Significance
Plant growth, the basis of agricultural production, is compromised when plants defend themselves against herbivores.Wound-induced growth reduction is coordinated between organs by hormones termed “jasmonates.” We developed a sensitive assay that marks tissues where wounding activates jasmonate function in seedlings. This assay showed that a key repressor of jasmonate responses is active mainly in roots where it permits normal growth. A deeper understanding of cell-size control is crucial in successfully engineering plants that display reduced growth restriction under stress.
Keywords: root growth, herbivory, plant fertility, mapping by sequencing
Abstract
Wound responses in plants have to be coordinated between organs so that locally reduced growth in a wounded tissue is balanced by appropriate growth elsewhere in the body. We used a JASMONATE ZIM DOMAIN 10 (JAZ10) reporter to screen for mutants affected in the organ-specific activation of jasmonate (JA) signaling in Arabidopsis thaliana seedlings. Wounding one cotyledon activated the reporter in both aerial and root tissues, and this was either disrupted or restricted to certain organs in mutant alleles of core components of the JA pathway including COI1, OPR3, and JAR1. In contrast, three other mutants showed constitutive activation of the reporter in the roots and hypocotyls of unwounded seedlings. All three lines harbored mutations in Novel Interactor of JAZ (NINJA), which encodes part of a repressor complex that negatively regulates JA signaling. These ninja mutants displayed shorter roots mimicking JA-mediated growth inhibition, and this was due to reduced cell elongation. Remarkably, this phenotype and the constitutive JAZ10 expression were still observed in backgrounds lacking the ability to synthesize JA or the key transcriptional activator MYC2. Therefore, JA-like responses can be recapitulated in specific tissues without changing a plant’s ability to make or perceive JA, and MYC2 either has no role or is not the only derepressed transcription factor in ninja mutants. Our results show that the role of NINJA in the root is to repress JA signaling and allow normal cell elongation. Furthermore, the regulation of the JA pathway differs between roots and aerial tissues at all levels, from JA biosynthesis to transcriptional activation.
In response to mechanical damage or herbivory, plants rapidly accumulate the lipidic prohormone jasmonic acid (JA) and biologically active jasmonoyl-l-isoleucine (JA-Ile), which inhibits growth and activates defense responses to promote plant fitness (1). In the current model of JA perception and signaling, JASMONATE ZIM DOMAIN (JAZ) proteins block the function of JA-responsive transcriptional activators in the absence of bioactive JA-Ile (2, 3). JAZ proteins form a repressor complex by recruitment of the general corepressor TOPLESS (TPL) and TPL-related proteins either directly or indirectly through interaction with the adaptor protein Novel Interactor of JAZ (NINJA) (4, 5). When JA-Ile accumulates, it acts as a bridging ligand between the F-box protein CORONATINE INSENSITIVE 1 (COI1) and JAZ proteins to form the JA coreceptor complex (6). This leads to ubiquitylation and degradation of JAZs, setting the JA-responsive transcriptional activators free to function (2, 3, 6).
Recent studies of the plant wound response and other JA-signaling processes have relied on measuring the accumulation of JAs and/or JA-dependent transcripts in entire tissue extracts (7, 8). Although such approaches have contributed to our current understanding of JA responses, a better spatiotemporal resolution could facilitate the study of pending questions on JA and wound signaling through novel cell biological and genetic means. Because reliable methods for in situ detection of jasmonates have rarely been implemented (9), transcriptional reporter lines based on robust JA-dependent promoters are one of the best tools to accurately localize JA signaling in planta. Another common feature of previous wound response studies in Arabidopsis is the use of adult rosettes, although experiments on the effects of exogenously applied JA have additionally used in vitro-grown seedlings (10). Small seedlings offer a convenient system to follow the spatiotemporal localization of JA signaling. Furthermore, visualizing JA signaling in all seedling tissues opens avenues to investigate how JA responses are temporally and spatially organized between organs in the plant body.
Arabidopsis seedlings have previously been the system of choice in many forward genetic screens that successfully identified JA-signaling components. These screens chiefly used the reduction in root growth caused by exogenously applied JA as a phenotypic output (10–12). However, specific screens for JA-mediated wound signaling have not been reported for Arabidopsis, and only one such screen was performed in tomato (13). Other genetic screens have used JA-responsive reporter lines to recover mutants with constitutive JA signaling (14), but in only two cases the corresponding mutant gene was identified and characterized (15, 16).
As part of a continuing effort to identify organ-specific activators or repressors of wound signaling, we screened seedlings from an EMS-mutagenized population of JAZ10-GUSPlus, a JA-responsive reporter, for either impaired activation after cotyledon wounding or constitutive expression without mechanical stimulation. The screen yielded mutant alleles of known JA biosynthesis and signaling components including three ninja alleles. Here we report the unexpected finding that NINJA is indispensable in repressing JA signaling in roots and in maintaining normal root growth. Even in a null JA synthesis background, these mutants display growth phenotypes normally observed when seedlings are wounded or treated with JAs. Our results provide evidence for a previously unrecognized organ specificity of JA responses, and we extend this observation to JAR1, which is involved in the synthesis of biologically active JA-Ile, and MYC2, a transcriptional activator of JA signaling.
Results
JA-Responsive Reporter for Spatial Localization of Wound-Induced Signaling.
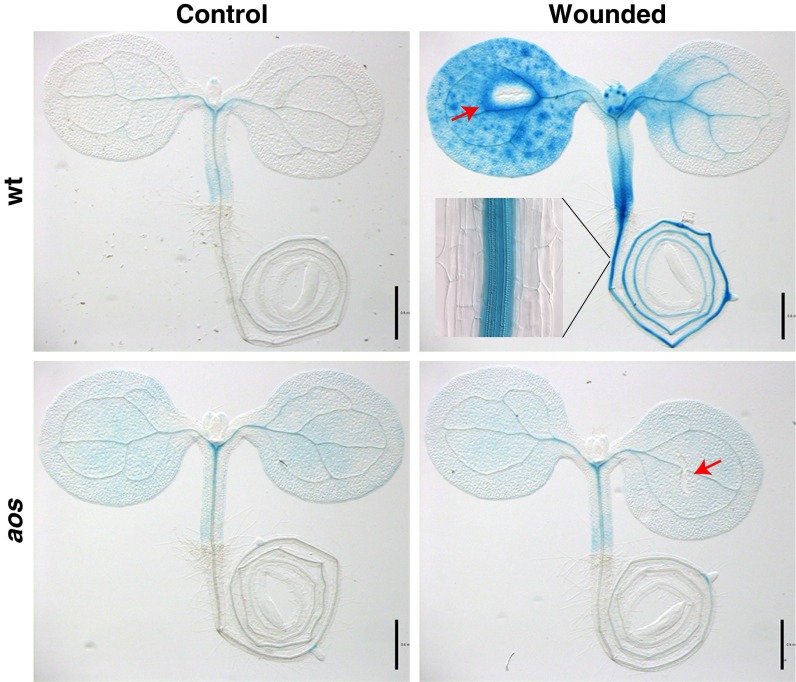
Mechanical wounding rapidly and strongly induces a well-characterized set of JA-dependent genes such as JAZ10 (17, 18). We used the promoter of this gene to drive the transcription of a secretable β-glucuronidase (GUS)–based reporter as a tool to visualize JA signaling in Arabidopsis. In untouched 5-d-old (d.o.) wild-type (WT) reporter seedlings, we detected weak JAZ10-GUSPlus (JGP) expression under our staining conditions (Fig. 1). However, piercing one cotyledon with a needle 2 h before GUS staining resulted in strong JGP activation not only in the wounded cotyledon but also in the shoots, the hypocotyl, and the root. The undamaged cotyledon displayed limited and more variable reporter staining, which suggests that long-distance wound signaling to this organ is weaker and/or takes more time to arrive. Wound-induced JGP expression was confined to the vasculature in the roots (Fig. 1, Inset) whereas in the aerial organs it spanned other tissues in addition to the vasculature. We have never observed JGP expression reaching the meristematic zone (MZ) of the primary root after cotyledon wounding, although cells of this zone respond strongly to exogenous methyl jasmonate (MeJA) (SI Appendix, Fig. S1), which is de-esterified in planta into the prohormone JA. Quantification of JAZ10 transcripts in roots and aerial organs of seedlings independently confirmed the strong induction of JAZ10 in both organ types after cotyledon wounding (SI Appendix, Fig. S2). We also crossed the JGP reporter into the JA-deficient mutant aos (19); wounding of this aos JGP line neither increased GUS activity (Fig. 1) nor induced high levels of JAZ10 transcripts (SI Appendix, Fig. S2), validating the JA dependency of the reporter line.
Fig. 1.
The JAZ10-GusPlus (JGP) reporter for JA-mediated wound signaling in 5-d.o. WT and aos mutant Arabidopsis seedlings. Detection of GUS activity was performed 2 h after wounding. Red arrows indicate cotyledon wounding sites. The reporter in the aos mutant background displays a constitutive faint blue staining in the aerial organs not observed in any other genotype and not attributable to a higher basal JAZ10 transcription (SI Appendix, Fig. S2). (Inset) Close-up of root. (Scale bars, 0.5 mm.)
coi1 and opr3 Mutant Alleles.
To search for genetically encoded activators of the wound response mediated by JA, we screened for mutant seedlings impaired in JGP reporter activation after cotyledon wounding. Five independent mutants (34A, 44B, 77A, 82B, 87A) were insensitive to wounding in the entire seedling, and four of them displayed insensitivity to MeJA treatment (SI Appendix, Fig. S1), suggesting that they were mutated in the JA receptor gene COI1. However, only lines 44B and 87A were completely male sterile as expected of coi1 null mutants, whereas in lines 34A and 82B male sterility was limited to the first few flowers (SI Appendix, Fig. S3). Complementation tests with the coi1-1 mutant (11) containing the JGP reporter and sequencing of the COI1 gene demonstrated that all four lines carried coi1 mutant alleles (SI Appendix, Table S1), hereafter designated coi1-32 and coi1-33 (fully male sterile) and coi1-8Lausanne (coi1-8L) and coi1-34 (partially male sterile). Note that the mutation in coi1-8L is identical to the previously reported coi1-8 (20).
The fifth wound-insensitive mutant (77A) was fully responsive to MeJA treatment (SI Appendix, Fig. S1) but displayed a slight sterility phenotype similar to that of coi1-8L and coi1-34 (SI Appendix, Fig. S3). The mutation causing this phenotype was identified by whole-genome sequencing and shown to lie in a splice donor site of the JA biosynthetic gene OXOPHYTODIENOATE-REDUCTASE 3 (OPR3) (21, 22). This mutant allele, designated opr3-2, expressed a longer-than-wild-type opr3 transcript that included an 81-nt intron (SI Appendix, Fig. S4 and Table S1). This does not disrupt the normal reading frame of the transcript but is predicted to add 27 amino acids between A116 and V117 to the final protein. The strongly impaired wound induction of JAZ10 transcripts in coi1-34 and opr3-2 seedlings was confirmed (SI Appendix, Fig. S5).
Roots Are the Main Activity Domain of JAR1.
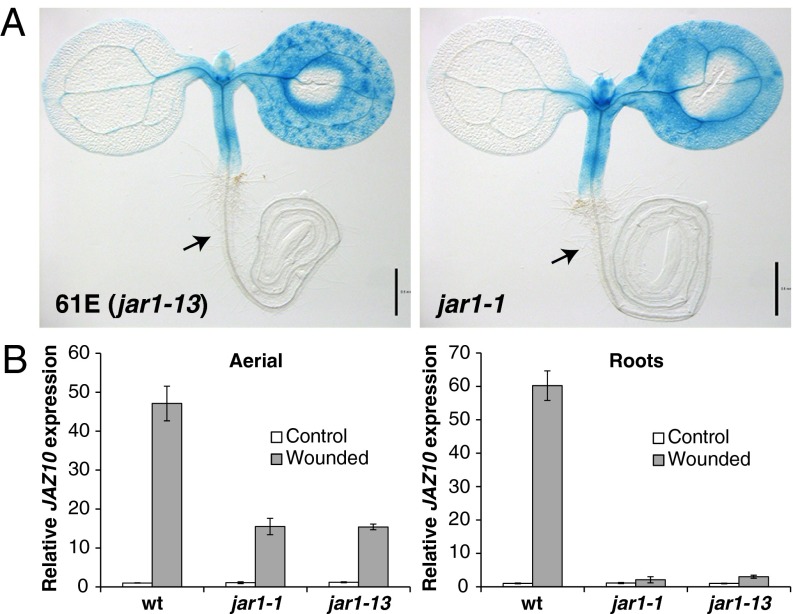
Next, we isolated mutant line 61E, where cotyledon wounding resulted in relatively normal JGP activity in the aerial organs but no or very little activity was observed in the roots (Fig. 2A; SI Appendix, Fig. S6). Concomitantly, we crossed the JGP reporter into the jar1-1 mutant (10) and observed a very similar phenotype, except that the lack of root response was fully penetrant (Fig. 2A; SI Appendix, Fig. S6). Crossing jar1-1 with line 61E did not complement the phenotype, indicating that 61E carried a mutant allele of JAR1, which we name jar1-13 (SI Appendix, Table S1). Quantification of JAZ10 transcripts showed that cotyledon wounding induced high JAZ10 levels in the aerial organs of the jar1 mutants, although they accumulated three times fewer transcripts than the WT (Fig. 2B). In contrast, roots of the wounded jar1 mutants did not induce JAZ10 transcription (jar1-1) or just barely (jar1-13). This phenotype could be due to a delay in long-distance JA signaling from the wounded aerial organs to the root. Therefore, we tested the JGP reporter at several late time points after wounding (up to 8 h) in WT and jar1-1 seedlings (SI Appendix, Fig. S7). In WT, JGP activity was saturated at 3 h postwounding in all responsive tissues. In jar1-1 aerial organs, reporter staining was weaker than WT at 2 h postwounding but also reached saturation after 3 h. Nevertheless, jar1-1 roots did not display reporter activity at any of the time points considered. Additionally, jar1-1 seedlings treated with MeJA for 2 h activated JGP in aerial organs but not in the root (SI Appendix, Fig. S8). In all these treatments jar1 mutants showed a relatively sharp separation of responsive vs. insensitive tissue around the hypocotyl–root interface.
Fig. 2.
jar1 mutations impair wound-induced JAZ10 expression mainly in roots. (A) JGP expression in 61E (jar1-13) and jar1-1 seedlings after cotyledon wounding. Black arrows indicate lack of staining in the root of wounded jar mutants. (Scale bars, 0.5 mm.) (B) Quantitative RT-PCR (qRT-PCR) of JAZ10 expression 1 h after wounding in aerial organs and roots of WT, jar1-1, and jar1-13 seedlings. JAZ10 transcript levels were normalized to those of UBC21 and displayed relative to the expression in the WT unwounded controls. Bars represent the means of three biological replicates (±SD), each containing a pool of organs from ∼60 seedlings.
Differential Effects of JA-Signaling Mutations in Resistance to a Generalist Herbivore.
To investigate whether jar1 mutations affect defense responses in aerial tissues, we tested the performance of our mutant set when challenged with larvae of the generalist lepidopteran Spodoptera littoralis. After 10 d of feeding, the average insect weights gained from the mutants impaired in JA signaling fell into two main categories (SI Appendix, Fig. S9). Insects gained the most weight in the full loss-of-function mutants coi1-1 and aos, as expected (23). This class included the coi1-34 mutant, which was as susceptible to the herbivores as aos and almost as susceptible as coi1-1.
The second group included jar1-1, jar1-13, and opr3-2, which were partially impaired in insect resistance. A similar intermediate phenotype was observed in Nicotiana attenuata lines silenced in JAR1 homologs (24), and it correlates with our observation that wound-induced JAZ10 transcription was decreased but still present in aerial tissues of jar1-1 and jar1-13 (Fig. 2B). Such clear correlation was not seen for opr3-2, where JAZ10 transcript induction upon wounding was almost completely suppressed (SI Appendix, Fig. S5), but herbivore resistance was only partially impaired.
NINJA Plays a Major Role as a Repressor of JA Signaling in Roots.
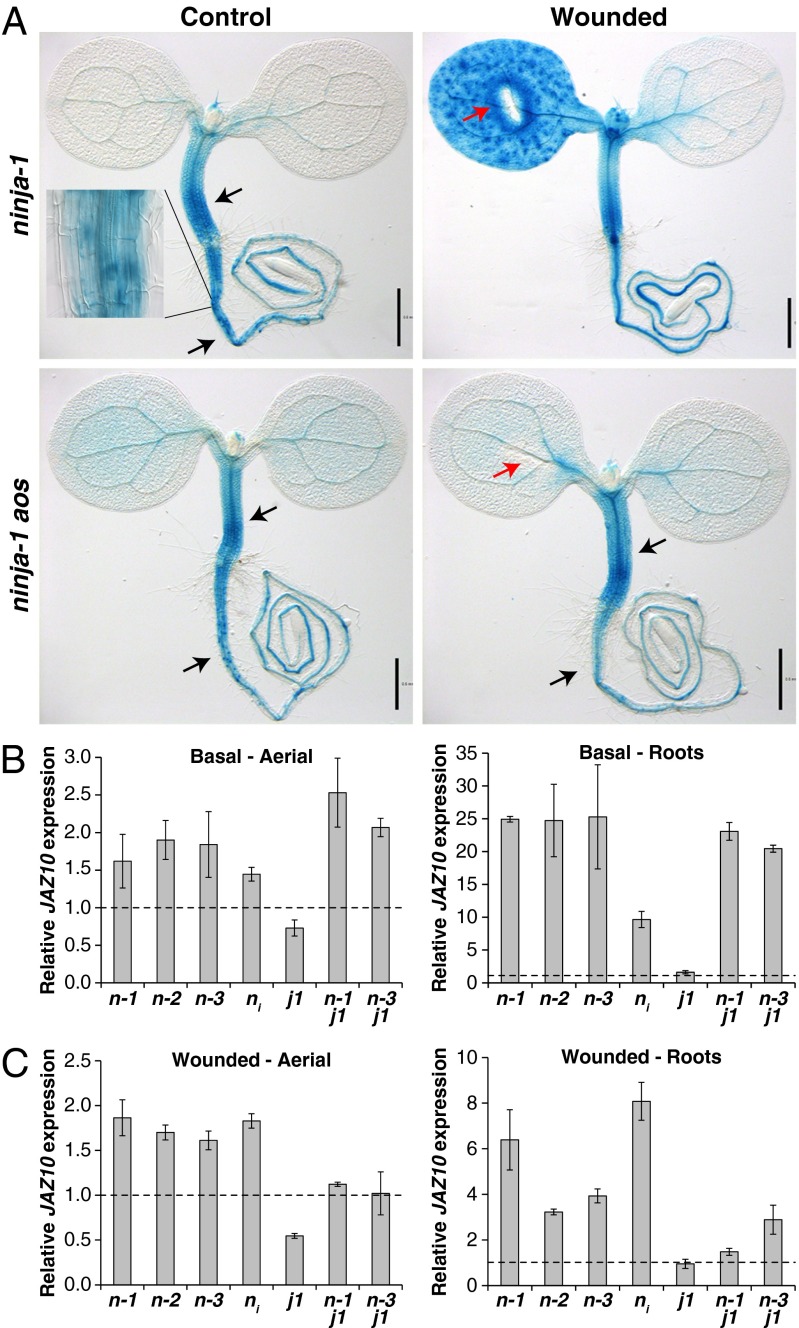
Next, we isolated three mutant lines displaying constitutive JGP activity in roots and hypocotyls without previous wounding (Fig. 3A; SI Appendix, Fig. S10). The staining in the mutant roots was present not only in the vasculature, which is the sole site of root JGP expression induced by cotyledon wounding in the WT (Fig. 1), but also in cortex cells of the collet and uppermost root regions. The constitutive activation also tended to reach farther down into the elongation zone (EZ), but it was not observed in the root MZ. Cotyledon wounding of these mutants induced JGP expression patterns similar to those in the WT (Fig. 3A; SI Appendix, Fig. S10). Whole-genome sequencing showed that all three mutants carried lesions in the gene encoding the corepressor NINJA and are designated ninja-1, ninja-2, and ninja-3 hereafter.
Fig. 3.
ninja mutants display constitutive JAZ10 expression. (A) JGP expression in control and wounded seedlings of ninja-1 and ninja-1 aos (compare with WT and aos in Fig. 1). Arrows indicate constitutive reporter activity in hypocotyl and roots (black) and cotyledon wounding sites (red). (Inset) Close-up of root. The ninja-2 aos double mutant displays the same expression pattern as ninja-1 aos. (Scale bars, 0.5 mm.) (B and C) qRT-PCR of JAZ10 expression basally (B) and 1 h after wounding (C) in ninja-1 (n-1), ninja-2 (n-2), ninja-3 (n-3), ninja RNAi (ni), jin1-7 (j1), ninja-1 jin1-7 (n-1 j1), and ninja-3 jin1-7 (n-3 j1). JAZ10 transcript levels were normalized to those of UBC21 and displayed relative to the expression of unwounded WT controls (B) or wounded WT samples (C); thus, WT levels are set to 1 in each plot and indicated with a dashed line. Bars represent the means of three biological replicates (±SD), except for n-2 and ni (two biological replicates), each containing a pool of organs from ∼60 seedlings. Complete qRT-PCR data are in Dataset S1.
JAZ10 expression was quantified in ninja mutants and a previously characterized RNAi line (4). Nontreated aerial organs of all four lines expressed ∼1.7 times more JAZ10 transcripts than the WT (Fig. 3B). These modest but significantly higher JAZ10 basal levels are in agreement with those reported for adult leaves of NINJA RNAi lines (4). In contrast, we observed a stronger effect in JAZ10 basal levels in the roots of the three ninja mutants, where 25 times more JAZ10 transcripts were detected and, in the NINJA RNAi line, with around 10 times more JAZ10 than WT (Fig. 3B).
One hour after wounding JAZ10 transcripts had accumulated to higher-than-WT levels in the ninja lines (Fig. 3C). Again, this effect was more prominent in roots (between 3 and 8 times more transcripts with respect to WT) than in aerial organs (∼1.8 times higher than the WT). A moderate up-regulation of multiple JA-regulated genes has been previously observed in leaves of a NINJA RNAi line (4). These include several defense genes such as VSP2, whose up-regulation we also confirmed in our ninja mutants (SI Appendix, Fig. S11). However, the mild increase in basal and induced JA signaling in ninja aerial organs did not result in enhanced resistance against S. littoralis larvae under our experimental conditions, even after prolonged feeding times (SI Appendix, Figs. S9 and S12).
A ninja-1 aos double mutant was mostly indistinguishable from the ninja-1 single mutant with respect to the constitutive JGP expression in hypocotyls and roots (Fig. 3A). Therefore, in those tissues loss of NINJA function is sufficient to abolish the repression on JA-responsive promoters, and JA is not required to activate them. The constitutive wound-like JGP expression in the hypocotyl and root of the ninja-1 aos double mutant did not change upon wounding, but cotyledons and shoots still failed to activate JGP (Fig. 3A). Moreover, ninja-1 aos and ninja-2 aos were as sensitive to herbivore attack as the aos mutant (SI Appendix, Fig. S13). Therefore, herbivory resistance in aerial organs of ninja mutants still required de novo JA biosynthesis and was not detectably influenced by the strong constitutive JA signaling of the roots.
The three ninja mutations that we characterized are predicted to cause partial or complete removal of NINJA’s C domain (SI Appendix, Figs. S14 and S15; Table S1), which is required for interaction with the JAZ repressors in yeast two-hybrid assays (4). To test if the ninja mutations impair JAZ repressor function in planta, we transformed the ninja mutants with constructs overexpressing JAZ10.3 and JAZ10.4, two truncated splice variants of JAZ10 that are resistant to COI1-mediated degradation in the presence of JA, effectively repressing JA responses (17, 25). As expected, a significant fraction of transformed (T1) WT seedlings overexpressing JAZ10.3 and JAZ10.4 displayed MeJA insensitivity in a root growth assay (SI Appendix, Fig. S16). However, T1 ninja seedlings remained more sensitive to MeJA than T1 WT, suggesting that the ninja mutations attenuated the repression mediated by JAZ10.3 and JAZ10.4. The extent of this attenuation remains to be assessed in independent T2 families.
The deregulated expression of JA response genes in the ninja mutants could be attributed to the activity of one or more JA-dependent transcription factors (TFs) normally subjected to JAZ-NINJA-TPL repression. Because the basic helix–loop–helix TF MYC2 is a central activator in several aspects of JA signaling (12, 26), we introgressed the ninja alleles into a myc2 mutant (jin1-7) (12). The constitutive JAZ10 expression was still present in aerial and root organs of ninja-1 jin1-7 and ninja-3 jin1-7 double mutants (Fig. 3B), suggesting that MYC2 has no role or is not the sole factor promoting deregulated JA signaling in ninja. For example, even if MYC2 has a major function in the root response to exogenously supplied JA (12, 26), we found that JAZ10 induction in roots after cotyledon wounding was not affected in jin1-2 and jin1-7 seedlings (Fig. 3C; SI Appendix, Fig. S17; Dataset S2). Instead, it was abolished only in a triple mutant between myc2 and two closely related TFs: myc3 and myc4 (SI Appendix, Fig. S17). On the other hand, myc2 single mutants displayed reduced JAZ10 expression in aerial organs after cotyledon wounding (Fig. 3C; SI Appendix, Fig. S17). This effect was also observed in ninja jin1-7 aerial organs, where the excess JAZ10 expression of ninja mutants after wounding was eliminated (Fig. 3C). A less clear effect was seen in roots of wounded double mutants, where the influence of jin1-7 on JAZ10 levels was only mild in ninja-3 but more prominent in ninja-1.
Primary Root Shortening in ninja as a Result of Reduced Cell Elongation.
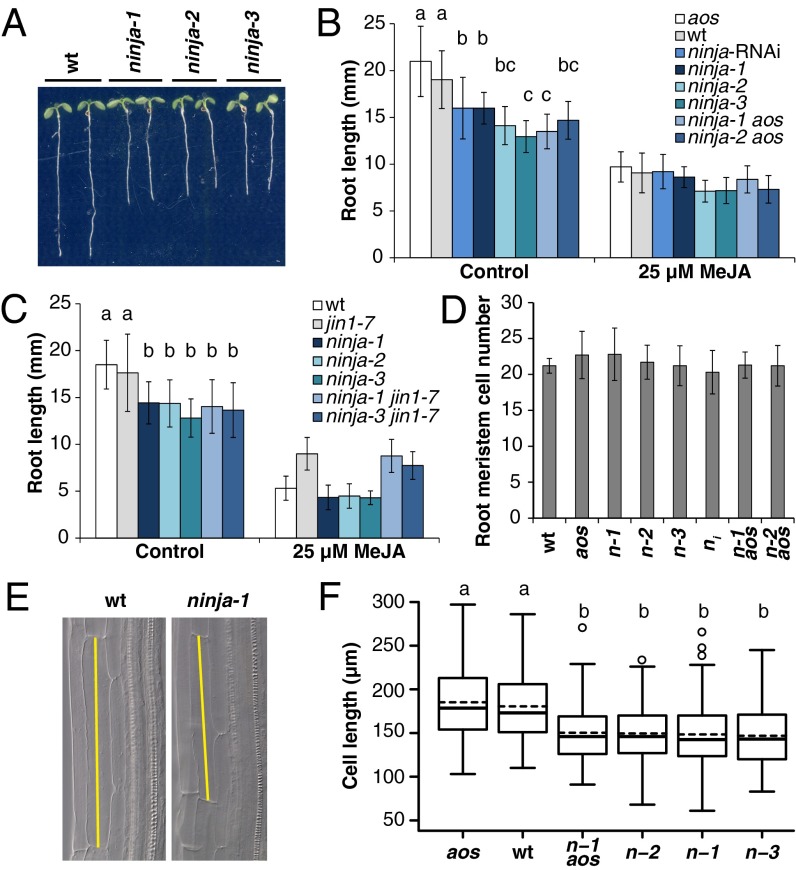
When grown in vertical plates, ninja mutant and RNAi lines had up to 30% shorter roots than WT (Fig. 4 A–C), indicating that lack of functional NINJA partly mimicked JA-mediated root growth inhibition. Remarkably, this short root phenotype was also present in ninja aos and ninja jin1-7 double mutants (Fig. 4 B and C), suggesting that loss of NINJA function bypasses the need of JA to repress root growth and that this may occur in the absence of MYC2. When grown in the presence of MeJA, ninja roots are as short as WT, and no hypersensitivity to MeJA was observed under the conditions tested (Fig. 4 B and C). Similarly, the roots of ninja jin1-7 were not different from the jin1-7 single mutant in media containing MeJA (Fig. 4C).
Fig. 4.
Reduced root growth and cell length in untreated ninja lines. (A) Representative 7-d.o. seedlings of WT and ninja mutants. (B and C) Quantification of root growth in WT and mutant lines grown in the absence or presence of 25 µM MeJA for 7 d. Data are the means (±SD) from at least 20 plants. Letters above bars indicate statistically significant differences between pairs of control samples as determined by Tukey’s honestly significant difference (HSD) test (P < 0.02). (D) Primary root meristem cell number in 5-d.o. seedlings of WT and several mutant lines. No statistically significant difference between genotypes was found by ANOVA (P = 0.475). (E) Representative cortex cells from WT and ninja-1 mutant roots. Yellow bars indicate cell length. (F) Box plot summary of cortex-cell length in 5-d.o. seedlings of WT and several mutant lines grown in the absence of MeJA. Medians and means are represented inside the boxes by solid and dashed lines, respectively. Circles depict outlier data points beyond ±1.5× the interquartile range defined by the whiskers. Letters indicate statistically significant differences between pairs as determined by Tukey’s HSD test (P < 2e-16).
Finally, we assessed if ninja root shortening was associated with a reduction in cell proliferation or cell elongation or, as when the WT is grown in the presence of exogenous MeJA, a combination of both (27). As an indicator of cell proliferation, we determined cell number in the root MZ of WT and ninja mutants, and no significant difference between them was detected (Fig. 4D). Conversely, cortex cell length in the differentiation zone (DZ) of ninja mutants was on average 20% shorter than in the WT (Fig. 4 E and F; SI Appendix, Table S2). Therefore, we attribute the root shortening in ninja primarily to a reduction in cell elongation.
Discussion
How is the specificity and diversity of JA responses determined throughout the plant? By spatially visualizing the effects of early JA signaling, we provide evidence that part of this diversity can be explained by organ specificity of NINJA, a member of the JA transcriptional repression complex, and of JAR1, the JA-Ile–conjugating enzyme. JAR1 and NINJA are indispensable to, respectively, activate and repress JA signaling in roots, providing a unique opportunity to study a complex network of interactions at an organ level. Roots also rely on a single member of the LIPOXYGENASE family, LOX6, for stress-induced JA accumulation (28). Together, these findings imply that the JA-signaling machinery is less redundant in the roots than in the aerial organs of Arabidopsis. How such arrangements evolved and what ecological consequences, if any, they have are open questions.
Additional coi1 and opr3 Alleles Compromised in JA-Mediated Wound Signaling.
The search for mutant seedlings impaired in JGP reporter activation after cotyledon wounding yielded one opr3, one jar1, and four coi1 mutant alleles, confirming the specificity of our screen. Arabidopsis COI1 has been independently identified in several forward genetic screens (11, 20) because it is a single-copy gene that is necessary for all JA-mediated processes known to date (29). Whereas all coi1 alleles identified herein showed abolished wound responses and JA sensitivity in seedlings, coi1-8L and coi1-34 were almost completely fertile. These two mutations cause amino acid substitutions at the end of β-sheets, forming the structurally important leucine-rich repeat (LRR) domains in COI1 (20). In coi1-8, this defect was previously reported to partially reduce COI1 protein stability while maintaining the ability to interact with recombinant JAZ1 protein (20). Our results show that this type of hypomorphic COI1 function is sufficient for JA-mediated fertility but insufficient for near-WT defense. Instead, full loss-of-function mutations such as coi1-33 affect the α-helices of the LRR domains as in the coi1-4, coi1-7, and coi1-9 mutants, which fail to accumulate the COI1 protein (20). On the other hand, the JA biosynthetic mutant opr3-2 is also hypomorphic, but the mutant enzyme is still partially functional in both fertility and defense responses. Even if JGP and JAZ10 expression are not properly induced in opr3-2 shortly after wounding, the partial herbivore resistance phenotype suggests that some JA signaling may still occur.
JAR1 Is Indispensable for Activating JA Signaling in Roots After Cotyledon Wounding.
It has been shown previously that JAR1 activity is not strictly required for the wound-induced expression of JA-responsive genes in rosette leaves, where other JA-amino synthetases may exist to account for the remaining JA-Ile levels in null jar1 mutants (8, 18, 30). Other reports indirectly support a primary role for JAR1 in the roots. First, JAR1 mRNA levels are highest in seedling roots (30). Second, jar1-1 displayed a root-specific impairment in the activation of a wound-induced FAD7p-LUC reporter (16). Third, jar1-1 is as susceptible as the JA-deficient mutant fad3-2 fad7-2 fad8 to Pythium irregulare, a soil-borne pathogen that infects roots (31). We found that jar1-1 and jar1-13 displayed an intermediate susceptibility to S. littoralis between that of the WT and coi1-1. This could be attributed to delayed or decreased JA-Ile accumulation that activates slower or reduced downstream defense responses (8, 30). In contrast, cotyledon wounding in jar1-1 and jar1-13 failed to activate JGP and JAZ10 expression in roots. JGP remained inactive in jar1-1 roots even at later cotyledon postwounding times as well as after short-term MeJA treatment. Taken together, our data indicate that the seedling wound response partly requires JAR1 in aerial organs whereas the roots are completely dependent on JAR1 activity. Nevertheless, when germinated and grown in medium supplied with exogenous MeJA, jar1 mutant seedlings retain partial sensitivity to JA (10). Thus, it is probable that exogenous JA overrides endogenous responses. Our results also favor the view that bioactive JA-Ile is not transported from wounded aerial tissues to basal organs. Instead, JA-Ile is most likely synthesized de novo in roots specifically through JAR1 action after the arrival of long-distance signals. This view is consistent with results from recent grafting experiments that demonstrated that roots produce JA and JA-Ile independently of leaves (28).
NINJA Is Essential for Repressing Basal JA Signaling and Promoting Cell Elongation in Roots.
Here, we report mutants in NINJA, a transcriptional corepressor of JA response genes that bridges target JAZ proteins to members of the TPL corepressor family (4). The lack of NINJA’s C domain in all three ninja mutant alleles most likely prevents its interaction with the ZIM domain of JAZ proteins and the recruitment of TPL corepressors to the JA transcriptional repression complex. This prediction is supported by our preliminary evidence that ninja mutants are less affected by the JA insensitivity caused by the nondegradable repressors JAZ10.3 and JAZ10.4. Although NINJA transcripts were detectable in both aerial organs and roots (SI Appendix, Fig. S14; Arabidopsis eFP browser, http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), loss of NINJA function caused ectopic JGP and JAZ10 expression predominantly in hypocotyls and roots, revealing a spatial specificity of the NINJA-dependent complex that represses basal JA signaling.
The major impact of NINJA on the basal repression of root JA signaling could be explained if other as-yet-unknown corepressors act redundantly with NINJA to constitutively inhibit JA signaling in aerial organs (and the root MZ), whereas NINJA is sufficient to repress early JA signaling in the older parts of the primary root. It is also possible that more direct mechanisms repress JA-dependent transcription in aerial organs. First, JAZ8 can act independently of NINJA because it carries an ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif capable of directly binding TPL (5). This feature is potentially shared by JAZ5, JAZ6, and JAZ7 (32). Second, JAZ1 may directly recruit the chromatin-modifying enzyme HISTONE DEACETYLASE 6 (HDA6) to reduce chromatin accessibility and repress gene expression (33). Alternatively, the tissue-specific differences in constitutive gene expression after losing NINJA-mediated repression may be a reflection of quantitative differences in the activity or basal concentration of specific JA-dependent transcriptional activators between organs.
Exogenously supplied JA is known to inhibit root growth by reducing both root meristem activity and cell length in the DZ (27). Our results, based on genetic approaches that did not require treatment with exogenous JA, indicate that root shortening in ninja mutants is primarily due to a reduction in cell length in the DZ. That is, a major role of NINJA in roots is in cell-size control. This agrees with the pattern of constitutive JGP expression observed in ninja roots, where derepression of the JAZ10 promoter encompasses the EZ and the DZ but not the MZ. The growth reduction in ninja mutant roots is most likely attributable to increased signaling through transcriptional activators of JA responses such as MYC2, which is normally repressed by a JAZ-NINJA-TPL complex (4, 26). However, a myc2 mutation did not suppress the ectopic JAZ10 expression and short root phenotypes in ninja. It is possible that, as in the wound response, MYC2 acts additively in the ninja mutants with MYC3 and MYC4, two related TFs that also form complexes with JAZ repressors and NINJA in vivo (4, 26). In this case, a combination of myc2, myc3, and/or myc4 mutants will potentially suppress the ninja phenotype. Alternatively, in seedlings grown under exogenous JA the major role of MYC2 is reducing root meristem cell number by directly repressing PLETHORA genes (27). When grown in the presence of JA, myc2 mutants showed normal root meristem size but retained partial sensitivity to JA-mediated root growth inhibition (27), which can be attributed to reduced cell elongation in the DZ independently of MYC2 function. In this respect, NINJA and MYC2 are likely to regulate different aspects (elongation vs. proliferation) of root growth.
Another possibility is that the ninja mutants derepress uncharacterized JA-specific growth factors that may explain the residual sensitivity of myc234 lines to JA-mediated root growth inhibition (26). The ninja mutants provide the opportunity to (i) uncover these factors and mediators of hormonal cross-regulation and (ii) study TF-specific effects on early JA signaling and cell elongation. However, it remains formally possible that the role of NINJA on root growth is independent of JA signaling.
Role of NINJA in Induced JA Signaling.
The higher-than-WT JAZ10 accumulation in both aerial and root organs of ninja mutants (Fig. 3C) (4) suggests an active role of NINJA in repressing JA-dependent TFs after cotyledon wounding. These excess JAZ10 levels were abolished in aerial organs of ninja jin1-7 double mutants, implying that NINJA corepressor function is required to attenuate MYC2 signaling after the wounding stimulus in aerial organs. However, unlike aerial organs where cotyledon wounding needs MYC2 to activate normal JA signaling, roots do not absolutely require MYC2 to induce WT JAZ10 levels because most likely MYC3 and MYC4 can also perform this function (SI Appendix, Fig. S17). This, and the finding that the constitutive JA signaling in ninja mutants can occur independently of MYC2, is in contrast with the major role attributed to this TF in root responses to exogenous JA (12, 26), emphasizing that the functional hierarchy of JA-signaling components varies according to both tissue and stimulus.
Upon strong and continuous stimulation of signaling in the presence of exogenous MeJA, the role of NINJA in suppressing JA responses becomes less evident. For example, we did not find ninja roots to be hypersensitive to JA, suggesting that NINJA exerts its corepression function mainly when JAZ proteins are most stable and not when they are rapidly degraded under a constant exogenous supply of JA. It is plausible that NINJA-independent repression mechanisms—mediated, for example, by JAZ8 (5) or JAZ1-HAD6 (33)—become predominant in such conditions. The increased sensitivity to JA of jaz10 loss-of-function plants (17, 34) further indicates that JAZ10 can act through NINJA-independent mechanisms to repress induced JA signaling.
This work describes the possibility of root growth inhibition by the JA-signaling machinery in the absence of JA synthesis. Our findings show that (i) it may be possible to manipulate what is normally JA-controlled growth inhibition in discrete parts of plants such as hypocotyls and roots, without the necessity of altering either JA synthesis or perception; and (ii) the ninja mutants will be powerful tools to dissect unexplored cellular events activated downstream of JA signaling to restrict root cell elongation. A deeper understanding of cell-size control will be crucial to successfully engineer plants that display reduced growth restriction under stress.
Materials and Methods
Arabidopsis thaliana accession Columbia was the WT background of the JGP reporter and all mutant and knock-down lines. Two different genetic screens were performed on EMS-mutagenized M2 seedlings that were evaluated for JGP activity by nondestructive GUS staining. In one screen, we searched for loss of GUS staining in the root after cotyledon wounding, and in the other we screened for constitutive GUS activity in the root of unwounded seedlings. Mutant alleles were identified by whole-genome next generation sequencing (NGS). Detailed experimental procedures of JGP reporter line generation, growth conditions, treatments, NGS, and measurements can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Christian Hardtke for bioinformatics support and comments on the manuscript, Niko Geldner for advice on reporter constructs, Philippe Reymond for S. littoralis eggs, and the Lausanne Genomic Technologies Facility for whole-genome sequencing. We also thank Alain Goossens for the NINJA RNAi line, Roberto Solano for the myc mutant combinations, and René Dreos for R expertise. This work was supported by Swiss National Science Foundation Grants 3100A0-122441 and 31003A-138235 and by EMBO Long-Term Fellowship ALTF 5-2009 (to I.F.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307910110/-/DCSupplemental.
References
- 1.Koo AJ, Howe GA. The wound hormone jasmonate. Phytochemistry. 2009;70(13–14):1571–1580. doi: 10.1016/j.phytochem.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 3.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 4.Pauwels L, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464(7289):788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shyu C, et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24(2):536–550. doi: 10.1105/tpc.111.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468(7322):400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glauser G, et al. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J Biol Chem. 2008;283(24):16400–16407. doi: 10.1074/jbc.M801760200. [DOI] [PubMed] [Google Scholar]
- 8.Koo AJ, Gao X, Jones AD, Howe GA. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009;59(6):974–986. doi: 10.1111/j.1365-313X.2009.03924.x. [DOI] [PubMed] [Google Scholar]
- 9.Mielke K, Forner S, Kramell R, Conrad U, Hause B. Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytol. 2011;190(4):1069–1080. doi: 10.1111/j.1469-8137.2010.03638.x. [DOI] [PubMed] [Google Scholar]
- 10.Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA. 1992;89(15):6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6(5):751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16(7):1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lightner J, Pearce G, Ryan CA, Browse J. Isolation of signaling mutants of tomato (Lycopersicon esculentum) Mol Gen Genet. 1993;241(5–6):595–601. doi: 10.1007/BF00279902. [DOI] [PubMed] [Google Scholar]
- 14.Lorenzo O, Solano R. Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol. 2005;8(5):532–540. doi: 10.1016/j.pbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14(7):1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda O, Sakamoto H, Nakao Y, Oda K, Iba K. CTD phosphatases in the attenuation of wound-induced transcription of jasmonic acid biosynthetic genes in Arabidopsis. Plant J. 2009;57(1):96–108. doi: 10.1111/j.1365-313X.2008.03663.x. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19(8):2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung HS, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146(3):952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JH, et al. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31(1):1–12. doi: 10.1046/j.1365-313x.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21(8):2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders PM, et al. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell. 2000;12(7):1041–1061. doi: 10.1105/tpc.12.7.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA. 2000;97(19):10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc Natl Acad Sci USA. 2001;98(22):12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Allmann S, Wu J, Baldwin IT. Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 2008;146(3):904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21(1):131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell. 2011;23(9):3335–3352. doi: 10.1105/tpc.111.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grebner W, Stingl NE, Oenel A, Mueller MJ, Berger S. Lipoxygenase6-dependent oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 2013;161(4):2159–2170. doi: 10.1104/pp.113.214544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry. 2009;70(13–14):1539–1546. doi: 10.1016/j.phytochem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Suza WP, Staswick PE. The role of JAR1 in Jasmonoyl-L: −Isoleucine production during Arabidopsis wound response. Planta. 2008;227(6):1221–1232. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 31.Staswick PE, Yuen GY, Lehman CC. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998;15(6):747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 32.Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152(3):1109–1134. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(30):12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno JE, et al. Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 2013;162(2):1006–1017. doi: 10.1104/pp.113.218164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.