Significance
A hemicatenane conjoins two DNA duplexes through a single-strand interlock. It has been proposed that hemicatenanes are important intermediates for replication, repair, and recombination. However, the biochemical analysis of hemicatenanes is hampered by the relative inaccessibility of such structures. We report here that a DNA topoisomerase III (Top3) from a hyperthermophilic archaeum can carry out synthesis and dissolution of hemicatenanes. We also show that a complex of human Top3α, Bloom helicase (Blm), and RecQ-mediated genome instability protein 1 and 2 has a biochemical function of reversing this hemicatenation. Our results demonstrate that type IA topoisomerases can regulate the formation of hemicatenane structures.
Keywords: DNA repair, DNA topology, genetic recombination, double Holliday junction
Abstract
Type IA DNA topoisomerases work with a unique mechanism of strand passage through an enzyme-bridged, ssDNA gate, thus enabling them to carry out diverse reactions in processing structures important for replication, recombination, and repair. Here we report a unique reaction mediated by an archaeal type IA topoisomerase, the synthesis and dissolution of hemicatenanes. We cloned, purified, and characterized an unusual type IA enzyme from a hyperthermophilic archaeum, Nanoarchaeum equitans, which is split into two pieces. The recombinant heterodimeric enzyme has the expected activities in its preference of relaxing negatively supercoiled DNA. Its amino acid sequence and cleavage site sequence analysis suggest that it is topoisomerase III, and therefore we named it “NeqTop3.” At high enzyme concentrations, NeqTop3 can generate high-molecular-weight DNA networks. Biochemical and electron microscopic data indicate that the DNA networks are connected through hemicatenane linkages. The hemicatenane formation likely is mediated by the single-strand passage through denatured bubbles in the substrate DNA under high temperature. NeqTop3 at lower concentrations can reverse hemicatenanes. A complex of human topoisomerase 3α, Bloom helicase, and RecQ-mediated genome instability protein 1 and 2 can partially disentangle the hemicatenane network. Both the formation and dissolution of hemicatenanes by type IA topoisomerases demonstrate that these enzymes have an important role in regulating intermediates from replication, recombination, and repair.
Type IA DNA topoisomerases are ubiquitous and highly conserved enzymes that have essential functions for life. They can efficiently alleviate DNA topological stress arising from unwinding and rewinding DNA during the processes of replication, transcription, recombination, repair, and chromatin remodeling (1–4). These enzymes accomplish the feat of topological transformation via strand passage through a reversible, enzyme-bridged, single-strand break. The transient break is generated by a reversible transesterification reaction: The active-site tyrosine serves as a nucleophile to create phosphodiester linkage to the nascent 5′-phosphoryl end; the 3′ end is bound noncovalently to the enzyme, and its reversal can reseal the break. This unique mechanism of strand passage through a tethered DNA gate allows type IA enzymes to carry out reactions other than solely serving as a swivel to relieve topological stress. The coordinated action of type IA enzymes with DNA helicases adds further versatility as a tool in regulating the production and resolution of intermediates generated during recombination and repair (5).
Type IA enzymes are present in all three domains of life, and they frequently exist in multiple forms in individual organisms. In eubacteria, there are two closely related isozymes of topoisomerase I (Top1) and III (Top3). Archaea have Top1 and/or Top3, and the hyperthermophiles have an additional type IA enzyme, reverse gyrase, which can help protect their genomes against duplex denaturation caused by the hostile temperatures where they inhabit. Multicellular eukaryotes have two structurally similar but functionally distinct isozymes, Top3α and Top3β. Top3α is essential for viability; mice and Drosophila with Top3α mutations die during early developmental stages, but animals with Top3β mutations can survive to adulthood (6). Interestingly Top3α has essential functions in two separate cellular compartments, mitochondria and nucleus. Besides a requisite function in the nucleus, Top3α also has a role in the maintenance of mitochondrial DNA and its functions (7).
Both genetic and biochemical experiments demonstrate that bacterial Top1 and Top3 have overlapping but distinct functions. Top1 along with gyrase plays an important role in the homeostasis of DNA supercoiling, with Top1 or gyrase removing either excessive negative or positive supercoiling, respectively (8). For example, plasmid DNA in bacteria with mutations inactivating Top1 is hypernegatively supercoiled (9), whereas the converse is true for bacteria with mutations inactivating gyrase (10). Because Top3 remains active in Top1-mutant cells, this result also suggests that Top1 is more efficient than Top3 in removing negative supercoiling, a result that is consistent with their in vitro biochemical properties (11). Although neither Top1 nor Top3 is required for viability in bacteria, the combined mutations in these isozymes are lethal (12, 13). However, the lethality of the double knockout can be rescued with a mutation in RecA, a gene with central importance in the homologous recombination pathway (12). Thus, Top1 or Top3 can resolve the intermediates generated during genetic recombination, failure of which can lead to cell death. The molecular nature of these cytotoxic recombination intermediates remains to be elucidated.
In addition to its overlapping functions with Top1, Top3 appears to have a unique biochemical role in chromosome segregation. Top3 has been shown to separate the late-stage replication intermediates such as catenated sister plasmid DNA with single-stranded gaps (14). In conjunction with RecQ helicase and single-strand binding proteins, Top3 also can decatenate theta-type replicating intermediates (15), and Top3/RecQ can catenate plasmid DNA (16). These diverse reactions are mechanistically related to the double Holliday junction dissolution reaction mediated by eukaryotic Top3 (or Top3α) and RecQ helicase. The convergent branch migration of the Holliday junctions will lead to the formation of a hemicatenane or a joint molecule with a limited number of single-strand, catenated interlocks, and additional single-strand passage reactions by Top3 can lead to the eventual separation of these topologically linked molecules (17).
We are interested in characterizing DNA topoisomerases from hyperthermophilic archaea, especially those from a parasitic archaeum, Nanoarchaeum equitans. It has a minimalistic genome of 0.49 Mb and encodes only the absolutely indispensable molecules for life (18, 19). The Nanoarchaeum genome codes for two type IA enzymes, reverse gyrase (20) and Top3, both of which are from split genes. Here we show that recombinant Top3 from coexpression of the split genes has the expected type IA enzymatic activities. We further show that it has a unique activity to generate and dissolve hemicatenane network structures. Because hemicatenane structures are potential intermediates from replication, recombination, and repair processes, the creation and characterization of hemicatenanes can be useful for providing important mechanistic insight into DNA transactions by type IA enzymes.
Results
Characterization of the Split Topoisomerase III in N. equitans.
Type IA topoisomerases, except for reverse gyrase, are present mostly as one single polypeptide chain. In N. equitans, however, this gene splits into two pieces, NEQ045 and NEQ324, according to its genomic sequence (18, 19), resulting in a hypothetical heterodimeric type IA topoisomerase, a structure which has not been reported before. Sequence alignment showed that NEQ045 corresponds to the N-terminal segment of Escherichia coli Top3 with 39% similarity (25% identity), and NEQ324 corresponds to the E. coli C-terminal segment with 32% similarity (18% identity) (Fig. S1). Domains I and III [topoisomerase-primase (TOPRIM) and winged helix domain (WHD) domains, respectively] of type IA core are encompassed in NEQ045, whereas domain IV [catabolic gene activator protein (CAP)-like] is in NEQ324. Interestingly, domain II (Hinge) is present in both subunits where the split site is located at the apex of the hinge, near the proposed break-point for gate opening (21). The residues essential for type IA enzyme activity, such as the catalytic tyrosine and the DxD TOPRIM motif, are conserved in NEQ045. Because the sequence homology is closer to that of Top3 and their biochemical activities are similar (as discussed in the following sections), we named the protein NeqTop3.
We constructed a vector for expressing NEQ045 and NEQ324 in E. coli and purified the heterodimeric protein to homogeneity (Materials and Methods and Fig. S2). NEQ045 and NEQ324 associate tightly throughout the purification steps, including gel filtration, indicating that they form a stable heterodimer.
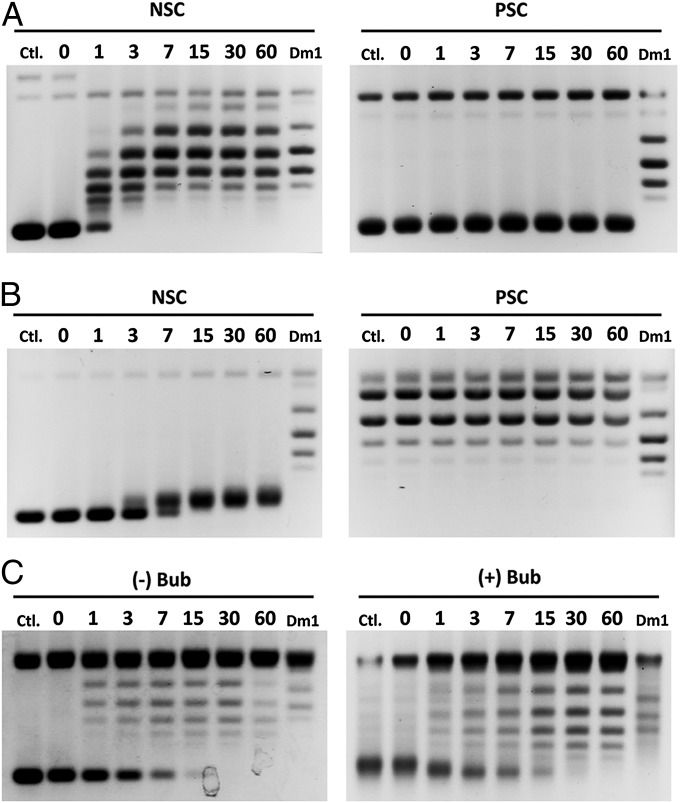
To test if the purified protein has the expected topoisomerase activities, we carried out supercoil relaxation assays. Type IA topoisomerase requires a single-stranded region for strand passage reactions, and thus it prefers relaxing negatively rather than positively supercoiled DNA. Unlike Drosophila topoisomerase IB, which relaxes positively and negatively supercoiled DNA with equal efficiency, NeqTop3 selectively relaxes the negatively supercoiled DNA with a reaction time of less than 7 min, whereas no relaxation for positively supercoiled DNA occurs after 60 min (Fig. 1A). The topology of DNA was confirmed further by analyzing the DNA products with gel electrophoresis in the presence of netropsin, which overtwists the DNA, thus reducing the positive supercoils or increasing the negative ones (Fig. 1B).
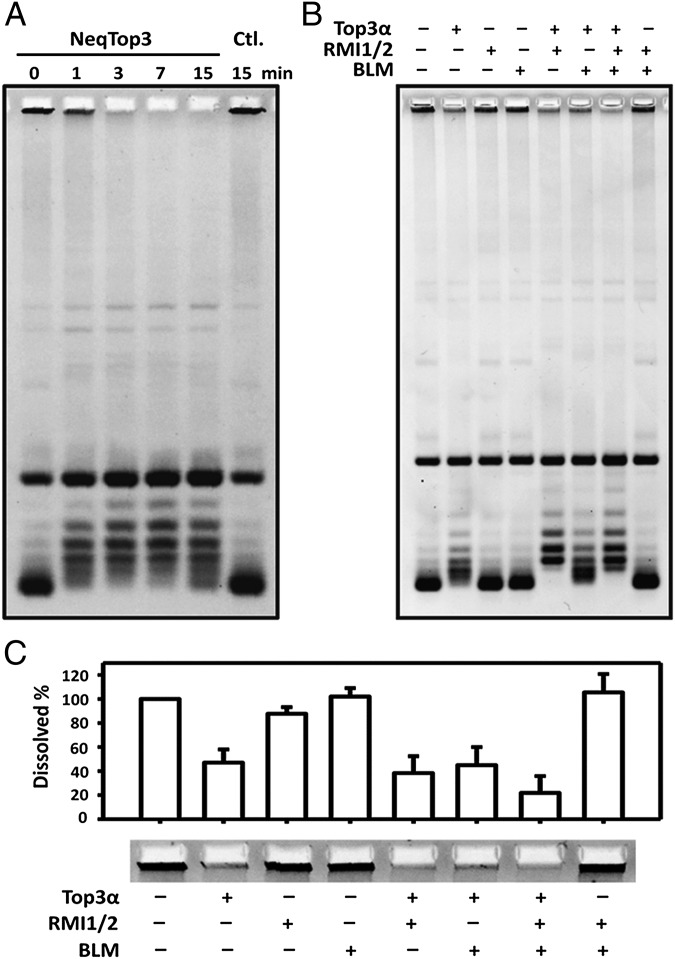
Fig. 1.
Type IA topoisomerase activity of NeqTop3. The relaxation assays were carried out with different DNA substrates. The reaction times (in min) are indicated on the top of each lane. (A) The preference for negatively supercoiled DNA over positively supercoiled DNA is shown by the relaxation reaction with NeqTop3. NeqTop3 (16 nM) completely relaxes negative supercoils (NSC) in 7 min but not positive ones (PSC). DmTop1 (Dm1) served as a control with which both negative and positive supercoils were relaxed. (B) The topology of DNA substrates was confirmed by netropsin gels (containing 5 μg/mL of netropsin). The reaction products shown in A were analyzed here to distinguish the negatively vs. positively supercoiled DNA and to demonstrate that the positively supercoiled substrate was refractory to relaxation by NeqTop3. (C) With DNA containing a single-stranded bubble, both positive and negative supercoils (+Bub or −Bub) can be relaxed.
We also used “bubbled” DNA substrates which contain a persistent single-stranded region to test the relaxation activity (22). NeqTop3 relaxes bubbled substrates in both positively and negatively supercoiled configurations (Fig. 1C); this result agrees with the notion that type IA topoisomerases can act on DNA regardless of its superhelical handedness as long as it harbors a single-stranded region.
Analysis of the Cleavage Sequence of NeqTop3.
Recent work showed that three highly conserved arginine and aromatic residues in domain I (R169, R173, and Y177 in E. coli Top1) can account for DNA cleavage site selection of Top1, known as “−4C specificity,” the fourth residue 3′ to the cleavage being a cytosine (23). These residues are absent in both eukaryotic and prokaryotic Top3. NEQ045 lacks these residues, and sequence comparison shows that it is more similar to prokaryotic Top3 than to Top1 (Fig. S3). To ascertain whether NeqTop3 has a DNA cleavage sequence specificity like that of Top1 and to examine the preferred binding site of NeqTop3, we determined its cleavage site sequences by primer extension using the NeqTop3-cleaved pUC19 as the DNA template. The cleavage hot spots are located mostly in AT-rich segments, and nucleotide sequence analysis shows that the −4C specificity is not observed (Fig. S4). This result indicates that NeqTop3 is more similar to prokaryotic Top3. Because AT-rich sequences are prone to denature at high temperature, it also demonstrates that this hyperthermophilic type IA topoisomerase preferentially interacts with a partially denatured single-stranded region when it is under the stress of negative supercoiling.
NeqTop3 Promotes DNA Network Formation.
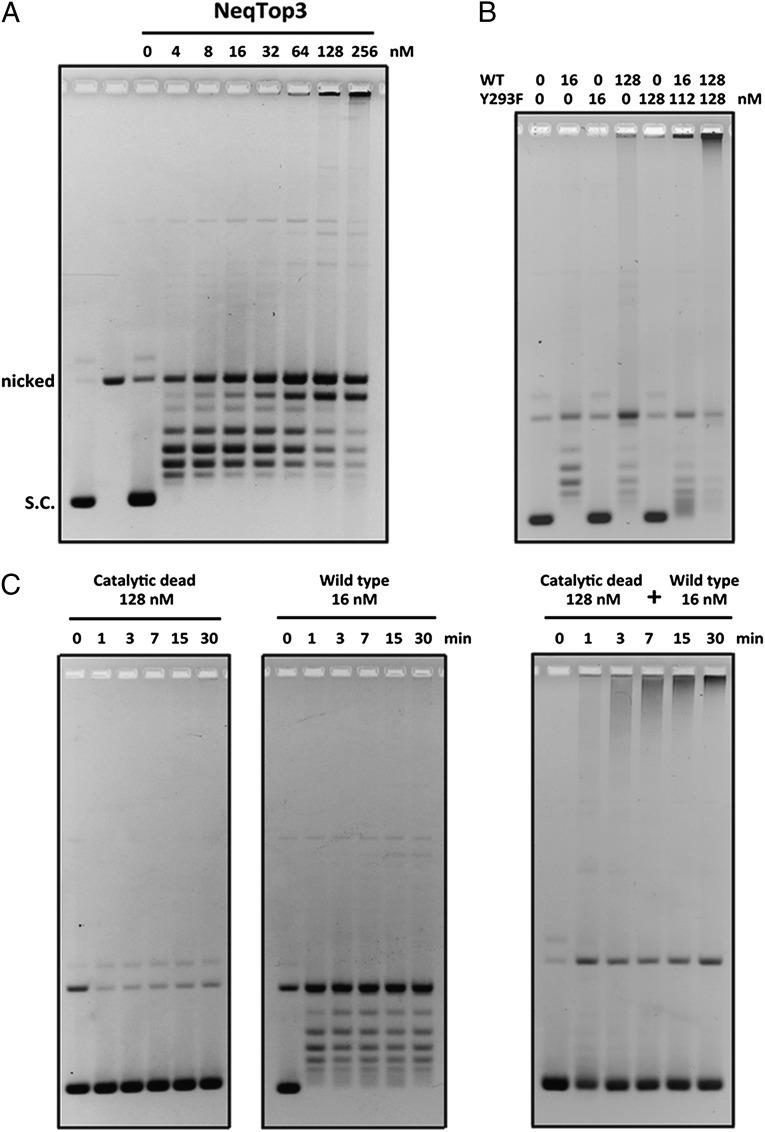
Although NeqTop3 is efficient in relaxing plasmid DNA at low concentrations, it generates an aggregated network product with limiting mobility in agarose gel electrophoresis when the enzyme is present at high concentrations (Fig. 2A). The requirement for an excess of enzyme could be caused by the need for the enzyme to serve a structural role in DNA condensation in addition to its function in carrying out intermolecular strand passage reaction. To test this hypothesis, we engineered and purified an active site Y293F mutant and showed that it can substitute for the wild-type protein in promoting network formation (Fig. 2B). NeqTop3-mediated network production is both concentration and time dependent. At a lower concentration of NeqTop3 (16 nM), DNA supercoils were relaxed quickly but networks were not produced even after prolonged incubation (Fig. 2C). With a high concentration of Y293F mutant protein neither supercoil relaxation nor network formation was observed. Interestingly, with both mutant and wild-type enzyme present, networks were produced in a time-dependent manner, suggesting that the network formation requires an excessive amount of NeqTop3 acting to condense DNA.
Fig. 2.
NeqTop3 induces the formation of a DNA network in a concentration-dependent manner. (A) Titration of NeqTop3 concentrations in relaxation assays. DNA networks failing to enter the gel appeared when high levels of NeqTop3 (≥64 nM) were present. (B) A low level of NeqTop3 (16 nM) also induced the formation of DNA networks when a high level of catalytic-dead NeqTop3 (128 nM) was included in the reaction. All the reactions were carried out at 80 °C for 30 min. (C) Time-course experiments of DNA network formation. A low concentration (16 nM) of active NeqTop3 showed only relaxation activity (Center), and a high concentration (128 nM) of catalytic-dead NeqTop3 had no effect on plasmid DNA (Left). When the two conditions were combined, aggregated networks could form at the 1-min time point and increased gradually with longer incubations (Right). DNA concentrations in all assays were 2.8 nM.
There are precedents showing that DNA topoisomerases can generate DNA networks. Aggregated DNA catenanes were observed in a reaction with an excess of either DNA Top2 or Top1 (24–26) and with Top3 assisted by a RecQ DNA helicase (16). However, the formation of catenated multimers mediated by Top1 requires the presence of a preexisting nick in the DNA substrate (25). Because the network formation with NeqTop3 was done with covalently closed plasmid DNA in the absence of any helicase, it is unclear if the DNA aggregates were made of catenanes.
Hemicatenane Structure of DNA Networks.
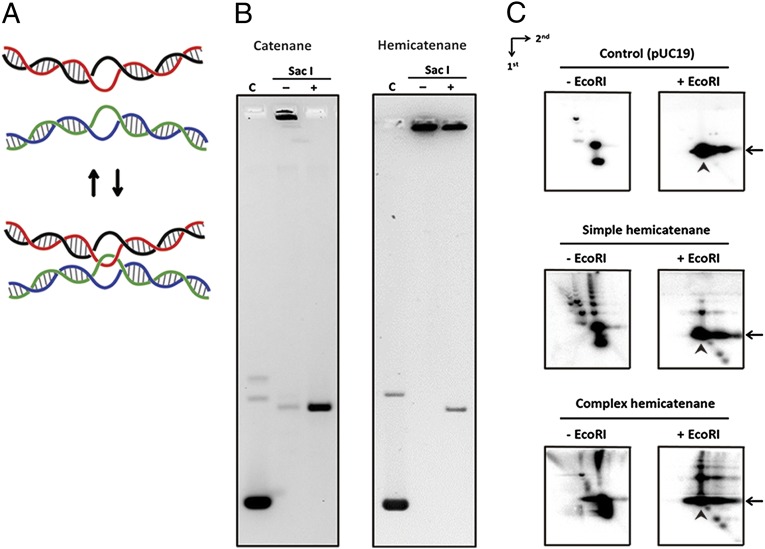
Because type IA topoisomerase mediates strand passage through a single-stranded region, if strand passage occurs between two bubbled regions in plasmid DNA, it can produce DNA networks containing the hemicatenane linkage (Fig. 3A). A simple diagnostic test for catenane vs. hemicatenane DNA linkage can be performed by linearizing DNA with a restriction enzyme: A catenane network can be unraveled completely by linearization, but a hemicatenane cannot be untangled readily. The aggregated network made by NeqTop3 can be converted to linear DNA only partially after restriction digestion, whereas in the control experiment the catenane network is converted efficiently to a linear form (Fig. 3B). We carried out similar experiments using network DNA products generated under less efficient conditions, so that only a smaller fraction of DNA was turned into high-molecular-weight species (as shown in Fig. 2A). To enhance the sensitivity of the detection, we used Southern blot analysis and probed with radio-labeled DNA. We obtained similar results under these conditions, showing that simpler hemicatenated networks cannot be unraveled efficiently by restriction digestion (Fig. S5). Interestingly, we can detect intermediate species with retarded mobilities compared with substrate DNA. We used 2D gel electrophoresis to confirm that neither the simpler nor the more complex network was joined through covalent linkage (Fig. 3C). In 2D gel analysis, the first dimensional run was under native conditions to resolve DNA according to size, and the second dimensional run was under alkaline conditions to disentangle the DNA not covalently joined. It is clear from these results that both simple and complex networks can be mostly converted to linear species after restriction digestion and under denaturing conditions, as is consistent with notion that they are hemicatenated species.
Fig. 3.
The hemicatenane structure of DNA networks. (A) A schematic model for generating and reversing hemicatenane linkage through single-strand passage between two DNA bubbles. (B) Drosophila topoisomerase II (DmTop2)-induced catenane was resolved by restriction enzyme treatment, indicating a topology of interlocked circular DNAs. The NeqTop3-induced network remained aggregated after restriction digestion, suggesting a distinct topology compared with catenanes. (C) In 2D neutral-alkaline agarose gels, aggregated networks without restriction digestion appeared either as a ladder of simple hemicatenanes or failed to enter the gel in complex hemicatenanes. After the linearization treatment by restriction digestion, most of the simple or complex hemicatenanes were converted to linear fragments after denaturation, confirming that the network formation did not involve covalent joining of DNA. Arrows indicate the position of linear monomer in the neutral dimension, and arrowheads indicate linear monomers under denaturing conditions. DNA fragments migrating faster than linear monomers were generated by the DNA cleavage activity from an excess of NeqTop3 present in the reaction mixtures.
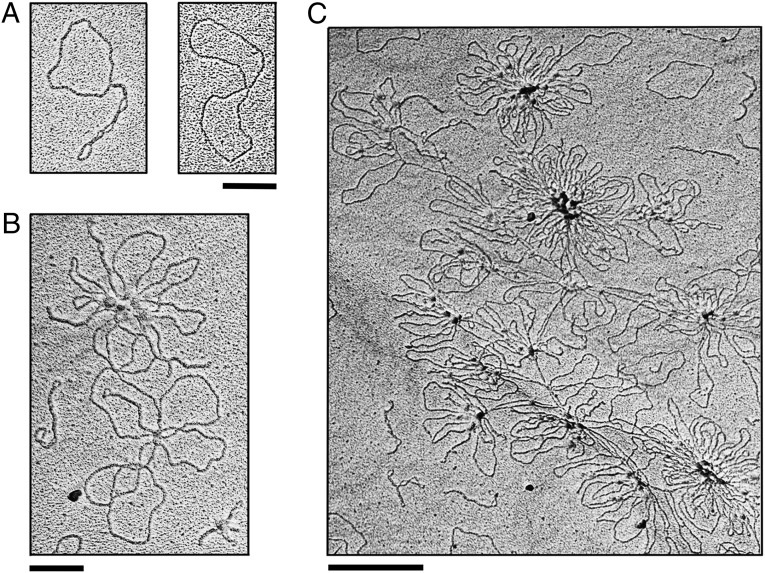
Two additional experiments were carried out to confirm that the networks are hemicatenated. The proposed hemicatenane can be considered as the simplest double Holliday junction with two DNA helices conjoined by a single topological linkage. Because the Holliday junction is sensitive to cleavage by a resolvase such as T7 endonuclease I (27), one would expect that NeqTop3-generated networks can be resolved by T7 endonuclease I digestion, and indeed this expectation is borne out by the experiments (Fig. S6). We also show that the resolution of hemicatenated networks is more efficient than the other two activities by T7 endonuclease I, linearization of nicked DNA and nicking/linearization of supercoiled DNA (Fig. S6). As a more direct way to monitor the formation of hemicatenated networks, we visualized hemicatenane species with different degrees of complexity by electron microscopy. At various time points after the reaction course and after reaction products had been purified of proteins, we observed the formation of dimer, multimer, and aggregated hemicatenanes with progressive complexity of hemicatenane linkage (Fig. 4).
Fig. 4.
Electron microscopy images of hemicatenane. (A) Plasmid DNAs form simple interlocked species in the NeqTop3 reaction. pUC19 DNA was incubated with active NeqTop3 (16 nM) and catalytic-dead NeqTop3 (128 nM) at 80 °C for 90 s. (B and C) A moderately complex network (B) and a large and extensively interlocked network (C), both from a 7-min reaction. All samples were purified free of proteins before electron microscopy analysis. (Scale bars: 200 nm in A and B; 500 nm in C.).
Reversal of Hemicatenation with NeqTop3.
The formation of hemicatenanes with high concentrations of NeqTop3 presumably results from the enzyme’s dual role in both bringing together DNA into a smaller hydrodynamic volume and mediating strand passage through the single-stranded region. This proposal also suggests that at a lower concentration of the enzyme the entropic factor would drive the reversal of hemicatenation (Fig. 3A). We tested this hypothesis by incubating the hemicatenane networks with a lower level of NeqTop3, and the reaction products were assayed at various time points. Under such conditions, NeqTop3 indeed can undo the network and regenerate the monomeric plasmid DNA (Fig. 5A). Therefore, the strand-passage activity of NeqTop3 can allow the interconversion between monomeric circles and hemicatenane networks depending on the presence or absence of DNA condensation.
Fig. 5.
NeqTop3 and human Top3α can reverse hemicatenation. (A) Hemicatenanes can be reversed rapidly by a low level of NeqTop3 (8 nM) treatment. The reversal is complete in 3 min, whereas the no-enzyme control (Ctl., rightmost lane) shows no reversal. (B) Top3α can partially dissolve the hemicatenane (lane 2). Rmi1/Rmi2 stimulates the activity of Top3α, both in relaxation and dissolution. The reactions containing Blm, Top3α, and Rmi1/Rmi2 showed the highest dissolution activity, with almost 80% of hemicatenane being dissolved. (C) Quantification of the hemicatenane DNA from experiments similar to B (n = 3).
The reversal of hemicatenation as shown here was carried out with a less complex hemicatenane structure. We also noticed that when such a reaction was performed with a highly complex, aggregated network, the reversal efficiency was greatly reduced. It is possible that multiple interlocked linkages in the complex hemicatenanes may exclude enzymes from efficiently untying the hemicatenane linkage.
The experiments presented here demonstrate that a type IA enzyme such as NeqTop3 can mediate the equilibrium of a hemicatenation reaction. However, it is unclear whether this reaction is uniquely associated with a special function of NeqTop3. Therefore we examined this question with another type IA enzyme, Top1 from Thermotoga maritima, and demonstrated that TmaTop1 can mediate both hemicatenation and its reversal, depending on the presence of DNA-condensing agents (Fig. S7). This result thus suggests that hemicatenation is a unique reaction mediated by a hyperthermophilic type IA enzyme.
Hemicatenane Can Be Dissolved by Human Dissolvasome.
DNA double-strand breaks can be repaired through a homologous recombination pathway in which the double Holliday junction is a key intermediate (28, 29). An important way to process a double Holliday junction is through convergent branch migration mediated through a coordinated action of Bloom syndrome helicase and Top3α, which results in exclusively noncrossover products (30, 31). The hemicatenane structure can be considered as a double Holliday junction with the least topological linkage before its final dissolution. Therefore we expect hemicatenane networks to be dissolved by the combined activities of Blm helicase and Top3α. Because human Blm, Top3α, and RecQ-mediated genome instability protein 1 and 2 (Rmi1/Rmi2) form a complex, the dissolvasome (32, 33), we prepared a complex of these four purified proteins and tested whether they can dissolve hemicatenane networks (Fig. 5 B and C). Top3α alone is capable of dissolving the network, and the presence of Blm/Rmi1/Rmi2 further stimulates this activity. Because only a single or a limited number of topological linkages connect two helices in a hemicatenane, it is reasonable to expect that the strand-passage activity of Top3α is responsible for the dissolution step, and the presence of Rmi1/Rmi2 or Rmi1/Rmi2 and Blm can stimulate this activity further in situations similar to previous observations regarding the relaxation activity of Top3α (34, 35).This result adds additional evidence that the network is conjoined through hemicatenane linkages and that it can be dissolved by a type IA enzyme from either a prokaryotic or eukaryotic source.
Discussion
In this study we demonstrated a reaction, DNA hemicatenane network formation, mediated by a type IA topoisomerase. The hemicatenated network is distinct from the catenated one in that it cannot be untangled readily by linearizing DNA circles. However, after linearization by restriction digestion, the hemicatenane network can be untangled under denaturing conditions, demonstrating that there is no covalent joining in the hemicatenane formation. We also have used both electron microscopy and resolution reaction by T7 endonuclease I treatment to confirm the hemicatenane structure. The mechanism of this efficient hemicatenation reaction by a hyperthermophilic type IA enzyme, such as NeqTop3, is presumably the result of strand passage through the single-stranded bubbles created under high temperature. Mapping of the DNA cleavage sites showed that they are located in regions with high AT content, suggesting that NeqTop3 acts on the regions that are prone to denaturation under high temperature. Because NeqTop3 can generate nicks in these AT-rich regions, it is possible that the fraying of single-strand breaks at high temperature also can enhance the single-strand character of DNA at NeqTop3 binding sites and facilitate the hemicatenation or catenation reaction.
Although both hemicatenation and catenation can produce aggregated DNA networks, the use of restriction digestion to linearize the plasmid DNA can serve as a diagnostic test to differentiate these DNA structures. The data presented here demonstrated that, although most of the DNA after linearization remained as interlocked networks, a fraction of DNA appeared as freed linear products. It is possible that NeqTop3 can mediate some extent of catenation. It is interesting that another mode of catenation, a double-stranded segment sandwiched between two complementary single strands, can be freed after restriction digestion. However, the hemicatenane networks are not entirely insensitive to liberation after linearization, because concerted unwinding and rewinding in the front and in the wake of the interlocked joint potentially can untangle the linkage, especially if the hemicatenation site is proximal to the linear ends. Despite this uncertainty in measuring the exact extent of catenation vs. hemicatenation, the results presented here demonstrated that most of the DNA networks produced by NeqTop3 are produced through the hemicatenated linkage (Fig. 2B). The hemicatenation and catenation reactions are related by the number of single-strand DNA passage events at a given site. It is unclear what controls the strand passage to stop at the hemicatenane stage because further strand passage reactions could lead to the formation of catenane. In some situations a type IA enzyme can mediate catenane formation of fully double-stranded DNA; bacterial type IA enzyme can mediate the formation of interlocked rings among plasmid DNA containing at least a single-strand break in one of the partner DNAs (25). With either a preexisting single-stranded bubble or a coordinated unwinding action by a yeast RecQ helicase, Sgs1, Top3 can catenate intact plasmid DNA (36). A mechanistically similar reaction is observed with bacterial RecQ helicase and Top3 (37). Four consecutive and coordinated strand-passage events are required to generate one catenane interlock, but only one single-strand passage is needed to create a hemicatenane linkage. The mechanism by which the extent of strand passage during each reaction cycle is regulated remains to be elucidated.
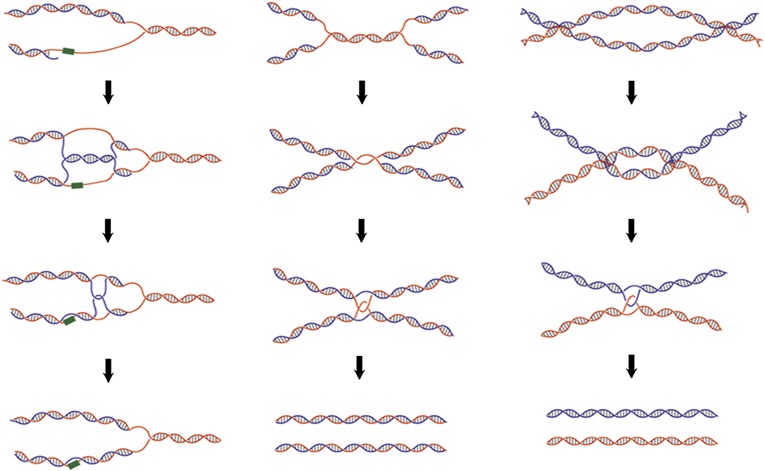
Hemicatenane DNA previously was synthesized with a pair of oligonucleotides containing dinucleotide repeats (38). In the process of annealing the oligonucleotide pairs, slippage in base pairing in the dinucleotide repeats allows the formation of hemicatenane loops in a fraction of the annealed products. The approach described in this paper provides a simple system for investigating the mechanistic insights into both the generation and dissolution of the hemicatenane. We can modulate the complexity of hemicatenanes readily by varying the amounts of topoisomerase, DNA condensation agents, and incubation time (Fig. 2). Hemicatenanes are an important intermediate in replication, repair, and recombination processes and must be resolved to maintain genome stability. Some of the proposed pathways that may lead to DNA structures related to hemicatenanes are shown in Fig. 6. Replication fork stalling caused by DNA damage can generate structures of “pseudo-double Holliday junction,” the processing of which can result in hemicatenane linkage between the replicated DNA strands (39). At a late stage of DNA replication, a theta-type late-replication intermediate can form in which two replicated DNA rings are linked by single-strand catenane in the unreplicated parental duplex (40). The maturation of this replication intermediate can lead to the formation of catenanes with single-strand breaks, which then can be processed by a type IA enzyme to generate hemicatenanes. Similar structures of late replication, presegregation sister chromatids with hemicatenane linkage, have been proposed also to exist in the archaeal chromosomes (41) and in the products from in vitro DNA replication of plasmid DNA using Xenopus egg extracts (42). Finally, as described in the previous section, the convergent branch migration of the double Holliday junction leads to the formation of a hemicatenane before the eventual dissolution of the conjoined DNA helices (17). In this case, only a single topological linkage is responsible for connecting the pair of DNA duplexes. The relative rate in the dissolution of the hemicatenane compared with the overall branch migration and the regulation in this final dissolution step may play a critical role in the pairing of sister chromatids and in the processing of recombination/repair intermediates.
Fig. 6.
Schematic models for the generation of hemicatenane structures during DNA transactions. Three examples of pathways to yield hemicatenane structures are shown here. (Left) Lagging strand synthesis encounters a damage site, and a pseudo-double Holliday structure can be produced by the pairing of the lagging strand with the complementary leading strand. The dissolution of the pseudo-double Holliday structure leads to hemicatenanes and allows replication to bypass the damage site. (Center) Convergence of two replication forks at the final stage of replication can lead to either single-strand catenane or hemicatenane conjoining the two replicated duplexes. Both single-strand catenane and hemicatenane can be resolved by a type IA topoisomerase, allowing the segregation of the daughter chromosomes. (Right) Convergent branch migration of a double Holliday junction can result in the formation of a hemicatenane just before the final separation of two helices undergoing recombination.
The relevance of the hemicatenated network generated by NeqTop3 to the double Holliday junction structure also is highlighted by the ability of the human dissolvasome, a protein complex of Top3α/Blm/Rmi1/Rmi2, to disentangle the network structure. The major enzymatic activity responsible for the dissolution activity is from the strand-passage action of Top3α, whereas Blm and Rmi1/Rmi2 play a role in facilitating the reaction, presumably because the hemicatenane structure as synthesized by NeqTop3 contains a single or a limited number of topological linkages. Therefore, the unwinding activity from the Blm helicase is not as important as it is in the dissolution of double Holliday junction structures containing a pair of duplexes with extensive base-paired regions that require considerable branch migration to bring together the Holliday junctions. The exact roles of Blm and Rmi1/Rmi2 in the disentanglement of hemicatenated networks now can be examined with the substrate described here.
Materials and Methods
Sequence Alignment.
Sequence alignment was performed using the AlignX function from Vector NTI Advance 11 (Life Technologies, Invitrogen).
Expression and Purification of N. equitans Split Top3.
The genes NEQ045 and NEQ324 were synthesized, and the sequence was optimized with the codon used for E. coli; a hexahistidine tag was introduced at the 3′ end of NEQ324. These genes were subcloned into the pETDuet-1 vector (Novagen) for coexpression.
The NeqTop3 protein was expressed in Rosetta (DE3, pLysS) cells (Novagen) by induction with 0.5 mM IPTG for 20 h at 20 °C. The cells were harvested and resuspended in lysis buffer consisting of 20 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, 1 mM PMSF, 5 mM 2-mercaptoethanol, and protease inhibitor mixture (Complete, EDTA-free tablets; Roche). After cell lysis by sonication, the cell debris was removed by centrifugation (20,000 × g, 30 min), and the supernatant of cell lysate (fraction I) was fractionated with four chromatographic steps (GE Healthcare). The first step was a HisTrap FF crude Nickel column; after loading, the column was washed successively with a lysis buffer containing 20 mM and then 75 mM imidazole. NeqTop3 eluted at the 250-mM imidazole step (fraction II) was purified further through the HiTrap Heparin HP affinity column. The pooled peak fractions (fraction III) were diluted two times with 20 mM Tris⋅HCl (pH 8.0) and adsorbed to Mono S 5/50 GL column, and the peak eluted fractions were pooled as fraction IV. The final step was gel filtration with HiLoad 16/60 Superdex 200, and fractions from a single homogeneous peak were pooled, dialyzed overnight in 50% (vol/vol) glycerol, 20 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, 1 mM PMSF, and 5 mM 2-mercaptoethanol, and stored at −20 °C (fraction V).
Fraction V NeqTop3 was more than 98% pure as judged by PAGE, and a purification table was constructed for all five purification steps (Fig. S3). The final specific activity of fraction V was 3.9 × 104 U/mg with one unit defined as the amount of enzyme required to relax 500 ng of negative-supercoiled pUC19 in a reaction volume of 30 μL in 30 min at 80 °C.
Enzymes.
Recombinant Drosophila Top1 (43) and Top2 (44) were purified following published procedures. Purification of Archaeoglobus fulgidus reverse gyrase followed the procedure of Rodriguez and Stock (45). Top1 from T. maritima was purified as described in ref. 46. All restriction endonucleases and T7 endonuclease I were from New England BioLabs.
DNA Substrates.
All DNA substrates except catenane/hemicatenane networks were purified by CsCl-ethidium double-banding ultracentrifugation. Positive- and negative-supercoiled bubbled DNA substrates were prepared as previously described (22). Nicked pUC19 DNA was made using DNase I with a saturated amount of ethidium bromide to generate single-nicked plasmid (47). DNA catenanes were prepared by published procedures (24). Reactions for preparing hemicatenane contained 22.5 μg/mL (12.7 nM) pUC19 DNA, 10 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 50 μg/mL gelatin, 16 nM active NeqTop3, and 128 nM catalytic-dead NeqTop3. Reactions were performed at 80 °C for 90 s and were stopped by 15 mM EDTA, 0.3% SDS, and 0.75 mg/mL proteinase K, incubated at 45 °C for 30 min. The DNA was purified by phenol-chloroform extraction and alcohol precipitation.
Relaxation Assays.
Reactions contained 500 ng (2.8 nM) of DNA substrate, 10 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 50 μg/mL gelatin, and an appropriate amount of NeqTop3 in a total volume of 30 μL, with a drop of mineral oil layered on top. The reactions were performed at 80 °C for 30 min, stopped by 15 mM EDTA, 0.3% SDS, and 0.75 mg/mL proteinase K, further incubated at 45 °C for 30 min, and analyzed by agarose gel electrophoresis.
Hemicatenane Dissolution Assays.
For dehemicatenation by NeqTop3, reactions contained 500 ng of hemicatenane substrate, 10 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 50 μg/mL gelatin, and 8 nM of NeqTop3 in a total volume of 20 μL. Reactions were carried out in 80 °C and stopped by adding a stop solution to 15 mM EDTA, 0.3% SDS, and 0.75 mg/mL proteinase K.
For hemicatenane dissolution by human enzymes, the reactions contained 40 mM Hepes⋅KOH (pH 7.5), 1 mM MgCl2, 50 μg/mL BSA, 1 mM DTT, and an appropriate amount of Blm helicase, Top3α, and Rmi1/Rmi2 and with 1 mM ATP in some of the reactions. Reactions were carried out at 37 °C for 1 h and were stopped by 15 mM EDTA, 0.3% SDS, and 0.75 mg/mL proteinase K. For the analysis of hemicatenane by 2D agarose gel electrophoresis, samples first were run in 1× Tris-phosphate electrophoresis buffer, 1 V/cm for 12 h at room temperature, and then were presoaked in alkaline buffer (30 mM NaOH, 1 mM EDTA) and run in 4 V/cm for 3 h at 4 °C under alkaline conditions. To enhance the minor signals, the DNA was transferred to a nitrocellulose membrane and detected by Southern blot.
T7 Endonuclease I Sensitivity Assays.
The T7 endonuclease I sensitivity assay was done in a 30-μl reaction containing 500 ng DNA substrates plus an appropriate amount of T7 endonuclease I (New England BioLabs) following the manufacturer’s recommendation.
Electron Microscopy.
The hemicatenane DNA product was purified to be free of proteins and was suspended in 10 mM Tris⋅HCl (pH 8.0) and 1 mM EDTA. Purified DNA was directly mounted on parlodion-coated 400-mesh copper grids (EM Sciences) and visualized by electron microscopy as described previously (48).
Supplementary Material
Acknowledgments
We thank Ming-yi Huang and Yushen Qian for technical assistance. This work was supported by National Institutes of Health Grant GM29006 (to T.-S.H.), by National Science Council, Taiwan, and Academia Sinica intramural funding (T.-S.H.), and by National Institutes of Health Grant GM031819 (to. J.D.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304103110/-/DCSupplemental.
References
- 1.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Chen SH, Chan N-L, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem. 2013;82(1):139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 3.Schoeffler AJ, Berger JM. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41(1):41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 5.Plank J, Hsieh TS. Helicase-appended topoisomerases: New insight into the mechanism of directional strand transfer. J Biol Chem. 2009;284(45):30737–30741. doi: 10.1074/jbc.R109.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen SH, Wu J, Hsieh TS. Essential Functions of Topoisomerase IIIα in the Nucleus and Mitochondria. In: Pommier Y, editor. DNA Topoisomerases and Cancer. New York: Springer; 2012. pp. 103–117. [Google Scholar]
- 7.Wu J, Feng L, Hsieh TS. Drosophila topo IIIalpha is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proc Natl Acad Sci USA. 2010;107(14):6228–6233. doi: 10.1073/pnas.1001855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H-Y, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 9.Pruss GJ. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J Mol Biol. 1985;185(1):51–63. doi: 10.1016/0022-2836(85)90182-2. [DOI] [PubMed] [Google Scholar]
- 10.Lockshon D, Morris DR. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res. 1983;11(10):2999–3017. doi: 10.1093/nar/11.10.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGate RJ, Marians KJ. Molecular cloning and DNA sequence analysis of Escherichia coli topB, the gene encoding topoisomerase III. J Biol Chem. 1989;264(30):17924–17930. [PubMed] [Google Scholar]
- 12.Zhu Q, Pongpech P, DiGate RJ. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc Natl Acad Sci USA. 2001;98(17):9766–9771. doi: 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stupina VA, Wang JC. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J Biol Chem. 2005;280(1):355–360. doi: 10.1074/jbc.M411924200. [DOI] [PubMed] [Google Scholar]
- 14.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988;263(26):13366–13373. [PubMed] [Google Scholar]
- 15.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30(6):779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol Cell. 1999;3(5):611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 17.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17(11):1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Giulio M. The split genes of Nanoarchaeum equitans are an ancestral character. Gene. 2008;421(1-2):20–26. doi: 10.1016/j.gene.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Waters E, et al. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA. 2003;100(22):12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capp C, Qian Y, Sage H, Huber H, Hsieh TS. Separate and combined biochemical activities of the subunits of a naturally split reverse gyrase. J Biol Chem. 2010;285(51):39637–39645. doi: 10.1074/jbc.M110.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg H, Lima CD, Mondragón A. Conformational changes in E. coli DNA topoisomerase I. Nat Struct Biol. 1999;6(10):918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 22.Plank JL, Chu SH, Pohlhaus JR, Wilson-Sali T, Hsieh TS. Drosophila melanogaster topoisomerase IIIalpha preferentially relaxes a positively or negatively supercoiled bubble substrate and is essential during development. J Biol Chem. 2005;280(5):3564–3573. doi: 10.1074/jbc.M411337200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Cheng B, Tse-Dinh YC. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc Natl Acad Sci USA. 2011;108(17):6939–6944. doi: 10.1073/pnas.1100300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh T, Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- 25.Tse Y, Wang JC. E. coli and M. luteus DNA topoisomerase I can catalyze catenation of decatenation of double-stranded DNA rings. Cell. 1980;22(1 Pt 1):269–276. doi: 10.1016/0092-8674(80)90174-9. [DOI] [PubMed] [Google Scholar]
- 26.Baldi MI, Benedetti P, Mattoccia E, Tocchini-Valentini GP. In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase. Cell. 1980;20(2):461–467. doi: 10.1016/0092-8674(80)90632-7. [DOI] [PubMed] [Google Scholar]
- 27.Dickie P, McFadden G, Morgan AR. The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem. 1987;262(30):14826–14836. [PubMed] [Google Scholar]
- 28.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 29.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464(7290):937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plank JL, Wu J, Hsieh TS. Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci USA. 2006;103(30):11118–11123. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 32.Raynard S, et al. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J Biol Chem. 2008;283(23):15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22(20):2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J Biol Chem. 2010;285(28):21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Hickson ID. The Bloom’s syndrome helicase stimulates the activity of human topoisomerase IIIalpha. Nucleic Acids Res. 2002;30(22):4823–4829. doi: 10.1093/nar/gkf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cejka P, Plank JL, Dombrowski CC, Kowalczykowski SC. Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: A mechanism for disentangling chromosomes. Mol Cell. 2012;47(6):886–896. doi: 10.1016/j.molcel.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harmon FG, Kowalczykowski SC. Biochemical characterization of the DNA helicase activity of the escherichia coli RecQ helicase. J Biol Chem. 2001;276(1):232–243. doi: 10.1074/jbc.M006555200. [DOI] [PubMed] [Google Scholar]
- 38.Gaillard C, Strauss F. DNA loops and semicatenated DNA junctions. BMC Biochem. 2000;1:1. doi: 10.1186/1471-2091-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberi G, et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19(3):339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurse P, Levine C, Hassing H, Marians KJ. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J Biol Chem. 2003;278(10):8653–8660. doi: 10.1074/jbc.M211211200. [DOI] [PubMed] [Google Scholar]
- 41.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26(3):816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas I, Hyrien O. Hemicatenanes form upon inhibition of DNA replication. Nucleic Acids Res. 2000;28(10):2187–2193. doi: 10.1093/nar/28.10.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown SD, Zhang CX, Chen AD, Hsieh TS. Structure of the Drosophila DNA topoisomerase I gene and expression of messages with different lengths in the 3′ untranslated region. Gene. 1998;211(2):195–203. doi: 10.1016/s0378-1119(98)00119-x. [DOI] [PubMed] [Google Scholar]
- 44.Hu T, Sage H, Hsieh TS. ATPase domain of eukaryotic DNA topoisomerase II. Inhibition of ATPase activity by the anti-cancer drug bisdioxopiperazine and ATP/ADP-induced dimerization. J Biol Chem. 2002;277(8):5944–5951. doi: 10.1074/jbc.M111394200. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez AC, Stock D. Crystal structure of reverse gyrase: Insights into the positive supercoiling of DNA. EMBO J. 2002;21(3):418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh TS, Capp C. Nucleotide- and stoichiometry-dependent DNA supercoiling by reverse gyrase. J Biol Chem. 2005;280(21):20467–20475. doi: 10.1074/jbc.M502739200. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh TS, Wang JC. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- 48.Griffith JD, Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.