Significance
Ribosomes are complex macromolecular machines universal to all domains of life that synthesize all cellular proteins. They consist of many different protein and RNA building blocks. Their biogenesis—a paradigm for the assembly of macromolecular machines in general—is a highly complex and tightly regulated process in eukaryotes requiring the concerted action of many ribosome assembly factors. In this study we analyze the cross-talk between two of these assembly factors and ribosomal building blocks with biophysical methods and show how it might contribute to the fidelity and efficiency of ribosome assembly.
Abstract
Factor activating Pos9 (Fap7) is an essential ribosome biogenesis factor important for the assembly of the small ribosomal subunit with an uncommon dual ATPase and adenylate kinase activity. Depletion of Fap7 or mutations in its ATPase motifs lead to defects in small ribosomal subunit rRNA maturation, the absence of ribosomal protein Rps14 from the assembled subunit, and retention of the nascent small subunit in a quality control complex with the large ribosomal subunit. The molecular basis for the role of Fap7 in ribosome biogenesis is, however, not yet understood. Here we show that Fap7 regulates multiple interactions between the precursor rRNA, ribosomal proteins, and ribosome assembly factors in a hierarchical manner. Fap7 binds to Rps14 with a very high affinity. Fap7 binding blocks both rRNA-binding elements of Rps14, suggesting that Fap7 inhibits premature interactions of Rps14 with RNA. The Fap7/Rps14 interaction is modulated by nucleotide binding to Fap7. Rps14 strongly activates the ATPase activity but not the adenylate kinase activity of Fap7, identifying Rps14 as an example of a ribosomal protein functioning as an ATPase-activating factor. In addition, Fap7 inhibits the RNA cleavage activity of Nob1, the endonuclease responsible for the final maturation step of the small subunit rRNA, in a nucleotide independent manner. Thus, Fap7 may regulate small subunit biogenesis at multiple stages.
Ribosomes are complex ribonucleoprotein particles responsible for the biosynthesis of all cellular proteins. Eukaryotic ribosomes consist of ∼80 ribosomal proteins and four RNA molecules. Their assembly also requires up to 200 different auxiliary ribosome assembly factor proteins and ∼70 small nucleolar RNAs (snoRNAs) (1–3). The assembly factors transiently bind to and act on the nascent ribosome in a temporally and spatially well-defined and highly regulated manner. Many of them belong to different classes of GTPases and ATPases (4, 5). One peculiar example for such a factor is a protein called Fap7 in Saccharomyces cerevisiae (6) and cofilin interacting protein (hCINAP) or adenylate kinase 6 (hADK6) in humans (7). Fap7 contains the Walker A and B sequence elements of an ATPase but structurally resembles an adenylate kinase (AK) (8,9). A weak NTPase and a nucleotide kinase activity have been demonstrated for the human protein in vitro (8, 9). Fap7 was shown to bind specifically to ribosomal protein Rps14 and to transiently associate with preribosomal complexes at multiple points during the biogenesis pathway (10). Depletion of Fap7 or mutations in its ATPase motifs lead to defects at different stages in small ribosomal subunit (SSU) biogenesis (10). Thus, the function of Fap7 seems to be closely associated with its ATP-binding and/or ATPase/kinase activity. The molecular basis for this connection and the functional consequences of the Fap7/Rps14 interaction have, however, not been established yet.
Eukaryotic ribosome assembly is initiated by the transcription of a single large precursor ribosomal RNA (pre-rRNA) in the nucleolus. This pre-rRNA contains the 18S rRNA of the SSU, as well as the 25S and 5.8S rRNA of the large subunit separated by internal and external spacer elements (1–3). Many ribosomal proteins and a defined set of early assembly factors bind cotranscriptionally to the pre-rRNA, leading to an early 90S assembly intermediate. A series of well-coordinated nucleolytic processing steps then leads to separate precursors for the large and small ribosomal subunits. These are then further processed and remodeled. Immediately before export into the cytoplasm the pre-SSU contains all but two (Rps10, Rps26) of the ribosomal proteins and seven additional ribosome assembly factors, including the endonuclease Nob1 (11). When Fap7 is depleted in yeast or mutated in its Walker B motif, the nascent SSU lacks a third ribosomal protein, Rps14 (12). It also contains a 20S pre-rRNA, which requires cleavage by Nob1 at site D to yield mature 18S rRNA (13,14). The pre-SSU binds Nob1 already in the nucleus, but D-site cleavage occurs only after its export into the cytoplasm (15). Depletion of Nob1 leads to the accumulation of 20S pre-rRNA containing SSU precursors (13, 14). Interestingly, depletion of Fap7, mutations in its Walker A and B motifs (10), or mutations in the arginine-rich C-terminal tail of Rps14 also lead to accumulation of 20S pre-rRNA (16). Furthermore, Rps14 has been reported to interact directly with Nob1 in vitro (17).
Before 20S pre-rRNA cleavage by Nob1 in the cytoplasm, the pre-SSU undergoes a quality control step by forming an 80S complex with mature 60S large ribosomal subunits in an eIF5b (fun12)-dependent manner (12, 18). GTP hydrolysis by eIF5b bound to this complex causes a conformational change in the SSU, which triggers the cleavage of the 20S pre-rRNA at the D-site by Nob1 (18). After formation of the 18S rRNA the remaining ribosome assembly factors are released while the 80S complex dissociates (12). Importantly, Fap7 depletion or mutations in its Walker A and B motifs prevent the dissociation of the 80S complex after this quality control checkpoint (12).
Archaeal homologs of eukaryotic ribosome assembly factors have served as valuable models for structural, functional, and biophysical studies (19–24). Here we use the homologs of Fap7, Rps14, and Nob1 from the hyperthermophilic archaeon Pyrococcus horikoshii (PhFap7, PhS11, and PhNob1, respectively) to delineate the functional consequences of their interactions in vitro. We find that PhFap7 binds PhS11 with extremely high affinity and blocks both of its RNA interaction motifs. PhS11 binding strongly activates the ATPase but not the AK activity of PhFap7 and also modulates its nucleotide-binding preferences. PhFap7 also binds PhNob1 and inhibits its D-site RNA cleavage activity. Taken together our data suggest that the essential ribosome biogenesis factor Fap7 performs a dual role in the biogenesis of the SSU in the nucleotide-dependent release of Rps14, as well as by regulating D-site cleavage.
Results
PhFap7 Binds PhS11 with Exceptionally High Affinity.
As in many other archaeal genomes a homolog of yeast Fap7 (ScFap7) and hADK6 is found in the genome of P. horikoshii (PhFap7). Importantly, PhFap7 shares exactly those sequential features with ScFap7 and hADK6 that distinguish these proteins from other AKs. These include an hhhD/ExH-type Walker B motif typical for NTPases but not for AKs (25), and a Walker A motif containing a threonine/serine residue with the capacity to complex a metal ion instead of the glycine common to all other classes of AKs at this position (SI Appendix, Fig. S1).
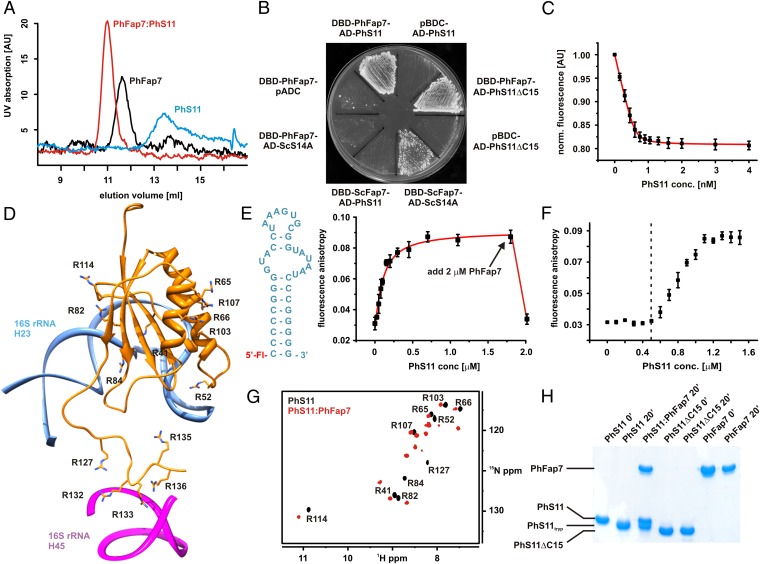
To test whether PhFap7 binds ribosomal protein PhS11—the archaeal homolog of Rps14—analytical gel filtration and NMR spectroscopy (Fig. 1A and SI Appendix, Fig. S2) were used. Both experiments revealed the formation of a stable 1:1 PhFap7:PhS11 complex. PhFap7:PhS11 complex formation was also observed in yeast two-hybrid experiments (Fig. 1B). A yeast two-hybrid interaction was also observed between ScFap7 and yeast Rps14 (ScS14A) but not between ScFap7 and PhS11 or PhFap7 and ScS14A (Fig. 1B). The PhFap7:PhS11 interaction was not abolished when the unstructured 15-aa C-terminal region was deleted in a PhS11ΔC15 mutant (Fig. 1B and SI Appendix, Fig. S2). Thus, the C terminus of PhS11 is not essential for PhFap7 binding.
Fig. 1.
PhFap7 binds PhS11 with high affinity and blocks both of its rRNA-binding elements. (A) Gel filtration profiles of PhFap7 (black), PhS11 (blue), and a 1:1 PhFap7/PhS11 (red) complex. (B) Yeast two-hybrid binding assays: PhS11 vs. PhFap7, ScS14a vs. ScFap7, PhFap7 vs. ScS14a, PhS11ΔC15 vs. PhFap7. (C) Fluorescence quenching of fl-PhFap7 upon titration with PhS11 for the determination of the KD of the PhFap7:PhS11 complex. (D) Model of PhS11 bound to ribosomal RNA based on the structure of the T. thermophila SSU X-ray structure (27). Arginine side chains are shown as sticks and labeled. (E) Changes in fluorescence anisotropy upon titration of fluoresceine-labeled h23-RNA (fl-h23, Left) with PhS11. Addition of PhFap7 (arrow) in an amount equimolar to PhS11 leads to a drop in fluorescence anisotropy to a value similar to that for free fl-h23. (F) Competitive binding of PhS11 to unlabeled PhFap7 and 5′ fluoresceine-labeled h23-RNA observed in fluorescence anisotropy measurements. Formation of the PhS11:fl-h23 complex is only detectable upon saturation of PhFap7 by PhS11 (dashed vertical line). (G) Overlay of 15N-TROSY-HSQC spectra of free 15N-Arg-labeled PhS11 (black, signal assignments are indicated) and bound to unlabeled PhFap7 (red). (H) Protection of the C terminus of PhS11 against trypsin digestion by PhFap7 binding analyzed by SDS/PAGE.
The affinity of the PhFap7:PhS11 interaction was quantified in fluorescence quenching experiments with PhFap7 fluorescein labeled at cysteine 40 (fl-PhFap7), yielding a KD of 22 ± 3 pM (Fig. 1C). A high-affinity fl-PhFap7:PhS11 interaction was also evident from fluorescence anisotropy measurements (SI Appendix, Fig. S2). The affinity of PhS11ΔC15 for fl-PhFap7 could not be measured by fluorescence quenching because the addition of PhS11ΔC15 did not cause a change in fl-PhFap7 fluorescence intensity. However, for the PhFap7:PhS11ΔC15 interaction, a KD of 6.7 ± 0.5 nM was derived from a competition experiment in which fl-PhFap7 was titrated with PhS11 in the presence of a 1,000-fold excess of PhS11ΔC15 (SI Appendix, Fig. S2), implying that the C-terminal tail of PhS11 contributes to the affinity for PhFap7.
The tight interaction between PhS11 and PhFap7 significantly stabilizes PhS11 against thermal unfolding in vitro (SI Appendix, Fig. S3).
PhFap7 Blocks Both RNA-Binding Elements of PhS11.
The S. cerevisiae and Tetrahymena thermophila SSU crystal structures revealed that Rps14 interacts with two parts of the 18S rRNA (26, 27): the globular body of Rps14 binds helix 23 (h23), and its basic tail contacts helix 45 (h45) (Fig. 1D). The majority of the basic residues involved in RNA binding, including those of the C-terminal tail, are conserved in PhS11 (SI Appendix, Fig. S4).
In vitro fluorescence anisotropy titration experiments showed that PhS11 bound to 5′-fluorescein labeled h23 of P. horikoshii 16S rRNA (fl-h23) with a dissociation constant of 78 ± 14 nM (Fig. 1E). Addition of PhFap7 to the fl-h23:PhS11 complex led to a reduction of the fluorescence anisotropy to the same value as observed for free fl-h23. Thus, PhFap7 is able to displace h23-RNA from its binding site on PhS11. Furthermore, when a mixture of fl-h23 and unlabeled PhFap7 was titrated with PhS11, an increase in fluorescence anisotropy due to RNA–protein complex formation occurred only after PhFap7 was saturated (Fig. 1F), indicating that PhFap7 and h23 compete for the same binding site on PhS11.
Five of the 14 arginine residues of PhS11 are located in close proximity to the h23 binding site in the body of the protein and five in the C-terminal tail of PhS11 (Fig. 1D). To identify the PhFap7 binding site, PhS11 was produced in a form whereby only the arginines are 15N-labeled. A 15N-TROSY-HSQC spectrum of this protein showed only 10 out of the expected 14 signals at 42 °C (Fig. 1G). Nine of these signals correspond to arginine residues in the globular body of PhS11 and one to R127 in the C-terminal tail. The backbone amide protons of the four remaining arginines of the C-terminal tail (R132, R133, R135, and R136) exchange rapidly with the solvent water, preventing their detection. Addition of unlabeled PhFap7 to 15N-arginine-labeled PhS11 led to two dramatic changes in the spectrum: the number of signals increased to the expected 14, and five out of nine arginine signals from the body of PhS11 (R41, R52, R82, R84, and R114) showed significant changes in their chemical shifts (Fig. 1G). In similar experiments with 15N-lysine-labeled PhS11, residue K129 in the C terminus also showed an altered chemical shift upon PhFap7 binding (SI Appendix, Fig. S5). Taken together, these data demonstrate that PhFap7 binds to both the h23-RNA binding site in the globular domain of PhS11 and the basic C-terminal tail of PhS11.
A direct interaction between the C-terminal tail of PhS11 and PhFap7 in solution was also observed in protease protection experiments. The high number of arginine residues in the C terminus of PhS11 renders it highly susceptible to degradation by trypsin. After trypsin digestion the C terminus of PhS11 was completely removed. In contrast, PhS11ΔC15 was not degraded by trypsin. Addition of PhFap7 to PhS11 significantly reduced tryptic cleavage of the C-terminal tail (Fig. 1H).
Binding of Nucleotides and PhS11 to PhFap7 Is Interdependent.
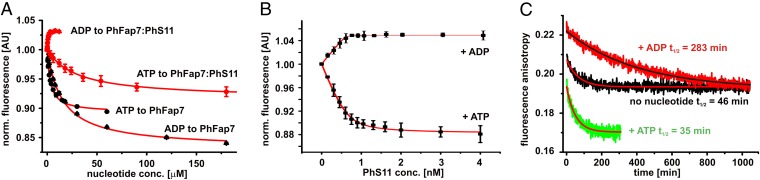
NMR titrations showed that both PhFap7 alone and the PhFap7:PhS11 complex bind ADP and ATP (SI Appendix, Fig. S6). The KDs for ATP and ADP in the presence of Mg2+ were measured in fluorescence quenching experiments using either free fl-PhFap7 or the fl-PhFap7:PhS11 complex (Fig. 2A). PhFap7 bound ATP with a KD of 3.5 ± 0.3 μM and ADP with a KD of 15.0 ± 0.6 μM. Binding of PhS11 lowered the KD of PhFap7 for ADP ∼33-fold to 0.46 ± 0.09 μM, whereas the KD for ATP increased ∼ninefold to 30.0 ± 1.1 μM compared with free PhFap7. Thus, although free PhFap7 had ∼fourfold higher affinity for ATP, the PhFap7:PhS11 complex bound ADP ∼67-fold better than ATP.
Fig. 2.
Nucleotides and PhS11 modulate each other’s affinities for PhFap7. (A) Fluorescence quenching experiments to determine the KDs of ADP and ATP for free PhFap7 and the PhFap7:Phs11 complex, respectively. (B) Fluorescence quenching experiments to determine the KD of PhS11 for PhFap7 in the presence of ADP and ATP, respectively. (C) Dissociation kinetics of the fl-PhFap7:PhS11 complex upon addition of a 100-fold excess of unlabeled PhFap7 in the absence of nucleotide (black) or in the presence of ADP (red) and ATP (green), observed by fluorescence anisotropy.
ATP and ADP also modulate the affinity of PhFap7 for PhS11 (Fig. 2B). In fluorescence quenching experiments with fl-PhFap7 in the presence of 2 mM nucleotide and 5 mM Mg2+ PhS11 bound the ADP–PhFap7 complex with a KD of <3.7 ± 4 pM, which is ∼sixfold lower than for free PhFap7. In contrast, the ATP–PhFap7 complex bound PhS11 with an increased KD of 70.4 ± 18 pM. The ATP analog ATPγS also diminished the affinity of PhFap7 for PhS11 (SI Appendix, Fig. S7). Thus, ADP and ATP inversely modulated the affinity of PhFap7 for PhS11, resulting in a >20-fold difference in the PhS11 affinity for PhFap7 in the presence of either nucleotide. ADP and ATP also inversely influenced the dissociation rates of the PhFap7:PhS11 complex, as observed in kinetic fluorescence anisotropy measurements. ATP, ATPγS, and the transition state analog ADP*AlFx increased the dissociation rate of the complex compared with the nucleotide-free state, whereas ADP significantly decreased the dissociation rate (Fig. 2C and SI Appendix, Fig. S7). Thus, ATP binding but not hydrolysis is required to destabilize the PhFap7:PhS11 complex.
Interestingly, the basic C terminus of PhS11 is important for modulating the affinity of the PhFap7:PhS11 complex for nucleotides because the nucleotide affinities of the PhFap7:PhS11ΔC15 complex were very similar to that of free PhFap7 (SI Appendix, Fig. S6).
A direct influence of nucleotide binding on the interaction of the C terminus of PhS11 with PhFap7 is evident from NMR experiments. Upon titration of 15N-arginine-labeled PhS11 bound to unlabeled PhFap7 with ATPγS the C terminus of PhS11 becomes solvent-exposed but is stably bound to PhFap7 in the presence of ADP. The C-terminal peptide of PhS11 is able to interact with an RNA corresponding to helix 45 of the P. horikoshii 16S rRNA (SI Appendix, Fig. S8).
PhS11 Stimulates the ATPase but Not the AK Activity of PhFap7.
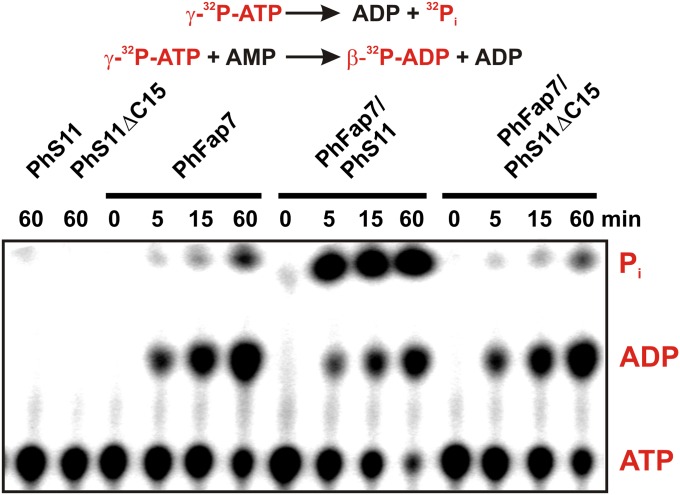
The influence of PhS11 and PhS11ΔC15 on the ATPase and AK activity of PhFap7 was analyzed with γ-32P-labeled ATP as a substrate in the presence of 10 mM unlabeled AMP, allowing both activities to be observed simultaneously (Fig. 3). Neither PhS11 nor PhS11ΔC15 significantly modulated the AK activity of PhFap7. In contrast, PhS11, but not PhS11ΔC15, strongly activated the ATPase activity of PhFap7. The activation of the ATPase activity of PhFap7 by full-length PhS11 was also seen under conditions where only the ATPase reaction can take place (SI Appendix, Fig. S9). In single turnover kinetic experiments PhS11 binding to PhFap7 increased the kcat for the ATPase reaction ∼170-fold (SI Appendix, Fig. S9), suggesting that PhS11 directly influences the chemical step of this reaction. PhFap7 was also able to use the other naturally occurring NTPs as substrates in PhS11-stimulated NTPase reactions (SI Appendix, Fig. S9).
Fig. 3.
Stimulation of the ATPase activity of PhFap7 by PhS11. Thin-layer chromatographic kinetic analysis of the reactions catalyzed by PhFap7, the PhFap7:PhS11 complex, and the PhFap7:PhS11ΔC15 complex with γ-[32P]ATP as substrate under conditions (10 mM AMP) where the ATPase reaction (upper line) and the kinase reaction (lower line) occur simultaneously. Radioactively labeled substrates and products are colored red.
PhNob1 Binds PhFap7 but Not PhS11.
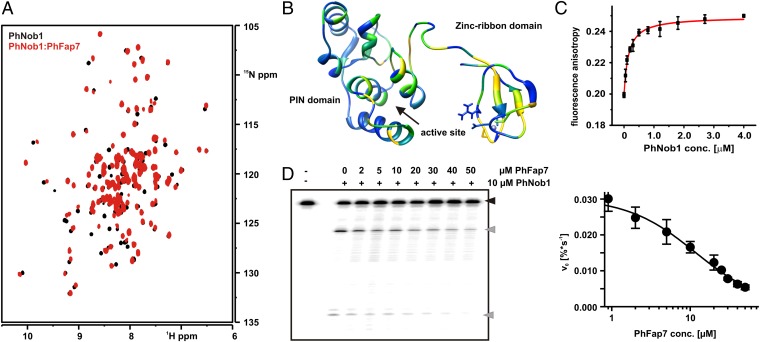
ScS14 has been reported to bind to Nob1 (17). However, no interaction was evident in NMR titration experiments using 15N-labeled PhNob1 and unlabeled PhS11 (SI Appendix, Fig. S10). In contrast, the addition of unlabeled PhFap7 to 15N-PhNob1 led to significant chemical shift changes for numerous PhNob1 backbone signals (28) (Fig. 4A). Mapping of these chemical shift changes on the structure of PhNob1 (24) showed that significant alterations were observed for residues in the zinc ribbon RNA-binding domain and in the linker connecting this domain to the catalytic PilT N-terminus (PIN) like domain (Fig. 4B). Interestingly, significant chemical shift changes were also observed for amino acids in spatial proximity to the active site on the PIN domain, whereas other areas of this domain were not influenced by PhFap7 binding (Fig. 4B). A stable interaction between PhFap7 and PhNob1 was also observed by analytical gel filtration (SI Appendix, Fig. S10). However, simultaneous addition of PhFap7 and PhS11 to PhNob1 did not lead to formation of a stable ternary complex, instead only the PhFap7:PhS11 complex was visible. This implies that the PhFap7:PhNob1 and the PhFap7:PhS11 interactions are mutually exclusive.
Fig. 4.
PhFap7 binds to PhNob1 and inhibits the D-site cleavage activity of PhNob1. (A) Overlays of HSQCs of 15N-labeled PhNob1 (black) and bound to PhFap7 (red). (B) Mapping of chemical shift changes induced by PhFap7 on the structure of PhNob1. Yellow corresponds to large, green to intermediate, and blue to no chemical shift changes. (C) Determination of the PhFap7:PhNob1 affinity by fluorescence anisotropy titrations. (D) D-site RNA cleavage by PhNob1 in the absence and presence of PhFap7 analyzed by denaturing PAGE (Left). A black arrow indicates uncleaved substrate, gray arrows cleavage products. D-site RNA cleavage velocity of PhNob1 as a function of PhFap7 concentration (Right).
Fluorescence anisotropy measurements using fl-PhFap7 yielded a KD of 141 ± 19 nM for the PhNob1:PhFap7 interaction (Fig. 4C). In contrast to the PhFap7:PhS11 interaction, the affinity of PhFap7 for PhNob1 did not change significantly upon addition of ADP, ATP, or ATPγS, and 5 mM Mg2+ and PhNob1 does not stimulate the ATPase activity of PhFap7 (SI Appendix, Fig. S10).
PhFap7 Inhibits Cleavage of D-Site RNAs by PhNob1.
The PhFap7-induced chemical shift changes for PhNob suggested that PhFap7 would inhibit the endoribonuclease activity of PhNob1 on substrates containing the D-site region of the pre-rRNA. D-site RNA cleavage assays (24) were carried out in the presence of increasing amounts of PhFap7. A significant reduction of specific RNA cleavage at the D-site was observed (Fig. 4D). In contrast, BSA was not able to inhibit PhNob1-induced D-site cleavage. In the presence of an excess of tRNAs, a Ki for the PhNob1 mediated D-site RNA cleavage of 12 ± 2 μM was estimated from kinetic analysis.
Discussion
A number of eukaryotic ribosome assembly factors are conserved in archaea. Here we used the P. horikoshii homologs of the essential ribosome assembly factors Fap7 and Nob1, as well as of the ribosomal protein Rps14, as a model system to elucidate their mutual interactions and the resulting functional consequences in vitro.
Similar to its eukaryotic homolog (10), PhFap7 binds to the ribosomal protein PhS11. It does so with an exceptionally high affinity, with a KD-value in the picomolar range. Furthermore, this interaction blocks both RNA interaction elements of PhS11. The very high affinity of PhFap7 for PhS11 suggests that Rps14 binding is likely to be the most important biological function of Fap7. Given the high affinity of these proteins for each other it is unlikely that Rps14 exists in its free form in the cell as long as the expression of the two proteins is balanced. Fap7 therefore presumably has an important function in protecting the highly basic Rps14 from premature or nonspecific interactions with cellular RNAs. Interaction with Fap7 might also prevent thermal denaturation of Rps14, as well as premature degradation of its unstructured and highly protease susceptible C terminus. Many other ribosomal proteins also have extended unstructured and positively charged N- or C-terminal tails or linkers prone to nonspecific interactions with cellular nucleic acids and/or degradation (26, 27). Indeed Fap7 is not the only ribosome assembly factor described to be responsible for protecting a ribosomal protein from such detrimental effects before incorporation into the nascent ribosome. The chaperones Sqt1, Rrb1, and Yar1 have been shown to bind to the ribosomal proteins Rpl10, Rpl3, and Rps3, respectively, which all contain long basic N- or C-terminal extensions (29–31). The 50-aa C-terminal extension of Rpl10, for example, has been shown to directly interact with its chaperone Sqt1. However, in contrast to Fap7, these chaperones have no enzymatic function. Instead the release of Rpl10 from Sqt1 requires the activity of an additional specialized GTPase, Lsg1 (29). Therefore, Fap7 likely represents an example of a ribosomal protein protection factor with a built-in ATPase-based release system.
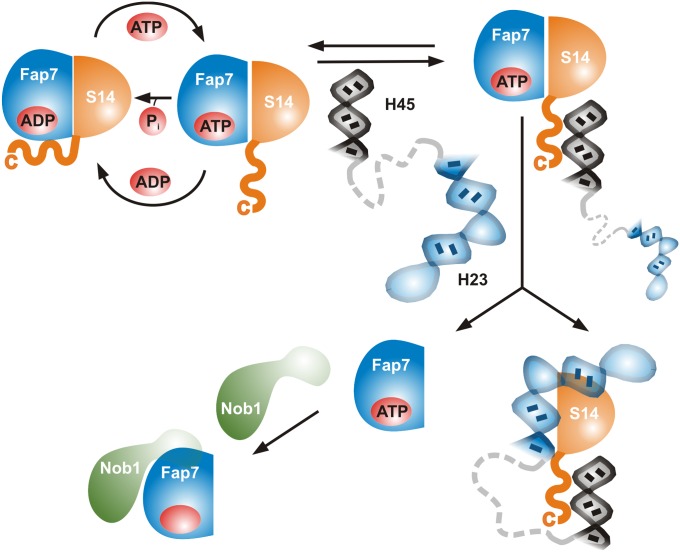
Although perfectly suitable for a protective function, the high affinity of PhFap7 for PhS11 calls for a mechanism for the release of PhS11 in ribosome biogenesis. Although ATP diminishes the affinity of PhFap7 for PhS11 in comparison with ADP, the KD is still in the picomolar range. At the same time, cross-talk between nucleotide and PhS11 affinity absolutely depends on the presence of the basic C-terminal tail of PhS11 because PhS11ΔC15 is unable to modulate nucleotide binding and ATPase activity. Furthermore, ATPγS binding to the PhS11:PhFap7 complex leads to the specific exposure of the C terminus of PhS11 to the solvent. We therefore propose a two-stage model for Rps14 release from Fap7 (Fig. 5). In the ADP-bound state both the body and the C-terminal tail of Rps14 interact strongly with Fap7, as indicated by the NMR data (SI Appendix, Fig. S8) and the higher ADP affinity of the PhFap7:PhS11 complex in comparison with free PhFap7 and the PhFap7:PhS11ΔC15 complex. Upon exchange of ADP to ATP the Rps14 C-terminal tail becomes exposed and thereby available for interactions with RNAs. Given the sequence of the C terminus these RNA interactions may initially be unspecific and transient but restrict the protein–protein interactions of Fap7 to the body of Rps14, resulting in a lowered affinity comparable to the situation in the PhFap7:PhS11ΔC15 complex. However, in the preribosomal complex, the RNA interactions of the Rps14 C terminus presumably involve helix 45, and this may bring the complex into the vicinity of helix 23. Helix 23, in its correctly folded form, may then displace Fap7 from the body of Rps14. In such a model the incorporation of Rps14 into the nascent ribosome would be highly dependent on the correct orientation of the two RNA substrates in the preribosome. This means that the Fap7:Rps14 affinity may be a key quality control element monitoring formation of the correct 3D preribosome structure.
Fig. 5.
Model for the hierarchy of Fap7 functions during ribosome biogenesis.
Upon integration of Rps14 into the preribosome Fap7 might become available to bind to and regulate the activity of Nob1. Although the measured affinity of PhFap7 for PhNob1 is in a biologically relevant range, it is significantly lower than the affinity of PhFap7 for PhS11, suggesting that Nob1 binding is a secondary function of Fap7 and is dependent on the prior release of Rps14 into the preribosomal particle. In this context it should be noted that Rps14 is a primary rRNA-binding protein, which is incorporated early into small subunit precursors. This implies that Fap7 would become available early in the ribosome biogenesis pathway for additional tasks at later stages of ribosome formation. In contrast to PhS11, PhNob1 does not stimulate nucleotide turnover by PhFap7, suggesting that the enzymatic functions of Fap7 are connected exclusively to its function in Rps14 incorporation into the pre-SSU. Furthermore, the PhFap7:PhNob1 interaction is not modulated by nucleotide binding. Therefore, other ribosome assembly factors and/or rRNA elements in the structural context of the nascent SSU may be required for the timed release and activation of Nob1. Although no ribosome assembly factors interacting with Fap7 have been described, Dim2 (Pno1) and the kinase Rio2 interact directly with Nob1 (11, 17) and may modify its structure, activity, and its interaction with Fap7 and RNA. In addition, elongation factor eIF5b and the RNA helicase Prp43 stimulate D-site cleavage by Nob1 in pre-40S particles (14, 18). Further studies of the Fap7:Nob1 interaction in the context of the nascent subunit are clearly required to clarify its physiological importance.
One particularly interesting finding of our study was that PhS11 specifically acts as an ATPase-activating protein for PhFap7 without influencing its AK activity. To our knowledge this is an unprecedented role for a ribosomal protein. Furthermore, the ATPase stimulation depends on the arginine-rich C terminus of PhS11. Interestingly, previously reported mutations of these arginines in ScS14 lead to defects in SSU biogenesis (16). The stimulation of the PhFap7 ATPase activity by PhS11 quickly resets the PhFap7:PhS11 complex into the inactive ADP-bound state and may therefore be important to restrict the lifetime of the interaction-prone ATP-containing complex. Functionally, this could limit premature integration of Rps14 in an RNA-rich environment, such as the preribosome, or Rps14 ribosome incorporation under conditions of ATP deprivation. The functional role of the PhS11-independent AK activity of PhFap7 is not clear yet.
Fap7 has recently been described as an important factor for a late cytoplasmic step of SSU biogenesis: the progression of the nascent small subunit through a translation-like quality control cycle involving a transient complex with the 60S subunit (12, 18). Fap7 depletion or mutations in its Walker motifs lead to an arrest of this complex, and it was proposed that the ATPase activity of Fap7 provides part of the energy to drive progression through this cycle (12). This proposal is not entirely consistent with our findings because the ATPase activity of free PhFap7 is strongly reduced in the absence of PhS11, and in the cytoplasm, PhFap7 is likely bound to PhS11. However, one prominent effect of Fap7 depletion is the absence of Rps14 from the nascent ribosomal subunit. This in turn probably has consequences for the correct positioning of helix 45 and the binding of rpS26, which is in direct contact with the 3′ end of the SSU RNA. Such structural defects are likely to be detected by a quality control system. It is therefore possible that the arrest of the nascent small subunit in the quality control complex previously described (12) arises as an indirect consequence of Fap7 depletion, causing the absence of Rps14.
In conclusion, we have shown that PhFap7 is able to modulate multiple RNA–protein interactions during ribosome biogenesis and that one of its binding partners, the ribosomal protein PhS11, plays a role as an ATPase-activating factor for PhFap7. This suggests a hierarchical model for Fap7 function during the biogenesis of the SSU. Additional experiments are necessary to verify the mechanism of ATPase activation by Rps14 and to ascertain the conservation of the properties of the Fap7:Rps14 and Fap7:Nob1 interactions between archaeal and eukaryotic systems.
Materials and Methods
PhFap7 and PhNob1 were produced as described previously (24, 32). PhS11 was overexpressed from a pET11a-derived vector and purified as described for PhNob1. RNAs were obtained commercially (Dharmacon). Protein–protein interactions were characterized using analytical gel filtration, yeast two-hybrid assays, and NMR spectroscopy as described in SI Appendix. Binding affinities were measured in fluorescence quenching and anisotropy experiments on a Fluorolog3 (HoribaJobinYvon). ATPase and AK assays were carried out at 50 °C with γ-[32P]-ATP as a substrate. Experimental details and data analysis are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Markus Bohnsack, Katherine E. Sloan, Sriram Srikant, and Elke Duchardt-Ferner for the critical reading of the manuscript and helpful discussions. This work was supported by an Aventis Foundation Endowed Professorship (J.W.), the Center for Biomolecular Magnetic Resonance, the Cluster of Excellence “Macromolecular Complexes” (K.-D.E., E.S., and J.W.), Johann-Wolfgang-Goethe-University Frankfurt, and the Collaborative Research Center (SFB) 902 “Molecular principles of RNA-based regulation,” funded by the Deutsche Forschungsgemeinschaft (E.S. and J.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306389110/-/DCSupplemental.
References
- 1.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13(5):255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 2.Henras AK, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65(15):2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803(6):673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15(12):2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juhnke H, Charizanis C, Latifi F, Krems B, Entian KD. The essential protein fap7 is involved in the oxidative stress response of Saccharomyces cerevisiae. Mol Microbiol. 2000;35(4):936–948. doi: 10.1046/j.1365-2958.2000.01768.x. [DOI] [PubMed] [Google Scholar]
- 7.Santama N, et al. Characterization of hCINAP, a novel coilin-interacting protein encoded by a transcript from the transcription factor TAFIID32 locus. J Biol Chem. 2005;280(43):36429–36441. doi: 10.1074/jbc.M501982200. [DOI] [PubMed] [Google Scholar]
- 8.Ren H, et al. The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus. Proc Natl Acad Sci USA. 2005;102(2):303–308. doi: 10.1073/pnas.0407459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drakou CE, et al. hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies. Proteins. 2012;80(1):206–220. doi: 10.1002/prot.23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol. 2005;25(23):10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strunk BS, et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333(6048):1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150(1):111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatica A, Oeffinger M, Dlakić M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol. 2003;23(5):1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pertschy B, et al. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284(50):35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581(15):2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Jakovljevic J, et al. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell. 2004;14(3):331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- 17.Campbell MG, Karbstein K. Protein-protein interactions within late pre-40S ribosomes. PLoS ONE. 2011;6(1):e16194. doi: 10.1371/journal.pone.0016194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebaron S, et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19(8):744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaRonde-LeBlanc N, Wlodawer A. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure. 2004;12(9):1585–1594. doi: 10.1016/j.str.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Ng CL, et al. Conformational flexibility and molecular interactions of an archaeal homologue of the Shwachman-Bodian-Diamond syndrome protein. BMC Struct Biol. 2009;9:32. doi: 10.1186/1472-6807-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia MZ, Horita S, Nagata K, Tanokura M. An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3′ end fragment of 16S rRNA. J Mol Biol. 2010;398(5):774–785. doi: 10.1016/j.jmb.2010.03.055. [DOI] [PubMed] [Google Scholar]
- 22.Taylor AB, et al. The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site. Nucleic Acids Res. 2008;36(5):1542–1554. doi: 10.1093/nar/gkm1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39(6):2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veith T, et al. Structural and functional analysis of the archaeal endonuclease Nob1. Nucleic Acids Res. 2012;40(7):3259–3274. doi: 10.1093/nar/gkr1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 27.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331(6018):730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 28.Veith T, et al. Backbone and side chain NMR resonance assignments for an archaeal homolog of the endonuclease Nob1 involved in ribosome biogenesis. Biomol NMR Assign. 2012;6(1):47–50. doi: 10.1007/s12104-011-9323-4. [DOI] [PubMed] [Google Scholar]
- 29.West M, Hedges JB, Chen A, Johnson AW. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol. 2005;25(9):3802–3813. doi: 10.1128/MCB.25.9.3802-3813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iouk TL, Aitchison JD, Maguire S, Wozniak RW. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol Cell Biol. 2001;21(4):1260–1271. doi: 10.1128/MCB.21.4.1260-1271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loar JW, et al. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 2004;168(4):1877–1889. doi: 10.1534/genetics.104.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellmich UA, Wöhnert J. Backbone resonance assignments for a homolog of the essential ribosome biogenesis factor Fap7 from P. horikoshii in its nucleotide-free and -bound forms. Biomol NMR Assign. 2012 doi: 10.1007/s12104-012-9423-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.