Significance
Transposons in higher eukaryotes are subject to epigenetic silencing. Some transposons have found residence in introns of genes. It is unclear how intronic transposon-containing genes are regulated. This paper reports a cellular factor, ANTI-SILENCING 1 (ASI1), which is required for proper expression of intronic transposon-containing genes. ASI1 plays an important role in regulating genome DNA methylation patterns. The work represents a major advancement in cellular antisilencing mechanisms. A previously undescribed mechanism for higher eukaryotes to cope with the collateral effects of silencing intronic transposon elements is discovered. This work has implications beyond the plant epigenetics field because studies in mammalian systems have suggested important roles of intronic heterochromatin on 3′ distal polyadenylation, although the underlying mechanism is not known.
Keywords: DNA methylome, ChIP, gene expression
Abstract
DNA methylation-dependent heterochromatin formation is a conserved mechanism of epigenetic silencing of transposons and other repeat elements in many higher eukaryotes. Genes adjacent to repetitive elements are often also subjected to this epigenetic silencing. Consequently, plants have evolved antisilencing mechanisms such as active DNA demethylation mediated by the REPRESSOR OF SILENCING 1 (ROS1) family of 5-methylcytosine DNA glycosylases to protect these genes from silencing. Some transposons and other repeat elements have found residence in the introns of genes. It is unclear how these intronic repeat elements-containing genes are regulated. We report here the identification of ANTI-SILENCING 1 (ASI1), a bromo-adjacent homology domain and RNA recognition motif-containing protein, from a forward genetic screen for cellular antisilencing factors in Arabidopsis thaliana. ASI1 is required to prevent promoter DNA hypermethylation and transcriptional silencing of some transgenes. Genome-wide DNA methylation analysis reveals that ASI1 has a similar role to that of the histone H3K9 demethylase INCREASE IN BONSAI METHYLATION 1 (IBM1) in preventing CHG methylation in the bodies of thousands of genes. We found that ASI1 is an RNA-binding protein and ensures the proper expression of IBM1 full-length transcript by associating with an intronic heterochromatic repeat element of IBM1. Through mRNA sequencing, we identified many genes containing intronic transposon elements that require ASI1 for proper expression. Our results suggest that ASI1 associates with intronic heterochromatin and binds the gene transcripts to promote their 3′ distal polyadenylation. The study thus reveals a unique mechanism by which higher eukaryotes deal with the collateral effect of silencing intronic repeat elements.
In higher eukaryotes including plants, DNA methylation is an important epigenetic mark that silences transposons and other repetitive elements. In Arabidopsis thaliana, DOMAINS REARRANGED METHYLASE 2 (DRM2) catalyzes de novo DNA methylation in all cytosine contexts including CG, CHG, and CHH (H represents A, T, or G) (1), through the RNA-directed DNA methylation pathway (RdDM) (2–9). At the same time, preexisting DNA methylation in plants can be pruned by enzymatic excision that is catalyzed by a subfamily of bifunctional DNA glycosylases represented by REPRESSOR OF SILENCING 1 (ROS1) and DEMETER (DME) (10–14). Following the enzymatic removal of 5-methylcytosine, the resultant single-nucleotide gap is filled with an unmodified cytosine through the DNA base excision repair pathway (15, 16).
Cytosine methylation and demethylation are both tightly linked with histone modifications. Increased DNA methylation was observed in an A. thaliana mutant defective in INCREASED DNA METHYLATION 1 (IDM1), an acetyltransferase that catalyzes acetylation of histone H3 lysine 18 (H3K18) and lysine 23 (H3K23) necessary for subsequent DNA demethylation and prevention of transcriptional silencing (17). In A. thaliana, the repressive histone mark H3K9me2 is established mainly by the histone methyltransferase KRYPTONITE (KYP) (18). KYP and CHROMOMETHYLASE 3 (CMT3) bind to CHG methylation and H3K9me2, respectively, thereby forming a self-reinforcing loop of DNA methylation and H3K9me2 (4, 19). In addition, the binding of SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) to methylated H3K9 helps recruit the DNA-DEPENDENT RNA POLYMERASE IV (Pol IV) for generation of heterochromatic siRNAs that mediate RdDM (20). Accumulation of H3K9 methylation is negatively regulated by the histone demethylase IBM1, the mutation of which results in elevated levels of H3K9me2 and concomitant CHG hypermethylation in thousands of genic loci (21, 22).
Although DNA methylation at promoters and intergenic transposon elements (TEs) generally correlates with transcriptional repression, the role of gene body methylation is mostly unclear. On a whole-genome scale, A. thaliana and human cytosine methylation show a preference for nucleosome-bound DNA over flanking sequences (23), indicating an interplay between DNA methylation and RNA biogenesis because nucleosomes are highly enriched on exons and are preferentially positioned at intron–exon and exon–intron boundaries (23). It has been proposed that exonic cytosine methylation may serve as a physiological barrier that slows Pol II during transcription elongation, thereby influencing cotranscriptional pre-mRNA splicing (24, 25). Indeed, a positive correlation between DNA methylation and inclusion of alternative exons was observed in human and honeybee (26, 27). DNA methylation can also be found in introns and show, in some cases, positive regulation of gene expression by unknown mechanisms. Gene expression of EARLY GROWTH RESPONSE 2 (EGR2), a mammalian tumor suppressor, is enhanced by intronic DNA methylation (28). Similarly, DNA methylation of an intronic repeat element was shown to be necessary for generating full-length transcripts of IBM1 in A. thaliana (29). Alternative polyadenylation was observed in mouse imprinted histocompatibility 13 (H13) and HECT AND RLD DOMAIN CONTAINING PROTEIN 3 (Herc3) genes, in which differential DNA methylation in an intron dictates RNA polyadenylation sites (30, 31). It follows that intronic DNA methylation can be a cis-element that affects alternative polyadenylation and regulates the expression levels of functional full-length transcripts. The transacting molecular factors involved in such epigenetic regulation, however, remain to be identified.
In this study, we identified and characterized ANTI-SILENCING 1 (ASI1), a previously undescribed epigenetic regulator that is important for genome DNA methylation patterns and prevention of gene silencing in A. thaliana. Through a forward genetic screen, we isolated the asi1-1 mutant that exhibited transcriptional silencing of reporter genes resulting from promoter DNA hypermethylation. We found that ASI1 is necessary for preventing gene body CHG hypermethylation across the genome. Our results show that a defect in the expression of IBM1 underlies the DNA hypermethylation patterns observed in the asi1-1 mutant. At IBM1 and other genic loci that contain heterochromatic elements in introns, ASI1 is required for the production of full-length transcripts by promoting distal polyadenylation downstream of the intronic heterochromatic elements. ASI1 associates specifically with heterochromatic sequences in introns and exhibits RNA-binding activity in vitro. Our research has thus uncovered a transacting factor that is critical for the proper expression of genes with intronic heterochromatic elements. These results increase our understanding of the mechanisms used by higher eukaryotes to deal with the collateral effect of silencing intronic repeat elements.
Results
ASI1 Is an Antisilencing Factor.
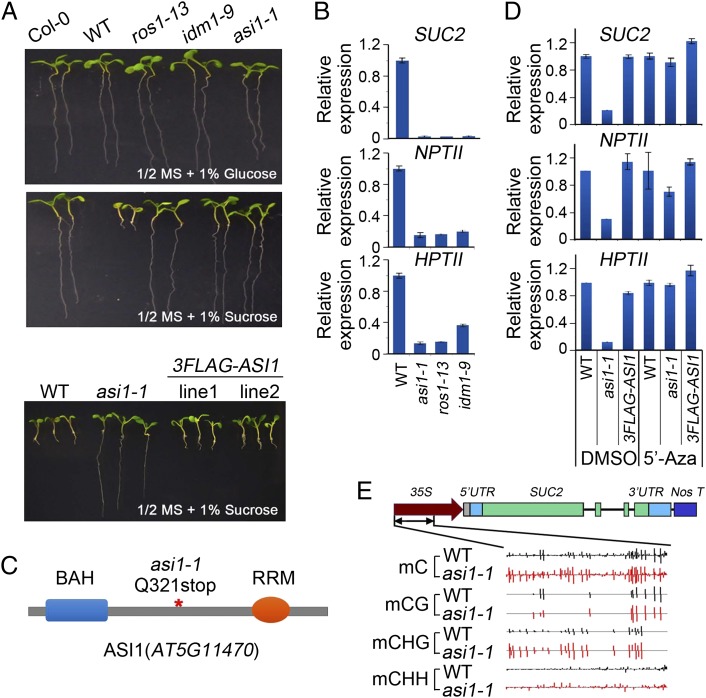
To identify antisilencing factors required for prevention of transcriptional gene silencing, we developed a unique transgene-based forward genetic screen in A. thaliana. In this system, the SUCROSE-PROTON SYMPORTER 2 (SUC2) gene encoding a sucrose transporter is overexpressed under the Cauliflower Mosaic Virus 35S promoter (32). When grown on a sucrose-containing culture medium, the 35S-SUC2 transgenic plants overaccumulate sucrose because of the high expression of SUC2, resulting in severe inhibition of root growth (Fig. 1A). The transgenic plants also contain two 35S promoter-driven marker genes, HYGROMYCIN PHOSPHOTRANSFERASE II (HPTII) and NEOMYCIN PHOSPHOTRANSFERASE II (NPTII). From an ethyl methanesulfonate-mutagenized 35S-SUC2 [hereinafter referred to as the wild type (WT)] population, we isolated a previously undescribed mutant, asi1-1, and mutant alleles of the known antisilencing factors ROS1 and IDM1 (10, 17), which show a derepression of root growth and silencing of the 35S-SUC2, 35S-HPTII, and 35S-NPTII transgenes (Fig. 1B). Map-based cloning revealed that asi1-1 is a recessive mutant allele of AT5G11470 that encodes a bromo-adjacent homology (BAH) domain and an RNA recognition motif (RRM)-containing protein (Fig. 1C). The asi1-1 mutation changed glutamine-321 to a premature stop codon (Fig. 1 and Fig. S1). Introduction of WT AT5G11470 genomic DNA rescued the short root and transgene silencing defects of the asi1-1 mutant (Fig. 1A and Fig. S2), suggesting that ASI1 is a previously undescribed antisilencing factor.
Fig. 1.
Characterization of the transgene silencing phenotypes of the asi1-1 mutant. (A) The root growth phenotype of asi1-1, ros1-13, and idm1-9 on 1/2 MS media supplemented with 1% sucrose or 1% glucose (Top and Middle) and of two complemented transgenic lines on medium containing 1% sucrose (Bottom). (B) Relative expression levels of SUC2, NPTII, and HPTII in the WT, asi1-1, ros1-13, and idm1-9. (C) Diagram of ASI1 (AT5G11470) protein domains. The red asterisk indicates the position of the asi1-1 mutation. (D) Relative expression of SUC2, NPTII, and HPTII in the WT, asi1-1, and FLAG-ASI1/asi1-1 complementation plants treated with DMSO (negative control) or 5′Aza. (E) A snapshot of IGB (33) shows the methylation level of the 35S promoter. MethylC sequencing data were mapped to the 35S-SUC2 transgene sequence to retrieve methylation information.
To test whether DNA methylation is associated with transgene silencing, we treated the asi1-1 mutant, the WT, and complementation plants with 5-Aza-2′-deoxycytidine (5′Aza), a DNA methylation inhibitor. We found that the silencing of transgenes in asi1-1 was suppressed by 5′Aza treatment (Fig. 1D), suggesting that the silencing in asi1-1 was associated with DNA methylation. We then determined the DNA methylation levels of the 35S promoter in asi1-1 by genome bisulfite sequencing. The data showed clear increases in CHG (H is A, T, or C) and CHH methylation in the 35S promoter in asi1-1 (Fig. 1E). These results suggested that the transgene silencing in asi1-1 was caused by hypermethylation of promoter DNA. Real-time PCR assays showed that the expression of ROS1 and IDM1 was not reduced in asi1-1 (Fig. S3), indicating that ASI1 may be a previously undescribed regulator of active DNA demethylation.
ASI1 Prevents CHG Hypermethylation in Gene Bodies Like IBM1.
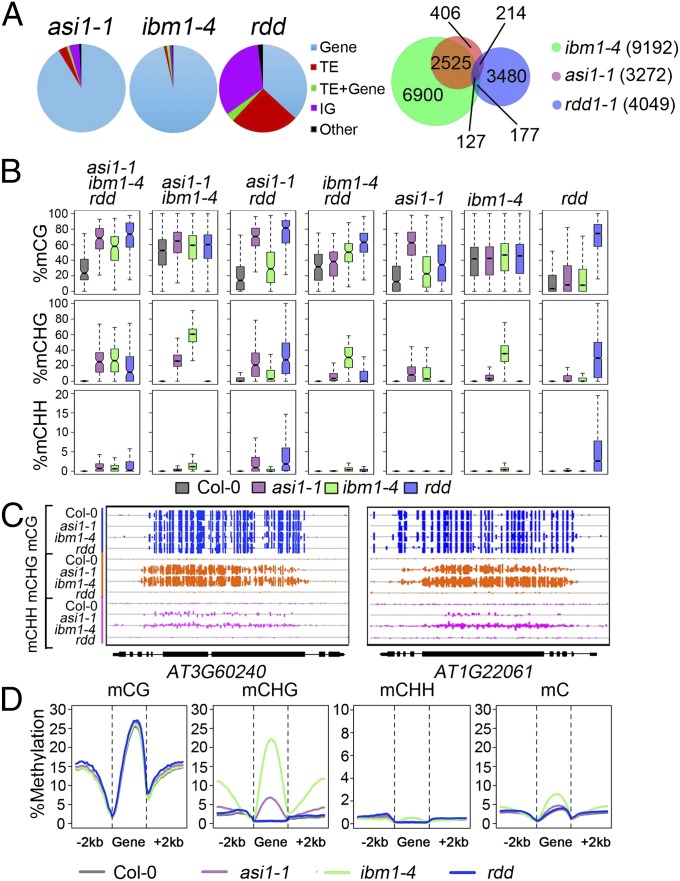
Based on the whole-genome bisulfite sequencing data, we found 3,767 differentially methylated regions (DMRs) in asi1-1 mutant plants, among which 3,272 DMRs are hypermethylated and 495 are hypomethylated (Fig. 2A and Fig. S4). We compared the DMRs in asi1-1 with those in rdd (ros1, dml2, and dml3 triple mutant) and ibm1-4. ROS1, DEMETER-LIKE 2 (DML2), and DML3 are 5-methylcytosine DNA glycosylases required for protecting transposon elements (TEs) and other repeat sequences from hypermethylation (12–14). IBM1 is a histone H3K9 demethylase that protects genes from CHG hypermethylation by CMT3 (21). As expected, most of the hyper-DMRs in rdd corresponded to TEs and intergenic sequences rather than to genes. However, most of the hyper-DMRs in asi1-1 were mapped to genes, which was very similar to the case with ibm1-4 (Fig. 2A). Approximately 81% (2,652/3,272) of the hyper-DMRs in asi1-1 overlapped with those in ibm1-4, whereas only 10% overlapped with those in rdd (Fig. 2A and Fig. S4). In DMRs that overlapped between asi1-1 and ibm1-4, CHG methylation was dramatically increased (Fig. 2 B and C). In contrast, the DMRs that were shared by asi1-1 and rdd or by asi1-1, ibm1, and rdd showed increases not only in CHG methylation but also in CG and CHH methylation (Fig. 2B).
Fig. 2.
Whole-genome methylation analysis of the asi1-1 mutant and comparison of the asi1-1 mutant with the ibm1-4 and rdd mutants. (A) Analysis of hyper-DMRs. (Left) Composition of the hyper-DMRs in asi1-1, ibm1-4, and rdd mutants. Gene, DMRs in protein-coding genes but not TEs; IG, intergenic regions; Other, DMRs that overlap with other features like pseudogenes, miRNAs, rRNAs, etc.; TE, DMRs in TEs but not protein-coding genes; TE + Gene, DMRs in both TEs and protein-coding genes. (Right) Numbers of hyper-DMRs that are overlapping among or unique to the asi1-1, ibm1-4, and rdd mutants. In a few cases where one region in one mutant overlaps with two regions in another mutant, we calculated the number of overlapping regions from the perspective of asi1-1, ibm1-4, and rdd, in that order. (B) Box plots displaying the distribution of average CG, CHG, or CHH methylation levels calculated for hyper-DMRs in the indicated subgroups. (C) Snapshots of two representative DNA hypermethylation regions that show dramatic increases in CHG methylation and slight increases in CHH methylation in asi1-1 and ibm1-4. (D) Average methylation levels in gene bodies and flanking 2-kb regions. Each gene was aligned from start to end and divided into 20 equal bins. Upstream and downstream 2-kb regions were each also divided into 20 equal bins. Weighted methylation level was calculated for each of the 60 bins across all of the corresponding regions and plotted. Only cytosines with fourfold or more coverage were used for this analysis.
To further characterize the DNA methylation profiles, we analyzed the distribution of DNA methylation along the gene bodies, TEs, and their 2-kb upstream and downstream flanking sequences in the three mutants. There was a significant increase in CHG methylation and a slight increase in CHH methylation but no change in CG methylation in gene bodies in asi1-1 and in ibm1-4 (Fig. 2 B–D). In contrast, the CHG and CHH methylation of TEs was slightly reduced in asi1-1 and ibm1-4 mutants but not in the rdd mutant (Fig. S5). Like the genic CHG hypermethylation in ibm1-4, the genic CHG hypermethylation in asi1-1 was strongly associated with gene length. Longer genes were more heavily hypermethylated than shorter ones in asi1-1 and ibm1-4 (Fig. S6). These results suggest that ASI1 may act in the same pathway as IBM1 in preventing genic CHG methylation.
ASI1 Controls the Expression of Full-Length IBM1 Transcript.
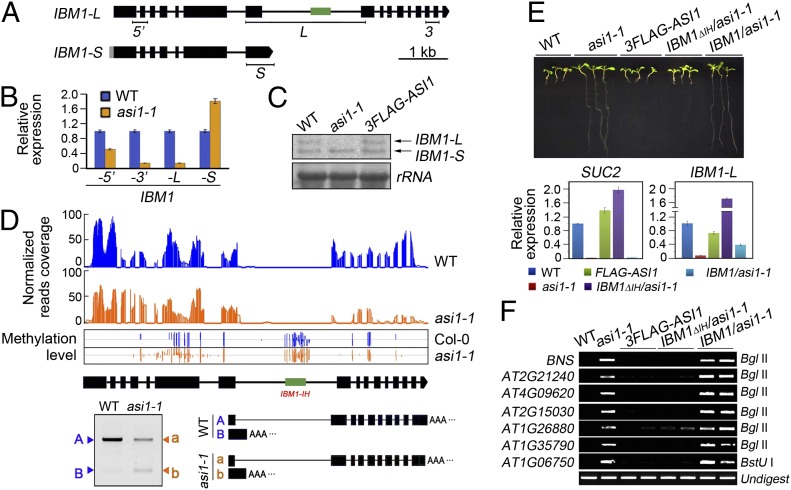
IBM1 is known to produce two transcripts (29), a long form (IBM1-L) containing the jmjC histone demethylase domain and a short form (IBM1-S) lacking the domain (Fig. 3A). Real-time RT-PCR assays showed that the IBM1-L transcript was substantially reduced in asi1-1 compared with the WT, whereas the IBM1-S transcript was slightly increased (Fig. 3B). The total amount of IBM1 transcripts (IBM1-5′) containing both IBM1-L and IBM1-S was slightly reduced in asi1-1 (Fig. 3B). Northern analysis supported these conclusions (Fig. 3C). Consistent with these findings, mRNA sequencing (mRNA-seq) experiments showed that the expression of the IBM1-3′ region was reduced (Fig. 3D). The 3′Rapid Amplification of cDNA Ends (RACE) assay revealed the presence of two major polyadenylated IBM1 transcripts (Fig. 3D). Polyadenylation occurred predominantly at the 3′ distal end in the WT but mainly in the seventh intron of the IBM1 gene in the asi1-1 mutant (Fig. 3D). These results showed that ASI1 is required for accumulation of the IBM1-L transcript.
Fig. 3.
Regulation of IBM1 expression by ASI1. (A) Schematic representation of IBM1-L and IBM1-S and positions of primers used for quantitative real-time PCR. (B) Relative expression of IBM1-5′, IBM1-3′, IBM1-L, and IBM1-S in the WT and the asi1-1 mutant. (C) Detection of the long and short forms of IBM1 transcripts in WT, asi1-1, and complemented asi1-1 by Northern blot analysis. (D) mRNA-seq 3′ RACE data. (Top) A snapshot of IGB (33) showing the read coverage of IBM1 transcripts and methylation levels. (Bottom) The 3′ RACE PCR and sequencing results. The uppercase blue letters and lowercase orange letters indicate the IBM1 transcripts in the WT and asi1-1, respectively. The green box in the IBM1 gene structure indicates the intronic heterochromatin region (IH). (E) Root growth and relative expression of SUC2 and IBM1-L transcript in the WT, asi1-1, 3FLAG-ASI1/asi1-1, IBM1Δ1H/asi1-1, and IBM1/asi1-1. (F) DNA methylation-sensitive PCR analysis of the WT, asi1-1, FLAG-ASI1/asi1-1, IBM1Δ1H/asi1-1, and IBM1/asi1-1 at the tested loci including BONSAI (BNS).
The IBM1 gene contains a very large (1,974-bp) seventh intron, in which a ∼500-bp repeat sequence (referred to as IBM1-IH) is methylated and enriched in H3K9 dimethylation (Fig. 3D) (29). The accumulation of IBM1-L transcript depends on CG and CHG methylation of this region (29). Our whole-genome bisulfite sequencing confirmed that the IBM-IH region was indeed heavily methylated and that the asi1-1 mutation did not affect this methylation (Fig. 3D). To determine the potential importance of IBM1-IH in ASI1 regulation of IBM1 expression, we introduced a WT IBM1 genomic DNA clone driven by its native promoter (pIBM1::IBM1) as well as a corresponding construct lacking only the IBM1-IH region (pIBM1::IBM1ΔIH) into the asi1-1 mutant. The results showed that IBM1-L expression was restored to the WT level in asi1-1 by pIBM1::IBM1ΔIH but not by pIBM1::IBM (Fig. 3E). DNA methylation-sensitive PCR assays showed that CHG hypermethylation in tested loci, including BONSAI, was suppressed in pIBM1::IBM1ΔIH but not in pIBM1::IBM1 transformants (Fig. 3F). The transgene silencing and root growth phenotypes of asi1-1 were also suppressed in pIBM1::IBM1ΔIH but not in pIBM1::IBM1 transformants (Fig. 3E). Furthermore, developmental defects of asi1-1 such as dwarf stature and short siliques, which were also observed in ibm1 mutant plants, were rescued by expression of pIBM1::IBM1ΔIH (Fig. S7). These results demonstrated that the function of ASI1 in preventing transgene silencing and CHG hypermethylation is explained by its regulation of IBM1 and that this regulation requires the intronic heterochromatin region.
ASI1 Controls the Alternative Processing of Intronic Heterochromatin-Containing Genes by Promoting 3′ Distal Polyadenylation.
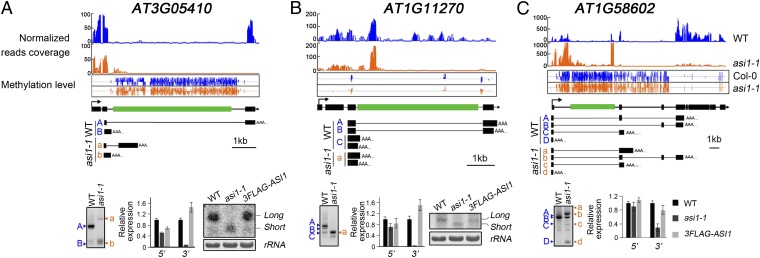
To investigate whether ASI1 plays a general role in regulating genes with a large and heterochromatic intron like that in IBM1, we searched and found 683 genes with intronic TEs; 45 of them possibly had a reduced expression in the 3′ region downstream of the TEs according to our cutoff (Table S1). The three candidate genes with the largest intronic TEs were selected for further analysis (Fig. 4 and Table S1). Our methylome data indicated the presence of a heavily methylated TE in a large intron in two of these three genes: COPIA8B in AT1G58602 and ATLINE2 in AT3G05410 (Fig. 4). The third gene, AT1G11270, contains a COPIA78 family TE in a large intron, and the TE is methylated near its two ends (Fig. 4). For all three of these genes, Northern blot and RT-qPCR analyses showed that there were lower levels of full-length transcripts with 3′ distal polyadenylation in asi1-1 than in the WT. The 3′ RACE assays showed the presence of multiple shorter transcripts with polyadenylation inside the TE or broader heterochromatic region for the three candidates and the absence of full-length transcripts with 3′ distal polyadenylation for two of the three genes in the asi1-1 mutant (Fig. 4). These results suggest that ASI1 is required for the proper expression of genes with heterochromatic introns.
Fig. 4.
Regulation of the expression of AT3G05410, AT1G11270, and AT1G58602 by ASI1. (Top) mRNA-seq data showing reduced transcript levels at the 3′ regions, genome bisulfite sequencing data showing DNA methylation levels, gene structures, and 3′ RACE results of (A) AT3G05410, (B) AT1G11270, and (C) AT1G58602. The green boxes indicate intronic TEs. (Bottom) Photographs of 3′ RACE PCR product gels, and real-time RT-PCR analysis of 5′ and 3′ transcript levels, and Northern blot analysis of the alternative transcripts in the WT, asi1-1, and complemented asi1-1.
ASI1 Is Enriched in the Intronic Heterochromatin Region of Target Genes.
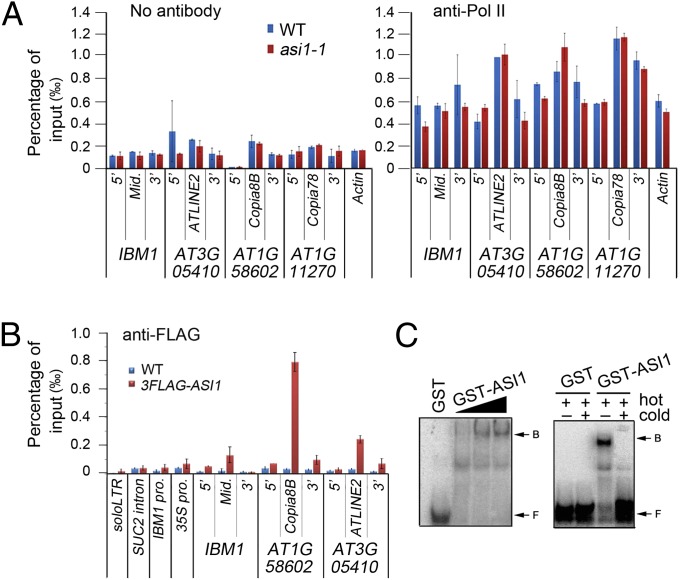
To investigate the underlying mechanism of ASI1-mediated mRNA processing of intronic heterochromatin-containing genes, we first performed chromatin immunoprecipitation (ChIP) assays to determine whether ASI1 affects Pol II elongation through the heterochromatic introns. The results show that there were no substantial differences between asi1-1 and the WT in Pol II occupancy in the heterochromatic intron or in the 5′ or 3′ flanking regions of IBM1, AT1G11270, AT3G05410, or AT1G58602 (Fig. 5A).
Fig. 5.
ASI1 is an RNA-binding protein and is enriched at intronic heterochromatin regions. (A) Pol II occupancy at different regions of the ASI1 target genes. Pol II occupancy at the 5′, intronic TE/heterochromatin, and 3′ regions of the target genes was determined by ChIP real-time RT-PCR. (B) Data from ChIP real-time RT-PCR using anti-FLAG antibodies on pASI1::3FLAG-ASI1 complementation lines showing ASI1 enrichment only at the TE/heterochromatin regions of the tested loci. WT plants were used as negative control. Data shown are from one of three independent experiments. (C) EMSAs showing ASI1 binding to single-stranded RNA in vitro. A 32P-labeled 60-nt single-stranded RNA oligo and an unlabeled RNA oligo were used as radioactive probe and unlabeled competitor, respectively. B, bound RNA band; F, free RNA band.
Because ASI1 has a BAH domain that is known to associate with chromatin (34), we determined whether ASI1 associates in vivo with the intronic heterochromatin of affected genes. ChIP experiments were carried out using anti-Flag antibodies on asi1-1 plants complemented with native promoter-driven 3FLAG-ASI1. WT plants without 3FLAG-ASI1 were used as the negative control. Real-time PCR assays showed that in comparison with the upstream and downstream regions, the intronic heterochromatic regions of IBM1, AT1G58602, and AT3G05410 were enriched with ASI1 (Fig. 5B). As expected, the IBM1 promoter; the SUC2 intron, which is a methylated intron (Fig. S8); and the solo LTR, which is a methylated transposon (35), were not enriched with ASI1 (Fig. 5B). These results suggest that ASI1 associates specifically with intronic heterochromatin.
ASI1 Is an RNA-Binding Protein.
Because the ASI1 protein has an RRM domain, which was predicted to have a single-stranded RNA binding function (36), we tested whether ASI1 may have RNA binding activity. Glutatione S-transferase (GST)–fused ASI1 protein was used for electrophoretic gel mobility shift assays. The results showed that GST-ASI1 but not GST alone could bind to a 60-nt single-stranded RNA. The binding signal increased as more GST-ASI1 protein was used, and the binding was competitively eliminated by addition of the unlabeled RNA of the same sequence (Fig. 5C). The results suggest that ASI1 is an RNA-binding protein, consistent with the presence of an RRM motif in the protein.
Discussion
DNA methylation at TEs and gene promoters is known to confer epigenetic silencing. This and several other studies (29–31) have found that DNA methylation at intronic TEs and other repeat elements is necessary for the proper expression of genes. It appears that organisms have evolved to take advantage of methylated introns to regulate the expression of genes harboring the methylated repeat elements. ASI1 is an important part of this unique mechanism of regulating genes via their intronic heterochromatic elements.
Mutation of ASI1 significantly decreases the level of full-length IBM1 transcripts. As a consequence, the asi1-1 mutant showed similar DNA hypermethylation patterns to those observed in ibm1. Both the asi1-1 and ibm1 mutants accumulate genome-wide DNA hypermethylation that mainly occurs in the CHG context within gene bodies. Comparative analyses of asi1-1 and ibm1 methylomes showed that the majority of hypermethylated loci in asi1-1 are also hypermethylated in ibm1. These overlapping loci include the BONSAI locus, the hypermethylation of which allowed the isolation of ibm1 mutants. A small percentage of asi1-1–induced hypermethylation also exists in the DNA demethylase triple mutant rdd. Some of this hypermethylation does not occur in ibm1, indicating that ASI1 may have IBM1-independent roles in regulating DNA demethylation and preventing transcriptional silencing. Nevertheless, transformation of pIBM1::IBM1ΔIH in the asi1-1 mutant restored the expression of full-length IBM1 transcripts and rescued not only the gene body CHG hypermethylation phenotype but also the developmental defects of asi1-1 mutant plants. These results suggest that many of the functions of ASI1 can be attributed to its regulation of IBM1 expression. Compared with asi1-1, ibm1 displayed greater levels of CHG hypermethylation. Such a pattern is consistent with the fact that asi1-1 mutation substantially reduces, but does not abolish, the full-length IBM1 transcript.
The seventh intron of the IBM1 gene contains a heterochromatic repeat sequence where DNA methylation and histone H3K9 methylation are necessary for accumulation of the full-length functional transcripts (29). IBM1, AT3G05410, and AT1G58602 maintain dense DNA methylation in the intronic heterochromatic element in the asi1-1 mutant, indicating that the heterochromatic feature alone is insufficient to promote the 3′ distal polyadenylation that generates full-length transcripts and that the transacting factor ASI1 is necessary to ensure the production of full-length transcripts.
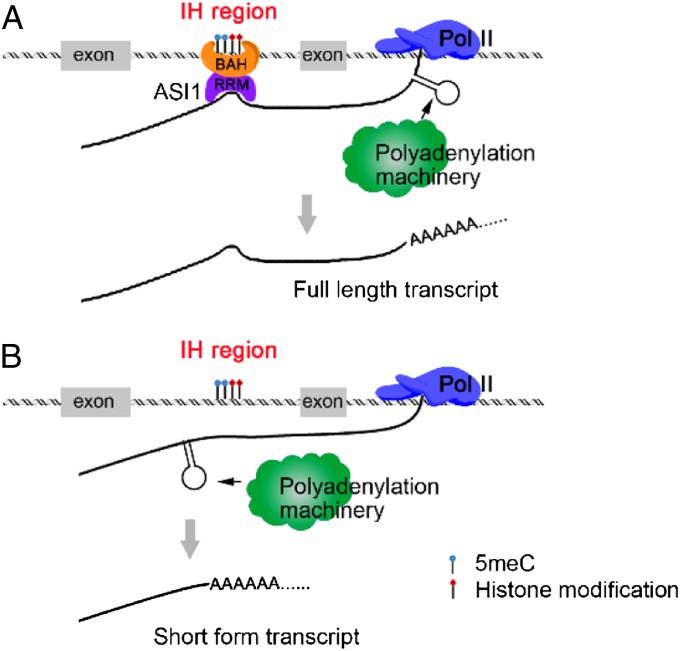
At IBM1 and several other genes with intronic heterochromatic elements, ChIP assays showed that occupancy of the elongating Pol II is not affected by the asi1-1 mutation, suggesting that transcription elongation is independent of ASI1 function. In addition, Pol II does not seem to exhibit greater occupancy at the intronic heterochromatic regions than at their corresponding 5′ and 3′ portions, indicating that ASI1 function does not involve alteration in transcriptional elongation kinetics. mRNA-seq and individual 3′-RACE analyses demonstrated that ASI1 facilitates distal polyadenylation downstream of the intronic heterochromatic elements or inhibits proximal polyadenylation, thereby promoting the generation of full-length transcripts. Highly used polyadenylation sites are positively associated with an mRNA structure that is energetically favorable and one that effectively exposes a critical polyadenylation cis-element (37). ASI1 contains a chromatin-binding BAH domain and an RNA-binding RRM motif. ChIP assays revealed that ASI1 associates with heterochromatic elements that reside within introns but not within gene promoters or intergenic regions, although it remains unclear how such specificity is achieved. One possibility is that unlike heterochromatic elements in gene promoters or intergenic regions, intronic heterochromatic elements are actively transcribed and thus must have various active chromatin marks in addition to their heterochromatic marks. Given that ASI1 also showed RNA-binding activities in vitro, we hypothesize that the intronic heterochromatin-associated ASI1 may bind to the pre-mRNAs in vivo and the RNA-binding subsequently induces an RNA structure that favors 3′ distal polyadenylation over proximal polyadenylation. Alternatively, ASI1 may impede the formation of an RNA structure that favors proximal polyadenylation. In either case, ASI1 promotes distal polyadenylation that generates full-length transcripts (Fig. 6).
Fig. 6.
A hypothetic model for ASI1-dependent regulation of alternative polyadenylation of intronic heterochromatin-containing genes. (A) ASI1 binds to the intronic heterochromatic region and the nascent pre-mRNA via the BAH domain and the RRM motif, respectively. The association of pre-mRNA with ASI1 induces formation of a structure that favors 3′ distal polyadenylation over proximal polyadenylation. (B) In the absence of ASI1, the pre-mRNA forms a structure that favors proximal polyadenylation, resulting in decreased accumulation of the full-length transcripts. Gray boxes represent the exons.
Our mRNA-seq analyses identified dozens of intronic heterochromatin-containing genes that require ASI1 for proper expression. Other intronic heterochromatin-containing genes may require the ASI1 paralog, AT3G15605, for proper expression. For some of these intronic heterochromatin-containing genes, proximal polyadenylation-generated shorter transcripts could be functional, and the alternative polyadenylation may even be subjected to regulation by developmental or environmental cues. It appears that the control of gene expression by intronic heterochromatic elements is a common biological feature and that plants have acquired an ASI1-dependent mechanism to ensure the proper expression of this type of genes. Alternative polyadenylation regulated by intronic DNA methylation has also been observed in the mouse imprinted genes H13 and Herc3, in which the methylated and unmethylated alleles use downstream and upstream polyadenylation sites, respectively (30, 31), supporting the conclusion that DNA methylation plays a positive role in determining the use of polyadenylation sites. It is likely that mammalian cells require transacting factors like ASI1 for the epigenetic regulation of alternative polyadenylation.
Materials and Methods
Details are provided in SI Materials and Methods, including plant materials and growth conditions, map-based cloning and identification of the ASI1 locus, plasmid construction and mutant complementation, DNA methylation-sensitive PCR, quantitative real-time PCR, Northern blot analysis, 3′RACE, ChIP assays, protein purification and EMSAs, whole-genome bisulfite sequencing and analysis, and mRNA-seq data analysis. Primers used in this study are listed in Table S2. The raw data of mRNA-seq of asi1-1, WT, and bisulfite sequencing of asi1-1 have been deposited in the GEO database (accession no. GSE48026). The bisulfite sequencing of ibm1-4 has also been deposited in the GEO database (accession no. GSE48053).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 (to J.-K.Z.) and by the Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
Data deposition: The raw data of mRNA sequencing of asi1-1, WT, and bisulfite sequencing of asi1-1; and the bisulfite sequencing of ibm1-4 have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE48026 and GSE48053).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315399110/-/DCSupplemental.
References
- 1.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12(13):1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126(6):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447(7143):418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21(3):367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12(8):483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012;15(5):517–522. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Pikaard CS, Haag JR, Pontes OM, Blevins T, Cocklin R. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb Symp Quant Biol. 2012;77:205–212. doi: 10.1101/sqb.2013.77.014803. [DOI] [PubMed] [Google Scholar]
- 10.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 11.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124(3):495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103(31):11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67(6):671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 14.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104(16):6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Zhu JK. Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol. 2012;77:161–173. doi: 10.1101/sqb.2012.77.014936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336(6087):1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 19.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151(1):167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498(7454):385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319(5862):462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 22.Miura A, et al. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28(8):1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberdoerffer S. A conserved role for intragenic DNA methylation in alternative pre-mRNA splicing. Transcription. 2012;3(3):106–109. doi: 10.4161/trns.19816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla S, Oberdoerffer S. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim Biophys Acta. 2012;1819(7):673–683. doi: 10.1016/j.bbagrm.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8(11):e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unoki M, Nakamura Y. Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett. 2003;554(1–2):67–72. doi: 10.1016/s0014-5793(03)01092-5. [DOI] [PubMed] [Google Scholar]
- 29.Rigal M, Kevei Z, Pélissier T, Mathieu O. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 2012;31(13):2981–2993. doi: 10.1038/emboj.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowley M, Wood AJ, Böhm S, Schulz R, Oakey RJ. Epigenetic control of alternative mRNA processing at the imprinted Herc3/Nap1l5 locus. Nucleic Acids Res. 2012;40(18):8917–8926. doi: 10.1093/nar/gks654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood AJ, et al. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22(9):1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei M, et al. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 2011;156(3):1116–1130. doi: 10.1104/pp.110.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25(20):2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446(1):189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 35.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25(12):2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cléry A, Blatter M, Allain FH. RNA recognition motifs: Boring? Not quite. Curr Opin Struct Biol. 2008;18(3):290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Khaladkar M, Smyda M, Hannenhalli S. Epigenomic and RNA structural correlates of polyadenylation. RNA Biol. 2011;8(3):529–537. doi: 10.4161/rna.8.3.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.