Significance
We have developed a quantitative Förster resonance energy transfer-based methodology using multiphoton fluorescence lifetime imaging microscopy to investigate in single live cells the interaction between the substrate adaptor protein Kelch-like ECH associated protein 1 (Keap1) and transcription factor NF-E2 p45-related factor 2 (Nrf2), the master regulator of the mammalian cell response to oxidants and electrophiles. The application of this methodology revealed that the system is much more dynamic than previously anticipated, whereby Keap1 utilizes a unique cyclic mechanism to target Nrf2 for ubiquitination and proteasomal degradation, which also leads to Keap1 regeneration. The nature of this mechanism allows rapid transcriptional responses to environmental changes and is capable of accommodating multiple modes of regulation.
Keywords: sulforaphane, FRET, FLIM, protein–protein interactions
Abstract
The transcription factor NF-E2 p45-related factor 2 (Nrf2), a master regulator of cytoprotective genes, is controlled by dimeric Kelch-like ECH associated protein 1 (Keap1), a substrate adaptor protein for Cullin3/RING-box protein 1 ubiquitin ligase, which normally targets Nrf2 for ubiquitination and degradation but loses this ability in response to electrophiles and oxidants (inducers). By using recombinant proteins and populations of cells, some of the general features of the regulation of Nrf2 by Keap1 have been outlined. However, how the two proteins interact at a single-cell level is presently unknown. We now report the development of a quantitative Förster resonance energy transfer-based system using multiphoton fluorescence lifetime imaging microscopy and its application for investigating the interaction between Nrf2 and Keap1 in single live cells. By using this approach, we found that under homeostatic conditions, the interaction between Keap1 and Nrf2 follows a cycle in which the complex sequentially adopts two distinct conformations: “open,” in which Nrf2 interacts with a single molecule of Keap1, followed by “closed,” in which Nrf2 binds to both members of the Keap1 dimer. Inducers disrupt this cycle by causing accumulation of the complex in the closed conformation without release of Nrf2. As a consequence, free Keap1 is not regenerated, and newly synthesized Nrf2 is stabilized. On the basis of these findings, we propose a model we have named the “cyclic sequential attachment and regeneration model of Keap1-mediated degradation of Nrf2.” This previously unanticipated dynamism allows rapid transcriptional responses to environmental changes and can accommodate multiple modes of regulation.
The transcription factor NF-E2 p45-related factor 2 (Nrf2) is a master regulator of the expression of large networks of genes that make up more than 1% of the human genome and encode detoxification, antioxidant, and anti-inflammatory cytoprotective proteins (1). Under homeostatic (basal) conditions, monomeric Nrf2 binds to two distinct sites on its major negative regulator, homodimeric Kelch-like ECH-associated protein 1 (Keap1) (Fig. 1A), which forms a RING E3-ubiquitin ligase with Cullin-3/Rbx1 and continuously targets the transcription factor for ubiquitination and proteasomal degradation (2–4). In response to electrophiles and oxidants (termed inducers), which recognize and chemically modify specific cysteine residues of Keap1 (5), ubiquitination of Nrf2 is inhibited; as a consequence, Nrf2 accumulates and activates gene transcription (6, 7). Functionally, the products of Nrf2-dependent genes are extraordinarily versatile and, via mechanisms that include direct antioxidant activity, obligatory 2-electron reduction reactions, conjugation with endogenous ligands, and the recognition, repair, and removal of damaged proteins, these cytoprotective proteins detoxify numerous endogenous and exogenous damaging agents and facilitate their elimination.
Fig. 1.
Cartoon representations and fluorescence lifetime data for Keap1-Nrf2 complexes. (A) Complex of wild-type Nrf2 with Keap1, in which Nrf2 is bound to Keap1 through both its high-affinity ETGE and low-affinity DLG motifs. (B) Complex of Nrf2ΔDLG mutant with Keap1, in which Nrf2 interacts with the Keap1 dimer only through the high-affinity ETGE motif. (C) Complex of Nrf2-doubleETGE mutant with Keap1, in which Nrf2 interacts with Keap1 through two high-affinity ETGE motifs. (D) Fluorescence lifetime data for cells transfected with EGFP-Nrf2 wild-type and mutant fusion proteins.
Dysregulation (either down- or upregulation) of Nrf2 activity is causally associated with susceptibility to chronic degenerative diseases, including cancer, cardiovascular disease, and neurodegenerative conditions, and Nrf2 is now widely recognized as a drug target (8). Indeed, induction of Nrf2-dependent genes is a highly effective strategy for protection against oxidants and carcinogens and has protective effects in numerous animal models of carcinogenesis of the stomach, bladder, lung, breast, prostate, pancreas, liver, colon, and skin; however, persistent activation of Nrf2 is frequently exploited by cancer cells and promotes their survival and resistance to chemotherapy (6). Mutations in KEAP1 or NRF2 that abrogate formation of the complex and lead to Nrf2 accumulation and constitutive activation of the pathway have been detected in several types of human cancer (9, 10). A recent comprehensive genomic characterization study conducted by the Cancer Genome Atlas Research Network has identified the occurrence of mutations in NRF2, KEAP1, or CUL3 in 34% of 178 human lung squamous cell carcinomas (11). Thus, a detailed mechanistic understanding of the intricate regulation of the activity of Nrf2 is critical.
There are more than 600 genes in the human genome that encode RING or RING-like E3 ligases, only a subset of which have been investigated in detail (12). Although studies to date using purified recombinant proteins or populations of cells have given insights into some general features of the regulation of Nrf2 by Keap1, understanding the precise details of how the two proteins interact at a single-cell level is currently lacking. Our aim was to investigate the dynamic interaction between Keap1 and Nrf2 in single live cells in real space and time. This type of analysis offers several advantages over techniques that analyze average cell populations at individual times, including detailed information on rapid and dynamic spatial and temporal changes that can be precisely quantified. By using this approach, we found that the Keap1-dependent E3-ligase uses a unique cyclic mechanism to target its substrate Nrf2 for ubiquitination and proteasomal degradation.
Results
Keap1-mCherry and EGFP-Nrf2 Fusion Proteins Faithfully Represent the Functional Endogenous Proteins.
We first designed constructs encoding fusion proteins of Nrf2 and Keap1 with the donor/acceptor Förster resonance energy transfer (FRET) pair EGFP and mCherry, respectively (Fig. 1A). The single-particle electron microscopy-generated structure of the Keap1 dimer (13) indicated that its C terminus is significantly closer to the Nrf2-binding site than the N terminus. Although there is currently no crystal structure available for Nrf2, it is known to bind to Keap1 through its Neh2 domain at the N terminus (14), suggesting that a fluorescent protein fused here may be in close proximity to the C terminus of Keap1. On the basis of these considerations, we engineered constructs encoding proteins of mCherry fused to the C terminus of Keap1 and of EGFP fused to the N terminus of Nrf2. To overcome the possibility that a fluorescent protein fused to the C terminus of Keap1 may interfere with its function, we added the sequence AEAAAKEAAAKA between the C terminus of Keap1 and mCherry, which forms an α-helix with two twists (15), thus separating the mCherry from Keap1 and allowing both proteins to maintain their function. Correct expression of the fusion proteins was verified by Western blotting after transfection of the plasmids into human embryonic kidney (HEK) 293 cells (Fig. S1A). Critically, addition of the fluorophores did not affect the interaction between Keap1 and Nrf2 (Fig. S1B) or the transcriptional activity of Nrf2 (Fig. S1C), nor did it influence the ability of Keap1 to target Nrf2 for degradation or to respond to inducers or proteasomal inhibition (Fig. S1D). The normal subcellular localization of Nrf2 and Keap1 was also unaffected by the presence of the fluorescent proteins (Fig. S1 E and F). Thus, Keap1-mCherry and EGFP-Nrf2 faithfully replicate the functional endogenous proteins.
In the Basal State, the Keap1-Nrf2 Complex Forms Two Distinct Conformations.
We then transfected the Keap1-mCherry and EGFP-Nrf2 expression vectors into HEK293 cells and looked for a FRET interaction. In control cells, in which Keap1-mCherry is not present, the fluorescence lifetime of EGFP is 2,375 ps (n = 13; Figs. 1D and 2A). When Keap1-mCherry is coexpressed with EGFP-Nrf2, the EGFP mean lifetime is reduced to 2,155 ps (n = 14; P < 0.0001; Figs. 1D and 2B), indicating that when EGFP is fused to the N terminus of Nrf2, and mCherry to the C terminus of Keap1, the fluorophores are sufficiently close to each other and in the correct orientation for FRET to occur. A system in which to study the direct interaction between Keap1 and Nrf2 in real space and time in single live cells was thus established.
Fig. 2.
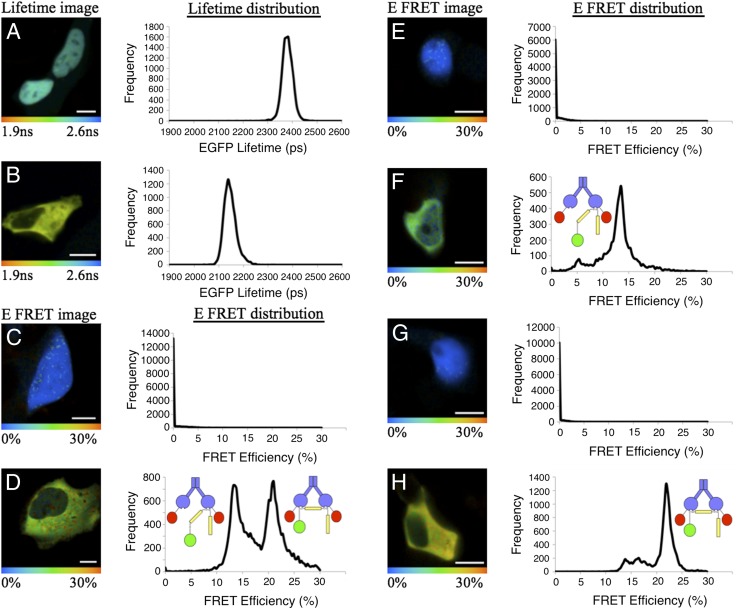
The Keap1-Nrf2 complex exists in two distinct conformations. HEK293 cells were transfected with wild-type EGFP-Nrf2 + mCherry (A and C), EGFP-Nrf2∆DLG + mCherry (E), EGFP-Nrf2-doubleETGE + mCherry (G), EGFP-Nrf2 + Keap1-mCherry (B and D), EGFP-Nrf2∆DLG + Keap1-mCherry (F), or EGFP-Nrf2-doubleETGE + Keap1-mCherry (H) and then imaged 24 h later. (Left) Pictorial representations of the EGFP lifetime (A and B) or the E-FRET (C–H), in which the color of the cell corresponds with the lifetime of EGFP, ranging from 1.9 to 2.6 ns, or to the E-FRET, ranging from 0% to 30%. (Right) Lifetime (A and B) or E-FRET (C–H) data from each pixel of the image, plotted on a graph. The lifetime of EGFP-Nrf2 in the presence of mCherry alone (A) is significantly longer than in the presence of Keap1-mCherry (B), indicating there is a FRET interaction between the two fusion proteins. This lifetime difference is seen in the images, in which A is blue and B is yellow. The E-FRET in wild-type or mutant EGFP-Nrf2 and mCherry cotransfected cells (C, E, and G) is 0, corresponding to the blue color of the cells. In EGFP-Nrf2 and Keap1-mCherry cotransfected cells (D), the E-FRET graph shows two distinct peaks (one centered at 13% and the other at 21%), suggesting there are two different FRET interactions between the EGFP and mCherry fluorophores within the Keap1-Nrf2 complex. These different E-FRET populations are shown pictorially (D, Left), where the green and yellow colors are distributed evenly across the cell. In EGFP-Nrf2∆DLG and Keap1-mCherry cotransfected cells (F), the E-FRET graph shows one peak at 13%, indicating there is a single FRET interaction between the EGFP and mCherry fluorophores within the Keap1-Nrf2∆DLG complex. This E-FRET population is shown pictorially in the image, in which the green color is distributed evenly across the cell. In EGFP-Nrf2-doubleETGE and Keap1-mCherry cotransfected cells (H), the E-FRET graph shows two peaks (a small one at 13% and a larger one at 21%), suggesting there is one major FRET interaction between the EGFP and mCherry fluorophores within the Keap1-Nrf2-doubleETGE complex. The distribution of the two E-FRET populations is shown pictorially in the image, in which the predominant color is yellow (corresponding to the 21% population), along with a small amount of green (corresponding to the 13% population). (Scale bar, 10 μm.)
Fluorescence lifetime changes can be indicative of alterations in the conformation of protein complexes, which in turn can be monitored by quantifying the FRET efficiency (E-FRET) (16). We quantified E-FRET for EGFP-Nrf2 in the presence and absence of Keap1-mCherry. As expected, in the absence of Keap1-mCherry, the value for E-FRET is close to 0 across the cell (n = 13; Fig. 2C). In contrast, in the cytoplasm of EGFP-Nrf2 + Keap1-mCherry cotransfected cells, E-FRET > 0 (n = 14). Surprisingly, the data for E-FRET revealed the presence of two distinct populations of interacting fluorophores; that is, two distinct peaks of E-FRET: one centered at 13%, and the other at 21% (Fig. 2D, graph). This pattern was highly reproducible, and although the relative areas of the peaks varied among cells, both populations were always present, and their distribution was uniform throughout the cytoplasm (Fig. 2D, image). This unexpected finding implied that in live cells, the Keap1-Nrf2 complex exists in two distinct conformations in the basal state and suggested that the system may be much more dynamic than previously anticipated.
Assignment of the Two Conformations of the Keap1-Nrf2 Complex.
Monomeric Nrf2 binds to a Keap1 dimer through two distinct motifs (Fig. 1A): a high-affinity ETGE motif and a low-affinity DLG motif (17, 18). Our first approach in determining the identity of the two E-FRET populations was to use an Nrf2-deletion mutant lacking the DLG motif (Fig. 1B). EGFP-Nrf2∆DLG was able to bind to Keap1-mCherry and generate a FRET signal. In the presence of free mCherry, the lifetime of EGFP-Nrf2∆DLG was 2,382 ps (n = 8), whereas when Keap1-mCherry was expressed in place of mCherry, the mean lifetime was significantly reduced to 2,227 ps (n = 11; P < 0.0001; Fig. 1D and Fig. S2 A and B). Interestingly, in the presence of the EGFP-Nrf2∆DLG mutant (n = 11), the mean fluorescence lifetime decrease is still significant (P < 0.0001) but is less pronounced than with the wild-type EGFP-Nrf2 (2,227 ps vs. 2,155 ps; Fig. 1D), suggesting that this difference in lifetime is correlated to a change in the interaction between Nrf2 and Keap1 in the absence of the DLG motif. Quantification of E-FRET for EGFP-Nrf2∆DLG + Keap1-mCherry (n = 11) revealed that compared with wild-type EGFP-Nrf2 + Keap1-mCherry, the interaction at 13% was maintained, whereas the 21% FRET efficiency population was absent (compare Fig. 2 D and F). Thus, the 13% E-FRET population represents Nrf2 bound to Keap1 through its ETGE motif alone. These data also indicate that the 21% E-FRET population represents Nrf2 bound to Keap1 through its DLG and ETGE motifs simultaneously, as this conformation cannot be formed by the Nrf2∆DLG mutant, and in Nrf2∆DLG-transfected cells, the 21% E-FRET population was not seen. As both the 13% and the 21% FRET efficiency populations are present in wild-type EGFP-Nrf2 + Keap1-mCherry–transfected cells, these data suggest that Keap1 and Nrf2 form two distinct complexes, both of which are present within the same cell at the same time in the basal state.
One caveat to this conclusion is that because the Keap1 dimer contains two mCherry fluorophores, it is possible for the EGFP-Nrf2, which is bound to Keap1 through the DLG and ETGE motifs, to “FRET” with both fluorophores simultaneously. Thus, a single conformation of the Keap1-Nrf2 complex may be able to give rise to two FRET interactions leading to two different E-FRET populations. To test this hypothesis, a second mutant of Nrf2 was used that binds to Keap1 more tightly as a result of the replacement of the DLG motif with a second ETGE motif (Fig. 1C). If the two E-FRET populations come from a single conformation of the Keap1-Nrf2 complex, then the tighter binding of EGFP-Nrf2-doubleETGE will have no effect on the E-FRET when transfected along with Keap1-mCherry. However, if the two E-FRET populations represent two distinct conformations of the Keap1-Nrf2 complex, one in which only the ETGE motif is bound to a Keap1 dimer and the other in which both the ETGE and DLG motifs are bound, then the addition of a second ETGE motif should increase the abundance of the complex in which both motifs are bound.
When EGFP-Nrf2-doubleETGE was transfected along with Keap1-mCherry, a FRET signal characterized by a reduction in EGFP fluorescence lifetime was measured (n = 13; P < 0.0001). Interestingly, the mean lifetime of this doubleETGE mutant was even shorter than the lifetime of the wild-type EGFP-Nrf2 (2,093 ps vs. 2,155 ps; Fig. 1D and Fig. S2 C and D) and corresponded to an increase in the protein interaction at 21% E-FRET (Fig. 2H). The subsequent use of a Keap1 mutant, which is unable to dimerize, termed Keap1-mono-mCherry, cotransfected with EGFP-Nrf2∆DLG resulted in the complete loss of the higher E-FRET population at 21% (n = 8; Fig. S2 G and H) and firmly established that the lower E-FRET population at 13% represents FRET between EGFP and the Keap1-mCherry protein, which is bound to the ETGE motif of Nrf2 (i.e., FRET in cis). Together, these experiments demonstrate that the Keap1-Nrf2 complex is found in two distinct states, one in which only the ETGE motif of Nrf2 is bound to Keap1 (the open conformation), corresponding to the lower 13% E-FRET population, and another in which both the DLG and ETGE motifs of Nrf2 are bound (the closed conformation), corresponding to the higher 21% E-FRET population (Fig. 2D, cartoons).
Inducers Promote the Closed Conformation of the Keap1-Nrf2 Complex.
To determine whether inducers altered the conformation of the Keap1-Nrf2 complex, we quantified the E-FRET in the induced state and compared it with the basal state by imaging the same cell under both basal and induced conditions. In response to inducers, Nrf2 accumulates within 1 h (19, 20), and we chose this as the time at which to study the Keap1-Nrf2 interaction in the induced state. In uninduced EGFP-Nrf2 + Keap1-mCherry cotransfected cells, the EGFP lifetime was stable during this period (n = 6; P > 0.05; Fig. S3 A, B, and E). Importantly, the E-FRET distribution was also unaffected by repeated imaging (Fig. S3 C and D). When EGFP-Nrf2 + Keap1-mCherry cotransfected cells were imaged before and 1 h after treatment with the inducer sulforaphane (SFN), the fluorescence lifetime of EGFP-Nrf2 was shorter compared with the basal state (n = 6; P = 0.009; Fig. S4A), corresponding to a reduced lifetime in the induced state. This means that the EGFP and mCherry fluorophores are closer together in the induced state. Although it has been previously shown biochemically that inducers do not cause dissociation of Nrf2 from Keap1 (21), our result was surprising because it suggested that SFN causes a conformational change in the complex, resulting in a tighter interaction between the two proteins. Most importantly, the reduction in lifetime on SFN treatment was coupled with a change in E-FRET that dramatically shifted from being equally distributed in the open (13% E-FRET) and closed (21% E-FRET) conformations at the basal state (Fig. 3A) to favoring almost exclusively the closed conformation in the induced state (Fig. 3B). This shift was not restricted to the Cys-151–targeting inducer SFN (22), as sulfoxythiocarbamate alkyne (STCA), which targets Cys-273, Cys-288, and Cys-613 of Keap1 (23), produced comparable results (n = 4; P = 0.019; Fig. 3 C and D and Fig. S4B). In contrast to cells transfected with EGFP-Nrf2 + wild-type Keap1-mCherry, addition of SFN did not cause a significant reduction in fluorescence lifetime in cells cotransfected with EGFP-Nrf2 + mutant Cys151Ser Keap1-mCherry (n = 7; P > 0.5; Fig. S5), confirming that Cys-151 is the primary target of SFN. The absence of change in fluorescence lifetime also implies there is no conformational change in the Cys151Ser Keap1-Nrf2 complex on treatment with SFN.
Fig. 3.
E-FRET in EGFP-Nrf2–transfected cells imaged before and after treatment with inducers, a proteasomal inhibitor (MG132), and an inhibitor of the protein synthesis (cycloheximide; CHX). HEK293 cells were transfected with EGFP-Nrf2 + Keap1-mCherry, and the E-FRET was quantified for individual cells that were imaged twice: once in the basal state (A, C, E, and G) and once again after a 1-h treatment with 5 μM SFN (B), 10 μM STCA (D), or 10 μM MG132 (F) or after a 15-min treatment with 10 μM CHX (H). (Left) Pictorial representations in which the color of the cell corresponds with the E-FRET, ranging from 0% to 30%. (Right) E-FRET from each pixel of the image plotted on a graph. The graphs show that the E-FRET distribution is altered by both inducers, as well as by inhibition of the proteasome or the protein synthesis, whereby there is an increase in the interaction at 21% E-FRET. This can be also seen in the images in the left columns, in which there is an increase in the amount of yellow relative to green in response to all treatments. (Scale bar, 10 μm.)
The Interaction Between Keap1 and Nrf2 Follows a Cycle.
The data so far show that in the basal state, the Keap1-Nrf2 complex is found in both an open and a closed conformation. However, it is unclear whether these two states are in equilibrium with one another or represent two phases of a cycle in which one conformation is formed first and then progresses to the second, thereby allowing ubiquitination of Nrf2. Inducers promote the formation of the closed conformation; however, this could occur by either a change in the equilibrium dynamics or an alteration in the binding cycle, both of which could manifest themselves as an increase at 21% E-FRET. To distinguish between these two possibilities, we used the proteasomal inhibitor MG132, which, to our knowledge, does not bind to Keap1 but stabilizes Nrf2 by blocking its degradation by the proteasome. We reasoned that if the two conformations are in equilibrium with one another, MG132 will not alter the relative ratio between them. However, if the two states of the complex represent two phases on a cycle, then blocking the cycle by inhibiting the degradation of Nrf2 will lead to the accumulation of the complex at the later point of the cycle. After treatment with MG132, there was a reduction in the lifetime of EGFP-Nrf2 (n = 7; P = 0.0003; Fig. S4C) coupled with an increase in the E-FRET distribution at 21% (Fig. 3 E and F). Similarly, blocking entry into the cycle by inhibiting new translation of Nrf2 should also lead to the accumulation of the complex at the later stage of the cycle. Indeed, inhibition of protein synthesis by cycloheximide had a similar effect to that of MG132 treatment (n = 3; P = 0.049; Fig. 4 G and H and Fig. S4D). Importantly, none of the inducer or inhibitor treatments affected the lifetime of EGFP in cells cotransfected with EGFP-Nrf2 and free mCherry (n = 5–13; P > 0.05; Fig. S4E). Together, these results strongly suggest that the interaction between Keap1 and Nrf2 follows a cycle in which the 21% E-FRET population (closed conformation) is the later phase in the cycle and the 13% population (open conformation) represents the initial phase of the cycle.
Fig. 4.
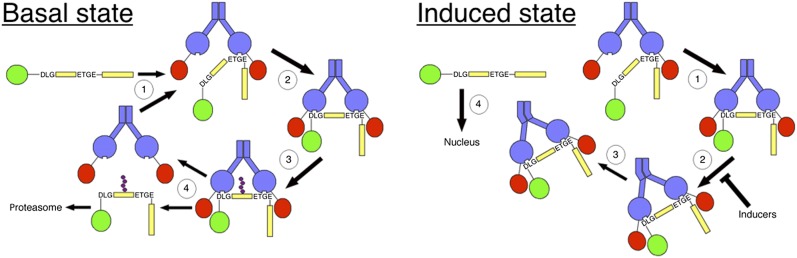
The cyclic sequential attachment and regeneration model of Keap1-mediated degradation of Nrf2. In the basal state, newly translated Nrf2 (yellow) binds sequentially to a free Keap1 dimer (blue), first through the ETGE motif to form the open conformation (1) and then through the DLG motif, to form the closed conformation (2). Once in the closed conformation, Nrf2 can be targeted for ubiquitination by the Keap1-dependent E3-ubiquitin ligase (3). Ubiquitinated Nrf2 is released from Keap1 and degraded by the proteasome. The free Keap1 dimer is regenerated and able to bind to newly translated Nrf2 (4), and the cycle begins again. In the induced state, the formation of the closed conformation is uncoupled from ubiquitination (2). As a consequence, Nrf2 is not released from Keap1 (3), free Keap1 is not regenerated, and newly translated Nrf2 accumulates and turns on the expression of cytoprotective genes (4).
Discussion
The cyclic model of Keap1-mediated degradation of Nrf2 is shown in Fig. 4. Newly translated Nrf2 binds first through its high-affinity ETGE motif to a single member of a free Keap1 dimer to form the open conformation. It has been shown that when the ETGE motif alone is bound to Keap1, Nrf2 is not targeted for ubiquitination by the E3-ubiquitin ligase (17), and therefore, while in the open conformation, Nrf2 is not ubiquitinated. After a period in the open conformation, the cycle progresses to form the closed conformation through the binding of the low-affinity DLG motif to the other member of the Keap1 dimer. In the closed conformation, Nrf2 is polyubiquitinated and then subsequently released for degradation by the proteasome. The regenerated free Keap1 dimer is then able to bind to newly synthesized Nrf2, and the cycle begins again. We have named this model the “cyclic sequential attachment and regeneration model of Keap1-mediated degradation of Nrf2.” Inhibition of the proteasomal degradation of Nrf2 blocks the cycle, leading to the accumulation of the Keap1-Nrf2 complex in the closed conformation. Inducers function to inhibit the ubiquitination of Nrf2, but crucially, in the absence of new translation, Nrf2 is not stabilized in the induced state, and Nrf2-target genes are not upregulated (24, 25), suggesting it is the de novo translated Nrf2 which translocates to the nucleus, whereas the existing population of Nrf2 remains bound to Keap1.
Our data show that, at first glance paradoxically, inducer activity is coupled with the formation of the closed conformation of the Keap1-Nrf2 complex, the same conformation that in the basal state is associated with the ubiquitination of Nrf2. How can these two seemingly contradictory facts be reconciled? We suggest that the closed conformation in the basal and induced states are not identical or, more accurately, they are identical with respect to the relationship between the Nrf2-binding (Kelch) domain of Keap1 and Nrf2 but not with respect to Nrf2 with the rest of the Keap1-dependent E3-ubiquitin ligase. In the basal state, the closed conformation orientates the lysine residues of Nrf2 so that they can be ubiquitinated by the E2 ubiquitin-conjugating enzyme that is bound to the E3. The binding of inducers to reactive cysteine residues of Keap1 leads to conformational changes in Keap1 (26) such that, although Nrf2 is still bound to Keap1 through both its ETGE and DLG motifs, it is no longer correctly aligned with the E2 ubiquitin-conjugating enzyme, and thus ubiquitination does not occur. This results in the Keap1-Nrf2 complex accumulating in the closed conformation, in which, without ubiquitination, Nrf2 is effectively trapped and cannot be released by the Keap1 dimer. Interestingly, in human tumors, gain-of-function mutations have been found in both the ETGE and DLG motifs of Nrf2 (9). This correlates well with our model, in which the loss of either motif will lead to stabilization of Nrf2 because of the inability of the complex to form the closed conformation and allow the cycle to proceed.
The model suggests that the Keap1-Nrf2 complex spends some of its time in the open conformation. This is consistent with other types of regulation of the pathway, such as by p21 in the case of p53 activation (27) or p62 in the case of autophagy inhibition (28). p21 and p62 bind to Nrf2 and Keap1, respectively, and, interestingly, to the same motifs exposed in the open conformation. Therefore, we propose that the Keap1-Nrf2 complex forms the open conformation to allow multiple modes of regulation, beyond electrophiles and oxidants. Consistent with our model, the responsiveness of the Keap1-Nrf2 pathway appears to be fine-tuned by the relative levels of Keap1 and Nrf2 proteins within the cell. Thus, in pancreatic cancer, a modest (<twofold) K-Ras–dependent upregulation in Nrf2 transcription causes an increase in Nrf2 protein levels and Nrf2-dependent transcription (29). This implies that in the basal state, the Nrf2 cycle operates at a level close to saturation and that any increase in Nrf2 overloads Keap1, which in turn allows newly translated Nrf2 to turn on expression of target genes. Conversely, a reduction in the level of Keap1 also leads to an increase in Nrf2 protein levels (30), which reinforces the idea that the system has little spare capacity. This “tipping point” state and lack of buffering capacity allows the pathway to respond exceedingly quickly to the cellular environment by the rapid upregulation of cytoprotective genes.
Materials and Methods
Chemicals and Inducers.
All chemicals were from Sigma unless specified. Inducers and compounds added to cells for the microscopy experiments were sourced as follows: SFN (LKT Laboratories Inc.), STCA (a gift from Young-Hoon Ahn and Philip A. Cole, Johns Hopkins University, Baltimore, MD), cycloheximide (Sigma), and MG132 (InSolution, Merck).
Cloning.
The pEGFP-C1 and pmCherry-N1 were from Clontech. The mouse Keap1, Nrf2, Nrf2ΔDLG, and Nrf2-doubleETGE cDNA vectors were kind gifts from Michael McMahon and John D. Hayes (University of Dundee, Dundee, United Kingdom). The Nrf2 mutant vectors contain the following mutations: Nrf2ΔDLG (D29A:L30G:G31E) and Nrf2-doubleETGE (I28E:D29E:L30T:V32E) (17). Both the Keap1-mono and Cys151Ser Keap1 plasmids were kind gifts from Masayuki Yamamoto (Tohoku University, Sendai, Miyagi, Japan). The Keap1-mono plasmid encodes full-length Keap1 with the following mutations: H96A:K97A:V98A:V99A:L100A (31). The Cys151Ser Keap1 plasmid also encodes full-length Keap1 with the following mutation: C151S (32). The general cloning strategy was the same for all constructs. The cDNA encoding the gene of interest was amplified using PCR to introduce the restriction enzyme consensus sequences. The primer sequences are shown in Table S1. The PCR product was gel purified and the DNA was extracted and digested, along with the destination vector. After gel purification, the two were ligated and transformed into competent bacteria. Transformed colonies were picked and grown overnight in 5-mL minicultures. From them, the plasmids were purified and sequenced.
Cell Culture.
HEK293 cells were grown in α-MEM supplemented with 10% heat-inactivated FBS (both from Gibco) on gelatin-coated plastic dishes (0.1% gelatin dissolved in water for 30 min). They were allowed to grow until they were confluent and then were split using a one-fifth dilution factor twice a week. For microscopy experiments, 200,000 cells were seeded onto Willco glass-bottom dishes. Cells were washed with PBS and imaged in 2 mL phenol red free CO2-independent DMEM supplemented with 10% FBS (Gibco). For experiments with inducers, 1 mL media was removed from the dish and transferred to an Eppendorf tube. To this, 2 μL of a 1,000× stock of inducer was added, and the solution was thoroughly mixed and then added back to the cells.
Transfections.
For each Willco dish, 100 μL opti-MEM was added to two Eppendorf tubes. To the first tube, DNA was added (0.75 μg EGFP-Nrf2 or 0.5 μg mCherry or Keap1-mCherry). To the second dish, 3.75 μL Lipofectamine 2000 (Invitrogen) was added. After a 12-min incubation at room temperature, the contents of the tubes were mixed and incubated for a further 15 min, and 200 μL of the final solution was added to the cells in 1.5 mL Opti-MEM (Invitrogen). After 4.5 h, the Opti-MEM was replaced with complete media.
Fluorescence Lifetime Imaging Microscopy/FRET Imaging.
Fluorescence lifetime imaging microscopy (FLIM)/FRET imaging was performed as described (16), using an inverted multiphoton laser-scanning microscope (Bio-Rad Radiance 2100MP) with a 60× oil immersion NA 1.4 Plan-Apochromat objective (Nikon) and Lasersharp 2000 software. The microscope was equipped with a Solent Scientific incubation chamber to maintain the cells at 37 °C. Two-photon excitation of EGFP was achieved using a Chameleon laser at 900 nm. The FLIM capability was provided by time-correlated single photon-counting electronics (SPC-830, Becker & Hickl). The two-photon laser power was adjusted to give a mean photon count rate of 105–106 photons per second. FLIM measurements were carried out over a 256 × 256 pixel area for 90 s.
FLIM Lifetime.
The .sdt file was imported into the SPCImage software (Becker & Hickl), and a region of interest was drawn around the area of the cell to be analyzed (corresponding to either the nucleus or the cytoplasm). The “bin” parameter was increased to 3, the “shift” parameter was fixed at 0, and all other parameters were left at the default unchecked positions (threshold = 5, scatter = unfixed, offset = unfixed). The image was analyzed using a single-component exponential decay model. The spatial distribution of the mean fluorescence lifetime of the EGFP donor is displayed using a continuous pseudocolor scale ranging from 1,900 to 2,600 ps.
FLIM/ FRET Efficiency.
The FRET efficiency was calculated using the SPCImage software (Becker & Hickl) for the same region of interest defined to calculate the mean fluorescence lifetime. The “bin” parameter was increased to 3, and all other parameters were left at the default unchecked positions. The image was analyzed using a two-component exponential decay model. To do this, the t2 (noninteracting proteins lifetime from control experiments in the absence of FRET) was fixed at 2,375 ps, whereas the t1 value (lifetime of the donor population that was interacting with the acceptor) was left unchecked. The spatial distribution of FRET efficiency (%) is displayed using a continuous pseudocolor scale ranging from 0% to 30%.
Statistical Analysis.
Averages and SD and P values were calculated using Excel (Microsoft). The P values for the EGFP lifetime data were calculated using two-tailed Student t test. For experiments in which the lifetime of EGFP was calculated in the same cell before and after addition of inducers, P values were calculated using two-tailed paired t test.
Supplementary Material
Acknowledgments
We thank Stephen M. Keyse, John D. Hayes, and Angus I. Lamond (University of Dundee) for helpful discussions and constructive criticism; Young-Hoon Ahn and Philip A. Cole (Johns Hopkins University) for gift of STCA; and Michael McMahon (University of Dundee) and Masayuki Yamamoto and Takafumi Suzuki (Tohoku University) for kind gifts of plasmids. We acknowledge the financial support of the Medical Research Council, Cancer Research UK (C20953/A10270), and Research Councils UK.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305687110/-/DCSupplemental.
References
- 1.Malhotra D, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38(17):5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25(1):162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99(18):11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler TW, Wakabayashi N. Nrf2: Friend or foe for chemoprevention? Carcinogenesis. 2010;31(1):90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13(11):1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 8.Crunkhorn S. Deal watch: Abbott boosts investment in NRF2 activators for reducing oxidative stress. Nat Rev Drug Discov. 2012;11(2):96. doi: 10.1038/nrd3655. [DOI] [PubMed] [Google Scholar]
- 9.Shibata T, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohta T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68(5):1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 13.Ogura T, et al. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci USA. 2010;107(7):2842–2847. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14(8):529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 16.Llères D, James J, Swift S, Norman DG, Lamond AI. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol. 2009;187(4):481–496. doi: 10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281(34):24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 18.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol Chem. 2006;387(10-11):1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 19.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem. 2003;278(7):4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 21.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci USA. 2005;102(29):10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23(22):8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn YH, et al. Electrophilic tuning of the chemoprotective natural product sulforaphane. Proc Natl Acad Sci USA. 2010;107(21):9590–9595. doi: 10.1073/pnas.1004104107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekhar KR, et al. Inhibition of the 26S proteasome induces expression of GLCLC, the catalytic subunit for gamma-glutamylcysteine synthetase. Biochem Biophys Res Commun. 2000;270(1):311–317. doi: 10.1006/bbrc.2000.2419. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi A, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44(18):6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34(6):663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 29.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taguchi K, et al. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol Cell Biol. 2010;30(12):3016–3026. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Maher J, Yamamoto M. Select heterozygous Keap1 mutations have a dominant-negative effect on wild-type Keap1 in vivo. Cancer Res. 2011;71(5):1700–1709. doi: 10.1158/0008-5472.CAN-10-2939. [DOI] [PubMed] [Google Scholar]
- 32.Takaya K, et al. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic Biol Med. 2012;53(4):817–827. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.