Significance
Zinc is an essential nutrient that is a cofactor in a wide range of enzymes and regulatory proteins. However, in excess, zinc is toxic to growth. As a consequence, all cells required mechanisms to maintain an optimal level of zinc. Despite a wealth of knowledge of proteins that use a structural or catalytic zinc cofactor, relatively little is know about the factors that sense and regulate intracellular zinc levels in eukaryotes. This report demonstrates that Loz1, a transcriptional repressor that contains a double C2H2-type zinc finger domain, plays a central role in zinc homeostasis in Schizosaccharomyces pombe. These studies provide important insight into the types of proteins and possible domains that can be used to sense zinc ions.
Keywords: metallosensor, metallothionein, ncRNA, noncoding RNA
Abstract
In Schizosaccharomyces pombe, alcohol dehydrogenase 1 (Adh1) is an abundant zinc-requiring enzyme that catalyses the conversion of acetaldehyde to ethanol during fermentation. In a zinc-replete cell, adh1 is highly expressed. However, in zinc-limited cells, adh1 gene expression is repressed, and cells induce the expression of an alternative alcohol dehydrogenase encoded by the adh4 gene. In our studies examining this zinc-dependent switch in alcohol dehydrogenase gene expression, we isolated an adh1Δ strain containing a partial loss of function mutation that resulted in higher levels of adh4 transcripts in zinc-replete cells. This mutation also led to the aberrant expression of other genes that are typically regulated by zinc. Using linkage analysis, we have mapped the position of this mutation to a single gene called Loss Of Zinc sensing 1 (loz1). Loz1 is a 55-kDa protein that contains a double C2H2-type zinc finger domain. The mapped mutation that disrupts Loz1 function leads to an arginine to glycine substitution in the second zinc finger domain, suggesting that the double zinc finger domain is important for Loz1 function. We show that loz1Δ cells hyperaccumulate zinc and that Loz1 is required for gene repression in zinc-replete cells. We also have found that Loz1 negatively autoregulates its own expression. We propose that Loz1 is a unique metalloregulatory factor that plays a central role in zinc homeostasis in S. pombe.
Zinc is an important trace metal element that has diverse biological roles. At the cellular level, zinc is an essential cofactor in a range of enzymes, including alkaline phosphatases, Cu-Zn superoxide dismutases, and alcohol dehydrogenases (1). Many regulatory proteins also contain small domains that are stabilized by zinc; these domains include the widely used C2H2-type zinc finger domain, the LIM domain, and the RING finger domain (2, 3). In addition to being a protein cofactor, a growing amount of evidence suggests that zinc can act as an intracellular signaling molecule (4, 5). Thus, zinc is essential for all life and has many cellular roles.
Although zinc is an essential nutrient, in excess, it is toxic to growth. Cells therefore rely on mechanisms to maintain a relatively constant intracellular level of zinc despite fluctuations in the extracellular environment. In eukaryotes, zinc-responsive transcription factors play a central role in zinc homeostasis by regulating the expression of genes required for zinc transport or zinc storage (6). In Saccharomyces cerevisiae, the transcription factor Zap1 binds to zinc-responsive elements located in target gene promoters and activates gene expression when cytoplasmic zinc levels are low (7). Zap1 target genes include ZRT1 and ZRT3, which are required for zinc uptake and the release of zinc from intracellular stores, respectively (7, 8). In mammals, fish, and flies, a different protein, metal responsive transcription factor 1 (MTF-1), prevents zinc from accumulating to toxic levels. In contrast to Zap1, MTF-1 activates target gene expression when zinc is in excess. MTF-1 regulates genes required for zinc efflux (ZnT-1) and zinc storage (MT-1) (9). Although studies with MTF-1 and Zap1 have revealed much about zinc homeostasis, zinc-dependent changes in gene expression are observed in organisms that lack homologs of MTF-1 and Zap1 (10, 11). Moreover, in mammals, zinc-dependent changes in gene expression occur in a manner that is independent of MTF-1 (12, 13). These observations suggest that other unidentified zinc-regulated factors play a critical role in zinc homeostasis in eukaryotes.
In the fission yeast Schizosaccharomyces pombe, ∼25 genes are induced in response to zinc deficiency (14, 15). These genes include, zinc regulated transporter 1 (zrt1), which encodes a high-affinity zinc uptake system, adh4, which encodes a putative mitochondrial iron-dependent alcohol dehydrogenase, and adh1AS, an antisense transcript generated at the adh1 gene locus. Other genes that are highly expressed under zinc-limited conditions include SPAC977.05c, SPBC1348.06c, and SPBPB2B2.15, which encode three highly related fungal proteins of unknown function. Genes that are repressed in response to zinc limitation include alcohol dehydrogenase 1 (adh1) and zinc binding metallothionein (zym1) (15, 16). Despite tight regulation of gene expression in response to zinc availability, there are no homologs of Zap1 or MTF-1 in fission yeast, suggesting that S. pombe uses a different factor(s) to sense and regulate gene expression in response to zinc. In our studies examining the regulation of adh1 gene expression by zinc, we isolated a second site mutation that led to increased expression of adh4, zrt1, SPBC1348.06c, and the adh1AS transcript in zinc-replete cells. This mutation also led to lower levels of adh1 and zym1 gene expression. In this report, we have mapped the position of this mutation to a single gene and demonstrate that this gene is essential for zinc-dependent regulation of gene expression. We have named this gene Loss Of Zinc sensing 1 (loz1).
Results
Spontaneous Mutation in adh1Δ Cells Leads to Aberrant Zinc-Dependent Regulation of Gene Expression.
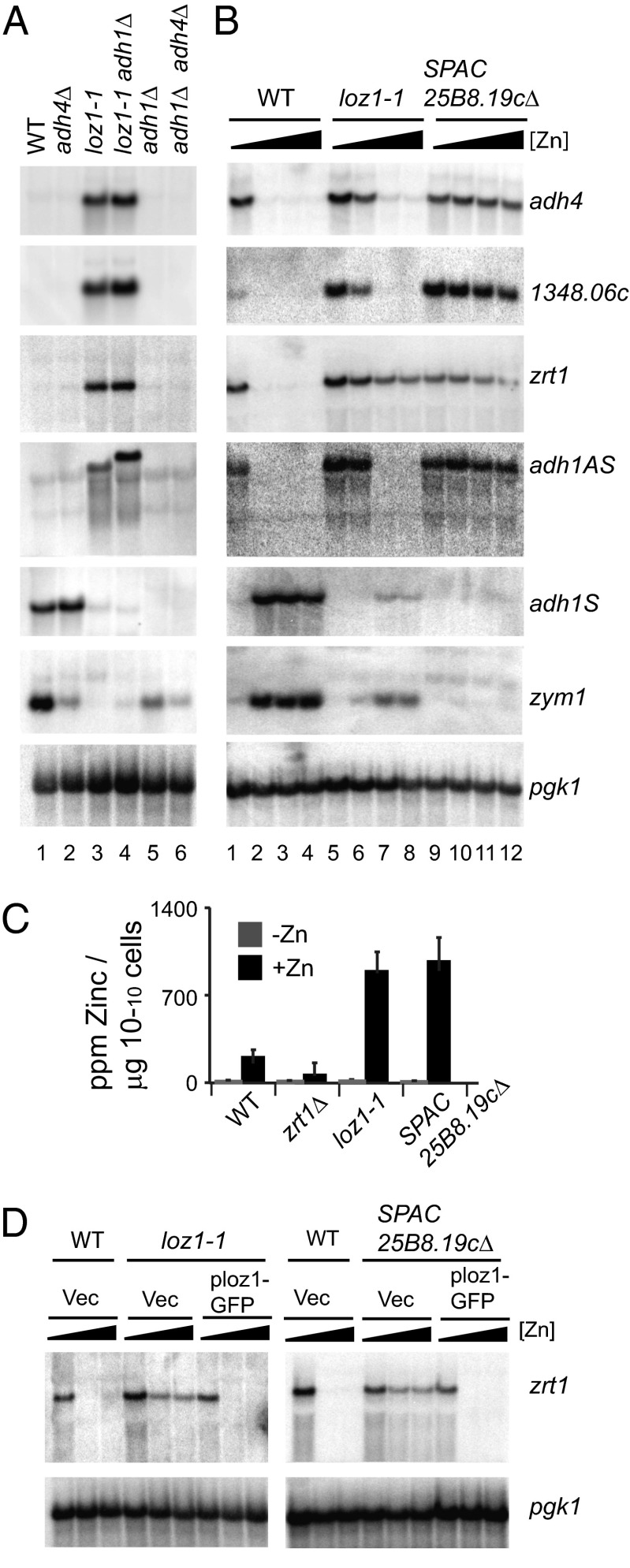
In our previous work, we found that adh1 and adh4 gene expression was regulated by zinc (15). During these studies, an adh1Δ colony was isolated that contained a spontaneous mutation resulting in aberrant zinc-dependent regulation of gene expression. The effects of this mutation (named loz1-1; Loss of Zinc Sensing 1 Allele 1) on gene expression are shown by RNA blot analysis in Fig. 1 A and B. In wild-type cells grown in the zinc-rich yeast extract + supplements (YES) medium, adh1 and zym1 transcripts accumulate to high levels, and zrt1, adh4, adh1AS, and SPBC1348.06c transcripts are not detected (Fig. 1A, lane 1). In cells lacking adh1 and/or adh4 grown under similar conditions, zrt1, adh1AS, and SPBC1348.06c transcripts were not detected (Fig. 1A, lanes 2, 5, and 6). Unexpectedly, zym1 transcripts levels were reduced relative to the wild-type control in the adh1Δ and adh4Δ cells. However, zym1 expression is tightly controlled in response to a number of environmental and stress pathways (16), suggesting that the changes of zym1 mRNA levels in these mutants could result from zinc-independent effects on gene expression. In contrast, in adh1Δ loz1-1 cells, zrt1, adh4, adh1AS, and SPBC1348.06c transcripts accumulated to high levels (Fig. 1A, lane 4). Zym1 transcripts levels were also lower than those observed in adh1Δ and wild-type cells. Thus, the gene expression profile of adh1Δ loz1-1 cells grown in YES medium was atypical and resembled that of a cell grown under zinc-limiting conditions.
Fig. 1.
A mutation in loz1 disrupts zinc-dependent regulation of gene expression. (A) Wild-type, adh4Δ, loz1-1, adh1Δ loz1-1, adh1Δ, and adh1Δ adh4Δ cells were grown to exponential phase in YES medium and total RNA purified for RNA blot analysis. RNA blots were incubated with strand-specific probes that hybridized to the adh4, zrt1, SPBC1348.06c, adh1AS, adh1, and zym1 transcripts. The adh1AS probe hybridizes to the extreme 5′ end of this transcript, a region that does not overlap with the adh1 ORF and is present in adh1Δ cells. Because of the high level of sequence conservation at the DNA level (>99% identity), the SPBC1348.06c probe can cross-hybridize to SPAC977.05c and SPBPB2B2.15 transcripts. For a loading control, blots were also probed for pgk1. (B) Wild-type, loz1-1, and loz1Δ cells were grown in ZL-EMM supplemented with 0, 50, 200, or 500 μM zinc. Total RNA was purified for RNA blot analysis. RNA blots were probed as described in A. (C) Wild-type, zrt1Δ, loz1-1, and loz1Δ cells were grown in ZL-EMM supplemented with 0 μM (-Zn) or 200 μM zinc (+Zn). Cells were harvested, and total cellular zinc levels measured by inductively coupled plasma mass spectrometry. (D) Wild-type, loz1-1, and loz1Δ cells containing the vector JK148 or plasmid pLoz1-GFP were grown in ZL-EMM supplemented with 0, 100, or 200 μM zinc, and total RNA was purified for RNA blot analysis.
The loz1-1 mutation spontaneously occurred in a strain lacking the adh1 gene. To determine whether the aberrant gene expression observed in adh1Δ loz1-1 cells depended on the loss of adh1 gene function, adh1Δ loz1-1 cells were crossed to a wild-type strain of the opposite mating type. When the resulting diploid was sporulated and individual asci isolated by tetrad dissection analysis, half of the ascospores accumulated high levels of adh4 transcripts in YES medium. This phenotype segregated independently from adh1Δ, and ascospores were isolated that had aberrant adh4 expression and the endogenous adh1 gene (Fig. 1A, lane 3 strain loz1-1). These results are consistent with the aberrant gene expression observed in adh1Δ loz1-1 being independent of adh1Δ.
To further examine the effects of the loz1-1 mutation on gene expression, total RNA was purified from wild-type and loz1-1 cells grown in a zinc-limited minimal medium (ZL-EMM), or in this medium with supplemented with 50, 200, or 500 μM zinc. In wild-type cells, high levels of adh4, zrt1, SPBC1348.06c, and adh1AS transcripts levels accumulated when cells were grown in ZL-EMM with no zinc supplement (Fig. 1B, lane 1). As expected, growth of wild-type cells in the presence of a zinc supplement led to a significant reduction in the levels of these transcripts, and an increase in zym1 and adh1 mRNA transcripts (Fig. 1B, lanes 2–4). In loz1-1 cells, high levels of adh4, zrt1, SPBC1348.06c, and adh1AS transcripts were detected in cells that had been grown in ZL-EMM with and without a 50 μM zinc supplement (Fig. 1B, lanes 5 and 6). Although there was a clear misregulation of zinc-dependent gene expression in loz1-1 cells grown in ZL-EMM + 50 μM zinc, at higher zinc levels (ZL-EMM + 200 or 500 μM zinc) there was a significant decrease in adh4, SPBC1348.06c, and adh1AS transcripts levels, and a modest reduction in zrt1 transcript levels. zym1 and adh1 mRNAs were also detected at these higher zinc levels in loz1-1 cells. Thus, the loz1-1 allele leads to impaired zinc-dependent gene expression that can be partially restored by the addition of zinc in excess.
The aberrant expression of zinc-regulated genes observed in loz1-1 cells could result from a mutation that impaired zinc uptake or a mutation that interfered with zinc-dependent regulation of gene expression. To distinguish between these possibilities, total cellular zinc levels were measured in wild-type and loz1-1 cells that had been grown in ZL-EMM or in this medium supplemented with 200 μM zinc. As a control, zinc levels were also examined in zrt1Δ cells that lack the high-affinity zinc uptake system. In zinc-replete medium, loz1-1 cells accumulated higher levels of zinc compared with the wild-type and zrt1Δ cells (Fig. 1C). Thus, under zinc-replete conditions, loz1-1 cells hyperaccumulate zinc, but “sense” that they are zinc limited. These results are consistent with a mutation in loz1 disrupting zinc-dependent regulation of gene expression.
Loz1 Is Required for Zinc-Dependent Regulation of Gene Expression.
The loz1-1 allele provided a tool to understand how S. pombe cells sense intracellular zinc levels. To determine how the loz1-1 mutation disrupted zinc sensing, the position of the mutation in loz1-1 cells was mapped by linkage analysis (Table S1). These experiments mapped the mutation to an ∼44-kb region on the right arm of chromosome 1. DNA sequencing through this region revealed a single C-G mutation at nucleotide position 1528 in the gene SPAC25B8.19c. To determine whether the mutation in SPAC25B8.19c was the cause of the impaired zinc-dependent regulation of gene expression, a plasmid that expressed a GFP-tagged SPAC25B8.19c gene from its native promoter (pLoz1-GFP) was integrated into loz1-1 cells. When loz1-1 cells containing the empty vector or pLoz1-GFP were grown in ZL-EMM with or without zinc supplements, pLoz1-GFP was able to restore zinc-dependent gene expression (Fig. 1D). To further examine the function of SPAC25B8.19c, a strain was constructed in which the entire SPAC25B8.19c ORF was deleted from the genome. In this cell type, adh4, SPBC1348.06c, zrt1, and adh1AS transcripts accumulated under all conditions, and no zym1 and adh1 transcripts were detected by RNA blot analysis (Fig. 1B, lanes 9–12). This aberrant gene expression could be fully restored by the addition of ploz1-GFP (Fig. 1D) and partially rescued by the addition of a plasmid expressing loz1-1 (Fig. S1). Taken together, these results indicate that SPAC25B8.19c plays a central role in zinc homeostasis and strongly suggest that the single base-pair mutation in SPAC25B8.19c is the cause of impaired zinc-dependent regulation of gene expression in loz1-1 cells. SPAC25B8.19c was therefore named Loz1.
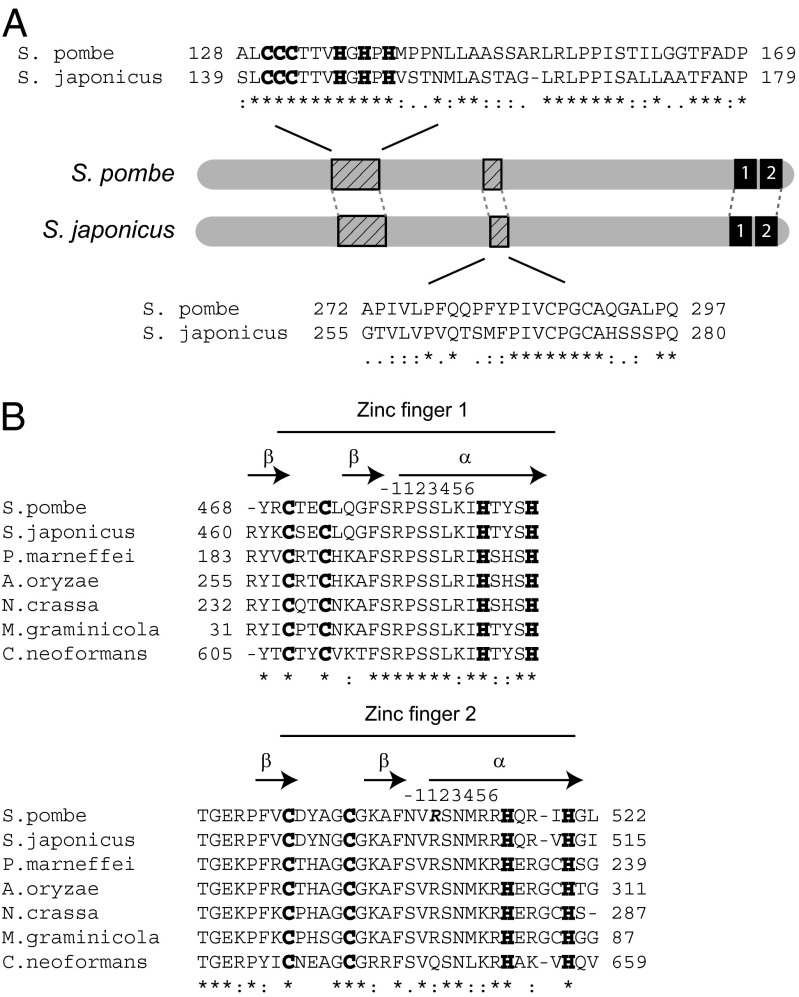
Loz1 is a 55-kDa protein that contains a double C2H2 type zinc finger domain at the extreme C terminus (Fig. 2A). The mutation that impairs Loz1 function results in an Arg510Gly substitution at position 1 of the alpha helix of the second zinc finger. In other fungal Loz1 homologs, the double zinc finger domain is highly conserved, consistent with it being important for Loz1 function (Fig. 2B). Outside of the double zinc finger domain, Loz1 shares little homology with other fungal proteins with the exception of a few smaller domains that are conserved in Schizosaccharomyces japonicus, a close relative of S. pombe (Fig. 2A).
Fig. 2.
Conserved Loz1 domains. (A) A schematic diagram of the Loz1 protein from S. pombe and S. japonicus. To identify conserved domains, the amino acid sequence of Loz1 was aligned with the Loz1 ortholog from S. japonicus by using CLUSTAL W. Black numbered boxes indicate the positions of the double zinc finger domains. The sequence alignment of other conserved domains has been shown and their positions indicated by striped rectangles on the schematic diagram. (B) An alignment of the Loz1 zinc finger domain with the corresponding region of Loz1 orthologs from S. japonicus, Penicillium marneffei, Aspergillus oryzae, Neurospora crassa, Mycosphaerella graminicola, and Cryptococcus neoformans. The cysteine and histidine residues that are predicted to coordinate zinc are shown in bold. The arginine altered to a glycine in loz1-1 is shown in bold and italics.
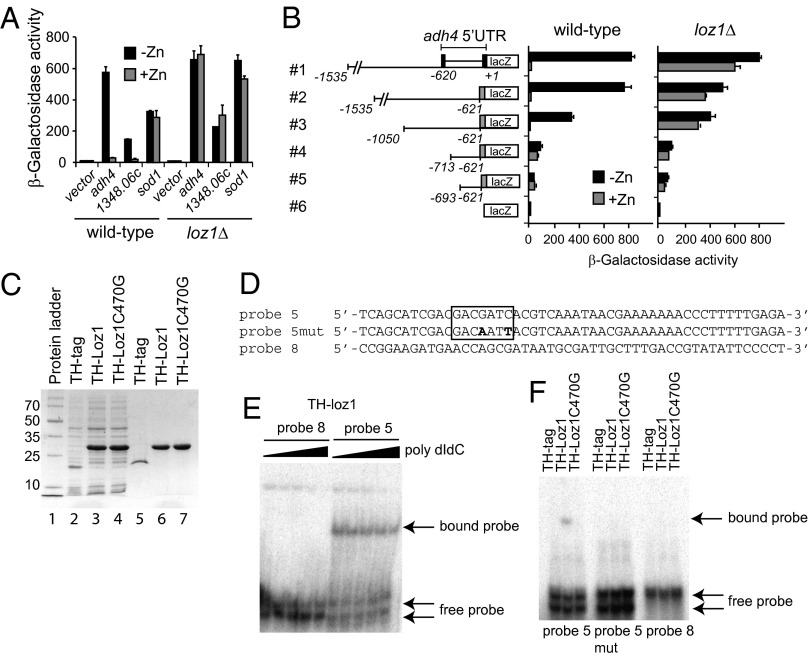
Because deletion of loz1 resulted in increased expression of adh4 and zrt1, we hypothesized that Loz1 played a central role in zinc homeostasis by acting as a transcriptional repressor in zinc-replete cells. To test whether Loz1 acted as a zinc-regulated transcriptional repressor, constructs were generated in which the adh4 and SPBC1348.06c promoters were fused to a lacZ reporter gene. When these constructs were integrated into wild-type or loz1Δ cells, β-galactosidase activity was robustly regulated by zinc in a manner that depended on loz1 (Fig. 3A). No zinc-dependent changes in β-galactosidase activity were observed in cells expressing a control sod1-lacZ reporter. When the adh4 and SPBC1348.06c reporters were introduced into loz1-1 cells, high levels of β-galactosidase activity were observed at intermediate zinc levels, consistent with the partial loss of gene expression in this strain (Fig. S2). Thus, Loz1 is necessary for the zinc-dependent regulation of the adh4-lacZ and SPBC1348.06c-lacZ reporter genes.
Fig. 3.
Loz1 is required for zinc-dependent transcriptional regulation of gene expression. (A) Wild-type and loz1Δ cells containing the empty vector, adh4-lacZ, or SPBC1348.06c-lacZ reporters were grown in ZL-EMM supplemented with 0 μM (-Zn) or 200 μM zinc (+Zn) before cells were harvested for β-galactosidase assays. (B) Wild-type and loz1Δ cells containing the indicated reporter constructs were grown as described in A. Cells were harvested and β-galactosidase activity was measured by standard procedures. (C) SDS/PAGE analysis of crude lysates from isopropyl β-D-1-thiogalactopyranoside-induced E. coli cells containing the Pet32a empty vector (TH tag), Pet32a-Loz1 (TH-Loz1), or Pet32a-Loz1Cys470Gly (TH-Loz1Cys470Gly) (lanes 2–4), or the Ni-NTA affinity-purified TH tag, TH-Loz1, or TH-Loz1Cys470Gly (lanes 5–7). Proteins were visualized by using Coomassie blue. The sizes of a molecular mass ladder (lane 1) are shown in kilodaltons on the left. (D) Sequence of the probes used for EMSA. All probes used were double-stranded; the sequence of the 5′ strand for each probe is shown. The GN(A/C)GATC element is boxed. Mutations in probe 5mut are highlighted in bold. (E) A representative EMSA using 0.1 ng of P32-labeled double-stranded probes T8 and T5 in the presence of increasing levels (0, 0.5, 1, 2.5, and 5 μg) of poly dIdC. (F) A representative EMSA using 0.1 ng of P32-labeled double-stranded probes T5, T5mut, and T8, and affinity-purified TH tag, TH-tagged Loz1, and TH-Loz1Cys470Gly in the presence of 1 μg of poly dIdC. Free probe and bound probe are indicated.
The adh4-lacZ reporter contained promoter and 5′ UTR sequences. A 5′ RACE revealed that the adh4 5′ UTR was 620 nt in length and contained a previously unannotated intron (Fig. S3). To test whether the adh4 5′ UTR was necessary for the Loz1-dependent regulation of adh4-lacZ reporter activity, a construct was generated in which the adh4 5′ UTR was exchanged for the nmt1 5′ UTR (Fig. S3). nmt1 (no message in thiamine 1) is a gene that is not regulated by zinc (15). When this hybrid reporter was expressed in wild-type cells, β-galactosidase activity was regulated by zinc (Fig. 3B, construct 2). In contrast, zinc responsiveness was severely reduced in loz1Δ cells expressing this reporter. Together these results indicate that the adh4 5′ UTR is not necessary for zinc-dependent regulation. To further map the region of the adh4 promoter required for Loz1-dependent regulation, additional truncated adh4-nmt1 hybrid reporters were introduced into wild-type and loz1Δ cells (Fig. 3B, constructs 3–6). These truncations resulted in a number of reductions in the total level of β-galactosidase activity, consistent with the loss of upstream activating sequences. When the zinc-dependent regulation was examined, the hybrid reporter containing adh4 nucleotides −1050 to −621 (construct 3) was not regulated by zinc in loz1Δ cells but retained full zinc responsiveness when expressed in a wild-type strain. In contrast, a shorter reporter containing adh4 nucleotides −713 to −621 (construct 4) was only weakly regulated by zinc in a wild-type strain. Together these results suggest that an element(s) critical to the loz1-dependent regulation of adh4 was located between nucleotides −1050 to −713.
Motif-based sequence analysis tools (17) revealed that three GN(A/C)GATC elements were located within nucleotides −1050 to −713 of the adh4 promoter. This element was also present in multiple copies in the promoters of other Loz1-regulated genes. To test whether Loz1 directly interacted with this candidate motif, in vitro DNA binding studies were performed by using a recombinant 96 aa C-terminal truncation of Loz1 and a 50-bp adh4 promoter fragment containing this motif. The C-terminal of Loz1 was chosen for these studies because this region contained the double zinc finger domain, a motif known to mediate site-specific interactions with DNA in other proteins (18). For the purification, the C-terminal of Loz1 was expressed in Escherichia coli as a thioredoxin-His tag fusion protein and was purified by using Ni-NTA affinity chromatography (Fig. 3C, TH-Loz1). The tag only (TH-tag) and a mutated form of the tagged Loz1 fusion protein containing a Cys470Gly substitution (TH-loz1C470G) were also expressed and purified as controls. The Cys470Gly substitution disrupts the first cysteine in zinc finger 1. Results showed that the radio-labeled adh4 promoter probe containing an GN(A/C)GATC element (Fig. 4D, probe 5) formed a complex in the presence of the TH-Loz1 and the nonspecific competitor poly dIdC (Fig. 4E). No complex was detected with this probe, and the thioredoxin-His tag only or the Cys470Gly mutant Loz1 protein (Fig. 4F, probe 5). In addition, no complex was detected when the GN(A/C)GATC element was mutated to GN(A/C)AATT (Fig. 4F, probe 5mut), or when TH-Loz1 was incubated with an adjacent adh4 promoter fragments lacking this motif (Fig. 4F, probe 8). These results suggest that the C-terminal 96 amino acids of Loz1 associate with a 50-bp region of the adh4 promoter that contains a GN(A/C)GATC element, and that the GATC core of this sequence is critical for this interaction.
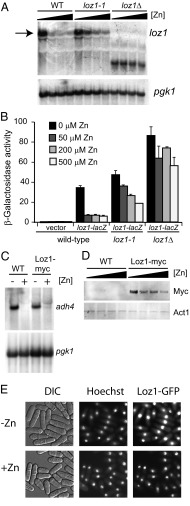
Fig. 4.
(A) Wild-type, loz1-1, and loz1Δ cells were grown in ZL-EMM supplemented with 0, 50, 200, or 500 μM zinc, cells were harvested, and total RNA was purified for RNA blot analysis. (B) Wild-type, loz1-1, and loz1Δ cells containing the loz1-lacZ reporter were grown in ZL-EMM supplemented with 0, 50, 200, or 500 μM zinc, and cells were harvested for β-galactosidase assays. (C) Wild-type and Loz1-myc cells were grown in ZL-EMM supplemented with 0 or 200 μM zinc, and total RNA was purified for RNA blot analysis. RNA blots were probed for adh4 and pgk1 mRNAs. (D) Wild-type and Loz1-myc cells were grown in ZL-EMM supplemented with 0, 50, 200, or 500 μM zinc, and crude protein extracts were isolated for immunoblot analysis. (E) loz1Δ cells expressing pLoz1-GFP were grown in ZL-EMM supplemented with 0 (-Zn) or 200 μM (+Zn) zinc. The cells were visualized for GFP by fluorescence microscopy. Hoechst stain was used to stain the nucleus, and differential interference contrast microscopy was used to confirm cell integrity.
Loz1 Negatively Autoregulates Its Own Expression.
Loz1 is required for gene repression in zinc-replete cells. To determine how Loz1 was regulated by zinc in S. pombe, we initially tested whether loz1 transcript levels depended on zinc levels. When wild-type were grown in ZL-EMM, an ∼3.1-kb loz1 transcript was detected by RNA blot analysis (Fig. 4A, see arrow). This band was absent from loz1Δ cells and was present at reduced levels in wild-type cells that had been grown in the presence of 50–500 μM zinc. A smaller band of unknown origin was also detected in loz1Δ cells. In loz1-1 cells, loz1 transcript levels were regulated by zinc, but to a lesser extent than the regulation observed in wild-type cells. These results suggested that Loz1 might negatively autoregulate its own expression. To test this hypothesis, the activity of a loz1-lacZ reporter was examined in wild-type, loz1-1, and loz1Δ cells that had been grown over a range of zinc levels (Fig. 4B). Consistent with Loz1 controlling its own expression, β-galactosidase activity was regulated by zinc in a manner that depended on Loz1.
The above results revealed that loz1 mRNA transcripts accumulate at lower levels in zinc-replete cells compared with zinc-limited cells. To determine whether a similar reduction was observed at the protein level, immunoblot analysis was used to examine Loz1 levels. To facilitate these studies, a yeast strain was generated in which a 13× myc epitope tag was fused to the C terminus of the endogenous Loz1 (Loz1-myc). In cells expressing the Loz1-myc fusion protein, adh4 transcript levels were regulated by zinc indicating that the Loz1-myc fusion is functional (Fig. 4C). When crude protein extracts were purified from Loz1-myc cells grown over a range of zinc levels, a major band of ∼75 kDa was detected by immunoblot analysis (Fig. 4D). This band was not detected in wild-type cells, suggesting that it was specific to the Loz1-myc strain. In addition, higher levels of Loz1-myc accumulated in zinc-limited cells, consistent with the higher expression of loz1 under this condition. Because Loz1 accumulated under all conditions, but plays a role in gene repression in zinc-replete cells, we hypothesized that the zinc-dependent changes in Loz1 function might result from zinc modulating the cellular distribution of Loz1. To test this hypothesis, loz1Δ cells expressing pLoz1-GFP were grown under zinc-limiting and zinc-replete conditions and the localization of Loz1-GFP visualized by immunofluorescence microscopy. Under all conditions, Loz1-GFP colocalized with a Hoechst DNA stain (Fig. 4E). Thus, Loz1 accumulates in the nucleus in zinc-limited and zinc-replete cells but plays a role in gene repression under zinc-replete conditions.
Increased adh4 Expression Confers a Growth Advantage to adh1Δ Cells.
Why would a spontaneous mutation that impaired zinc-dependent regulation of gene expression have occurred in adh1Δ cells? In S. cerevisiae, gene duplications leading to increased ADH4 copy number confer a significant growth advantage to adh1Δ cells (19). Because adh4 expression is tightly regulated by zinc in S. pombe, a straightforward explanation for the occurrence of the loz1-1 mutation is that it leads to increased adh4 expression that, in turn, compensates for the absence of adh1. To test this hypothesis, we investigated whether the loz1-1 mutation or increased adh4 expression would confer a growth advantage to adh1Δ cells. adh1Δ cells have a severe growth defect on YES medium and are unable to grow in the presence of the respiration inhibitor Antimycin A (Fig. S4). In contrast, adh1Δ loz1-1 cells had no growth defect in YES medium and were able to grow in the presence of antimycin A (Fig. S4). To determine whether increased adh4 expression would enhance growth of adh1Δ, a plasmid expressing adh4 from the constitutive pgk1 promoter (pgk1-adh4) was introduced in adh1Δ cells, and control wild-type and adh4Δ cells. The addition of pgk1-adh4 conferred a significant growth advantage to adh1Δ in both the presence and absence of antimycin A (Fig. S4). Thus, one potential explanation for the occurrence of the loz1-1 mutation is that it results in increased adh4 gene expression that, in turn, confers a growth advantage to adh1Δ.
Discussion
In S. pombe, zrt1, adh4, SPBC1348.06c, and adh1AS transcripts are specifically expressed in zinc-limited cells. In this study, we show that loz1-1 and loz1Δ mutations result in the expression of zrt1, adh4, SPBC1348.06c, and adh1AS genes in zinc-replete cells and lead to the hyperaccumulation of zinc. We have also found that Loz1 is a nuclear protein that associates with a GN(A/C)GATC element located in the adh4 promoter. Based on the above results, we propose that Loz1 plays a central role in zinc homeostasis in S. pombe.
The defects observed in loz1-1 cells are consistent with a loss-of-function mutation in a transcriptional repressor or a gain-of-function mutation in a transcriptional activator. We have found that the loz1-1 mutation is recessive (Fig. S5) and that a wild-type copy of loz1 can complement the aberrant gene expression observed in loz1-1 cells (Fig. 1D). We also show that deletion of the loz1 ORF results in the expression of zrt1, adh4, SPBC1348.06c, and the adh1AS transcript in high zinc (Fig. 1B). From these observations, we propose that loz1-1 is a loss-of-function mutation in a gene required for transcriptional repression.
If Loz1 functions in transcriptional repression, then why are adh1 and zym1 mRNA levels reduced in loz1Δ and loz1-1 cells? In S. pombe, adh1 gene expression is inhibited by increased expression of the adh1AS transcript (15). In this study, we have found that the expression of the adh1AS transcript depends on Loz1 (Fig. 1B). Together, these results suggest that Loz1 indirectly controls adh1 gene expression by regulating the levels of the adh1AS transcript. A potential clue to the regulation of zym1 by zinc is that low levels of a larger zym1 transcript are detected on RNA blots in loz1Δ cells and in wild-type cells grown under zinc-limiting conditions (Fig. 1B and Fig. S6). Intergenic transcripts that transverse through promoter regions can inhibit the recruitment of transcriptional activators, leading to decreased expression of the downstream gene (20, 21). Therefore, one explanation for the Loz1-dependent regulation of zym1 is that Loz1 regulates the expression of an intergenic transcript that transcriptionally interferes with zym1 expression. Supporting this hypothesis, probes that bind to the upstream region zym1 hybridize to a larger Loz1-regulated transcript (Fig. S6). Thus, the Loz1-dependent regulation of adh1 and zym1 genes is potentially indirectly mediated through the regulation of noncoding RNA transcripts at these loci.
A number of lines of evidence suggest that Loz1 is regulated at multiple levels by zinc. At a transcriptional level, Loz1 negatively regulates its own expression, leading to lower levels of loz1 mRNA transcript in high zinc (Fig. 4A). This transcriptional regulation, in turn, leads to lower levels of Loz1 protein accumulating in zinc-replete cells compared with zinc-limiting cells (Fig. 4D). Despite modest changes in Loz1 protein levels by zinc, Loz1 is required for gene repression in zinc-replete cells, suggesting that Loz1 activity is also modulated by zinc. In the two characterized zinc-responsive transcription factors, Zap1 and MTF1, C2H2-type zinc fingers have been implicated in zinc sensing (22–24). In our study, we find that an Arg510Gly substitution in zinc finger 2 partially disrupts Loz1 function and that the zinc finger region is necessary for DNA-binding function. Thus, one possibility is that the occupancy of the Loz1 zinc finger domains with zinc depends on cellular zinc levels that, in turn, controls Loz1 DNA binding function. Alternately, the N-terminal region of Loz1 contains small domains rich in cysteine and histidine residues. Although these domains do not match any known zinc-binding motif, cysteine and histidine residues are commonly involved in the coordination of zinc ions. Further studies with Loz1 should provide insight into whether Loz1 activity is directly controlled by zinc ion binding to one or more of these domains.
A number of domains have been implicated in zinc sensing in eukaryotes, some of which closely resemble commonly found zinc structural motifs. In Chlamydomonas reinhardtii, deletion of a C-terminal metallothionein-like domain from the copper sensing transcription factor, CRR1, leads to increased expression of genes required for zinc uptake and zinc hyperaccumulation (10). In Arabidopsis thaliana, bZIP19 and bZIP23 are required for the adaptation to zinc (11). Although it is unclear how bZIP19 and bZIP23 are regulated by zinc, both contain a small conserved Cys/His-motif that might be involved in zinc sensing. Zap1 contains multiple domains that are independently regulated by zinc (25). These domains include AD1, a transactivation domain that is rich in histidine and cysteine residues, and AD2, a transactivation domain that contains a unique regulatory zinc finger pair motif (26). In mammals, MTF-1 also contains multiple zinc-regulated domains including a zinc-responsive DNA binding domain containing six C2H2-type zinc fingers (9). The growing number of unique eukaryotic zinc-regulated factors that have been identified suggests that novel zinc-sensing domains may have evolved independently in different species. Intriguingly, studies with Zap1 have shown that AD1 is plays a primary role in activating genes required to maintain zinc homeostasis, whereas AD2 plays a more major role in gene regulation when zinc deficiency is combined with other environmental stresses (26). Thus, one potential explanation for the diversity in zinc-regulated factors is that each unique factor is able to optimally sense changes in zinc levels under normal and specialized environmental conditions. Future studies with Loz1 and other zinc-regulated factors should therefore provide important insight into how unique domains have evolved to sense zinc and the role that different zinc sensors play in maintaining zinc homeostasis and adapting to changes in zinc levels.
Materials and Methods
Additional details are found as SI Materials and Methods. All of the yeast strains used in this study are shown in Table S2. All primers used for plasmid generation are listed in Table S3.
Supplementary Material
Acknowledgments
We thank Andrea McCue for help with strain generation, Mike Carge for assistance with linkage analysis, Dr. Jian-Qiu Wu for kindly providing plasmids and yeast strains that were used in these studies, and Dr. R. Michael Townsend for critically reading the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300853110/-/DCSupplemental.
References
- 1.Andreini C, Bertini I. A bioinformatics view of zinc enzymes. J Inorg Biochem. 2012;111:150–156. doi: 10.1016/j.jinorgbio.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C, Banci L, Bertini I, Rosato A. Counting the zinc-proteins encoded in the human genome. J Proteome Res. 2006;5(1):196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 3.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 4.Kim AM, et al. Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem Biol. 2011;6(7):716–723. doi: 10.1021/cb200084y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukada T, Yamasaki S, Nishida K, Murakami M, Hirano T. Zinc homeostasis and signaling in health and diseases: Zinc signaling. J Biol Inorg Chem. 2011;16(7):1123–1134. doi: 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrensberger KM, Bird AJ. Hammering out details: Regulating metal levels in eukaryotes. Trends Biochem Sci. 2011;36(10):524–531. doi: 10.1016/j.tibs.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Eide DJ. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2009;284(28):18565–18569. doi: 10.1074/jbc.R900014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CY, et al. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics. 2008;9:370. doi: 10.1186/1471-2164-9-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Günther V, Lindert U, Schaffner W. The taste of heavy metals: Gene regulation by MTF-1. Biochim Biophys Acta. 2012;1823(9):1416–1425. doi: 10.1016/j.bbamcr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Sommer F, et al. The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell. 2010;22(12):4098–4113. doi: 10.1105/tpc.110.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assunção AG, et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA. 2010;107(22):10296–10301. doi: 10.1073/pnas.1004788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coneyworth LJ, et al. Identification of the human zinc transcriptional regulatory element (ZTRE): A palindromic protein-binding DNA sequence responsible for zinc-induced transcriptional repression. J Biol Chem. 2012;287(43):36567–36581. doi: 10.1074/jbc.M112.397000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu MS, Langkamp-Henken B, Chang SM, Shankar MN, Cousins RJ. Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc Natl Acad Sci USA. 2011;108(52):20970–20975. doi: 10.1073/pnas.1117207108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dainty SJ, Kennedy CA, Watt S, Bähler J, Whitehall SK. Response of Schizosaccharomyces pombe to zinc deficiency. Eukaryot Cell. 2008;7(3):454–464. doi: 10.1128/EC.00408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrensberger KM, et al. Zinc-dependent regulation of the Adh1 antisense transcript in fission yeast. J Biol Chem. 2013;288(2):759–769. doi: 10.1074/jbc.M112.406165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borrelly GP, et al. Surplus zinc is handled by Zym1 metallothionein and Zhf endoplasmic reticulum transporter in Schizosaccharomyces pombe. J Biol Chem. 2002;277(33):30394–30400. doi: 10.1074/jbc.M203145200. [DOI] [PubMed] [Google Scholar]
- 17.Chakravarty A, Carlson JM, Khetani RS, Gross RH. A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinformatics. 2007;8:249. doi: 10.1186/1471-2105-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366(6454):483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 19.Walton JD, Paquin CE, Kaneko K, Williamson VM. Resistance to antimycin A in yeast by amplification of ADH4 on a linear, 42 kb palindromic plasmid. Cell. 1986;46(6):857–863. doi: 10.1016/0092-8674(86)90067-x. [DOI] [PubMed] [Google Scholar]
- 20.Moseley JL, et al. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell. 2002;14(3):673–688. doi: 10.1105/tpc.010420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429(6991):571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 22.Bird AJ, et al. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 2003;22(19):5137–5146. doi: 10.1093/emboj/cdg484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae. J Mol Biol. 2006;357(4):1167–1183. doi: 10.1016/j.jmb.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1) Arch Biochem Biophys. 2007;463(2):201–210. doi: 10.1016/j.abb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 25.Frey AG, et al. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS ONE. 2011;6(7):e22535. doi: 10.1371/journal.pone.0022535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frey AG, Eide DJ. Roles of two activation domains in Zap1 in the response to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2011;286(8):6844–6854. doi: 10.1074/jbc.M110.203927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.