Abstract
The activation of immune cells is mediated by a network of signaling proteins that can undergo post-translational modifications critical for their activity. Methylation of nucleic acids or proteins can have major effects on gene expression as well as protein repertoire diversity and function. Emerging data indicate that indeed many immunologic functions, particularly those of T cells, including thymic education, differentiation and effector function are highly dependent on methylation events. The critical role of methylation in immunocyte biology is further documented by evidence that autoimmune phenomena may be curtailed by methylation inhibitors. Additionally, epigenetic alterations imprinted by methylation can also exert effects on normal and abnormal immune responses. Further work in defining methylation effects in the immune system is likely to lead to a more detailed understanding of the immune system and may point to the development of novel therapeutic approaches.
Keywords: Autoimmunity, Transmethylation, T cells, Lupus, EAE
1. Introduction
Nucleic acids and proteins can be fine-tuned by a wide spectrum of modifications, which are essential for regulating gene transcription and amplifying protein repertoires. Major post-translational modifications include phosphorylation, methylation, acetylation, nitration, citrullination and glycosylation. Phosphorylation, because of its ubiquitous role in cell signaling, has been the most studied with regard to immunity, but accumulating evidence indicates that methylation also plays an important role in immune cell function. Interestingly, post-translational modifications may trigger a break in tolerance and provoke an anti-self response [1], suggesting that inhibition of methylation may be a means to intervene in inflammatory or autoimmune processes. A past hurdle in such an approach was that transmethylation inhibitors were irreversible and somewhat toxic, but less toxic reversible inhibitors have now been developed, and their applicability to a variety of abnormal immunological responses has been tested. We summarize herein recent findings pertaining to methylation and its role in immune system function, and discuss the potential therapeutic utility of methylation inhibitors in immune-related diseases.
2. The transmethylation pathway
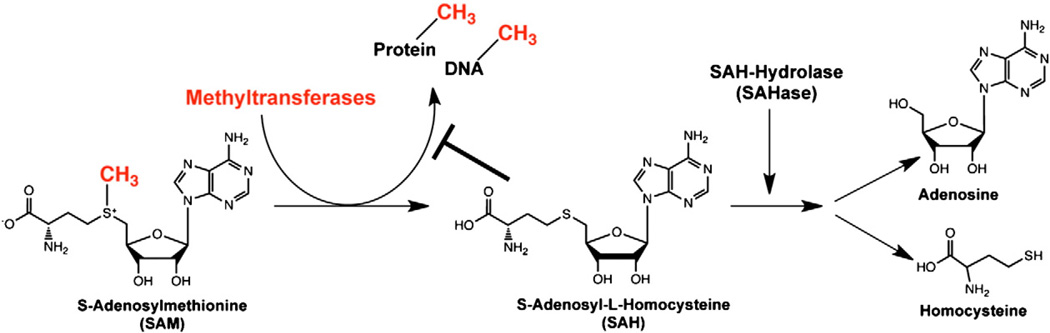
The major players controlling transmethylation are S-adenosylmethionine (SAM) and the methyltransferase (MT) superfamily, which includes DNA methyltransferases (DNMTs), protein arginine methyltransferases (PRMTs), and protein lysine methyltransferases (PKMT), among others. The terms methylation and transmethylation are used interchangeably to describe the general donation of methyl groups from SAM to various macromolecules [2–4]. MTs remove a methyl group from SAM, thereby converting it to S-adenosyl-L-homocysteine (SAH), which can act as a potent feedback inhibitor of upstream transmethylation reactions by blocking most MT activity (Fig. 1). Since the reactions are reversible, the ratio of SAM to SAH influences whether SAH is converted back into SAM or, more likely, hydrolyzed by S-adenosyl-L-homocysteine hydrolase (SAHase) into adenosine and homocysteine. Thus, blockade of SAHase activity raises intracellular SAH levels, reduces MT activity, and indirectly inhibits transmethylation reactions.
Figure 1.
Biochemical pathway of protein and DNA transmethylation. Methyltransferases (MT) catalyze the transfer of methyl groups from SAM to either proteins or DNA, resulting in conversion of SAM to SAH. SAH is then hydrolyzed into adenosine and homo-cysteine by SAHase. If the balance between SAM and SAH concentrations favor SAH, SAH can then provide negative feedback inhibition of transmethylation activity allowing restoration of SAM concentrations. Thus, pharmacological blockade of SAHase increases intracellular levels of SAH, thereby indirectly blocking MT activity.
In mice with the non-agouti mouse ax-mutation lacking the gene (Ahcy) encoding SAHase [5], the failure of embryo implantation suggests a critical role for SAHase in some cellular functions. Indeed, exposure of the inner cell mass of non-mutant embryos to an irreversible SAHase inhibitor in vitro results in abnormal proliferation and differentiation (Table 1). These findings are consistent with the observed high Ahcy mRNA expression throughout embryonic development [GNF SymAtlas database (http://symatlas.gnf.org)]. Correspondingly, in humans with decreased SAHase activity levels (5%–20% of normal) associated with elevated levels of serum creatine kinase, SAH, and SAM, severe defects, including myopathy and developmental delay, are seen [6–8]. Interestingly, dietary restriction of methionine and supplementation with creatine and phosphatidylcholine appreciably reduced such manifestations, suggesting that a minimum threshold of SAHase activity is sufficient to sustain normal cellular function.
Table 1.
Impact of transmethylation pathway on embryonic and disease development.
| Pathology/affected field | Treatment/model | Observation | References |
|---|---|---|---|
| Embryonic development | |||
| Mouse model | Ahcy (encoding SAHase) lacking mice | -Embryo implantation failure -High Ahcy mRNA expression during embryonic development |
[5] |
| SAHase inhibition | -Abnormal proliferation and differentiation of embryo inner cell mass | ||
| Human deficiency | Gene encoding SAHase 2 missense mutations |
-Myopathy-developmental delay -5–20% of normal SAHase activity level -High levels of creatine kinase, SAH and SAM |
[6–8] |
| Methionine dietary restriction | -Reduced symptoms | ||
| Adenosine deaminase deficiency | -57% reduction of cellular SAHase -DeoxyATP accumulation -T cell dysfunction as a hallmark |
[25–28] | |
| Autoimmunity | |||
| Rheumatoid arthritis | |||
| T cells | -Normal level of DNA methyltransferase activity and DNMT1 mRNA | [67,94] | |
| Synovial fibroblasts | -Decrease in global DNA methylation -Hypomethylation of CpG island in LINE-1 promoter |
||
| Monocytes | -Unmethylated CpG island within IL-6 promoter -Methylation changes in CpG island promoter of DR-3 |
||
| Type II collagen-induced | SAHase inhibition | -Reduced disease development -Reduced anti-collagen type II antibody level -Reduced T cell proliferative response -Fewer bone erosions -Amelioration of established disease |
[50] |
| Peptidoglycan-polysaccharide-induced | SAHase inhibition | -Reduced disease severity -Inhibition of IL-1β production (spleen+joint) |
[49] |
| Systemic lupus erythematosus (SLE) | Active SLE | -Global DNA hypomethylation -Reduced DNMT-1 mRNA levels |
[67,94,98] |
| CD4+ T cells | -CD70–CD40L (in women)–CD11a–perforin– IL-6–IL-4 demethylation→genes overexpressed | ||
| B cells | -CD70–perforin–CD40L–CD6 demethylation | ||
| Multiple sclerosis | White matter cells | -30% reduction in CpG island methylation -Hypomethylation of PAD2 region promoter |
[67] |
| CD4+ T cells | -No consistent variation in epigenetics between twins | ||
| EAE (MOG35–55) | SAHase inhibition | -Reduced disease incidence and severity -Reduced anti-MOG35–55 T cell proliferative response -Reduced Th1 cytokines response -Downregulation of cyclin D3 and cyclin-dependent kinases CDK4–CDK6 -Upregulation of p27 |
[35,36] |
| EAE (PLP139–151) | SAHase inhibition | -Blockade of disease induction -Reduced relapse incidence and severity -Inhibition of autoreactive CD4+ T cells |
|
| Epigenetic modification | |||
| Mouse lupus | DNA methyltransferase inhibitor | -Lupus-like disease development -Autoreactive potential acquired by CD4+ T cells -Possible role of CD4+ T cells in B cell IgG production and macrophage killing |
[91] |
| Human SLE | DNA methyltransferase inhibitor | -Demethylation of the silenced CD154 (CD40L) gene on the X-chromosome -Potential role for methylation in female predisposition to SLE |
[92,93] |
| Post-translational modifications | Bone marrow transfer from PCMT-KO mice | -Lupus-like disease development -Anti-DNA autoantibody production -Kidney disease -Isoaspartyl protein modification- associated pathogenesis |
[57] |
| Allergy | |||
| Delayed hypersensitivity reaction | SAHase inhibition | -Significant reduction in ear swelling | [37,51] |
| Transplantation | |||
| Mouse skin allograft | SAHase inhibition | -Significantly prolonged graft survival -Blockade of in vitro cytotoxic -T cell generation |
[29] |
3. Protein arginine methyltransferases
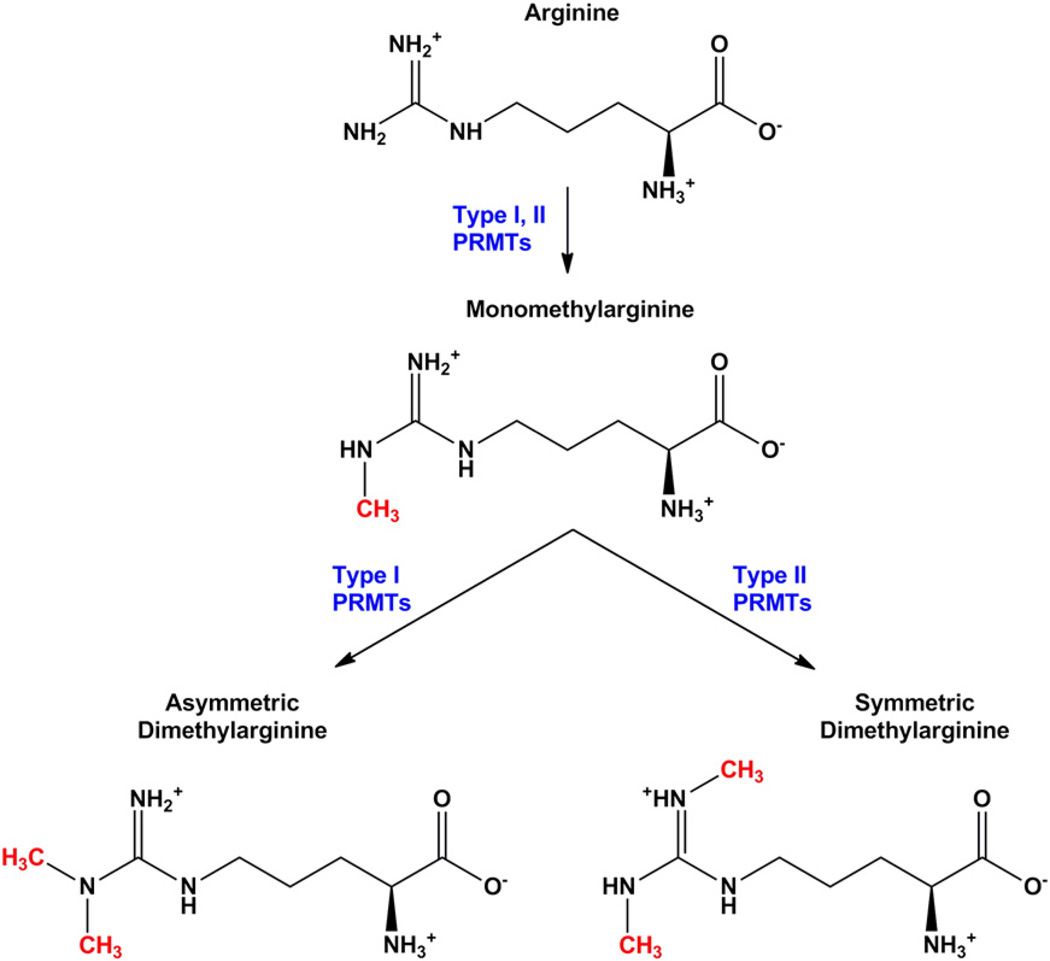
As mentioned above, the large family of MTs that catalyze the transfer of methyl-groups from SAM (~250 members) includes a sub-family of protein arginine methyltransferases (PRMTs) that have recently been shown to play key roles in many biological processes. The PRMT-mediated transfer of methyl groups yields three products: monomethyl, asymmetric dimethyl, and symmetric dimethyl arginine (Fig. 2). The eight PRMT members currently characterized (PRMT1–8) all generate monomethylarginine residues. PRMTs are further classified as type I (PRMT 1, 3, 4, 6, and 8) if they produce asymmetric dimethylarginines or type II (PRMT 5 and 7) if they produce symmetric dimethylarginines (46); PRMT 2 has not yet been classified. PRMT1 is thought to exert more than 90% of all type I PRMT activity. Three additional enzymes show PRMT activity and have been denoted as PRMTs 9, 10, and 11, but their classification and substrates have not yet been defined [9].
Figure 2.
Arginine methylation. Monomethylarginine is generated upon transfer of a methyl group from SAM to the guanidino amino group of arginine. Type I protein arginine methyltransferases (PRMTs 1, 3, 4, 6 and 8) then transfer a second methyl group to the same nitrogen, resulting in an asymmetric dimethylarginine. Type II PRMTs (PRMTs 5 and 7) transfer the additional methyl group to the opposite terminal nitrogen, creating symmetric dimethylarginine. PRMT2 is not yet classified.
Arginine methylation can increase steric hindrance and hydrophobicity but, in contrast to phosphorylation, does not affect protein charge. Little is known about PRMT regulation, but homodimerization [10] and association with other cellular proteins appear to augment and/or inhibit their activity [11–13]. Arginine methylation occurs in a wide variety of substrates, including histones, transcription factors, cytoskeletal proteins, cytoplasmic signaling proteins, and apoptosis proteins (reviewed in [14] and [15]). Additional substrates of PRMT methylation have recently been identified and include the estrogen receptor ERα [16], splicing and elongation factors SAP49, UIC, and CA150 [17], FGF-2 [18], and Ewing Sarcoma Oncoprotein [19,20]. Further work is needed to define the sub-cellular distribution, potential redundancy, and expression profiles of these enzymes in specific cell types, particularly immunocytes.
4. The role of transmethylation in immunity
Initial studies showed that transmethylation in monocytes was necessary for chemotactic responsiveness, as well as the morphologic changes, arachidonic acid release from membrane phospholipids, and second messenger signaling via phosphoinositide metabolism [21–23]. Subsequently, when the turnover rate of SAM was measured in many cell types, the amount of SAM consumed was found to be 4–10 times higher in activated vs. resting lymphocytes, and 3–5 times higher in resting lymphocytes vs. most other cell types, suggesting that lymphocytes may be more sensitive than most other cells to transmethylation inhibition (Table 2) [24].
Table 2.
Transmethylation effects in immune cell types.
| Type of immunocytes | Studied aspect | Consequences | References |
|---|---|---|---|
| Immunocyte differentiation | -Selective demethylation or de novo methylation of genes in tissues or lineage-specific manner -Demethylation of Lck gene in T cells -Demethylation of Pou2af1 in B cells -Methylation of dachshund homologue 1 (Dach1) in common lymphoid progenitors and DN thymocytes -Histone modification as pre-priming marks of lineage differentiation |
-Myeloid/lymphoid commitment -T cell/B cell commitment |
[79] |
| Thymocyte development | -Demethylation of CD8α and CD8β genes | -Transition of double negative to double positive stage | [80,81] |
| -Retention of demethylated CD8 genes | -Single-positive CD4+ T cells | ||
| -SAHase inhibition | -Arrested development (CD8lo and CD4+CD8+ double positive stages) -Not due to increased apoptosis -T cell-specific inhibition of co-receptor CD4 and CD8 mRNA |
[31] | |
| B cell/T cell | -Demethylation of TCRβ and Igκ loci -IgH locus: preference for D element pre-marked with histone modification -Plant homeodomain (PHD) of Rag2 |
-Increased gene accessibility allowing V(D)J rearrangement | [79–82] |
| CD4+ T cells | |||
| Th1 | -Demethylation of the IFN-γ gene -Methylation of the IL-4 gene |
-Control of IFN-γ gene accessibility by transcription factor T-bet | [80,84–87] |
| Th2 | -Methylation of the IFN-γ gene -Demethylation of the IL-4 gene |
-Control of IL-4 gene accessibility by transcription factor GATA-4 | |
| Th17 | -Acetylation of histone H3 in the IL-17/IL-17F promoter region -Trimethylation of H3K4 in IL-17/IL-17F promoter region |
-Transcription of IL-17 and IL-17F cytokines | [88,89] |
| nTreg | -Increased acetylation of histone H3 in FOXP3 promoter region -Trimethylation of H3K4 in FOXP3 promoter region -Complete DNA demethylation in the FOXP3 promoter |
-nTreg phenotype -Requires complete DNA demethylation of FOXP3 for a permanent regulatory state |
[90] |
| Human monocytes | -Adenosine deaminase inhibitor | -Increased intracellular SAH levels -Decreased chemotactic responsiveness and attendant morphologic changes |
[21,22] |
| -SAHase inhibition | -Decreased arachidonic acid release from membrane phospholipids -Decreased second messenger activation via phosphoinositide metabolism |
||
| Macrophages | -SAHase inhibition | -Normal antigen processing and presentation -Significant reduction of TNF-α |
[33] |
| B cells | -SAHase inhibition | -No inhibition of B cell proliferation | [29] |
| T cells Mouse | -S-adenosyl-L-methionine burst | -High sensitivity of transmethylation inhibition | [24] |
| -Irreversible SAHase inhibition | -Inhibition of conA stimulated-T cell proliferation and IL-2 production -Reduced OVA-specific T cell responses -Reduced anti-OVA antibody levels |
[29,30] | |
| -Reversible SAHase inhibition | -No inhibition of conA stimulated-T cell proliferation or IL-2 production -Did inhibit IL-12p40 and TNF-α from monocytes |
[37] |
As a corollary, adenosine deaminase deficiency, an inborn error of purine metabolism that results in severe combined immunodeficiency with characteristic T cell dysfunction, is associated with a 57% reduction in cellular SAHase activity [25] and accumulation of deoxyATP [26–28]. In this condition, adenosine is not deaminated to inosine, but rejoins with endogenous homocysteine to resynthesize SAH. As the SAH accumulates, it indirectly inhibits transmethylation reactions. Combined with the above findings, the adenosine deaminase deficiency phenotype led to the hypothesis that SAHase is required for proper immune cell function.
Indeed, when Wolos and co-workers studied the effects of the irreversible SAHase inhibitor (Z)-5′-fluoro-4′,5′-didehydro-5′-deoxyadenosine (MDL 28842) in murine systems, they found that it blocked generation of cytotoxic T cells in vitro and significantly prolonged skin allograft survival in vivo (Table 1) [29]. When they stimulated T cells with concanavalin A and B cells with lipopolysaccharide in the presence and absence of this compound, they found that it inhibited IL-2 production and the proliferation of T, but not B, cells despite SAH accumulation in both cell types (Table 2). Moreover, mice treated with this SAHase inhibitor showed reduced OVA-specific T cell responses and anti-OVA antibody levels [30].
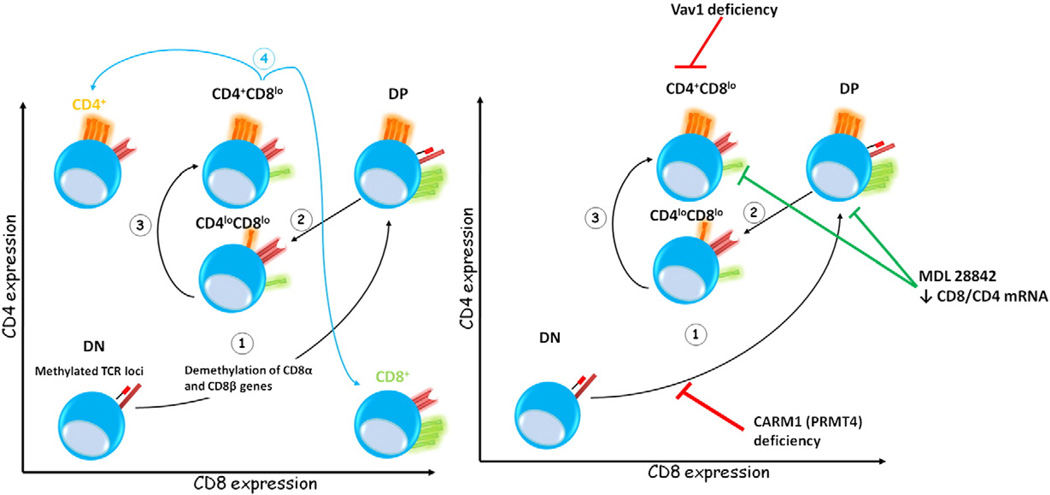
A possible mechanistic explanation for the T cell-specific effects following SAHase blockade was provided by Cohen and colleagues. Working with a murine adenosine deaminase model, they showed that treatment with MDL 28842 arrested thymocyte development at the CD8lo and CD4+CD8+ double-positive stages, an effect not due to increased apoptosis, but to T cell-specific inhibition of mRNA for the CD4 and CD8 co-receptor molecules (Fig. 3 and Table 2) [31]. These results suggest that a SAM-mediated methylation event(s) is required for the signal that regulates transcription of the T cell-specific molecules during intrathymic T cell development. Interestingly, mice deficient in the protein arginine methyltransferase CARM1 (PRMT4) exhibited partial developmental arrest of CD4−CD8− double-negative thymocyte progenitors (Fig. 3) [32]. Finally, mouse peritoneal macrophages treated with MDL 28842 and subsequently stimulated with LPS showed normal antigen processing and presentation, but significantly reduced TNF-α production, suggesting that SAHase inhibitors perhaps preferentially block proinflammatory responses (Table 2) [33].
Figure 3.
Methylation events during thymocyte development. Gene methylation status fluctuates during thymocyte differentiation. Double-negative (DN) thymocytes exhibit methylated TCR loci and expression of TCRβ is closely associated with signaling through the IL-7Rα and demethylation of the TCRβ locus, leading to locus accessibility and VDJ recombination. The first step (1) in thymocyte differentiation is the transition from DN to double-positive (DP) CD4+CD8+ cells and is accompanied by demethylation of CD8α and CD8β. The second (2) and third (3) steps lead to intermediate thymocyte subpopulations, e.g., CD4loCD8lo and CD4+CD8lo, and correspond to acquisition of a functionally rearranged TCR, again implying DNA and protein methylation modifications. Finally, CD4+ or CD8+ T cells are produced (4), but whether methylation status affects CD4 or CD8 expression remains to be clearly elucidated. However, retention of demethylated CD8 genes in CD4+ single cells was demonstrated, and de novo remethylation of CD8α genes was associated with apoptosis of misselected thymic CD8+ T cell emigrants that do not correctly recognize MHC class I in the periphery, inhibiting development of autoimmune disease. Interestingly, inhibition of SAHase activity by MDL28842 leads to arrested thymocyte differentiation at the CD8lo and CD4+CD8+ DP stages associated with decreased CD8 and CD4 mRNA. This effect may be a result of MDL28842 inhibition of PRMT4 and/or Vav1 activity, as both have been implicated in thymocyte developmental arrest.
We recently published a series of papers detailing the immunosuppressive characteristics of a novel, potent, and reversible SAHase inhibitor, methyl 4-(adenine-9-yl)-2-hydroxy-butanoate (DZ2002) [34–37], which unlike traditional irreversible inhibitors is not cytotoxic at working concentrations, thus allowing treatment for up to 9 months. DZ2002 blocked neither concanavalin A-induced T cell proliferative responses nor IL-2 production, but in monocytes inhibited IL-12p40 and TNF-α production as well as expression of costimulatory molecules (Table 2) [37]. Moreover, DZ2002 reduced delayed-type hypersensitivity reactions, pointing to potentially important roles for SAHase in both macrophages and T cells.
We also examined the role of DZ2002 in T-dependent antigen responses [34] in DZ2002-treated mice and found suppression of OVA-specific lymphocyte proliferation, anti-OVA IgG levels, and IL-2 and IFN-γ production. However, IL-4 production and anti-OVA IgG1 antibody levels were less affected, suggesting that SAHase inhibition preferentially targets type 1 helper T cell (Th1)-specific responses. Mice treated with SAH alone showed similar reductions in T cell activity and OVA-specific responses, confirming that, at least in part, the mode of action of DZ2002 involves increases in intracellular SAH levels. These data indicate that in vivo transmethylation inhibition has a profound suppressive effect on T cells.
5. The role of transmethylation in cell signaling
One of the first reported observations of the direct involvement of methyltransferases in receptor-mediated signaling pathways was the specific binding of PRMT1 to interferon-α receptor I (IFNAR1) in a two-hybrid screening analysis (Table 3) [38]. Binding of type I IFNs to its receptor (IFNAR1) signals through the Jak/Stat family of kinases and transcription factors, which leads to anti-viral, antiproliferative, and immunoregulatory activities. Indeed, when PRMT1 expression was blocked by antisense oligonucleotides in a myeloma cell line, these cells become less responsive IFN-β. This novel and unexpected observations led to a reassessment of the role of protein arginine methylation in cellular signaling. Further evidence for the importance of transmethylation in this pathway comes from a study detailing the requirement of STAT1 arginine methylation by PRMT1 for type I IFN signaling [39]. Although STAT1 phosphorylation was unaffected by a transmethylation inhibitor, its ability to bind to DNA response elements was reported to be severely compromised. This finding, however, has not been replicated in other studies, perhaps due to technical reasons [40–42].
Table 3.
Signaling pathways of methylation and transmethylation.
| Treatment/model | Signaling pathway | References | |
|---|---|---|---|
| Immunocytes | |||
| T cells | -CD28-activated | -Vav1 R-methylation and translocation into the nucleus | [44,45] |
| CD4+ T cells (but not CD8+ T cells) | -SAHase inhibition | -Reduced Vav R-methylation -Reduced Akt, Erk 1/2, NF-κB phosphorylation |
[36] |
| Protein methyltransferase | |||
| PRMT-1 | -Arginine methyltransferase | -Binding to IFNAR1 -Inhibition of antiproliferative effects of IFN-β by blocking PRMT-1 -STAT1 R-methylation by PRMT1? |
[38] [39–42] |
| SETD6 | -Lysine methyltransferase | -Monomethylation of RelA at Lysine 310 -Basal condition: RelAK310me1 bound to H3K9me2 by recognition of the GLP domain of SETD6 -Pro-inflammatory stimulus: PKCζ-mediated phosphorylation of RelAK310me1 blocks GLP-RelAK310me1 recognition→chromatin relaxation and RelA target genes expression |
[43] |
| Autoimmunity | |||
| Lupus | -Transgenic mouse that inducibly expresses a dominant-negative MEK in T cells -Hydralazine/ Erk inhibitor-treated murine T cells -Human T cells from SLE patient |
-Decreased Erk 1/2 pathway signaling -Impaired PKCδ -Decreases DNA methylation -Modification of gene expression rendering T cells autoreactive |
[95–97] |
Interestingly, another protein methyltransferase, SETD6, has been implicated in NF-κB signaling (Table 3). This methyltransferase is a PKMT that allows monomethylation of RelA at lysine 310 [43]. This form of monomethylated RelA (RelAK310me1) can be found bound to chromatin under basal conditions, and its binding is mediated via recognition of its GLP domain (ankyrin domain), which anchors RelA310me1 to the dimethylated lysine 9 of H3. Phosphorylation by protein kinase Cζ of RelAK310me1 at serine position 311 blocks GLP-RelAK310me1 recognition, thereby leading to chromatin relaxation and expression of RelA target genes, including IL-8, IL-1A, MYC and CCND1. This mechanism appears to be a protective cellular event that illustrates synergistic cross-talk between phosphorylation and methylation reactions, which are often studied separately.
Recently, it was shown that Vav1, a guanine nucleotide exchange factor important in T cell signaling, is methylated following CD28 ligation [44]. In CD28-activated cells, methylated Vav1 translocates to the nucleus, while unmethylated Vav1 remains in the cytoplasm. Moreover, indirect inhibition of transmethylation reactions by irreversible blocking of SAHase activity demonstrates a functional defect in IL-2 production that directly links methylation with T cell effector function. These findings suggest that methylation either directs Vav1 to the nucleus or, following phosphorylation, allows it to act as a transcription factor [45]. We recently confirmed and extended those observations [36] by showing that in the presence of DZ2002, activated CD4, but not CD8, T cells had reduced Vav1 methylation, suggesting that the two cell types differ in their post-activation methylation requirements (Table 3). CD4 T cells treated with DZ2002 also showed reduced phosphorylation of several key signaling molecules, including Akt, Erk1/2, and NF-κB, which led to a downstream reduction in calcium mobilization.
Similar CD4 T cell signaling abnormalities are also seen in mice carrying an arginine-to-glycine mutation in the pleckstrin homology domain of Vav1 [46], suggesting that methylation at that particular arginine residue is essential for Vav1 function in this subset. Furthermore, Vav1-deficient mice showed defective positive and negative selection in the DP thymocytes and reduced T cell receptor-mediated proliferation and IL-2 secretion, demonstrating the broader role of Vav1 in T cell function (Fig. 3) [47–49]. No clear explanation for the lineage-specific effect of arginine methylation of the pleckstrin homology domain was provided but, in conjunction with our findings, it appears that CD4 and CD8 T cells have differing requirements of arginine methylation in this particular domain. The findings suggest that agents that block or inhibit methylation reactions may be exploited when devising treatments for CD4-mediated autoimmune diseases.
6. SAHase inhibition and autoimmune models
Initially, Wolos and colleagues reported a dramatic block in the development of type II collagen-induced arthritis in MDL 28842-treated mice, directly demonstrating that transmethylation inhibition could be an effective treatment for autoimmune diseases [50]. At the highest dosage, none of the treated mice developed disease, while 87% of untreated controls did. Moreover, this treatment was associated with reduced anti-collagen type II antibody levels, decreased T cell proliferative responses to this antigen, and fewer bone erosions (Table 1). Even more striking was the amelioration of established disease. In addition, Saso et al., using a separate irreversible SAHase inhibitor, DHCaA, showed in vitro inhibition of T cell proliferation and in vivo inhibition of delayed type hypersensitivity reactions as well as a reduction in severity of peptidoglycan polysaccharide-induced arthritis [51]. Of interest, disease in this arthritis model is mediated by IL-1β, suggesting that methylation inhibitors may interfere with IL-1β processing, which encompasses an initial TLR-mediated induction of pro-IL-1β followed by inflammasome-mediated conversion and secretion of mature IL-1β [52,53]. Indeed, our preliminary data support the idea that methylation inhibitors broadly block TLR-signaling and production of proinflammatory cytokines (unpublished data).
In light of our previous findings with DZ2002, we used a similar approach to establish the efficacy of transmethylation inhibition in the treatment of experimental autoimmune encephalomyelitis (EAE), a classic Th1-type autoimmune disease [35]. EAE can be induced in mice by immunization with several myelin antigens, such as peptide 35–55 of myelin oligodendrocyte glycoprotein (MOG), which leads to chronic paralysis and demyelination of the central nervous system, and peptide 139–151 of proteolipid protein (PLP), which results in a relapsing/remitting type of disease [54]. Administration of the reversible SAHase inhibitor DZ2002 reduced the incidence and severity of MOG35–55-induced EAE in the C57BL/6 mouse, and also decreased anti-MOG35–55 T cell responses as well as other Th1 cytokine production. Inhibition of proliferation was associated with downregulation of several cyclins and cyclin-dependent kinases (CDKs) (e.g., cyclin D3, CDK4, and CDK6), as well as upregulation of the CDK inhibitor p27 (Table 1).
In a more recent study, we reexamined the effects of DZ2002 in the PLP139–151-induced relapsing/remitting model of EAE, which closely reflects the course of human MS. Impressively, we not only blocked induction of disease, but reduced the severity and incidence of relapses [36]. Moreover, the effectiveness of this treatment appears to be primarily mediated by its inhibitory effects on autoreactive T cells, since the ability of encephalitogenic CD4+ T cells to induce disease upon adoptive transfer was greatly reduced in DZ2002-treated mice (Table 1).
Finally, it should be noted that global transmethylation inhibitors, reversible or otherwise, likely exert many unanticipated off-target effects on a diverse array of genes/ proteins since there are >250 known methyltransferases. These unintended effects could potentially lead to adverse events. Therefore, efforts to develop more specific transmethylase inhibitors, such as PRMT1-specific inhibitors, are highly warranted.
7. Post-translational protein modifications and autoimmunity
On a different note, tolerance to self-proteins can be compromised by post-translational protein modifications. This possibility has been supported by several experiments, particularly those conducted by Mamula and colleagues, which showed that 1) intracellular isoaspartyl (isoAsp) residues accumulate with age in T cells from autoimmune mice and are elevated in hyperproliferating T cells [55], 2) immunization with in vitro isoAsp-modified tyrosinase-related protein (TRP)-2 rendered it immunogenic and a target for CD8 T cells [56], 3) transfer of bone marrow from mice lacking protein carboxyl methyltransferase (PCMT), the isoAsp repair enzyme, into wild-type recipients resulted in development of lupus-like disease associated with anti-DNA autoantibodies and kidney disease (Table 1) [57], and 4) mice immunized with the isoAsp form of mouse cytochrome c mounted a strong B and T cell response, and anti-isoAsp cytochrome c antibodies cross-reacted with native cytochrome c [1]. Other post-translational protein modifications have also been implicated as playing a role in the pathogenesis of autoimmune diseases, for example, in RA, citrullination (conversion of arginine to citrulline) [58–63] and glycosylation [64–66]. Finally, DNA methylation was demonstrated to indirectly interfere with post-translational modifications implicated in autoimmunity. For example, promoter regions of peptidyl arginine deiminase type II (PAD2), an enzyme involved in the citrullination of myelin basic protein (MBP), have been shown to be hypomethylated and overexpressed in white matter from MS patients (Table 1). The citrullination of MBP by PAD2 is thought to promote protein autocleavage, resulting in the potential creation of neoepitopes for which tolerance has not been established [67]. Thus, epigenetic events (as detailed below) and post-translational protein modifications appear intimately intertwined.
8. Epigenetics, immunity, and autoimmunity
Because DNA can also be transmethylated similar to proteins, as noted above, blockade of transmethylation should significantly affect the epigenetics of immunocytes. Epigenetic modifications (heritable changes in gene expression that leave DNA sequences unaffected) are fundamental to tissue differentiation [68], gene imprinting [69], X chromosome inactivation [70], and suppression of foreign DNA acquired by mammalian cells during evolution [71]. DNA methylation and histone modifications alter chromatin structure in a way that affects DNA accessibility to transcription factors and gene expression [72,73].
DNA methylation, which occurs on cytosine bases in CpG-dinucleotides widely distributed in the genome, are mediated by three DNA methyltransferases (DNMT1, responsible for heritable maintenance of CpG-methylations during replication; DNMT3a and DNMT3b, responsible for de novo methylation). These enzymes catalyze the transfer of methyl groups from SAM to the 5 position of the cytosine ring. Methylation of CpG islands in or near a promoter region contributes to the silencing of genes by promoting chromatin remodeling and preventing the binding of transcriptional factors.
Gene expression can also be controlled by methylation of certain lysine or arginine residues within the histone tail. For example, methylation of lysine 9 or 27 on histone H3 (H3K9 or H3K27), or lysine 20 on histone H4 (H4K20), is associated with transcriptional repression by formation of heterochromatin (dense form of chromatin) [74]. In fact, the presence of both H3K9me3/H3K9me2 and H3K27me3 in the same region of the genome marks the local CpG islands for DNMT-mediated methylation [75]. In contrast, di- and trimethylation of lysine 4 on histone H3 (H3K4me3 or H3K4me2) is found in actively transcribed euchromatin (less compact form of chromatin) and may promote transcription, notably by association with RNApol II in actively transcribed genes [74,76]. Indeed, methylation of arginine residue 3 on histone H4 (H4R3) or arginine 17 on histone H3 (H3R17) was associated with gene expression [67]. Epigenetic regulation bequeaths gene expression changes to subsequent cell generations. Thus, it is not surprising that the mechanism is ubiquitous in the immune system [77,78] wherein the preservation of adaptations developed in various cell subsets permits and transmits a rapid and robust defense against specific pathogens.
Hematopoietic cell lineages nicely illustrate epigenetic regulation mediated either by selective demethylation or de novo methylation at the DNA level or by histone modifications as a pre-priming mark for lineage specific differentiation (reviewed in [79]). For example, demethylation of the Lck gene (encoding a Src family kinase essential in TCR receptor signaling) or POU domain class 2-associating factor (Pou2af1 encoding a B-cell specific co-activator) has been demonstrated in T and B cells, respectively (Table 2). Similar demethylation events were observed in CD8α and CD8β genes during thymocyte transition from the double-negative (DN) to the double-positive (DP) stage (Fig. 3 and Table 2) (reviewed in [80]). Moreover, during lineage development, many genes are marked with H3K4me2 either close to their transcription start points or in transcription factor binding regions. In some cell types, activation of these genes is characterized by an additional methylation event (H3K4me3), while other genes are silenced by demethylation (H3K4). The latter is the case for recombination activating gene-2 (Rag-2) and GATA-binding protein-1 (GATA-1), which both exhibit the H3K4me2 dimethylation priming mark in hematopoietic stem cells (HSC), but are either trimethylated (H2K4me3), as is the case with Rag2, or demethylated, as is the case with GATA-1, upon differentiation to B cells and inversely in erythroid cells (Table 2) (reviewed in [79]).
V(D)J rearrangement of B and T cell receptor genes is also regulated by epigenetic events that promote the accessibility of certain gene loci, such as Igκ and TCRβ (Table 2) [81]. For example, a single methylated CpG island in the heptamer of the Dβ element recombination signal sequence (RSS) can block RAG-mediated cleavage, suggesting methylation plays a role in recombinant regulation [82]. Indeed, hypomethylation of CpG dinucleotides may be induced by the action of a V(D)J element promoter (Pβ1), since hypermethylation of CpG dinucleotides across Dβ1–Jβ1 clusters was demonstrated after Pβ1 deletion [83]. Furthermore, as previously discussed, histone modifications can also predict a V(D)J rearrangement. The D element of either the IgH or Igκ loci can be targeted for histone modifications (acetylation and methylation) that later induce rearrangement. In this context, the Rag-2 enzyme recognizes the H3K4me3 motif via its specific plant homeodomain (PHD) finger, thus directly activating its cleavage activity (Table 2). Therefore, it is likely that epigenetic modifications are necessary in orchestrating recombination events during immunoglobulin or TCR gene rearrangement [79,82].
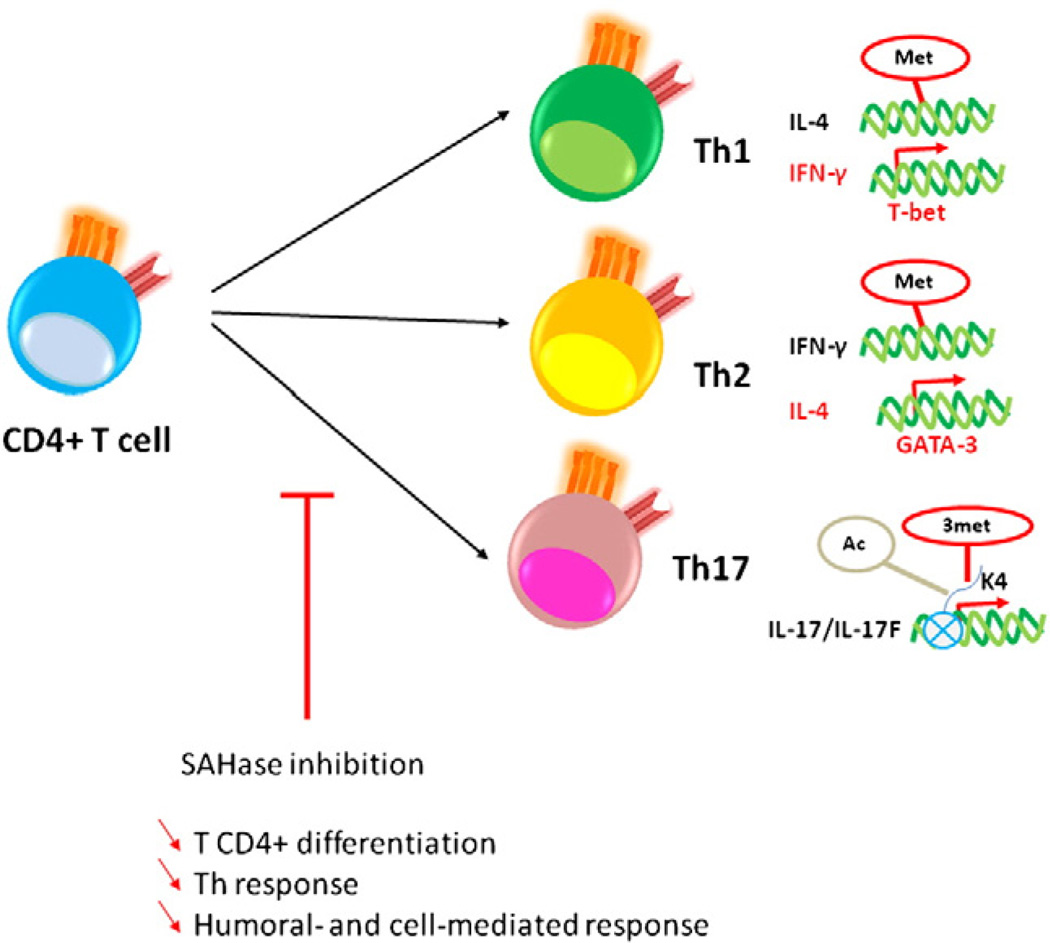
Naïve CD4+ T cell differentiation is also a well-studied example of epigenetic regulation primarily mediated by two transcriptional factors, T-bet and GATA-3, which, through chromatin modifications, enable access of transcription factors to the IFN-γ gene or to the IL-4 gene and subsequent differentiation into Th1 or Th2 CD4+ T cells, respectively (Table 2) [84,85]. This is accomplished by demethylation of the IFN-γ gene and methylation of the IL-4 gene for Th1 cells [86], and the converse for Th2 cells (Fig. 4) [80,87].
Figure 4.
Putative role of methylation during CD4+ T cell and global immune responses. DNA or histone methylation allows DNA structural changes and transcription factor accessibility. These events occur during CD4+ T cell differentiation and direct CD4+-mediated helper responses. Th1 and Th2 differentiationismediatedbymethylationofthe IL-4 gene and demethylationofthe IFN-γ gene,ormethylation of the IFN-γ gene and demethylation of the IL-4 gene, respectively. Moreover, histone modifications in the IL-17 and IL-17F promoter regions (e.g. H3K4 trimethylation and H3 acetylation) lead to the expression of these cytokines and a Th17 response. Thus, SAHase inhibition may block T helper responses by ineffective epigenetic events (DNA or protein) leading to defective humoral- and cell-mediated responses, [38,51,52]. H3K4, lysine 4 of histone 3; 3met, trimethylation; ac, acetylation; met, metylation).
Epigenetic modifications play similarly significant roles in the differentiation of naïve CD4+ T cells into Th17 and regulatory T cells. Th17 cells are produced in the presence of IL-6 and TGF-β [88], which induce histone H3 acetylation and H3K4me3 in the IL-17 and IL-17F promoter regions, allowing transcription of the respective cytokines (Fig. 4 and Table 2) [89]. Natural regulatory T cells of the CD4+CD25+FOXP3+ phenotype are characterized by complete DNA demethylation in the FOXP3 promoter and increased histone acetylation and H3K4me3 in associated chromatin. Of note, complete demethylation of FOXP3 is required for a permanent regulatory functional status [90]. Considering the effects of methylation on several CD4+ T cell subtypes, it can be surmised that epigenetic changes likely play a significant role in autoimmunity.
Indeed, the role of DNA methylation has been extensively investigated in autoimmunity, especially in lupus. Early studies showed that CD4+ T cells treated in vitro with the DNMT inhibitor 5-azadeoxycytidine acquired autoreactive potential, as indicated by the ability of these cells to induce a lupus-like disease upon adoptive transfer (Table 1) [91]. In further support of the role of epigenetic changes in lupus, it was found that T cells from patients with active SLE had decreased deoxymethylcytosine content and DNMT1 mRNA (Table 1) [92], suggesting that abnormal demethylation may lead to overexpression of certain genes leading to a break in tolerance. In this regard, particular emphasis has been placed on genes affecting adhesion molecules (LFA-1), apoptosis-related proteins (perforin), and costimulatory molecules (CD70 and CD154). Of interest, female-derived CD4+ T cells treated with 5-azadeoxycytidine demethylate the normally silenced copy of methylated CD154 (CD40L) on the X-chromosome, leading to a double-dose of this cost-imulatory molecule and thus providing a potential role for methylation in the female predominance of this disease [93]. These susceptibility genes are found overexpressed in CD4+ T and B cells, but not in CD8+ T cells from SLE patients. Further investigations using 5-azadeoxycytidine defined a putative model in which drug-induced autoreactive CD4+ T cells eliminated macrophages by perforin secretion or signal delivered through T cell FasL, TWEAK and TRAIL promote B cell activity by overexpression of CD70 and/or IFN-γ and IL-4. The release of nucleosomal material following macrophage apoptosis may induce anti-nucleosomal T-cell responses, leading to an anti-nucleic acid antibody response [91,94].
Erk signaling pathways have also been implicated in DNA methylation modifications by regulating DNMT activity. Inhibition of Erk was investigated in T cells by using hydralazine, an Erk-inhibiting drug, or T cells from transgenic mice that inducibly express a dominant negative form of mitogen-activated protein kinase (MEK) (Table 3). Both of these approaches led to decreased DNA methylation associated with overexpression of LFA-1 and CD70 [67,95,96]. Interestingly, this decreased Erk signaling was also observed in T cells from SLE patients and appeared related to impaired protein kinase C δ activation [97]. However, this remains a matter of conjecture derived from in vitro studies and requires further work. Epigenetic changes have also been considered a potential explanation for the incomplete concordance of SLE in monozygotic twins, although this “incompleteness” may be explained by many other stochastically-imposed factors, including, in particular, environmental factors such as viral infections that can act as precipitating events in genetically-predisposed individuals. Interestingly, in contrast to lupus, T cells from RA patients show no defects in DNMT1 or LFA-1 activity, but instead show defects in global DNA hypomethylation in synovial fibroblasts, especially in CpG islands in long interspersed nuclear element-1 (LINE-1, an intragenomic parasitic DNA), and in monocytes, within the IL-6 promoter and the death receptor-3 inducing hyper-inflammation and apoptosis resistance gene (Table 1) [67,94].
9. Conclusions
Transmethylation is a key pathway common to both post-translational protein modification and gene regulation. By modifying protein functionality or generating novel structures, post-translational modifications can contribute to the pathogenesis of autoimmunity. Additionally, epigenetic mechanisms that disturb immune system gene expression can profoundly influence self-tolerance. Intervention in those processes with either small-molecule chemical inhibitors or agonist/antagonist biologicals could ultimately lead to novel autoimmune disease therapies.
Acknowledgments
This is manuscript number 20,949 from the Department of Immunology & Microbial Science of The Scripps Research Institute. We would like to thank Kat Occhipinti-Bender and Sara Benson, for editorial assistance. The work reported herein was supported by NIH grant 5R01AI076396.
Footnotes
10. Conflict of interest statement
The author(s) declare that there are no conflicts of interest.
References
- 1.Mamula MJ, Gee RJ, Elliott JI, Sette A, Southwood S, Jones PJ, Blier PR. Isoaspartyl post-translational modification triggers autoimmune responses to self-proteins. J. Biol. Chem. 1999;274:22321–22327. doi: 10.1074/jbc.274.32.22321. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee AK. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 4.Ueland PM, Helland S, Broch OJ, Schanche JS. Homocysteine in tissues of the mouse and rat. J. Biol. Chem. 1984;259:2360–2364. [PubMed] [Google Scholar]
- 5.Miller MW, Duhl DM, Winkes BM, Arredondo-Vega F, Saxon PJ, Wolff GL, Epstein CJ, Hershfield MS, Barsh GS. The mouse lethal nonagouti (a(x)) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. EMBO J. 1994;13:1806–1816. doi: 10.1002/j.1460-2075.1994.tb06449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baric I, Cuk M, Fumic K, Vugrek O, Allen RH, Glenn B, Maradin M, Pazanin L, Pogribny I, Rados M, Sarnavka V, Schulze A, Stabler S, Wagner C, Zeisel SH, Mudd SH. Sadenosylhomocysteine hydrolase deficiency: a second patient, the younger brother of the index patient, and outcomes during therapy. J. Inherit. Metab. Dis. 2005;28:885–902. doi: 10.1007/s10545-005-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baric I, Fumic K, Glenn B, Cuk M, Schulze A, Finkelstein JD, James SJ, Mejaski-Bosnjak V, Pazanin L, Pogribny IP, Rados M, Sarnavka V, Scukanec-Spoljar M, Allen RH, Stabler S, Uzelac L, Vugrek O, Wagner C, Zeisel S, Mudd SH. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buist NR, Glenn B, Vugrek O, Wagner C, Stabler S, Allen RH, Pogribny I, Schulze A, Zeisel SH, Baric I, Mudd SH. S-adenosylhomocysteine hydrolase deficiency in a 26-year-old man. J. Inherit. Metab. Dis. 2006;29:538–545. doi: 10.1007/s10545-006-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchet F, Schurter BT, Acuto O. Protein arginine methylation in lymphocyte signaling. Curr. Opin. Immunol. 2006;18:321–328. doi: 10.1016/j.coi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 12.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh V, Miranda TB, Jiang W, Frankel A, Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S, Newsham IF. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene. 2004;23:7761–7771. doi: 10.1038/sj.onc.1208057. [DOI] [PubMed] [Google Scholar]
- 14.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 15.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 16.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol. Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 17.Chan JY, Hsieh TY, Liu ST, Chou WY, Chung MH, Huang SM. Physical and functional interactions between hnRNP K and PRMT family proteins. FEBS Lett. 2009;583:281–286. doi: 10.1016/j.febslet.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Bruns AF, Grothe C, Claus P. Fibroblast growth factor 2 (FGF-2) is a novel substrate for arginine methylation by PRMT5. Biol. Chem. 2009;390:59–65. doi: 10.1515/BC.2009.001. [DOI] [PubMed] [Google Scholar]
- 19.Kim JD, Kako K, Kakiuchi M, Park GG, Fukamizu A. EWS is a substrate of type I protein arginine methyltransferase, PRMT8. Int. J. Mol. Med. 2008;22:309–315. [PubMed] [Google Scholar]
- 20.Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:1125–1137. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- 21.Pike MC, DeMeester CA. Inhibition of phosphoinositide metabolism in human polymorphonuclear leukocytes by S-adenosylhomocysteine. J. Biol. Chem. 1988;263:3592–3599. [PubMed] [Google Scholar]
- 22.Pike MC, Kredich NM, Snyderman R. Requirement of S-adenosyl-L-methionine-mediated methylation for human monocyte chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 1978;75:3928–3932. doi: 10.1073/pnas.75.8.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pike MC, Snyderman R. Transmethylation reactions are required for initial morphologic and biochemical responses of human monocytes to chemoattractants. J. Immunol. 1981;127:1444–1449. [PubMed] [Google Scholar]
- 24.German DC, Bloch CA, Kredich NM. Measurements of S-adenosylmethionine and L-homocysteine metabolism in cultured human lymphoid cells. J. Biol. Chem. 1983;258:10997–11003. [PubMed] [Google Scholar]
- 25.Tsuchiya S, Nakae S, Konno T, Tada K. S-adenosylhomocysteine hydrolase activity in a lymphoblastoid cell line from a patient with adenosine deaminase deficiency disease. J. Inherit. Metab. Dis. 1981;4:197–201. doi: 10.1007/BF02263651. [DOI] [PubMed] [Google Scholar]
- 26.Cohen A, Hirschhorn R, Horowitz SD, Rubinstein A, Polmar SH, Hong R, Martin DW., Jr. Deoxyadenosine triphos-phate as a potentially toxic metabolite in adenosine deaminase deficiency. Proc. Natl. Acad. Sci. U. S. A. 1978;75:472–476. doi: 10.1073/pnas.75.1.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullman B, Cohen A, Martin DW. Characterization of a cell culture model for the study of adenosine deaminase- and purine nucleoside phosphorylase-deficient immunologic disease. Cell. 1976;9:205–211. doi: 10.1016/0092-8674(76)90111-2. [DOI] [PubMed] [Google Scholar]
- 28.Ullman B, Gudas LJ, Cohen A, Martin DW., Jr. Deoxyadenosine metabolism and cytotoxicity in cultured mouse T lymphoma cells: a model for immunodeficiency disease. Cell. 1978;14:365–375. doi: 10.1016/0092-8674(78)90122-8. [DOI] [PubMed] [Google Scholar]
- 29.Wolos JA, Frondorf KA, Babcock GF, Stripp SA, Bowlin TL. Immunomodulation by an inhibitor of S-adenosyl-L-homocysteine hydrolase: inhibition of in vitro and in vivo allogeneic responses. Cell. Immunol. 1993;149:402–408. doi: 10.1006/cimm.1993.1165. [DOI] [PubMed] [Google Scholar]
- 30.Wolos JA, Frondorf KA, Davis GF, Jarvi ET, McCarthy JR, Bowlin TL. Selective inhibition of T cell activation by an inhibitor of S-adenosyl-L-homocysteine hydrolase. J. Immunol. 1993;150:3264–3273. [PubMed] [Google Scholar]
- 31.Benveniste P, Zhu W, Cohen A. Interference with thymocyte differentiation by an inhibitor of S-adenosylhomocysteine hydrolase. J. Immunol. 1995;155:536–544. [PubMed] [Google Scholar]
- 32.Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, Richie E, Bedford MT. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J. Biol. Chem. 2004;279:25339–25344. doi: 10.1074/jbc.M402544200. [DOI] [PubMed] [Google Scholar]
- 33.Lambert LE, Frondorf KA, Berling JS, Wolos JA. Effects of an S-adenosyl-L-homocysteine hydrolase inhibitor on murine macrophage activation and function. Immunopharmacology. 1995;29:121–127. doi: 10.1016/0162-3109(94)00051-g. [DOI] [PubMed] [Google Scholar]
- 34.Fu YF, Wang JX, Zhao Y, Yang Y, Tang W, Ni J, Zhu YN, Zhou R, He PL, Li C, Li XY, Yang YF, Lawson BR, Zuo JP. S-adenosyl-L-homocysteine hydrolase inactivation curtails ovalbumin-induced immune responses. J. Pharmacol. Exp. Ther. 2006;316:1229–1237. doi: 10.1124/jpet.105.093369. [DOI] [PubMed] [Google Scholar]
- 35.Fu YF, Zhu YN, Ni J, Zhong XG, Tang W, Re YD, Shi LP, Wan J, Yang YF, Yuan C, Nan FJ, Lawson BR, Zuo JP. A reversible S-adenosyl-L-homocysteine hydrolase inhibitor ameliorates experimental autoimmune encephalomyelitis by inhibiting T cell activation. J. Pharmacol. Exp. Ther. 2006;319:799–808. doi: 10.1124/jpet.106.107185. [DOI] [PubMed] [Google Scholar]
- 36.Lawson BR, Manenkova Y, Ahamed J, Chen X, Zou JP, Baccala R, Theofilopoulos AN, Yuan C. Inhibition of transmethylation down-regulates CD4 T cell activation and curtails development of autoimmunity in a model system. J. Immunol. 2007;178:5366–5374. doi: 10.4049/jimmunol.178.8.5366. [DOI] [PubMed] [Google Scholar]
- 37.Wu QL, Fu YF, Zhou WL, Wang JX, Feng YH, Liu J, Xu JY, He PL, Zhou R, Tang W, Wang GF, Zhou Y, Yang YF, Ding J, Li XY, Chen XR, Yuan C, Lawson BR, Zuo JP. Inhibition of S-adenosyl-L-homocysteine hydrolase induces immunosuppression. J. Pharmacol. Exp. Ther. 2005;313:705–711. doi: 10.1124/jpet.104.080416. [DOI] [PubMed] [Google Scholar]
- 38.Abramovich C, Yakobson B, Chebath J, Revel M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 40.Komyod W, Bauer UM, Heinrich PC, Haan S. I. Behrmann. Are STATS arginine-methylated? J. Biol. Chem. 2005;280:21700–21705. doi: 10.1074/jbc.C400606200. [DOI] [PubMed] [Google Scholar]
- 41.Meissner T, Krause E, Lodige I, Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–589. doi: 10.1016/j.cell.2004.11.024. discussion 589–590. [DOI] [PubMed] [Google Scholar]
- 42.Weber S, Maass F, Schuemann M, Krause E, Suske G, Bauer UM. PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes Dev. 2009;23:118–132. doi: 10.1101/gad.489409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, Tennen RI, Kuo AY, Tanjing S, Cheung R, Chua KF, Utz PJ, Shi X, Prinjha RK, Lee K, Garcia BA, Bedford MT, Tarakhovsky A, Cheng X, Gozani O. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat. Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanchet F, Cardona A, Letimier FA, Hershfield MS, Acuto O. CD28 costimulatory signal induces protein arginine methylation in T cells. J. Exp. Med. 2005;202:371–377. doi: 10.1084/jem.20050176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tybulewicz VL. Vav-family proteins in T-cell signalling. Curr. Opin. Immunol. 2005;17:267–274. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Prisco A, Vanes L, Ruf S, Trigueros C, Tybulewicz VL. Lineage-specific requirement for the PH domain of Vav1 in the activation of CD4+ but not CD8+ T cells. Immunity. 2005;23:263–274. doi: 10.1016/j.immuni.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Fischer KD, Zmuldzinas A, Gardner S, Barbacid M, Bernstein A, Guidos C. Defective T-cell receptor signalling and positive selection of Vav-deficient CD4+ CD8+ thymocytes. Nature. 1995;374:474–477. doi: 10.1038/374474a0. [DOI] [PubMed] [Google Scholar]
- 48.Tarakhovsky A, Turner M, Schaal S, Mee PJ, Duddy LP, Rajewsky K, Tybulewicz VL. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 49.Zhang R, Alt FW, Davidson L, Orkin SH, Swat W. Defective signalling through the T- and B-cell antigen receptors in lymphoid cells lacking the vav proto-oncogene. Nature. 1995;374:470–473. doi: 10.1038/374470a0. [DOI] [PubMed] [Google Scholar]
- 50.Wolos JA, Frondorf KA, Esser RE. Immunosuppression mediated by an inhibitor of S-adenosyl-L-homocysteine hydrolase. Prevention and treatment of collagen-induced arthritis. J. Immunol. 1993;151:526–534. [PubMed] [Google Scholar]
- 51.Saso Y, Conner EM, Teegarden BR, Yuan CS. S-adenosyl-L-homocysteine hydrolase inhibitor mediates immunosuppressive effects in vivo: suppression of delayed type hypersensitivity ear swelling and peptidoglycan polysaccharide-induced arthritis. J. Pharmacol. Exp. Ther. 2001;296:106–112. [PubMed] [Google Scholar]
- 52.Baccala R, Gonzalez-Quintial R, Lawson BR, Stern ME, Kono DH, Beutler B, Theofilopoulos AN. Sensors of the innate immune system: their mode of action. Nat. Rev. Rheumatol. 2009;5:448–456. doi: 10.1038/nrrheum.2009.136. [DOI] [PubMed] [Google Scholar]
- 53.Theofilopoulos AN, Gonzalez-Quintial R, Lawson BR, Koh YT, Stern ME, Kono DH, Beutler B, Baccala R. Sensors of the innate immune system: their link to rheumatic diseases. Nat. Rev. Rheumatol. 2010;6:146–156. doi: 10.1038/nrrheum.2009.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu. Rev. Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 55.Yang ML, Doyle HA, Gee RJ, Lowenson JD, Clarke S, Lawson BR, Aswad DW, Mamula MJ. Intracellular protein modification associated with altered T cell functions in autoimmunity. J. Immunol. 2006;177:4541–4549. doi: 10.4049/jimmunol.177.7.4541. [DOI] [PubMed] [Google Scholar]
- 56.Doyle HA, Zhou J, Wolff MJ, Harvey BP, Roman RM, Gee RJ, Koski RA, Mamula MJ. Isoaspartyl post-translational modification triggers anti-tumor T and B lymphocyte immunity. J. Biol. Chem. 2006;281:32676–32683. doi: 10.1074/jbc.M604847200. [DOI] [PubMed] [Google Scholar]
- 57.Doyle HA, Gee RJ, Mamula MJ. A failure to repair self-proteins leads to T cell hyperproliferation and autoantibody production. J. Immunol. 2003;171:2840–2847. doi: 10.4049/jimmunol.171.6.2840. [DOI] [PubMed] [Google Scholar]
- 58.Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S, Yamamoto K. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 2005;44:40–50. doi: 10.1093/rheumatology/keh414. [DOI] [PubMed] [Google Scholar]
- 59.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Mechin MC, Vincent C, Nachat R, Yamada M, Takahara H, Simon M, Guerrin M, Serre G. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 60.Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P, Moscarello MA. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J. Neurosci. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubin B, Sonderstrup G. Citrullination of self-proteins and autoimmunity. Scand. J. Immunol. 2004;60:112–120. doi: 10.1111/j.0300-9475.2004.01457.x. [DOI] [PubMed] [Google Scholar]
- 62.Vossenaar ER, Despres N, Lapointe E, van der Heijden A, Lora M, Senshu T, van Venrooij WJ, Menard HA. Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res. Ther. 2004;6:R142–R150. doi: 10.1186/ar1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vossenaar ER, Radstake TR, van der Heijden A, van Mansum MA, Dieteren C, de Rooij DJ, Barrera P, Zendman AJ, van Venrooij WJ. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann. Rheum. Dis. 2004;63:373–381. doi: 10.1136/ard.2003.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Backlund J, Treschow A, Bockermann R, Holm B, Holm L, Issazadeh-Navikas S, Kihlberg J, Holmdahl R. Glycosylation of type II collagen is of major importance for T cell tolerance and pathology in collagen-induced arthritis. Eur. J. Immunol. 2002;32:3776–3784. doi: 10.1002/1521-4141(200212)32:12<3776::AID-IMMU3776>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 65.Buzas EI, Gyorgy B, Pasztoi M, Jelinek I, Falus A, Gabius HJ. Carbohydrate recognition systems in autoimmunity. Autoimmunity. 2006;39:691–704. doi: 10.1080/08916930601061470. [DOI] [PubMed] [Google Scholar]
- 66.Opdenakker G, Dillen C, Fiten P, Martens E, Van Aelst I, Van den Steen PE, Nelissen I, Starckx S, Descamps FJ, Hu J, Piccard H, Van Damme J, Wormald MR, Rudd PM, Dwek RA. Remnant epitopes, autoimmunity and glycosylation. Biochim. Biophys. Acta. 2006;1760:610–615. doi: 10.1016/j.bbagen.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Meda F, Folci M, Baccarelli A, Selmi C. The epigenetics of autoimmunity. Cell. Mol. Immunol. 2011;8:226–236. doi: 10.1038/cmi.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 69.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 70.Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 71.Bird AP. Functions for DNA methylation in vertebrates. Cold Spring Harb. Symp. Quant. Biol. 1993;58:281–285. doi: 10.1101/sqb.1993.058.01.033. [DOI] [PubMed] [Google Scholar]
- 72.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 73.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 74.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 75.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Esteller M. The coming of age of DNA methylation in medicine in the genomics and postgenomics era. Clin. Immunol. 2002;103:213–216. doi: 10.1006/clim.2002.5216. [DOI] [PubMed] [Google Scholar]
- 78.Teitell M, Richardson B. DNA methylation in the immune system. Clin. Immunol. 2003;109:2–5. doi: 10.1016/s1521-6616(03)00224-9. [DOI] [PubMed] [Google Scholar]
- 79.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat. Rev. Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 80.Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin. Immunol. 2003;109:37–45. doi: 10.1016/s1521-6616(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 81.Inlay M, Xu Y. Epigenetic regulation of antigen receptor rearrangement. Clin. Immunol. 2003;109:29–36. doi: 10.1016/s1521-6616(03)00199-2. [DOI] [PubMed] [Google Scholar]
- 82.Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat. Immunol. 2003;4:624–630. doi: 10.1038/ni0703-624. [DOI] [PubMed] [Google Scholar]
- 83.Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PD beta 1 from the TCR beta locus causes hyper-methylation that impairs D beta 1 recombination by multiple mechanisms. Immunity. 2000;13:703–714. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]
- 84.Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 86.Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, Reiner SL. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat. Immunol. 2002;3:652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- 87.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 88.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 89.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 90.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum. Immunol. 1986;17:456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 92.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 93.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 94.Richardson B. DNA methylation and autoimmune disease. Clin. Immunol. 2003;109:72–79. doi: 10.1016/s1521-6616(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 95.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 96.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J. Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 98.Renaudineau Y, Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J. Med. 2011;60:10–16. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]