kinetic analysis to determine the KD. However, the shape of the binding curves and a weak interaction with a KD above 20 μM, as estimated from the highest concentration tested are both essentially similar to those observed in measurements of ILT2/4 and HLA-G binding (14). As additional control for the SPR experiments, we tested refolded KIR2DL2 as analyte over surfaces used for both the KIR2DS4-stem and KIR3DL2-D1D2stem experiments, with the addition of the natural HLA-C ligand as a positive control for the refolded KIR2DL2-stem. These experiments demonstrated the expected specificity of the recombinant KIR2DL2-stem for its HLA-C ligand as complex and not OC. The absence of binding to other MHC-I, including HLA-F as either complex or OC, provided additional evidence against nonspecific interactions with the refolded KIR and MHC-I proteins tested (Fig. 4B).
HLA-F and KIR3DL2 function in the presence or absence of other KIR receptors
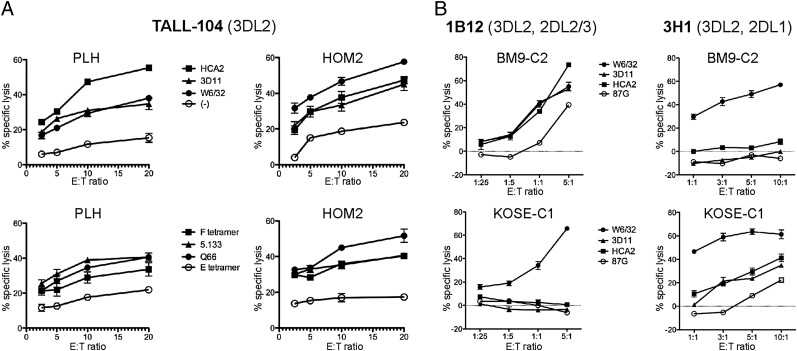
The interaction between KIR3DL2, HLA-F, and MHC-I was tested functionally, using the KIR3DL2+, CD16− T cell line TALL-104 as effector against B-LCL targets, which are known to express both HLA-F and free forms of MHC-I. Blocking was performed with tetramers and specific mAb to test for interactions between KIR3DL2 and MHC-I or HLA-F. Minimal cytolysis was observed over a wide range of E:T ratios for B-LCL targets PLH (HLA-A*03, B*47) and HOM2 (HLA-A*03, B*27), and inhibition of lysis was significantly reversed in the presence of specific mAbs used to block MHC-I or HLA-F on the target. Furthermore, reversal of inhibition also occurred in the presence of HLA-F tetramer and KIR3DL2-reactive mAb used to directly block KIR3DL2 on the effector (Fig. 5A). Reactivity of KIR3DL2 with HLA-F could also be observed using two KIR3DL2+ NK clones, coexpressing either KIR2DL1 or KIR2DL2/3. Target B-LCL expressing either HLA-C1 (BM9, HLA-A2/-B35) or HLA-C2 (KOSE HLA-A2/-B35) was used to assay KIR3DL2 function in the presence or absence of a cognate KIR2DL–HLA-C interaction, to examine the effect of KIR3DL2/HLA-F in the absence of cognate KIR–HLA interaction on licensed NK cells. In the absence of ligand for KIR2DL on the target, lysis by the respective effector was enhanced by blocking MHC-I or HLA-F, although to a degree much milder than the classical KIR2DL–HLA-C interaction (Fig. 5B). Alternatively, when the corresponding KIR2DL receptor was present, lysis was controlled by the interaction between HLA-C ligand and KIR2DL receptor and was reversed only in the presence of W6/32, which binds HLA-C1 or C2 complex. The choice of HLA-A*02+ targets in this instance demonstrated that the effect of KIR3DL2 inhibition by HLA-F can occur independently of HLA-A*03.
FIGURE 5.
Functional interactions between KIR3DL2 and HLA-F. (A) TALL-104 lysis of LCL targets is inhibited by HLA-F and KIR3DL2. B-LCLs (indicated above each graph) were subjected to lysis at varying E(TALL-104):T ratios. B-LCLs were incubated prior to lysis with MHC-I–specific (upper) or KIR3DL2-specific mAb and HLA tetramers (lower). Conditions are indicated in the legends presented between each set of profiles. (B) Functional stimulation of KIR3DL2+, 2DL+ NK clones is inhibited through HLA-F and MHC-I when KIR2DL ligand is absent. NK clones expressing KIR3DL2 and either KIR2DL1 or KIR2DL2/3 were used as effectors against B-LCL targets expressing either the C1 or the C2 ligand, as indicated in the box between graphs. Targets were subjected to lysis at varying E:T ratios in the presence of different mAbs, as indicated in the legend. 87G, an HLA-G–specific IgG2a Ab, was used as control.
HLA-F and MHC-I OC modulate KIR3DL2 reactivity toward target and between effectors
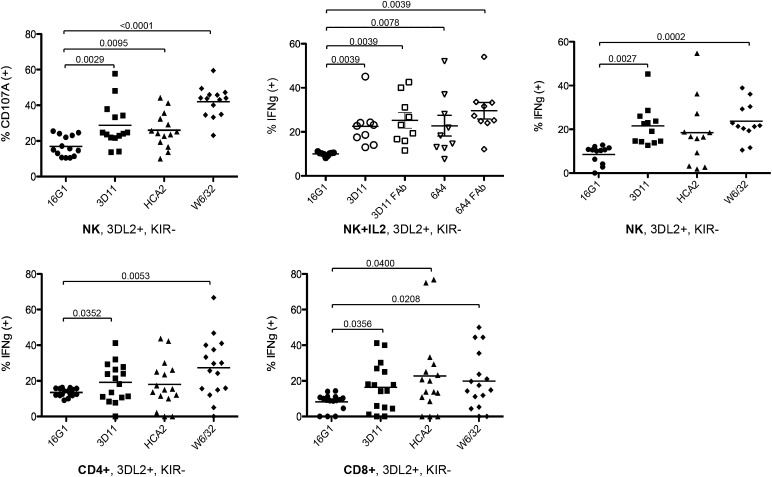
Extending these functional findings, we used IL-2–stimulated purified whole NK and T cell populations gated for the KIR3DL2 single-positive subset (KIR2DL1, -S1, -L2/3 and KIR3DL1− and NKG2A−) to measure cytokine production after exposure to HLA-F+ targets. NK cells were exposed to target cell line T2, which, similar to other B-LCL, expresses surface HLA-F and MHC-I OC but has reduced levels of surface MHC-I complex, including HLA-E, owing to deficiency for TAP and Tapasin. Accordingly, IL-2–activated NK cells from random healthy donors were exposed to T2 cells with and without addition of MHC-I and HLA-F mAbs and tested for the presence of CD107A or intracellular IFN-γ. Both CD107A and intracellular IFN-γ were significantly increased when either HLA-F (mAb 3D11) or MHC complex (mAb W6/32) was blocked (Fig. 6). Blocking by W6/32 suggests that the MHC complex also acts as an inhibitory ligand, but this interpretation is confounded by the fact that W6/32 also binds to different conformational forms of peptide-bound and peptide-free MHC (29). To control for artifactual responses due to Ab-dependent cell-mediated cytotoxicity, we performed a subset of the experiments with Fab produced from two HLA-F mAbs. The results were essentially similar for HLA-F blocking with whole mAb, supporting the specificity of the observed responses to the HLA-F and KIR interactions (Fig. 6).
FIGURE 6.
Regulation of activated KIR3DL2 single-positive NK and T cells by ligands MHC-I OC and HLA-F. Upper, IL-2–activated NK cells were generated from multiple random individuals, as described in Materials and Methods, and incubated with LCL target T2 in the presence or absence of blocking mAb or mAb Fab fragment, as indicated beneath each set. Both 3D11 and 6A4 are anti–HLA-F–specific mAbs, and HCA2 and W6/32 mAbs detect polymorphic MHC-I, as described in Materials and Methods. KIR3DL2 single-positive NK cells from random healthy donors were gated and analyzed by FACS for percentage of CD107A+ cells (upper left: n = 14) and percentage of IFN-γ+ cells (upper middle: n = 9 and right: n = 12) from two separate experiments. Lower, HLA-F+ CD4+ and CD8+ T cell lines (n = 16) were generated through stimulation with CD3 mAb. KIR3DL2 single-positive cells were analyzed by FACS for IFN-γ expression, with and without prior incubation with mAbs, as indicated beneath respective sets of data points. Relative p values are indicated with bars above each pairwise comparison where values were significant (<0.05).
Given that activated lymphocytes express both HLA-F and MHC-I OC, similar experiments were performed with IL-2/CD3–activated CD4+ or CD8+ T cells from random healthy donors to assess whether expression of these ligands during immune activation influences receptor–ligand cross-talk responses between effector cells. T cells were stimulated for 4 d with IL-2 and CD3, at which time the expression of HLA-F and MHC-I OC is at a peak (see Fig. 2). Even in the absence of target, significant shifts in IFN-γ responses between activated effectors were obtained when blocking HLA-F (mAb 3D11) and MHC (mAb W6/32) between the effector cells alone, whereas broader variation in responses was seen when blocking MHC-I OC (mAb HCA2). The latter result should be considered within the context that HCA2 reacts with only a subset of MHC-I alleles and the individuals used for these experiments expressed different levels of HCA2-reactive MHC-I.
Discussion
Functional interaction between KIR and MHC-I has been implicated in a range of immunological roles, from pregnancy to transplantation (30) and from autoimmunity to infectious disease (31). However, a complete picture of KIR–ligand function has yet to be fully elucidated. Our limited understanding of the expression and function of HLA-F, which goes hand in hand with the expression of free forms of MHC class I, reveals an underlayer of possible KIR–MHC interactions that may be particularly relevant to the inflammatory response in which upregulation of free forms of MHC-I occurs in its most dramatic fashion. HLA-F is unique among HLA class I in being surface expressed exclusively as an OC without peptide and β2m (19, 32)—possibly dependent on coexpression of other MHC-I OCs (21). Indeed, coexpression of MHC-I OC and HLA-F on activated cells and their potential ability to physically interact suggests overlapping or interdependent functions. Starting with these considerations, this study set out to obtain evidence for HLA-F as a ligand for immune receptors and to possibly relate the findings to other MHC-I OCs. The novel receptor–ligand relationship between KIR3DL2 and KIR2DS4 and HLA-F and other MHC-I OCs suggests a potentially broader definition of KIR function that increases in depth with the onset of an activated immune response. KIR–HLA interaction potentially extends beyond the detection of missing self to affect the dynamics of an ongoing inflammatory response as new receptor–ligand interactions come into play.
Although several receptor–ligand relationships between MHC-I OCs have been suggested (33), most of these have been envisioned as acting in cis, possibly playing a role in stabilizing the MHC-I OC and directing MHC-I internalization. Indeed, HLA-F has been proposed for such a role, including both stabilization and internalization, either as a coreceptor for MHC-I ligands acquired extracellularly or through HLA-F–specific internalization signals or both (21, 22). The trans receptor LILRB2 (ILT4) has been found to be a receptor for peptide-free MHC-I, binding both folded and free HC forms, and the activating receptor LILRA1, which displayed a preference for binding to HLA-C HC (34).
Early work defined a specific receptor–ligand pairing between KIR3DL2 and HLA-A*03/-A*11, presumably as complex with bound peptide, and not other MHC-I allotypes (35–37). In addition, studies with HLA-A*03 tetramers and KIR3DL2 transfectants suggested that tetramer binding was dependent on the peptide used to form the tetramer (38). The data presented in this article would appear to conflict with those reports, in particular that not only HLA-A*03 and HLA-A*11 but also HLA-B*07 bind to KIR3DL2 and do so in the OC form and not as complex. However, it may be important to consider that historically most functional studies used to define KIR specificity have been carried out using B-LCL cell lines as targets, which express an activated phenotype including the expression of HLA-F and MHC-I OCs (36, 37, 39). In these in vitro experiments, restriction of KIR3DL2 to the allotypes defined might be influenced by the differential ability of allelic MHC to interact with HLA-F. An alternative interpretation of peptide-dependent tetramer binding is that refolding with peptides of varying binding affinities affects the stability of tetramer reagents. Certain peptides may confer stable structures, thus predominantly favoring the complex form of MHC-I, whereas others confer less stability leading to higher proportion of peptide free MHC-I as tetramers, as we have observed. In this case, what is being considered peptide dependent binding may in fact be OC binding, reflecting instead the pool of peptide free MHC-I in specific peptide-MHC-I tetramer preparations.
Although the functional data previously reported and presented in this article support an inhibitory function for KIR3DL2, evidence suggests alternative functions are also likely (40). Previous studies reported that unlike classical KIR interactions, KIR3DL2 single-positive cells are hyporesponsive in individuals with the classical HLA-A*03 and/or -A*1l (41). This finding would contradict the idea that KIR3DL2 encounters its putative ligand as a peptide-bound complex under normal resting conditions and supports the concept that KIR3DL2+ cells may instead encounter their respective ligand under inflammatory conditions. Whether the function of KIR3DL2 and that of classical KIR overlap at all—even without considering the roles played by HLA-F and other MHC-I OCs—is therefore questionable. The expression of KIR3DL2 increases upon activation from basal levels present on resting cells (42), and the majority of functional evidence gathered for KIR3DL2 has been performed with activated effectors such as IL-2–activated NK cell populations or KIR3DL2+ NK cell clones (36, 37). Further, KIR3DL2 can bind CpG oligodeoxynucleotides (ODNs), which are subsequently cointernalized with KIR3DL2 and shuttled to TLR9, resulting in cytokine release (43). Therefore, the ability of KIR3DL2 to respond to the expression of HLA-F and free forms of MHC-I, combined with its downregulation by ODN, suggests that functional responses mediated by KIR3DL2 may be of greater influence during the inflammatory response than the detection of missing self under homeostatic conditions. Downregulation by ODN could serve a dual purpose in provoking a TLR-based proinflammatory signal while also reducing a functional interaction between KIR3DL2 and HLA-F/MHC-OC.
The coincident upregulation of KIR3DL2, HLA-F, and MHC-I OC under inflammatory conditions implies communication may occur between KIR3DL2+ NK and T cells, with activated HLA-F+ lymphocytes directed through KIR3DL2 and HLA-F–MHC-I OC interactions. Although the function of KIR2DS4 has so far been inferred largely from genetic studies, our findings, combined with the structural similarities with KIR3DL2 previously noted (28), suggest that activating KIR receptors also function in such capacity. Such communication has the potential to affect the magnitude and nature of the inflammatory response, and indicates the possibility that KIR3DL2+ or KIR2DS4+ cells become transiently “licensed” or experience an increase in functional capacity upon encounter with HLA-F and peptide-free MHC under proinflammatory conditions. Licensing of NK raises the question of why we have inhibitory receptors specific for self–MHC-I that do not have a ligand pair in certain individuals, as is often the case with the KIR2DL1/2 and HLA-C1, C2 pairings. Similarly, licensing also poses the question of why we have activating receptors that are specific for self–MHC-I that through licensing render those NK unresponsive, as in the case of KIR2DS1 and HLA-C1 individuals (44). As explanations, alternative possibilities for ligands have been suggested, including pathogen-encoded or other stress-activated signals that might provide high-affinity ligands (44, 45). The finding of HLA-F and MHC-I OC as ligands expressed on activated cells fits precisely among such possibilities.
A potentially relevant example of KIR/MHC-I OC pairing, and the only one reported outside the work presented in this article, is that measured between KIR3DL1/KIR3DL2 and HLA-B*27. HLA-B*27:05 is unusual among MHC-I alleles in being expressed constitutively not only as complex but also as a dimer without peptide or β2m. Specifically, KIR3DL1 interacts with HLA-B*27 complex, whereas KIR3DL2 interacts with HLA-B*27 expressed as a homodimer without peptide (46). Considering the strong association of HLA-B*27 with ankylosing spondylitis and related autoimmune conditions and that KIR3DL2, expressed as a homodimer, has the potential to sense HLA-F and MHC-I OC, raises the possibility that KIR3DL2 recognition of HLA-B*27 may represent an aberrant encounter that does not typically occur under resting conditions, leading to immune dysregulation. This possibility may relate to the expression of KIR3DL2 on Th17 CD4 T cells and their apparent increase in responsiveness in patients with ankylosing spondylitis (25).
HLA-F may also play a potential role in the immunology of pregnancy through modification of, or interaction with, specific HLA-E and HLA-G receptors. HLA-F, -E, -G, and -C are coexpressed in extravillous trophoblasts that have invaded the maternal decidua in contact with decidual NK cells (dNKs) (47). The ability of HLA-F to associate with MHC-I and KIR3DL2 and KIR2DS4 raises the possibility that HLA-F may be involved in stabilizing receptor–ligand interactions between extravillous trophoblasts and dNK during pregnancy, where dNK responses likely contribute to the immune regulation of pregnancy (48). We know that HLA-F and HLA-E physically interact as OCs, suggesting the possibility that HLA-F might modify the recognition of HLA-E by CD94/NKG2 heterodimers or, alternatively, that HLA-E may modify HLA-F interaction with KIR3DL2.
These findings may also be relevant to MHC-I and KIR genetics and disease, possibly extending previous knowledge of KIR ligands to include specifically HLA-F as the prototypical MHC-I OC alongside all MHC-I OCs. The evidence implicating KIR with disease is primarily derived from genetic association data (31), suggesting that HLA-F should be considered in studies of KIR in complex disease in at least two respects. First, HLA-F coding sequences are conserved but expression levels of HLA-F vary, presumably based on HLA-F genotype in which substantial noncoding variation has been identified (49). Second, because MHC-I OC and HLA-F are coexpressed and the affinities between HLA-F and different MHC-I alleles vary, the potential exists for HLA-F expression levels to be modified by allelic MHC-I OC and vice versa. Thus, not only HLA-F but also coordinated MHC-I variation may be important considerations in obtaining a precise interpretation of MHC-I–KIR genetic associations.
Acknowledgments
We thank Fujio Magaki for valuable advice.
This work was supported by National Institute of Child Health and Human Development Grant HD45813 (to D.E.G.); the University of Washington Center for AIDS Research, a National Institutes of Health–funded program (Grant P30 AI027757) supported by the following National Institutes of Health Institutes and Centers: National Institute of Allergy and Infectious Diseases; National Cancer Institute; National Institute of Mental Health; National Institute on Drug Abuse; National Institute of Child Health and Human Development; National Heart, Lung, and Blood Institute; and National Center for Complementary and Alternative Medicine (to J.P.G. and D.E.G.); and interim funding from the Clinical Research Division at the Fred Hutchinson Cancer Research Center (to D.E.G.).
The online version of this article contains supplemental material.
- dNK
- decidual NK cell
- KIR
- killer Ig-like receptor
- β2m
- β2-microglobulin
- MHC-I
- class I MHC
- OC
- open conformer
- ODN
- oligodeoxynucleotide
- SPR
- surface plasmon resonance.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Gumperz J. E., Valiante N. M., Parham P., Lanier L. L., Tyan D. 1996. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J. Exp. Med. 183: 1817–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litwin V., Gumperz J., Parham P., Phillips J. H., Lanier L. L. 1993. Specificity of HLA class I antigen recognition by human NK clones: evidence for clonal heterogeneity, protection by self and non-self alleles, and influence of the target cell type. J. Exp. Med. 178: 1321–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier L. L., Phillips J. H. 1996. Inhibitory MHC class I receptors on NK cells and T cells. Immunol. Today 17: 86–91 [DOI] [PubMed] [Google Scholar]
- 4.Parham P., Norman P. J., Abi-Rached L., Guethlein L. A. 2012. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velardi A., Ruggeri L., Mancusi A. 2012. Killer-cell immunoglobulin-like receptors reactivity and outcome of stem cell transplant. Curr. Opin. Hematol. 19: 319–323 [DOI] [PubMed] [Google Scholar]
- 6.Cooley S., Weisdorf D. J., Guethlein L. A., Klein J. P., Wang T., Le C. T., Marsh S. G., Geraghty D., Spellman S., Haagenson M. D., et al. 2010. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116: 2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Male V., Trundley A., Gardner L., Northfield J., Chang C., Apps R., Moffett A. 2010. Natural killer cells in human pregnancy. Methods Mol. Biol. 612: 447–463 [DOI] [PubMed] [Google Scholar]
- 8.Bashirova A. A., Thomas R., Carrington M. 2011. HLA/KIR restraint of HIV: surviving the fittest. Annu. Rev. Immunol. 29: 295–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhrberg M., Valiante N. M., Shum B. P., Shilling H. G., Lienert-Weidenbach K., Corliss B., Tyan D., Lanier L. L., Parham P. 1997. Human diversity in killer cell inhibitory receptor genes. Immunity 7: 753–763 [DOI] [PubMed] [Google Scholar]
- 10.Colonna M., Borsellino G., Falco M., Ferrara G. B., Strominger J. L. 1993. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc. Natl. Acad. Sci. USA 90: 12000–12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cella M., Longo A., Ferrara G. B., Strominger J. L., Colonna M. 1994. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 180: 1235–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern M., Ruggeri L., Capanni M., Mancusi A., Velardi A. 2008. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood 112: 708–710 [DOI] [PubMed] [Google Scholar]
- 13.Shiroishi M., Tsumoto K., Amano K., Shirakihara Y., Colonna M., Braud V. M., Allan D. S., Makadzange A., Rowland-Jones S., Willcox B., et al. 2003. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. USA 100: 8856–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiroishi M., Kuroki K., Rasubala L., Tsumoto K., Kumagai I., Kurimoto E., Kato K., Kohda D., Maenaka K. 2006. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc. Natl. Acad. Sci. USA 103: 16412–16417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajagopalan S., Bryceson Y. T., Kuppusamy S. P., Geraghty D. E., van der Meer A., Joosten I., Long E. O. 2006. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 4: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pietra G., Romagnani C., Manzini C., Moretta L., Mingari M. C. 2010. The emerging role of HLA-E-restricted CD8+ T lymphocytes in the adaptive immune response to pathogens and tumors. J. Biomed. Biotechnol. 2010: 907092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee N., Llano M., Carretero M., Ishitani A., Navarro F., López-Botet M., Geraghty D. E. 1998. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 95: 5199–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llano M., Lee N., Navarro F., García P., Albar J. P., Geraghty D. E., López-Botet M. 1998. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur. J. Immunol. 28: 2854–2863 [DOI] [PubMed] [Google Scholar]
- 19.Lee N., Geraghty D. E. 2003. HLA-F surface expression on B cell and monocyte cell lines is partially independent from tapasin and completely independent from TAP. J. Immunol. 171: 5264–5271 [DOI] [PubMed] [Google Scholar]
- 20.Lee N., Ishitani A., Geraghty D. E. 2010. HLA-F is a surface marker on activated lymphocytes. Eur. J. Immunol. 40: 2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodridge J. P., Burian A., Lee N., Geraghty D. E. 2010. HLA-F complex without peptide binds to MHC class I protein in the open conformer form. J. Immunol. 184: 6199–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodridge J. P., Lee N., Burian A., Pyo C. W., Tykodi S. S., Warren E. H., Yee C., Riddell S. R., Geraghty D. E. 2013. HLA-F and MHC-I open conformers cooperate in a MHC-I antigen cross-presentation pathway. J. Immunol. 191: 1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cella M., Colonna M. 2000. Cloning human natural killer cells. Methods Mol. Biol. 121: 1–4 [DOI] [PubMed] [Google Scholar]
- 24.Myszka D. G. 1999. Improving biosensor analysis. J. Mol. Recognit. 12: 279–284 [DOI] [PubMed] [Google Scholar]
- 25.Bowness P., Ridley A., Shaw J., Chan A. T., Wong-Baeza I., Fleming M., Cummings F., McMichael A., Kollnberger S. 2011. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J. Immunol. 186: 2672–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khakoo S. I., Geller R., Shin S., Jenkins J. A., Parham P. 2002. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J. Exp. Med. 196: 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivian J. P., Duncan R. C., Berry R., O’Connor G. M., Reid H. H., Beddoe T., Gras S., Saunders P. M., Olshina M. A., Widjaja J. M., et al. 2011. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graef T., Moesta A. K., Norman P. J., Abi-Rached L., Vago L., Older Aguilar A. M., Gleimer M., Hammond J. A., Guethlein L. A., Bushnell D. A., et al. 2009. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 206: 2557–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giacomini P., Beretta A., Nicotra M. R., Ciccarelli G., Martayan A., Cerboni C., Lopalco L., Bini D., Delfino L., Ferrara G. B., et al. 1997. HLA-C heavy chains free of beta2-microglobulin: distribution in normal tissues and neoplastic lesions of non-lymphoid origin and interferon-gamma responsiveness. Tissue Antigens 50: 555–566 [DOI] [PubMed] [Google Scholar]
- 30.Parham P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5: 201–214 [DOI] [PubMed] [Google Scholar]
- 31.Kulkarni S., Martin M. P., Carrington M. 2008. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 20: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wainwright S. D., Biro P. A., Holmes C. H. 2000. HLA-F is a predominantly empty, intracellular, TAP-associated MHC class Ib protein with a restricted expression pattern. J. Immunol. 164: 319–328 [DOI] [PubMed] [Google Scholar]
- 33.Arosa F. A., Santos S. G., Powis S. J. 2007. Open conformers: the hidden face of MHC-I molecules. Trends Immunol. 28: 115–123 [DOI] [PubMed] [Google Scholar]
- 34.Jones D. C., Kosmoliaptsis V., Apps R., Lapaque N., Smith I., Kono A., Chang C., Boyle L. H., Taylor C. J., Trowsdale J., Allen R. L. 2011. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J. Immunol. 186: 2990–2997 [DOI] [PubMed] [Google Scholar]
- 35.Brando C., Mukhopadhyay S., Kovacs E., Medina R., Patel P., Catina T. L., Campbell K. S., Santoli D. 2005. Receptors and lytic mediators regulating anti-tumor activity by the leukemic killer T cell line TALL-104. J. Leukoc. Biol. 78: 359–371 [DOI] [PubMed] [Google Scholar]
- 36.Döhring C., Scheidegger D., Samaridis J., Cella M., Colonna M. 1996. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 156: 3098–3101 [PubMed] [Google Scholar]
- 37.Pende D., Biassoni R., Cantoni C., Verdiani S., Falco M., di Donato C., Accame L., Bottino C., Moretta A., Moretta L. 1996. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J. Exp. Med. 184: 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansasuta P., Dong T., Thananchai H., Weekes M., Willberg C., Aldemir H., Rowland-Jones S., Braud V. M. 2004. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 34: 1673–1679 [DOI] [PubMed] [Google Scholar]
- 39.Storkus W. J., Salter R. D., Alexander J., Ward F. E., Ruiz R. E., Cresswell P., Dawson J. R. 1991. Class I-induced resistance to natural killing: identification of nonpermissive residues in HLA-A2. Proc. Natl. Acad. Sci. USA 88: 5989–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivori S., Falco M., Carlomagno S., Romeo E., Soldani C., Bensussan A., Viola A., Moretta L., Moretta A. 2010. A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood 116: 1637–1647 [DOI] [PubMed] [Google Scholar]
- 41.Fauriat C., Andersson S., Björklund A. T., Carlsten M., Schaffer M., Björkström N. K., Baumann B. C., Michaëlsson J., Ljunggren H. G., Malmberg K. J. 2008. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group A KIR haplotype. J. Immunol. 181: 6010–6019 [DOI] [PubMed] [Google Scholar]
- 42.Chrul S., Polakowska E., Szadkowska A., Bodalski J. 2006. Influence of interleukin IL-2 and IL-12 + IL-18 on surface expression of immunoglobulin-like receptors KIR2DL1, KIR2DL2, and KIR3DL2 in natural killer cells. Mediators Inflamm. 2006: 46957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcenaro E., Carlomagno S., Pesce S., Moretta A., Sivori S. 2011. Bridging innate NK cell functions with adaptive immunity. Adv. Exp. Med. Biol. 780: 45–55 [DOI] [PubMed] [Google Scholar]
- 44.Fauriat C., Ivarsson M. A., Ljunggren H. G., Malmberg K. J., Michaëlsson J. 2010. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood 115: 1166–1174 [DOI] [PubMed] [Google Scholar]
- 45.Long E. O., Rajagopalan S. 2002. Stress signals activate natural killer cells. J. Exp. Med. 196: 1399–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kollnberger S., Chan A., Sun M. Y., Chen L. Y., Wright C., di Gleria K., McMichael A., Bowness P. 2007. Interaction of HLA-B27 homodimers with KIR3DL1 and KIR3DL2, unlike HLA-B27 heterotrimers, is independent of the sequence of bound peptide. Eur. J. Immunol. 37: 1313–1322 [DOI] [PubMed] [Google Scholar]
- 47.Ishitani A., Sageshima N., Lee N., Dorofeeva N., Hatake K., Marquardt H., Geraghty D. E. 2003. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J. Immunol. 171: 1376–1384 [DOI] [PubMed] [Google Scholar]
- 48.Erlebacher A. 2013. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 31: 387–411 [DOI] [PubMed] [Google Scholar]
- 49.Pyo C. W., Williams L. M., Moore Y., Hyodo H., Li S. S., Zhao L. P., Sageshima N., Ishitani A., Geraghty D. E. 2006. HLA-E, HLA-F, and HLA-G polymorphism: genomic sequence defines haplotype structure and variation spanning the nonclassical class I genes. Immunogenetics 58: 241–251 [DOI] [PubMed] [Google Scholar]