Significance
During protein synthesis, ribosomes catalyze peptide-bond formation between amino acids with differing efficiency. We show that two or more consecutive prolines induce ribosome stalling, and that stalling strength is influenced by the amino acid preceding and following the prolines. In bacteria, the elongation factor EF-P efficiently rescues the ribosome stalling irrespective of the XPP or PPX motif.
Abstract
Ribosomes are the protein synthesizing factories of the cell, polymerizing polypeptide chains from their constituent amino acids. However, distinct combinations of amino acids, such as polyproline stretches, cannot be efficiently polymerized by ribosomes, leading to translational stalling. The stalled ribosomes are rescued by the translational elongation factor P (EF-P), which by stimulating peptide-bond formation allows translation to resume. Using metabolic stable isotope labeling and mass spectrometry, we demonstrate in vivo that EF-P is important for expression of not only polyproline-containing proteins, but also for specific subsets of proteins containing diprolyl motifs (XPP/PPX). Together with a systematic in vitro and in vivo analysis, we provide a distinct hierarchy of stalling triplets, ranging from strong stallers, such as PPP, DPP, and PPN to weak stallers, such as CPP, PPR, and PPH, all of which are substrates for EF-P. These findings provide mechanistic insight into how the characteristics of the specific amino acid substrates influence the fundamentals of peptide bond formation.
Protein synthesis in the cell occurs on macromolecular machines called ribosomes. The ribosome synthesizes polypeptide chains by providing a platform upon which peptide-bond formation can occur between a peptidyl-tRNA located at the ribosomal P-site and an aminoacyl-tRNA in the A-site. However, the ribosome cannot form peptide bonds between all amino acids with the same efficiency; this is exemplified by the amino acid proline, which has an imino group instead of a primary amino group in other amino acids. Proline has been shown to be a particularly poor substrate for peptide-bond formation, both as a donor in the P-site and as an acceptor in the A-site (1–4). In fact, ribosomes stall when attempting to incorporate three or more consecutive proline residues (PPP) into the polypeptide chain (5–7). In this case, ribosome stalling results from the slow rate of peptide-bond formation between the peptidyl-Pro-Pro-tRNA located in the P-site and the Pro-tRNA in the A-site (6). In bacteria, the translational arrest is relieved by the translation elongation factor P (EF-P), which binds to the stalled ribosomes and stimulates peptide bond formation (5, 6). In Escherichia coli, EF-P is posttranslationally modified by YjeA, YjeK, and YfcM (EpmA, EpmB, and EpmC) (8–10) and the resulting lysinylation modification has been shown to be critical for the rescue activity of EF-P in vivo and in vitro (5, 6). EF-P homologs exist in all archaea and eukaryotes, termed aIF-5A and eIF-5A, respectively (11). Yeast eIF-5A has recently been shown to also rescue translational stalling at polyproline-stretches (12). Like EF-P, a/eIF-5A is also posttranslationally modified, but via hypusinylation rather than lysinylation (11). In addition to PPP, PPG triplets also induce ribosome stalling in bacteria, which is rescued by EF-P (6). Moreover, a recent proteomics study identified APP, YIRYIR, and GSCGPG motifs as conferring EF-P dependent translation (13). Ribosome stalling has also been observed at PPA, PPD, PPE, PPN, PPW, APP, and WPP triplets during in vitro translation (7), however it was not shown whether EF-P can relieve the translational arrest at these triplets. Ribosome profiling data indicate ribosome accumulation at PPP, PPG as well as PPD and PPE triplets, despite the profiling being performed with wild-type cells containing EF-P or eIF-5A (7, 14, 15).
To address the role of EF-P during translation in E. coli in vivo, we used SILAC (stable isotope labeling by amino acids in cell culture) coupled with high-resolution mass spectrometry (MS) to monitor the changes in expression levels of proteins in E. coli strains lacking either the efp gene, or one of the genes (yjeK, yjeA, yfcM) encoding the modification enzymes, relative to the parental wild-type E. coli strain. We found that in the absence of EF-P, YjeA, or YjeK, the majority of PPP-containing proteins are strongly down-regulated, whereas only specific subsets of XPP- and PPX-containing proteins are down-regulated. A systematic analysis of each of the 39 XPP/PPX combinations (where X means any amino acid) reveals the hierarchy of EF-P dependence. Moreover, we show in vitro and in vivo that the combinations of strong XPP with strong PPX motifs lead to XPPX quadruplets with the strongest effects, which are nevertheless efficiently relieved by EF-P. Collectively, our findings broaden the substrate range for EF-P activity from ∼100 PPP-containing proteins in E. coli to encompass the >1,300 additional XPPX-containing proteins. Eukaryotic proteomes, such as that of Homo sapiens contain >7,000 PPP-containing proteins and >15,000 XPPX-containing proteins that are all potential substrates for eIF-5A.
Results
Proteomic Analysis of Δefp, ΔyjeK, ΔyjeA, and ΔyfcM Strains.
SILAC was performed by growing the E. coli K-12 mutant (Δefp, ΔyjeK, ΔyjeA, or ΔyfcM and ΔlysA/ΔargA) strains in minimal medium containing the “light” forms of lysine (K0–12C614N2) and arginine (R0–12C614N4), and the wild-type E. coli parental strain (MG1655 ΔlysA ΔargA) in the “heavy” forms of lysine (K8–13C615N2) and arginine (R10–13C615N4). Cells were harvested at exponential growth phase, and the heavy-labeled (H) wild-type sample was mixed with each light-labeled (L) mutant sample in 1:1 ratio based on their total protein concentration. The relative changes in protein levels were then determined by calculating the inverted heavy/light (H/L) ratios for each mutant strain using MaxQuant (16). Our experimental setup resulted in 96 LC-MS measurements and 1,559,931 acquired MS/MS spectra. The database search led to identification of between 9,131 and 12,875 unique peptide sequences in individual replicate experiments, with an average absolute precursor ion mass accuracy of 0.33 ppm and SD of 0.45 ppm. Identified peptides mapped to 2,098 protein groups at 1% false discovery rate, comprising more than 48% of gene products in our sequence database (E. coli K-12 MG155). This result is well in line with a recent work where a total of 2,118 proteins were detected, of which 1,984 were quantified in E. coli grown in minimal medium (17). Of the 2,098 proteins identified across all experiments, between 1,418 and 1,687 were quantified (i.e., had two or more ratio counts in MaxQuant analysis) in a single experiment, and these were therefore used for all subsequent analysis (Dataset S1). The correlation between the two biological replicates for each mutant strain ranged between 87–95%, indicating high reproducibility of the data (Fig. S1). Furthermore, a high correlation (80–87%) also existed between the Δefp, ΔyjeK, and ΔyjeA datasets, consistent with the critical role that lysinylation of EF-P by YjeK and YjeA plays for EF-P activity (5, 6). Conversely, the low correlation (42–56%) observed between the Δefp/ΔyjeK/ΔyjeA and ΔyfcM data supports the observation that hydroxylation of EF-P by YfcM is not essential for EF-P activity (5, 6). The division between the Δefp/ΔyjeK/ΔyjeA and ΔyfcM data are also seen in the distribution of the protein ratios: Although the majority of normalized protein ratios was distributed around log2 = 0 for each dataset (demonstrating little or no change), the SD of the Δefp/ΔyjeK/ΔyjeA datasets from the median was larger, particularly for proteins with negative fold-change ratios (Fig. 1 A–C), compared with the ΔyfcM dataset (Fig. 1D). Similar trends were also obtained for the replicate samples (Fig. S2). Gene Ontology (GO) analysis indicates that the absence of active EF-P leads to down-regulation of proteins related to translation and ATP synthesis, and up-regulation of proteins related to chemotaxis, motility, and amino acid biogenesis (Fig. S3).
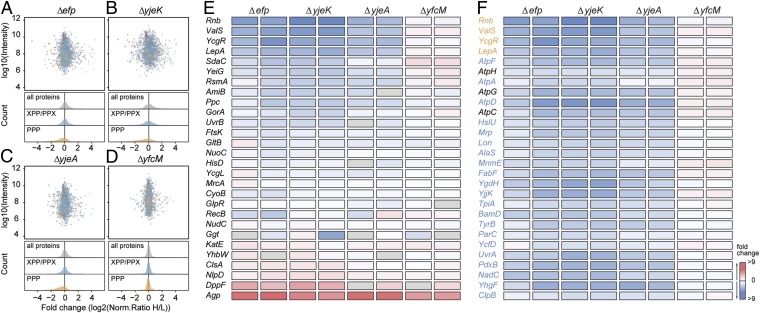
Fig. 1.
Proteomic analysis of Δefp, ΔyjeK, ΔyjeA, and ΔyfcM strains. (A–D) Scatter-plots of inverted normalized H/L ratios (log2-transformed) relative to the summed up protein intensity for a biological replicate of SILAC data from the Δefp (A), ΔyjeK (B), ΔyjeA (C), and ΔyfcM (D) strains, including density plots showing distribution of PPP- (gold) and XPP/PPX- (blue) containing proteins relative to all proteins (gray). (E) Heat map representation of selected PPP-containing proteins that were up- (red) and down- (blue) regulated in the biological replicates of Δefp, ΔyjeK, ΔyjeA, and ΔyfcM strains relative to wild-type strain (complete hierarchical clustering representation is available in Fig. S4). Gray boxes indicate that the protein was not identified or quantified. (F) As in E, but for selected proteins (PPP, gold; XPP/PPX, blue; non-PPP/PPX/XPP, black) that are down-regulated in Δefp, ΔyjeK, and ΔyjeA strains, but up-regulated or unchanged in ΔyfcM strains.
Down-Regulation of PPP-Containing Proteins in Δefp, ΔyjeK, and ΔyjeA Strains.
Because EF-P was shown to enhance translation of polyproline-containing proteins (5, 6), we addressed whether down-regulation of polyproline-containing proteins due to the absence of active EF-P was evident in the Δefp/ΔyjeK/ΔyjeA datasets. In total, we detected 44 of the 96 possible polyproline-containing proteins in our dataset (Dataset S1), 28 of which were quantified in at least one replicate experiment and 21 of which were quantified in both replicate experiments (Fig. 1E). As expected, we observed a marked shift in the distribution of protein ratios for the PPP-containing proteins in the Δefp/ΔyjeK/ΔyjeA, but not in the ΔyfcM data (Fig. 1 A–D): The median values for the PPP-containing protein normalized ratios were between −0.29 and −0.71 for Δefp/ΔyjeK/ΔyjeA compared with −0.04 and 0.04 for the ΔyfcM replicates. Consistently, hierarchical clustering reveals that most of the PPP-containing proteins are down-regulated in Δefp/ΔyjeK/ΔyjeA (blue in Fig. 1E), whereas in the ΔyfcM they remain unchanged or up-regulated (white or red, respectively, in Fig. 1E). The most strongly (and differentially) down-regulated polyproline-containing proteins are ribonuclease II (Rnb), Val-tRNA synthetase (ValS), the c-di-GMP-regulated flagellar velocity braking protein (YcgR), and translation elongation factor LepA (Fig. 1E), with ∼ninefold decreases in the protein ratios. Surprisingly, a number of PPP-containing proteins exhibit increased fold changes, such as DppF and Agp, indicating these proteins are up-regulated despite the lack of active EF-P. In these cases, we note that up-regulation is also observed in ΔyfcM, suggesting that the response is likely to be more general rather than directly related to EF-P. Hierarchical clustering of the entire set of 1,149 proteins quantified in each replicate of the Δefp/ΔyjeK/ΔyjeA/ΔyfcM strains (Fig. S4) reveals that many non-PPP–containing proteins cluster with the PPP-containing proteins (such as Rnb, ValS, YcgR, and LepA) i.e., also being down-regulated in Δefp/ΔyjeK/ΔyjeA, yet remaining unchanged or up-regulated in ΔyfcM (Fig. 1F). These non-PPP–containing proteins are assigned to diverse biological processes, cellular components, and molecular functions, and do not exhibit significant overlap with the assignments of the PPP-containing proteins. Therefore, we do not believe that the down-regulation of these non-PPP–containing proteins results from their being in pathways or processes directly related to the down-regulated PPP-containing proteins.
Because EF-P also relieves translational stalling at PPG triplets (6), we next examined whether these non-PPP–containing proteins contained PPG motifs. Although the PPG triplet was present in some of the non-PPP–containing proteins, such as AtpD, HslU, Mrp, Lon, AlaS, and MnmE, the majority of proteins lacked both PPP and PPG (Fig. 1F). Because proteomics studies identified APP as conferring EF-P dependence (13) and translational stalling has been shown to occur at other PPX triplets, namely PPA, PPD, PPE, PPN, and PPW (7, 15), we examined whether the differentially regulated non-PPP–containing proteins do in fact contain any XPPX motifs. Our analysis indicates that the vast majority (∼80%) does indeed contain at least one XPPX motif, for example FabF, YgdH, TpiA, and the protease Lon have XPPE, whereas BamD, YgdH, and TyrB have XPPN. Multiple XPPX motifs are also observed for some proteins, such as FabF (SPPE, VPPT), YgdH (NPPE, EPPN), YjjK (VPPK, IPPG), and Lon (GPPG, IPPE). The topoisomerase subunit ParC and the ribosomal protein L16 hydroxylase YcfD (also called RoxA) have four motifs each, IPPH/LPPG/MPPV/LPPQ and IPPG/VPPR/APPE/EPPY, respectively. In total, we detected 734 proteins containing 747 XPPX motifs, excluding PPP. Nevertheless, we observed proteins that lacked both PPP and XPPX motifs and yet still cluster with the PPP/XPPX-containing proteins, implying that they are coregulated in some manner (Fig. S4). This finding is exemplified by AtpH, AtpG, AtpC, which lack XPPX motifs, yet display the same regulation trends as the XPPX-containing proteins AtpF, AtpA, and AtpD (Fig. 1F), presumably because they are located together in the operon atpIBEFHAGDC and interact to form the ATP synthase complex. Surprisingly, the density plots showed no shift in the overall distribution of protein ratios for the XPPX-containing proteins in the Δefp/ΔyjeK/ΔyjeA compared with ΔyfcM (blue in Fig. 1 A–D): The median normalized H/L values for XPPX-containing protein ratios were between −0.06 and −0.01 for Δefp/ΔyjeK/ΔyjeA compared with −0.03 and 0.01 for the ΔyfcM replicates, suggesting that only distinct XPP- and/or PPX-containing proteins are strongly down-regulated in the absence of active EF-P.
Down-Regulation of Specific XPP- and PPX-Containing Proteins in Δefp, ΔyjeK, and ΔyjeA Strains.
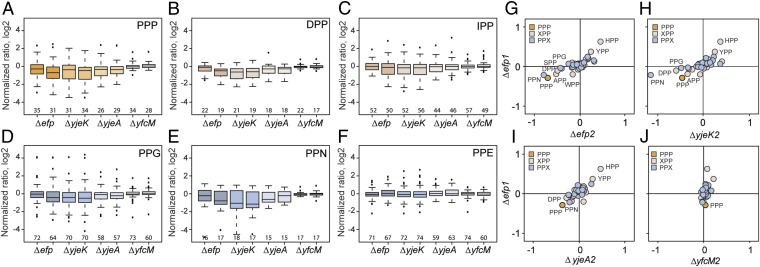
To analyze which distinct XPP- and/or PPX-containing proteins were down-regulated in the absence of active EF-P, we generated box plots to illustrate the fold-change distribution for proteins containing each of the 20 PPX and 20 XPP combinations in both replicates of the Δefp, ΔyjeK, ΔyjeA, and ΔyfcM strains (Figs. S5 and S6). Consistent with the density plot (Fig. 1 A–D), the box plots also show a significant shift in distribution of the PPP-containing proteins in the Δefp, ΔyjeK, and ΔyjeA strains, but not the ΔyfcM strain (Fig. 2A), as evident from the Krustal–Wallis test (Table S1). Similar trends were also observed for DPP (Fig. 2B), IPP (Fig. 2C), and APP (Fig. S5), as well as PPG (Fig. 2D) and PPN (Fig. 2E). Surprisingly, no change in distribution was observed for proteins containing PPD or PPE (Fig. 2F), which had been reported previously to induce translational stalling (7, 15). To obtain an approximate ranking of the influence that the different XPP and PPX motifs have on the distribution of the respective proteins, we plotted the median fold-change determined for each motif in replicate 1 of Δefp against the median fold change calculated from replicate 2 of the Δefp, ΔyjeK, ΔyjeA, and ΔyfcM strains (Fig. 2 G–J). A similar trend was observed in each plot where PPP and PPN produced the strongest down-regulation, followed by a clustering of DPP, APP, SPP, and PPG. HPP and YPP appear to correlate with up-regulation; however, we note that YPP and, in particular, HPP are relatively scarce in our dataset, occurring only in 18–24 and 5–7 proteins, respectively, identified across the four knockout strains (Figs. S5 and S6) and are not significant according to the Krustal–Wallis test (Table S1). We note that some other XPP and PPX motifs were also very scarce, such as PPW, PPC, and WPP, occurring only in 5–8, 3–7, and 4–6 proteins, respectively, identified across the four knockout strains (Figs. S5 and S6). The low occurrences of particular XPP and PPX motifs in our dataset correlates well with the general scarcity and underrepresentation of these triplets within the E. coli proteome (Fig. 3A): For example, PPW, PPC, WPP, HPP, and YPP are encoded 22, 12, 18, 27, and 59 times in the E. coli genome, all lower or equal to their expected frequencies of 39, 30, 39, 27, and 73, respectively. By comparison, PPP and PPG occur 101 and 183 times, similar to their expected frequencies of 114 and 190, respectively (Fig. 3A). Unfortunately, the low numbers of XPP/PPX motifs within our dataset prohibited us from performing similar box plot analysis of individual quadruplet XPPX motifs.
Fig. 2.
Proteins containing particular XPP/PPX-motifs are down-regulated in Δefp, ΔyjeK, and ΔyjeA strains. (A–F) Selected box-plot representations of inverted normalized H/L ratios (log2-transformed) for proteins containing PPP (gold) (A), XPP (yellow) motifs DPP (B), IPP (C), and PPX (blue) motifs PPG (D), PPN (E), and PPE (F) for Δefp, ΔyjeK, ΔyjeA, and ΔyfcM strains. Black center line, median value; box, lower quartile to upper quartile (25th to 75th percentile); whiskers, data points within 1.5× IQR (interquartile range, distance from median to lower or upper quartile); individual points, points at a greater distance from the median than 1.5× the IQR. (G–J) Scatter-plots of the median values of the normalized H/L ratios (log2-transformed) for all XPP/PPX-containing proteins comparing Δefp biological replicate-1 with replicate-2 of Δefp (G), ΔyjeK (H), ΔyjeA (I), and ΔyfcM (J) strains.
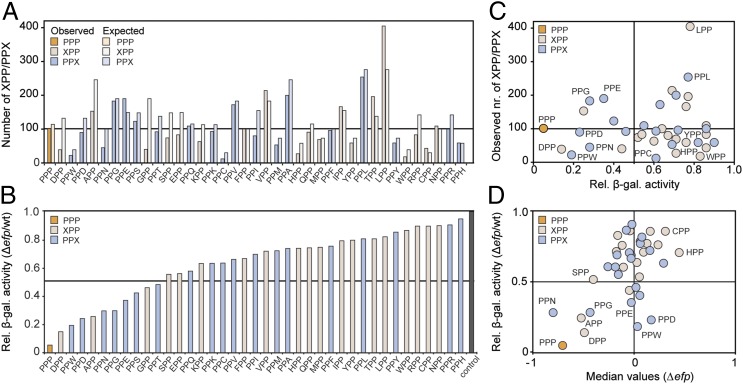
Fig. 3.
Systematic analysis of EF-P dependence of XPP/PPX-protein expression. (A) Bar graph showing the expected versus observed frequency of occurrence for PPP (orange), XPP (salmon), and PPX (blue) triplets within proteins encoded in the E. coli genome. (B) β-galatosidase activities of LacZ constructs containing PPP (orange), XPP(H) (salmon), or (F)PPX (blue) motifs were normalized in wild-type E. coli strains relative to Δefp strain. (C) Scatter plot of normalized β-galactosidase activities from B against the observed frequency of occurrence for PPP, XPP, and PPX triplets within the E. coli proteome from A. (D) Scatter plot of median values of the inverted H/L normalized ratio (log2) for all PPP-, XPP-, and PPX-containing proteins from Fig. 2G versus normalized β-galactosidase activities from B.
Systematic Analysis of the EF-P Dependence of XPP- and PPX-Containing Proteins.
To systematically analyze the influence of EF-P on the translation of proteins bearing XPP- or PPX triplets, we fused (E)PPP(H) and all 38 XPP/PPX motifs in front of the LacZ reporter protein, and monitored the effect on expression by comparing the β-galactosidase activity in wild-type E. coli (efp+) strain relative to that obtained in the Δefp strain (efp−) (Fig. 3B). Importantly, all 19 XPP motifs were followed by histidine (XPPH) and all 19 PPX motifs were preceded by glutamate (EPPX), because PPH and EPP motifs did not appear to be significantly affected by EF-P (Figs. S5 and S6) (5). The results show that, in the absence of EF-P, the introduction of any XPP or PPX motif leads to a reduction in β-galactosidase activity compared with the control lacking any XPP/PPX triplet. However, the extent of the reduction depended on the nature of the specific XPP or PPX motif; at one extreme, only a slight reduction was observed with PPH, whereas at the other extreme, PPP led to the largest reduction of >20-fold. The majority (∼70%) of XPP/PPX motifs (PPH to PPT in Fig. 3B) reduced the β-galactosidase activity by less than twofold, whereas the remaining 10 motifs ranged from threefold (GPP) to eightfold (DPP). These motifs include PPG and PPN (fourfold) as well as PPD (fivefold) and PPW (sixfold), which have been shown to induce translational stalling (7, 15). As for the proteomics study, the occurrence of XPP/PPX motifs encoded in the E. coli genome and their influence on EF-P dependent β-galatosidase activity do not correlate (Fig. 3B). For example, the majority (68%, 19 of 28) of the XPP/PPX motifs that cause less than twofold reduction in β-galactosidase activity are encoded less often in the E. coli genome than PPP which leads to >20-fold reduction. By contrast, we found a good correlation between the reduction in β-galactosidase activity caused by specific XPP/PPX motifs and the median values of the protein fold-change from the proteomic data, namely, PPP, PPN, APP, DPP, and PPG (Fig. 3D, Lower Left). However, no correlation was observed for PPE, PPD, and PPW (Fig. 3D, Lower Right). This latter finding is surprising given the previous reports that these three motifs strongly induce translational stalling in the absence of EF-P (7, 15).
EF-P Dependence of XPP- and PPX-Containing Proteins During in Vitro Translation.
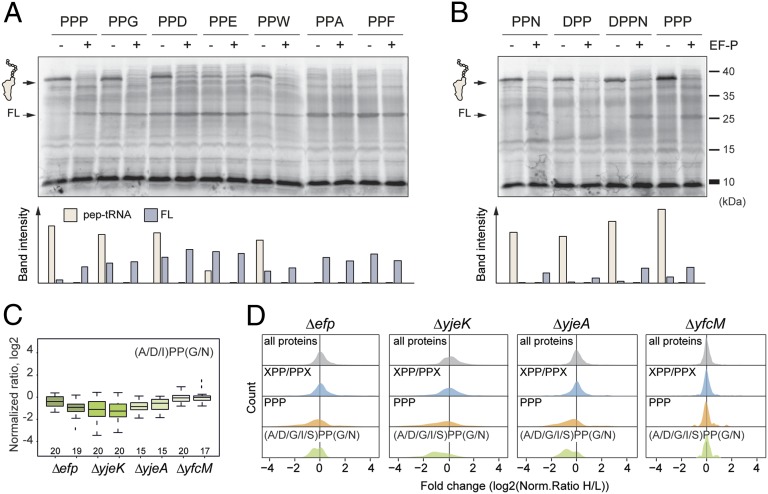
Therefore, we reassessed whether the PPD, PPE, and PPW triplets can induce translational stalling in vitro and, if so, whether the stalling can be alleviated by EF-P. To do this, we used an E. coli in vitro translation system reconstituted from recombinantly purified components (18) to translate reporter constructs containing the PPD, PPE, or PPW triplet motifs, in the absence and presence of EF-P (Fig. 4A). Translation of reporter constructs bearing triplet motifs predicted to have varying stalling capabilities, such as strong stallers, like PPP and PPG, as well as weak stallers, such as PPA and PPF, were also included as controls (Fig. 4A). In the absence of EF-P, accumulation of a stalled peptidyl-tRNA band was observed at the expected size of ∼38 kDa (corresponding to the ∼18-kDa nascent chain translated up to triplet motif, but remaining attached to ∼20 kDa tRNA) for the reporters containing PPP, PPG, PPD, PPW, and, to a lesser extent, PPE. In contrast, the low level of stall product prevented quantitation for PPA and PPF. Consistently, for these weak-stalling motifs, the full-length product (∼26 kDa) was much more abundant than at the strong stalling motifs (Fig. 4A). As expected, the presence of EF-P led to relief of ribosome stalling, as indicated by the loss of the peptidyl-tRNA band, as well as a corresponding increase in the amount of full-length product. We note however that for the weak-stalling motifs, addition of EF-P appeared to cause a slight decrease in the amount of full-length product, perhaps indicating that in the absence of significant stalling, the presence of additional EF-P can interfere with translation. In general, the in vitro results correlate well with the results from the in vivo β-galactosidase assays (Fig. 3B). In both cases, translation of reporters containing the PPP, PPW, PPD, and PPG triplets was strongly EF-P dependent and intermediate for PPE, whereas less effect was observed for the PPA and PPF triplets. Collectively, these findings suggest that the PPW, PPD, and PPE triplets do indeed confer EF-P dependence, suggesting that the lack of down-regulation of PPD/E/W-containing proteins in the absence of active EF-P (Fig. 2F and Fig. S6) may result from the low number of proteins identified with these motifs (as PPW) and/or be masked by competing regulatory effects that up-regulate these proteins in the absence of EF-P.
Fig. 4.
EF-P rescues translational stalling at XPPX motifs in vitro. (A and B) Autoradiographs of SDS–polyacrylamide gels indicating [35S]Met-labeled in vitro translation of reporters containing PPP, PPG, PPD, PPE, PPW, PPA, and PPF (A) or PPN, DPP, DPPN, or PPP (B). All reactions were performed in the absence (−) and presence (+) of active EF-P. (C) Box plot representation of inverted normalized H/L ratios (log2-transformed) for proteins containing (A/D/I)PP(G/N) motifs, for Δefp, ΔyjeK, ΔyjeA, and ΔyfcM strains. (D) Density plots showing distribution of proteins containing PPP (gold), XPP/PPX (blue), and (A/D/I)PP(G/N) (green) motifs relative to all proteins (gray). Both XPP/PPX and (A/D/I)PP(G/N) subsets have PPP-containing proteins excluded.
In the proteomics data, proteins containing PPN were more strongly down-regulated than PPP and DPP, whereas in the β-galatosidase assay, PPP and DPP exerted a stronger EF-P dependence than PPN. Closer examination of the PPN-containing proteins in the proteomics data reveals that in 30% of cases the amino acid preceding PPN was alanine, isoleucine, or aspartate, i.e., generating APP, IPP, and DPP, which are strong stalling motifs. In addition, 50% of the PPN-containing proteins also contained an additional XPPX motif. Thus, to reassess XPPN, we again used the reconstituted E. coli in vitro translation system to directly compare translation of reporter constructs containing PPP, (E)PPN, DPP(H), as well as DPPN, in the absence and presence of EF-P (Fig. 4B). In the absence of EF-P, peptidyl-tRNA was observed in all cases, with more stalling observed for DPPN than DPP(F) and (H)PPN, although stalling at DPPN was still slightly lower than for PPP (Fig. 4B). These findings indicate that although the quadruplet DPPN motif induced stronger stalling than the individual DPP and PPN motifs, the effects were not cumulative. To examine whether the stalling quadruplet motifs also exerted an affect in vivo, we generated box plots for a subset of proteins identified from the proteomics data that contain any of the (A/D/I)PP(G/N) motifs (excluding PPP-containing proteins) (Fig. 4C). As mentioned, the limited number of proteins precluded an analysis of proteins containing distinct XPPX motifs. The median normalized H/L ratio values for (A/D/I)PP(G/N)–containing protein ratios were between −0.30 and −0.99 for Δefp/ΔyjeK/ΔyjeA, which was comparable to the range observed for PPP (−0.29 and −0.71). As expected, we also observed a marked shift in the distribution of protein ratios for the (A/D/I)PP(G/N)–containing proteins in the Δefp/ΔyjeK/ΔyjeA, but not the ΔyfcM data (Fig. 4D).
Discussion
Collectively, our in vivo and in vitro data indicate that ribosome stalling occurs not only at PPP triplets, but also at XPP and PPX triplets with the efficiency of the stalling dictated by the nature of the amino acid preceding and following the diprolyl moiety. Nevertheless, in all cases, the efficiency of stalling induced by the XPP or PPX triplets was less than for PPP. However, the combination of specific XPP and PPX motifs to form XPPX quadruplets was shown to increase the efficiency of stalling, both in vivo and in vitro, to levels paralleling the PPP triplets. In all cases, we were able to show that EF-P was able to efficiently relieve the translation stalling. Consistent with our findings was the identification of proteins containing PPP, PPG, or APP as being down-regulated when the genes encoding EF-P, YjeA, or YjeK were deleted in Salmonella (8, 13). However, we did not observe any significant stalling in the absence of EF-P at non-diprolyl containing motifs, such as RME, YIR, PFF, YIRYIR, nor at the GSCGPG motif found in PoxB (Figs. S7 and S8 and Table S1). Moreover, although PoxB was down-regulated in the Δefp, ΔyjeK, ΔyjeA strains, it was also down-regulated in the ΔyfcM strain (Fig. S4). Thus, we believe the major role for EF-P in the cell is the relief of translational stalling at proline-containing motifs, predominantly at PPP as well as a specific subset of XPPX motifs.
Previous studies have shown that in the case of PPX motifs, translation stalling occurs with a peptidyl-Pro-Pro-tRNA in the P-site and an aminoacyl-tRNA in the A-site bearing the amino acid X (6, 7) (Fig. 5A). Kinetic studies have shown that when X is proline (P) or glycine (G), formation of the peptide bond between the A-site proline or glycine with the P-site proline is very slow in the absence of EF-P (6) (Fig. 5B). This result is consistent with the findings that Pro- and Gly-tRNA in the A-site act as poor acceptors (3, 4) and that Pro-tRNA in the P-site acts as a poor donor (1, 2). Additionally, stalling is enhanced when peptidyl-Pro-Pro-tRNA is situated in the P-site, presumably because proline in the −1 position imparts additional conformational constraints on the P-site proline that are unfavorable for peptide bond formation (Fig. 5B). Our studies indicate that, in addition to proline and glycine, tryptophan (W), aspartate (D), asparagine (N), glutamate (E), serine (S), and threonine (T) act as poor A-site acceptors that impair peptide bond formation and induce translational stalling (Fig. 5B), consistent with previous toeprinting studies (7). In contrast, peptide bond formation with the P-site diprolyl residue appears to occur efficiently when the A-site amino acid X is histidine (H), arginine (R), tyrosine (Y), leucine (L), or phenylalanine (F), suggesting that the nature of these amino acids enables them to act as efficient acceptors, even within the poor donor context of the diprolyl P-site substrate (Fig. 5B). This phenomenon is reminiscent of the selective A-site stalling that can occur at the ErmAL1 leader peptide (19), although there are clear differences with respect to the A-site specificities observed here.
Fig. 5.
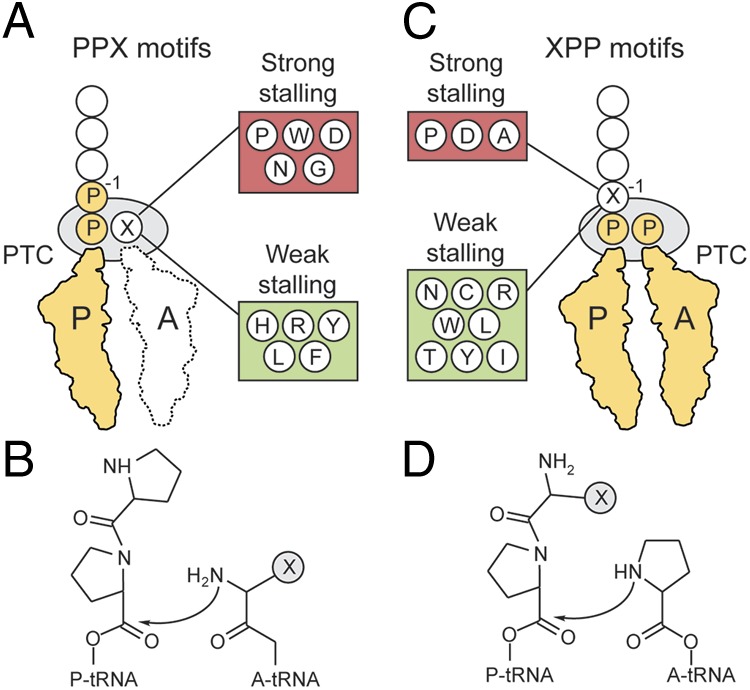
Model for ribosome stalling at PPX and XPP motifs. (A) Translation stalls at PPX motifs with the peptidyl-Pro-Pro-tRNA in the P-site and aminoacyl-tRNA bearing the amino acid X in the A-site. (B) Peptide bond formation between NH2 of amino acid X in the A-site with the carbonyl carbon of the Pro-tRNA in the P-site. (C) Translation stalls at XPP motifs with the peptidyl-X-Pro-tRNA in the P-site and Pro-tRNA in the A-site. (D) Peptide bond formation between imino group of Pro in the A-site with the carbonyl carbon of the peptidyl-X-Pro-tRNA in the P-site.
Translational stalling at XPP triplets occurs with the peptidyl-X-Pro-tRNA in the P-site and Pro-tRNA in A-site, as evidenced by toeprinting for WPP and APP (7) (Fig. 5C). In addition to proline, alanine (A), and glycine (G), our studies indicate that aspartate (D) and serine (S) in the −1 position also lead to translational stalling. By analogy with PPP, we propose that these amino acids (P, A, D, G, S) also constrain the P-site proline such that its donor capabilities are further diminished (Fig. 5D). Amino acids located deeper in the tunnel (−2, −3, −4… positions) could also influence stalling at both XPP and PPX motifs by modulating the donor capabilities of the P-site proline. Indeed, known regulatory stalling sequences, such as the bacterial TnaC and SecM as well as the uORF2 of human cytomegalovirus gp48 stall with Pro-tRNA in the P-site (20, 21). In both the PPX and XPP motifs, we detect no obvious characteristic of the amino acids, such as hydrophobicity, size, or charge, which correlates with either strong or weak stalling. Nevertheless, we note that proline, glycine, and aspartate induce strong stalling, whereas arginine, leucine, and tyrosine induce poor stalling, in the context of both XPP and PPX. High-resolution structures of stalled ribosomes should provide insight into the conformation of the P-site proline and how it is influenced by the local environment provided by the neighboring amino acids in the nascent polypeptide chain. Moreover, it will be interesting to see whether similar hierarchies of translational stalling at XPPX motifs are observed in archaeal and eukaryotic cells in the absence of IF-5A.
Methods
Proteomics.
SILAC (ΔargA, ΔlysA) wild type, and subsequently mutant strains, were generated using P1 transduction (22) from Keio strains (23, 24) as described (10), but with an E. coli MG1655 background. The strains were grown in MOPS medium, supplemented with 50 µg/mL heavy arginine and lysine (Cambridge Isotope Laboratories) for wild-type MG1655 SILAC strain and light arginine and lysine (Sigma) for Δefp, ΔyjeK, ΔyjeA, and ΔyfcM deletion strains. Cells were grown to midlog, harvested by centrifugation, and lysed. Cell lysates were mixed in 1:1 ratio and proteins digested as described (25). Resulting peptides were fractionated as described (26) and analyzed via LC-MS/MS using 120-min gradients (10). Data analysis was performed using MaxQuant v1.3.0.5 (16), with default settings against E. coli K-12 MG1655 protein sequence database from UniProtKB (09.09.2011).
β-Galactosidase Assays.
Cells producing CadC-LacZ hybrids TL30- XPP/PPX under control of the cadC promoter were grown in Lysogeny Broth (LB) to exponential growth phase (OD600 = 0.3–0.5). β-galactosidase activities were then determined as described (5) for at least three independent experiments.
In Vitro Translation Assays.
In vitro translation assays with EF-P were conducted using an Fluc-based reporter system as described (5), with indicated XPPX motifs substituted at positions 172–175 and the Fluc reporter shortened by removal of residues 183–539.
Genome Composition Analyses.
The tripeptide composition of the E. coli K-12 MG1655 and H. sapiens proteomes (from National Center for Biotechnology Information, ftp://ftp.ncbi.nih.gov/) and expected composition was based on single amino acid frequencies. The expected frequency of a PPX or XPP motif was calculated using (p2x)g, where p is the fraction of proline in the genome, x is the fraction of the amino acid X, and g is the genome size in amino acids.
Supplementary Material
Acknowledgments
We thank Liisa Arike, Triin Tammsalu, Ingrid Weitl, and Heidemarie Sieber for technical assistance. This research was supported the Deutsche Forschungsgemeinschaft WI3285/2-1 and a European Molecular Biology Organization Young Investigator grant (to D.N.W.), the Cluster of Excellence (Exc114/2) (K.J.), and the Estonian Science Foundation Grant 9289 (to J.R.). L.P. and G.C.A. received support from European Social Fund Program Mobilitas Grants MJD144 and MJD99, respectively. L.P. is funded by a Marie Curie FP7-PEOPLE-2011-IEF Postdoctoral Fellowship and A.L.S. is funded by an AXA Research Fund Postdoctoral Fellowship. MS analyses were partly supported by the European Regional Development Fund via the Center of Excellence in Chemical Biology (T.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310642110/-/DCSupplemental.
References
- 1.Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem Biophys Res Commun. 2008;366(4):1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- 2.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283(47):32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 3.Pavlov MY, et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106(1):50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson M, et al. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci USA. 2011;108(1):79–84. doi: 10.1073/pnas.1012612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 6.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 7.Woolstenhulme CJ, et al. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110(10):E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarre WW, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39(2):209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol. 2010;17(9):1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 10.Peil L, et al. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8(8):695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- 11.Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38(2):491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez E, et al. eIF5A Promotes Translation of Polyproline Motifs. Mol Cell. 2013;51(1):35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hersch SJ, et al. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio. 2013;4(2):e00180–13. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484(7395):538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 17.Soares NC, Spät P, Krug K, Macek B. Global dynamics of the Escherichia coli proteome and phosphoproteome during growth in minimal medium. J Proteome Res. 2013;12(6):2611–2621. doi: 10.1021/pr3011843. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19(8):751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 19.Ramu H, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41(3):321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Ito K, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun. 2010;393(1):1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol. 2011;21(2):274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Taylor AL, Trotter CD. Revised linkage map of Escherichia coli. Bacteriol Rev. 1967;31(4):332–353. doi: 10.1128/br.31.4.332-353.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto N, et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 26.Wiśniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8(12):5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.