Significance
We developed reagents and approaches to pull down ribosome-associated RNAs from Cre-labeled cells. We show that this strategy is useful to probe cell type-specific gene expression and the extent of transcript binding to ribosomes.
Keywords: heart, pressure overload
Abstract
Transcriptional profiling is a useful strategy to study development and disease. Approaches to isolate RNA from specific cell types, or from specific cellular compartments, would extend the power of this strategy. Previous work has shown that isolation of genetically tagged ribosomes (translating ribosome affinity purification; TRAP) is an effective means to isolate ribosome-bound RNA selectively from transgene-expressing cells. However, widespread application of this technology has been limited by available transgenic mouse lines. Here we characterize a TRAP allele (Rosa26fsTRAP) that makes this approach more widely accessible. We show that endothelium-specific activation of Rosa26fsTRAP identifies endothelial cell-enriched transcripts, and that cardiomyocyte-restricted TRAP is a useful means to identify genes that are differentially expressed in cardiomyocytes in a disease model. Furthermore, we show that TRAP is an effective means for studying translational regulation, and that several nuclear-encoded mitochondrial genes are under strong translational control. Our analysis of ribosome-bound transcripts also shows that a subset of long intergenic noncoding RNAs are weakly ribosome-bound, but that the majority of noncoding RNAs, including most long intergenic noncoding RNAs, are ribosome-bound to the same extent as coding transcripts. Together, these data show that the TRAP strategy and the Rosa26fsTRAP allele will be useful tools to probe cell type-specific transcriptomes, study translational regulation, and probe ribosome binding of noncoding RNAs.
Genome-wide and unbiased measurement of RNA transcript levels using microarrays and RNA-seq (1) has powered fundamental advances in biology over the past decade. However, when used to study tissues composed of multiple cell types, RNA expression profiling faces two fundamental limitations. First, whole-tissue transcript levels represent the average of the distinct cell lineages in the tissue, and this averaging process can result in loss of important information or misassignment of gene expression changes in one type of cell to another. Second, RNA profiling measures transcript abundance, but translational regulation is also an important determinant of gene expression (2, 3).
To overcome these limitations, approaches have been developed to isolate RNAs from selected cell types and/or selected transcript fractions (4–7). Often these approaches involve tissue dissociation followed by FACS, but this is slow and the dissociation procedure itself likely alters expression profiles. Translating ribosome affinity purification (TRAP) permits isolation of transcripts from selected cell types of intact tissues, without dissociation (4). In this approach, ribosomes of selected tissues are genetically labeled by transgenic expression of GFP fused to L10a, an integral component of the 60S ribosomal subunit. To collect RNA from the transgene-expressing subpopulation of a tissue, whole-cell lysates are prepared under conditions that stabilize ribosomes on RNA and block translation (Fig. 1A). Immunoprecipitation of GFP then selectively coprecipitates RNAs from the target cell population. Moreover, the coprecipitated RNAs (TRAP RNAs) are those actively engaged by ribosomes, so that the method also enriches for RNAs that are “actively translating.” Heiman et al. used this approach to profile actively translating RNAs in rare cell populations in the central nervous system (4). This study was based on tissue-selective expression of GFP-L10a using BAC transgenes, so that each lineage of interest required construction of a new transgenic mouse line.
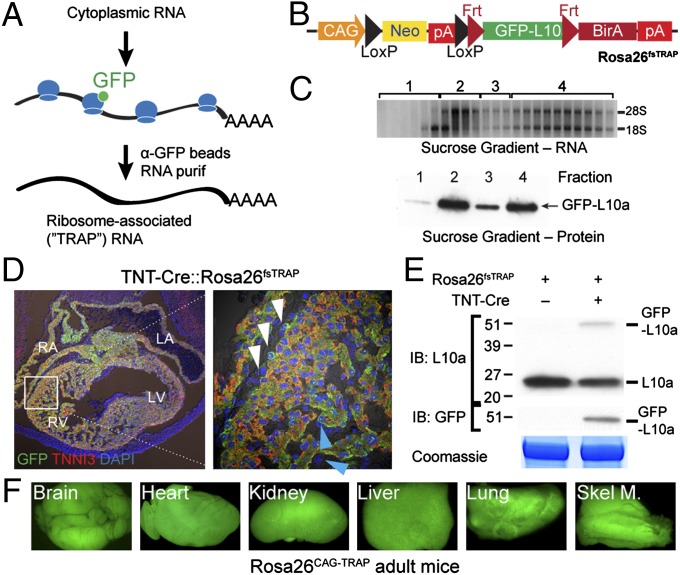
Fig. 1.
Generation and characterization of a Cre-activated TRAP allele. (A) TRAP strategy to pull down transcripts bound to GFP-tagged ribosomes. (B) Structure of targeted Rosa26fsTRAP allele. (C) EGFP-L10a incorporates into assembled ribosomes and polysomes. Equal volumes of the indicated fractions (1–4) from sucrose gradient polysome fractionation of Rosa26CAG-TRAP ES cells were assayed for GFP-L10a by Western blot. (D) Section of E12.5 TNT-Cre::Rosa26fsTRAP heart. TNNI3-positive cardiomyocytes coexpressed GFP. (E) Western blot showing Cre-dependent expression of GFP-L10a fusion protein in neonatal Rosa26fsTRAP heart. (F) Germ-line Cre recombination yielded Rosa26CAG-TRAP mice, which showed widespread GFP-L10a expression.
Here we report generation of a mouse line in which lineage-selective Cre activates expression of GFP-L10a. The resulting mouse line permits selective isolation of ribosome-bound RNA from Cre-marked cell lineages, and at the same time is useful as a Cre-activated fluorescent reporter. In validation experiments, we show that this mouse line in combination with endothelial-selective Tie2-Cre (8) effectively isolates endothelial lineage-selective genes, and we use this property to identify additional putative endothelial-selective transcripts. In combination with cardiomyocyte-selective cardiac troponin T (TNT)-Cre, we show that cell type-specific transcriptional profiling is helpful to define gene expression changes that occur in a specific cell type. Finally, we use this unique tool to analyze ribosome binding to transcripts. We show that ribosome binding to mitochondrial protein-coding transcripts is regulated in a tissue-specific manner, with high ribosome binding occurring in heart and liver. For noncoding transcripts, we show that a subset of long intergenic noncoding RNAs (lincRNAs) display low ribosome binding, consistent with their function in the absence of translation. However, most noncoding transcripts, including the majority of lincRNAs, display ribosome binding comparable to that seen for protein-coding transcripts. Collectively, these experiments indicate that the Cre-activated TRAP allele will be a useful tool for studying regulation of transcript ribosome binding and lineage-specific gene expression.
Results
Rosa26fsTRAP Allele.
Rpl10a encodes ribosomal protein L10a. Immunoprecipitation of GFP-tagged L10a using a GFP-specific antibody selectively isolates transcripts from GFP-L10a–expressing cells (Fig. 1A) (4). We targeted the Rosa26 locus with a construct containing a strong CAG promoter followed by a floxed neomycin resistance cassette, a transcriptional stop signal, and GFP-L10a cDNA (Fig. 1B and Fig. S1A). Incidentally, the GFP-L10a cDNA was flanked by Frt sequences, allowing its excision by Flp recombinase and subsequent expression of the Escherichia coli BirA enzyme, which permits selective biotinylation of appropriately tagged proteins. Cre-activated expression of BirA from the post-Flp recombined allele will be described in a subsequent manuscript.
Successful targeting of the Rosa26 locus in murine embryonic stem (ES) cells yielded Rosa26-flox-stop-TRAP (Rosa26fsTRAP). Correct homologous recombination was verified by Southern blotting (Fig. S1B). Cre-mediated recombination in ES cells yielded constitutively activated TRAP (Rosa26CAG-TRAP), in which the CAG promoter drives expression of the GFP-L10a protein (Fig. S1C). To examine the distribution of tagged protein in ribosomes, we fractionated ES cell lysates on sucrose gradients (Fig. 1C). As expected, GFP-L10a protein was distributed primarily in the polysome fraction (fraction 4, containing actively translating transcripts) and the free-ribosome fraction (fraction 2).
Cre-Mediated, Tissue-Specific Activation of Rosa26fsTRAP in Mice.
We used Rosa26fsTRAP ES cells to generate gene-targeted mice. Consistent with the allele design, we did not detect GFP fluorescence in the absence of Cre. To evaluate the Cre activation of GFP-L10a expression in specific cell lineages, we crossed in the TNT-Cre transgene, which is selectively active in cardiomyocytes. TNT-Cre activated GFP-L10a expression selectively in the heart (Fig. S1 D and E), where it was colocalized with the cardiomyocyte marker TNNI3 (Fig. 1D). GFP-L10a expression was not detected in epicardial (white arrowheads) or endocardial cells (blue arrowheads), which are not labeled by TNT-Cre. We confirmed Cre-dependent expression of GFP-L10a in neonatal heart by Western blotting (Fig. 1E). GFP-L10a was found only in lysates of Rosa26fsTRAP hearts that were also positive for the TNT-Cre transgene. Importantly, TNT-Cre::Rosa26fsTRAP and control hearts did not differ significantly upon echocardiographic analysis (Fig. S1F), indicating that the GFP-L10a protein does not interfere with heart function.
We also globally activated the GFP-L10a expression by excising the floxed stop cassette in the germ line. The resulting Rosa26CAG-TRAP mice were viable and fertile as homozygotes. Homozygotes survived to weaning at the expected Mendelian frequency [from heterozygous intercrosses, we obtained 25% (11/44) homozygotes], had normal heart function by echocardiography, and lived without overt phenotype or premature demise for over 8 mo. GFP-L10a was widely expressed, as determined by observation of robust GFP fluorescence (Fig. 1F) and by immunoblotting (Fig. S1G). GFP immunoprecipitation under conditions that stabilize ribosomes and block translational elongation (TRAP) pulled down about 40% of total RNA in heart and brain (heart 42 ± 7%, brain 38 ± 6%, n = 4, P > 0.05). These data indicate that the CAG promoter drives widespread GFP-L10a expression and that this is not overtly detrimental to mouse development or survival, and that the Rosa26fsTRAP allele will likely be useful for lineage tracing and TRAP analysis in multiple organs.
TRAP Identification of Endothelial-Enriched Transcripts.
TRAP has been used to measure cell-specific transcriptional profiles from whole tissues in which the cell population of interest selectively expresses GFP-L10a (4). To further characterize the Rosa26fsTRAP allele and to identify transcripts enriched in endothelial and endocardial cells, we generated Tie2-Cre::Rosa26fsTRAP mice. The endothelial cell-specific Tie2-Cre transgene selectively activated GFP-L10a expression in endothelial cells, as demonstrated by costaining with the endothelial marker PECAM1 (Fig. 2A). Next, we used GFP immunoprecipitation under conditions that stabilize ribosomes and block translational elongation to isolate total cytoplasmic RNA (input) and GFP-L10a–bound RNA (TRAP) from Tie2-Cre::Rosa26fsTRAP and TNT-Cre::Rosa26fsTRAP heart. We refer to the TRAP fraction from Tie2-Cre::Rosa26fsTRAP and TNT-Cre::Rosa26fsTRAP as “Tie2-TRAP” and “TNT-TRAP,” respectively. Quantitative RT-PCR (qRT-PCR) for canonical endothelial transcripts Cdh5 (VE-cadherin) and Tek (Tie2) showed that these transcripts were enriched in Tie2-TRAP by over 10-fold compared with the housekeeping gene Gapdh, whereas they were depleted in TNT-TRAP (Fig. 2B). On the other hand, cardiomyocyte-specific transcripts Myh6 (myosin heavy chain α) and Tnnt2 (TNT) were depleted in Tie2-TRAP (Fig. 2B). These cardiomyocyte transcripts were not substantially enriched by TNT-TRAP, likely because the preponderance of heart RNA arises from cardiomyocytes, so that TRAP cannot substantially further enrich the cardiomyocyte-specific transcripts.
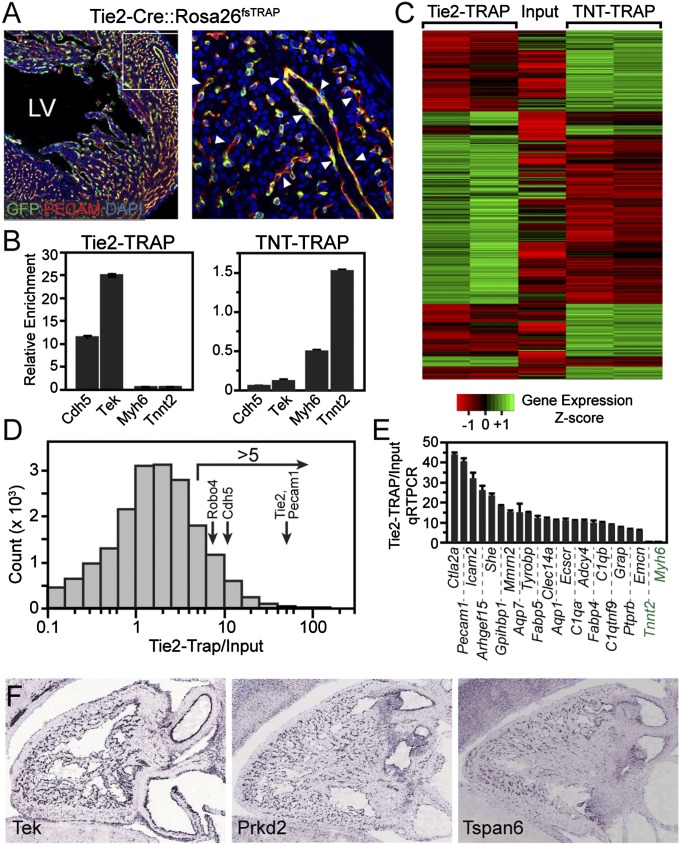
Fig. 2.
Capture of endothelial transcripts from heart with Tie2-TRAP. (A) Tie2-Cre–activated Rosa26fsTRAP expression in endothelial cells. Boxed area at Left is magnified at Right. LV, left ventricle. (B) Transcript enrichment by qRT-PCR in Tie2-TRAP or TNT-TRAP isolated from heart. (C) Heatmap showing an overview of genes differentially expressed between Tie2-TRAP and TNT-TRAP of heart RNA by RNA-seq. (D) Histogram of Tie2-TRAP:input ratios, showing high values (>5) for known endothelial-restricted transcripts. (E) qRT-PCR measurement of Tie2-TRAP:input ratio in heart RNA of arbitrarily selected genes with an RNA-seq ratio greater than 5. Tnnt2 and Myh6 were included as negative controls. (F) Eurexpress high-throughput in situ hybridization images of selected genes with high Tie2-TRAP:input ratio. The classic endothelial gene Tek (also known as Tie2) has a typical endothelial/endocardial staining pattern. Prkd2 and Tspan6 are less-characterized genes with endothelial-restricted expression identified by high Tie2-TRAP:input ratios. See also Fig. S2. Data in B and E are displayed as mean ± SEM.
Having established that Tie2-TRAP and TNT-TRAP selectively capture endothelial and cardiomyocyte transcripts, we next evaluated the global gene expression profile of endothelial and cardiomyocyte cells by RNA-seq (Datasets S1 and S2). There were 1,524 genes differentially expressed (P < 0.0001) between Tie2-TRAP and TNT-TRAP in biological duplicate samples (Fig. 2C). The Tie2-TRAP:input ratio is a measure of endothelial cell enrichment. For instance, the signature endothelial genes Pecam1, Tie2, Cdh5, and Robo4 all had Tie2-TRAP:input ratios higher than 5 (Fig. 2D). We randomly selected several additional transcripts with Tie2-TRAP:input ratios of greater than 5 and validated the RNA-seq data by qRT-PCR (Fig. 2E). In all 21 genes examined, the endothelium-biased signal found by RNA-seq was confirmed by qRT-PCR, whereas negative control genes Tnnt2 and Myh6 had low Tie2-TRAP:input ratios, as expected.
We identified putative endothelial/endocardial cell-enriched transcripts by Tie2-TRAP:input ratios higher than five and strong endothelial expression (Tie2-TRAP level > 10 RPKM). There were 908 genes that matched these criteria (Dataset S2), including known endothelial cell-specific transcripts Pecam1, Tie2, Cdh5, and Robo4. From these, we selected 290 genes with a range of Tie2-TRAP:input ratios and evaluated their expression pattern in Eurexpress, a public gene expression database containing high-throughput embryonic day (E)14.5 whole-embryo in situ hybridization data (9). Of these, sixty-four (47%) were expressed selectively in endothelial cells or endothelial cells plus the thymus (Fig. 2F and Fig. S2A), which contains blood cells that are also labeled by Tie2-Cre. Among these were known endothelial cell transcripts (e.g., Tek and Hdac7), as well as transcripts with previously less well characterized expression patterns (e.g., Prkd2 and Tspan6; Fig. 2F).
Collectively, these data indicate that Cre-activated TRAP and RNA-seq are unbiased means to interrogate cell type-specific transcriptomes.
Analysis of Cardiomyocyte Gene Expression.
Although many studies have reported on cardiac gene expression profiling, few have looked at gene expression selectively in cardiomyocytes from intact tissue. In part, this is due to the difficulty in obtaining purified cardiomyocyte populations without lengthy isolation procedures, which may themselves alter gene expression profiles. Although cardiomyocytes constitute a large proportion of cardiac mass and RNA, noncardiomyocytes also contribute to overall gene expression profiles. Disease models such as aortic banding alter cardiomyocyte and nonmyocyte gene expression, as well as the relative proportion of each cell type. Thus, selective measurement of gene expression in cardiomyocytes might yield different results in disease models compared with whole-heart expression profiling.
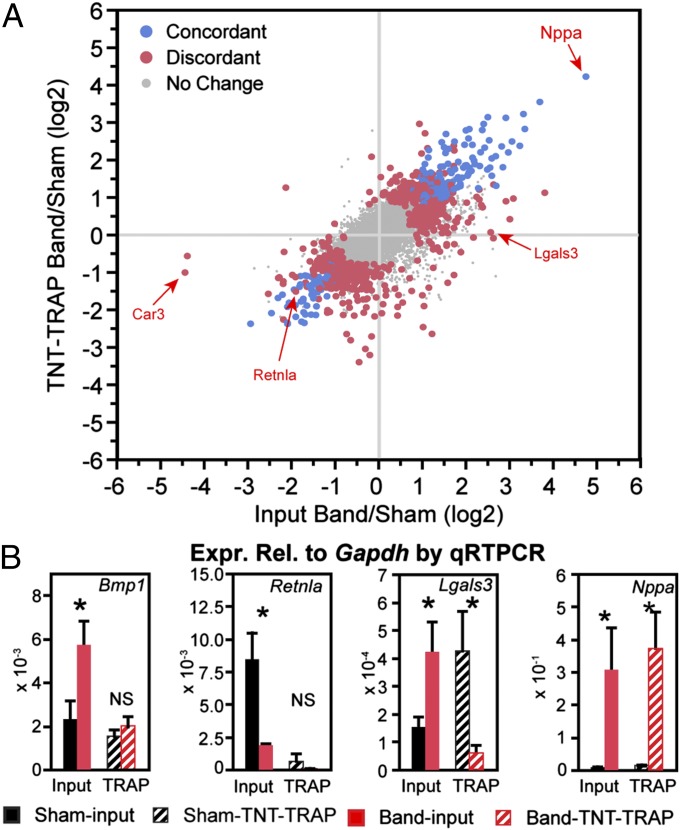
To investigate the usefulness of TNT-TRAP in transcriptional analysis of differential gene expression in heart disease models, we performed aortic banding or sham operation of TNT-Cre::Rosa26fsTRAP mice. We used moderate banding conditions that caused substantial hypertrophy without inducing systolic dysfunction 2 wk after the operation (Fig. S3). At these time points, we isolated total heart cytosolic RNA (input) or RNA bound to TNT-Cre–activated GFP-L10a (TNT-TRAP) from sham or band groups. Biological triplicate RNA samples were prepared for each of the resulting four groups and analyzed by qRT-PCR or RNA-seq (Datasets S1 and S3). We identified differentially expressed genes between band and sham (P < 0.05; expression level over 5 RPKMs in at least one sample) in input and TRAP samples. There were 521 and 548 genes differentially expressed between band and sham in input or TRAP fractions, respectively. Of these, 231 genes were differentially expressed in both input and TRAP fractions, 290 were changed in input but not TRAP, and 317 were changed in TRAP but not input (Fig. 3A). We evaluated 14 genes by qRT-PCR (Fig. 3B and Fig. S3 D and E). There was overall good correlation between fold change measured by RNA-seq compared with qRT-PCR (R = 0.9), and statistical comparisons agreed between RNA-seq and qRT-PCR in 18 of 25 cases (Fig. S3 D and E). These data indicate that cell type-selective Cre-activated Rosa26fsTRAP is a useful means to hone in on differential expression in the cell type of interest, even in cases where those cells constitute the majority of the tissue mass. Furthermore, the TRAP fraction constitutes actively translating RNAs and hence may more closely reflect gene product function than total RNA.
Fig. 3.
Analysis of differential gene expression using TRAP. (A) Gene expression was measured 2 wk after aortic banding. Fold change was calculated from RNA-seq of input and TNT-TRAP (n = 3). Blue, differentially expressed between band and sham in both input and TRAP RNA. Red, differentially expressed between band and sham in either input or TRAP but not both. Gray, genes that were not differentially expressed in either RNA fraction. Coding transcripts with RPKM >5 are shown. A subset of genes selected for qRT-PCR validation is indicated. (B) qRT-PCR validation of RNA-seq data. Expression of the indicated genes was measured by qRT-PCR. n = 3–6 samples per group. *P < 0.05. NS, not significant. Data are displayed as mean ± SEM.
Translational Regulation of Coding Transcripts.
Gene expression is regulated at the level of transcription and translation, and recent studies have suggested that translational regulation is at least as important as transcriptional regulation in determining cellular protein abundance (2). The predominant described use of TRAP has been to study tissue-specific gene expression. We tested the hypothesis that TRAP enrichment of ribosome-bound RNAs would allow it to measure one aspect of translational regulation, the extent of ribosome loading onto gene transcripts, herein termed “ribosome occupancy,” a measure of translational efficiency.
Sucrose gradient sedimentation has been the standard technique to measure ribosome occupancy. This method was used to show that in ES cells, actin (Actb) was predominantly associated with actively translating ribosomes, whereas roughly half of Atf5 and little of the noncoding RNA H19 were found in this fraction (10). TRAP may be an alternative, more streamlined method to assess ribosome binding and active translation. To benchmark TRAP’s effectiveness in measuring a transcript’s ribosome occupancy, we directly compared the ribosome binding of these transcripts by TRAP versus sucrose gradient sedimentation from ES cell lysates. The TRAP and sucrose gradient methods yielded similar results, in that Actb was predominantly found in the TRAP fraction, whereas only a minority of Atf5 and a small fraction of H19 transcripts were present in TRAP RNA (Fig. S4 A and B).
We further compared TRAP and sucrose gradient sedimentation by using each technique to measure translation of Actb transcripts when ES cells were cultured with increasing concentrations of des-methyl, des-amino-pateamine A (DMDA-PatA), a potent inhibitor of translational initiation (11). DMDA-PatA reduced Actb transcript abundance in both polysomal RNA and TRAP RNA, and relative inhibition measured by TRAP and sucrose gradient methods paralleled one another (Fig. S4C), although the sucrose gradient measurement was consistently shifted toward more potent DMDA-PatA inhibition. Likely this is because DMDA-PatA blocked polysome formation but left some monosomes intact (Fig. S4D), and RNA assembled on tagged monosomes will coprecipitate during the TRAP procedure. Collectively, these data show that the TRAP procedure pulls down ribosome-associated RNAs and can be used to assess mRNA ribosome occupancy.
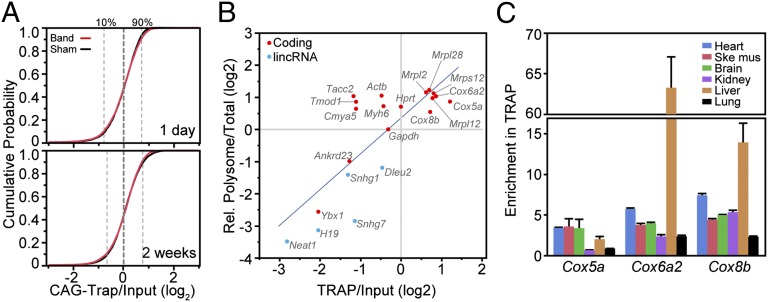
We next used the Rosa26 TRAP allele to investigate ribosome binding in tissues in vivo. In a complex tissue, the TRAP:input ratio is influenced by both GFP-L10a expression pattern and GFP-L10a ribosome binding. To focus on GFP-tagged ribosome binding to transcripts and minimize the effect of cell type-restricted GFP-L10a expression, we used Rosa26CAG-TRAP mice, in which the constitutively activated GFP-L10a is widely expressed by the CAG promoter. We generated four sets of RNA-seq data for sham and banded hearts, collecting heart ventricles 1 d and 2 wk after the operation (Datasets S1 and S4). We initially focused on coding transcripts, and used the TRAP:input ratio from Rosa26CAG-TRAP heart as a measure of ribosome occupancy. In the sham 2-wk sample, 80% of transcripts had CAG-TRAP:input ratios within an ∼1.4 log2 scale range (2.6-fold linear scale), suggesting that ribosome binding of most transcripts occurs within a limited dynamic range (Fig. 4A). The coding transcripts with the lowest ribosome binding (lowest 500) were enriched for functional annotations related to actin cytoskeleton organization and regulation of actin filaments [false discovery rate (FDR) = 4.8 × 10−5; Table 1]. In contrast, transcripts with the highest ribosome binding (highest 500) were strongly enriched for mitochondria (FDR = 1.6 × 10−35; Table 1) and oxidative phosphorylation (FDR = 5.5 × 10−20). Transcripts encoding ribosomal proteins were also highly overrepresented (FDR = 2.1 × 10−13); interestingly, the overrepresented ribosomal proteins were largely components of mitochondrial ribosomes. These overall patterns were also found in the other three (2-wk band and 1-d band and sham) datasets (Fig. 4A).
Fig. 4.
Analysis of ribosome binding using Rosa26CAG-TRAP. RNA-seq was performed on Rosa26CAG-TRAP heart 2 wk after sham or band operation. (A) Cumulative probability plot of the strength of ribosome binding (TRAP:input) for coding transcripts with RPKM >5 in sham or band. Most transcripts had TRAP:input ratios within a narrow range, and the distribution did not change with banding. (B) Comparison of sucrose gradient fractionation and TRAP measurement of translational activity. TRAP:input values are from RNA-seq, whereas polysome and total RNA measurements are from qRT-PCR and normalized to Gapdh. The best-fit line is shown. (C) qRT-PCR measurement of TRAP enrichment relative to Gapdh of selected mitochondrial protein transcripts in multiple tissues. These transcripts had high ribosome binding in heart and liver and lower ribosome binding in kidney and lung. Data are displayed as mean ± SEM.
Table 1.
Gene ontology analysis of genes with high or low ribosome binding
| Term | Adjusted P value |
| Low ribosome binding (bottom 500 ranked by TRAP:input) | |
| Actin binding | 8.4 × 10−5 |
| Actin filament-based process | 3.7 × 10−3 |
| High ribosome binding (top 500 ranked by TRAP:input) | |
| Mitochondrion | 1.6 × 10−35 |
| Oxidative phosphorylation | 5.5 × 10−20 |
| Ribonucleoprotein | 2.1 × 10−13 |
| Mitochondrial ribosome | 2.8 × 10−5 |
| NADH dehydrogenase activity | 7.9 × 10−3 |
| Protein targeting | 4.3 × 10−2 |
All functional terms with significant enrichment (Benjamini–Hochberg–corrected P value < 0.05) for genes with the strongest or weakest ribosome binding are shown.
We examined the relationship of ribosome occupancy measured by TRAP:input and by sucrose gradient sedimentation in Rosa26CAG-TRAP heart ventricles. We selected 21 genes that spanned the gamut of TRAP:input values and measured the fraction that partitioned into the polysome fraction, compared with GAPDH. Overall, TRAP:input and normalized polysome:total were moderately correlated (R = 0.78, P < 0.001; Fig. 4B). Moreover, these data also validated the high ribosome occupancy of mitochondrial proteins (seven out of the seven tested).
Cardiomyocytes are highly enriched for mitochondria. To determine whether high ribosome occupancy of these transcripts is ubiquitous or is regulated between different tissues, we used qRT-PCR to measure TRAP:input ratios for mitochondrial transcripts Cox5a, Cox6a2, and Cox8b in multiple tissues (Fig. 4C). Interestingly, their TRAP:input ratios varied substantially between tissues. For instance, Cox5a, encoding a cytochrome c subunit, showed strong enrichment in TRAP from heart but not in kidney or lung. Cox6a2 was strongly ribosome-bound in liver and heart and more weakly ribosome-bound in kidney and lung. These data indicate that translation of some mitochondrial transcripts is under strong tissue-specific regulation.
Increased translation has been identified as a hallmark of cardiac hypertrophy (12), although this has not been studied at a genome-wide scale. We used RNA-seq and the CAG-TRAP:input ratio to provide an unbiased assessment of changes in ribosome occupancy that occur following pressure overload. We found that the overall distribution of the TRAP:input ratio of coding transcripts was not significantly different in the band compared with the sham group at 1 d or 2 wk after banding (Fig. 4A), suggesting there is no significant change in overall ribosome occupancy after banding. Considering protein-coding genes with altered ribosome binding at 24 h between band and sham, the top 2.5 percentile (143 genes) was significantly enriched for genes related to ribosomes and translation, mitochondria, and sarcomeres (Dataset S4). These sarcomere genes included tropomodulin 1, myotilin, myozenin 2, PDZ, and limb domain 3.
Many Noncoding RNAs Exhibit Significant Ribosome Binding.
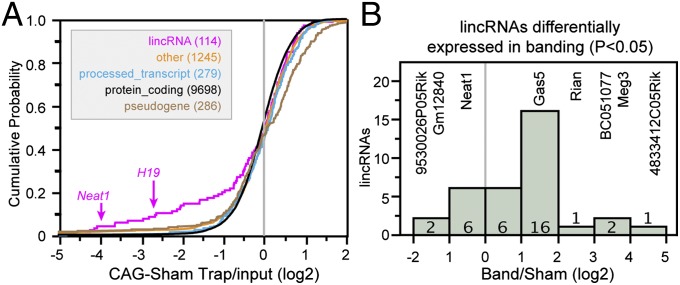
Unbiased transcriptional profiling has revealed many noncoding RNA classes, such as long intergenic noncoding RNAs, antisense transcripts, and pseudogenes, as well as truncated transcripts presumably generated by nonsense-mediated decay. The expression pattern and function of most of noncoding transcripts remain unknown. Using TRAP, we assayed ribosome binding to noncoding transcripts, subgrouped by their Ensembl transcript classification (SI Materials and Methods). Interestingly, the large majority of annotated noncoding transcripts bind to ribosomes with a similar distribution as coding transcripts (Fig. 5A). However, the lincRNA subclass showed a distribution that was significantly enriched in low ribosome binding (Fig. 5A). Among these transcripts were two of the better-characterized noncoding RNAs, H19 and Neat1 (Fig. 5A; see also Fig. 4B). These data indicate that a subset of lincRNAs is not significantly bound by ribosomes, consistent with their presumed function as RNAs rather than proteins.
Fig. 5.
lincRNA ribosome binding. (A) Noncoding RNA ribosome binding profiles. Cumulative probability plot of ribosome binding strength by different noncoding RNA classes shows that lincRNAs are enriched for weak ribosome binding. Other noncoding RNA classes share ribosome binding profiles similar to coding transcripts. (B) Histogram showing that expression of a subset of lincRNAs changed significantly in input RNA between band and sham groups 2 wk after the operation. Transcripts with extreme up- or down-regulation are indicated by name.
Surprisingly, the majority of lincRNAs (∼65%) showed a profile of stronger ribosome binding that was comparable to protein-coding transcripts (Fig. 5A). We used a coding probability score (13) to assess whether inaccurate annotation of coding transcripts as noncoding was likely to account for this unexpected ribosome binding. Among lincRNA transcripts expressed above threshold, only 10% had a score suggestive of coding potential. Thus, inaccurate classification of coding transcripts as lincRNAs is unlikely to account for the unexpectedly high frequency of substantial ribosome binding. Because ribosome-bound RNAs isolated by TRAP include both transcripts present in polysomes and monosomes (Fig. S4C), we performed sucrose density sedimentation to further characterize three selected ribosome-bound RNAs. Two of the three, Snhg1 and Dleu2, were found in the polysome fraction in the approximate proportion projected based on the TRAP:input value, whereas Snhg7 was relatively underrepresented in the polysome fraction relative to the TRAP:input value (Fig. 4B). These data suggest that a substantial fraction of lincRNAs is bound by ribosomes, including those assembled into polysomes.
We evaluated whether or not banding had a significant effect on ribosome binding of lincRNAs by analyzing RNA-seq data on the input and TRAP RNAs of sham or band-operated Rosa26CAG-TRAP mice. The overall distribution of lincRNA binding to ribosomes did not change significantly with banding (Fig. S5). We also examined individual lincRNAs for differential expression in banding. Out of 120 lincRNAs above the expression threshold, 34 were differentially expressed in input RNA with nominal P < 0.05 (n = 3), including the better-characterized lincRNAs Gas5 and Neat1 (Fig. 5B and Dataset S5).
Discussion
The TRAP approach improves expression profiling of whole tissues by isolating ribosomal RNAs from selected cell types (4). Our study advances the TRAP approach by providing a generic allele that can be combined with numerous Cre drivers to make this technology widely available. Moreover, we show that this technology can be effectively deployed to study translational regulation and noncoding RNA binding to ribosomes.
The Cre-activated TRAP allele will be useful to profile the ribosome-bound transcriptome of selected cell types. Using Tie2-Cre–activated TRAP, we identified a number of transcripts enriched in endothelial and endocardial cells. One of these was Prkd2, encoding a protein kinase that interacts with HDAC7 (14), a regulator of endothelial cell transcription (15). The biological significance of enriched expression of Prkd2 and other endothelium-enriched transcripts identified in this study requires further investigation.
A known limitation of gene expression profiling of whole tissues is that gene expression changes in disease models may occur as a result of a change in the proportion of cell types, or in cells that are not the primary interest of the study. Even when the cell type of interest constitutes the majority of the tissue mass, cell type-targeted expression profiling is important to isolate changes that occur in the cells of interest. We observed this in the heart, where less than half of the genes that were differentially expressed between band and sham in the whole heart were also differentially expressed in cardiomyocyte TRAP RNA. Importantly, TRAP RNAs are also the actively translated fraction, and hence might be expected to be more closely related to protein-coding gene product expression than total RNA.
Regulation of translation is an important mechanism for gene expression control (2), but genome-wide, unbiased analyses of translational regulation have been limited. We show that TRAP is an effective and relatively straightforward means to assess ribosome occupancy, one determinant of translational efficiency. Using TRAP, we provide genome-wide assessment of ribosome occupancy in pressure overload. Although studies of selected transcripts suggested that increased protein synthesis is a hallmark of cardiac hypertrophy (12), we did not detect globally increased transcript binding to ribosomes at 1 d or 2 wk after banding. However, other factors that we did not measure, such as total ribosome number and elongation rate, also influence protein synthesis rate. We also did not directly measure the number of ribosomes loaded onto transcripts, although TRAP efficiency is likely to correlate with ribosome loading under the conditions that we used.
We identified transcripts with relatively low or high ribosome binding in normal heart. Transcripts with low binding were overrepresented for genes involved in actin binding and cytoskeletal organization, suggesting that these genes are likely regulated at the translational level. Interestingly, highly ribosome-bound transcripts were highly enriched for mitochondrial proteins, and efficient ribosome binding of these mitochondrial transcripts is regulated, as it differed markedly between tissues.
There is growing interest in the function of noncoding RNAs, particularly lincRNAs. Whether lincRNAs are substantially translated into proteins has been debated (16–19). We used TRAP to show that ∼35% of lincRNAs above expression threshold showed low ribosome binding. Some of these RNAs, such as Neat1, localize to cellular compartments where they are inaccessible to the tagged ribosome (20, 21). Others such as H19 are present in the cytoplasm (22), but appear to be excluded from ribosomes. The signals and mechanisms that permit these RNAs to avoid ribosome binding remain to be elucidated, but may involve their binding by specific proteins (22). The remaining ∼65% of annotated lincRNAs did show ribosome binding comparable to protein-coding transcripts. This finding is consistent with recently reported results based on ribosome footprinting (16) and with prior studies showing ribosome association of individual noncoding RNAs such as Gas5 (23). The significance of this ribosome binding requires further investigation, but one possibility is that a subset of noncoding RNAs may regulate translation of other transcripts.
Recently, a complementary approach to isolating RNA from cells labeled by Cre was reported, in which Cre-activated transgenic expression of uracil phosphoribosyltransferase (UPRT) permits selective RNA labeling by thiouracil (TU tagging) (7). A strength of the TU-tagging strategy is the enhanced temporal resolution for active transcription. In contrast, TRAP provides more information on translational regulation and ribosome binding. The extent of enrichment of endothelial transcripts by Tie-Cre–driven TRAP also appeared to be higher than achieved by TU tagging, potentially due to baseline labeling by thiouracil in UPRT-negative cells (7).
In conclusion, we have established a Cre-activated Rosa26fsTRAP mouse line and highlighted areas in which this reagent may enhance studies of gene expression regulation and disease mechanisms. This mouse line can be combined with a diverse array of Cre drivers to provide new insights into gene expression regulation.
Materials and Methods
Animals were used according to protocols approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital. Rosa26fsTRAP mice were generated by homologous recombination in J1 ES cells. Poly(A) RNA was analyzed by RNA-seq or quantitative RT-PCR. Primer sequences can be found in Dataset S6. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
This study was supported by funding from the American Heart Association and the National Heart, Lung, and Blood Institute (U01HL098166, U01HL098188, and R01HL095712), and by charitable donations from Gail Federici Smitt, Edward Marram, and Karen Carpenter.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) Cardiovascular Development Consortium Server (accession no. GSE45152).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304124110/-/DCSupplemental.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MR, Robinson KJ, Cleary MD, Doe CQ. TU-tagging: Cell type-specific RNA isolation from intact complex tissues. Nat Methods. 2009;6(6):439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gay L, et al. Mouse TU tagging: A chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev. 2013;27(1):98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kisanuki YY, et al. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 9.Diez-Roux G, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9(1):e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampath P, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Low WK, et al. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell. 2005;20(5):709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Hannan RD, Jenkins A, Jenkins AK, Brandenburger Y. Cardiac hypertrophy: A matter of translation. Clin Exp Pharmacol Physiol. 2003;30(8):517–527. doi: 10.1046/j.1440-1681.2003.03873.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra M, Kasler H, McKinsey TA, Olson EN, Verdin E. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J Biol Chem. 2005;280(14):13762–13770. doi: 10.1074/jbc.M413396200. [DOI] [PubMed] [Google Scholar]
- 15.Chang S, et al. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126(2):321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bánfai B, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22(9):1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9(1):59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esakova O, Krasilnikov AS. Of proteins and RNA: The RNase P/MRP family. RNA. 2010;16(9):1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zieve G, Penman S. Small RNA species of the HeLa cell: Metabolism and subcellular localization. Cell. 1976;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- 22.Runge S, et al. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem. 2000;275(38):29562–29569. doi: 10.1074/jbc.M001156200. [DOI] [PubMed] [Google Scholar]
- 23.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.