Significance
The Cedars, in coastal northern California, is an active serpentinization site. The spring waters emerging from this system feature very high pH (≈11.5), low redox potential (≈−550 mV), and low ionic concentrations, making it an exceptionally challenging environment for life. The microbial communities are different in different springs, strongly correlated with the source of the serpentinizing groundwater feeding the springs (shallow or deep). The shallow groundwater community was similar to those described in other terrestrial serpentinizing sites, while the deep community was distinctly different from any other previously described terrestrial serpentinizing community. These communities have the potential to yield important insights into survival mechanisms in these challenging, early-earth analog environments.
Keywords: biodiversity, extremophile, alkaliphile, small subunit rRNA, hydrogen
Abstract
The Cedars, in coastal northern California, is an active site of peridotite serpentinization. The spring waters that emerge from this system feature very high pH, low redox potential, and low ionic concentrations, making it an exceptionally challenging environment for life. We report a multiyear, culture-independent geomicrobiological study of three springs at The Cedars that differ with respect to the nature of the groundwater feeding them. Within each spring, both geochemical properties and microbial diversity in all three domains of life remained stable over a 3-y period, with multiple samples each year. Between the three springs, however, the microbial communities showed considerable differences that were strongly correlated with the source of the serpentinizing groundwater. In the spring fed solely by deep groundwater, phylum Chloroflexi, class Clostridia, and candidate division OD1 were the major taxa with one phylotype in Euryarchaeota. Less-abundant phylotypes include several minor members from other candidate divisions and one phylotype that was an outlier of candidate division OP3. In the springs fed by the mixture of deep and shallow groundwater, organisms close to the Hydrogenophaga within Betaproteobacteria dominated and coexisted with the deep groundwater community members. The shallow groundwater community thus appears to be similar to those described in other terrestrial serpentinizing sites, whereas the deep community is distinctly different from any other previously described terrestrial serpentinizing community. These unique communities have the potential to yield important insights into the development and survival of life in these early-earth analog environments.
Serpentinization is the process whereby water reacts with ultramafic minerals (i.e., olivine and pyroxenes) to produce a new suite of minerals (i.e., serpentine, magnetite, and brucite), as well as hydrogen, methane, and highly alkaline (ultrabasic) fluids (1, 2). This is a common process at oceanic tectonic plate boundaries, where seawater interacts with deeply buried ultramafic rocks (e.g., the Lost City site located on the Mid-Atlantic Ridge) (3, 4). In contrast, terrestrial active serpentinization sites are relatively rare, reported only in California (5–7), Portugal (8, 9), Canada (10, 11), Turkey (12), Oman, and Yugoslavia (6). Thus, despite the difficulties inherent in accessing the oceanic sites, most of our geobiological knowledge of serpentinization systems stems from studies of the Lost City complex (3, 4, 13–16).
One of the first terrestrial active serpentinization sites that has been described is The Cedars, located north of San Francisco near the Russian River, within the Franciscan Subduction Complex (FSC) (5) (Fig. S1). As at Lost City, macroscopic (i.e., centimeter to meter scale) carbonate structures of various types are found at the air/water interfaces (Fig. S1) resulting from carbonate precipitation upon interaction of CO2 with the calcium in the ultrabasic waters (i.e., pH 11.4–11.9). In contrast to Lost City, however, the waters that feed The Cedars peridotite are not of marine origin, and thus contain low concentrations of sodium and sulfate (7). The low sodium is notable because known alkaliphiles typically pump sodium instead of protons, and use sodium-based ATPases to generate ATP, thus avoiding the problem of pumping protons at high pH (17, 18). This option is unlikely to be available at The Cedars, and so alternative—as yet undescribed—mechanisms may be at play in ATP generation. Low sulfate concentration is also important because there are no obvious electron acceptors in abundance at the very low Eh (electrode potential) of The Cedars (∼−550 mV), a stark contrast to marine serpentinizing systems, with abundant electron acceptors in the form of sulfate (∼25 mM). Thus, although there is plenty to “eat” (i.e., electron donors) in the form of hydrogen and methane produced by serpentinization at The Cedars, there is nothing obvious to “breathe” (i.e., electron acceptors). In addition, after interaction with peridotite, the water is Ca2+-rich and CO2-poor (7), adding further complications for microbial life.
Microbial community and metagenomic analyses have begun to reveal the diversity of microbial life at the Lost City oceanic serpentinizing site (13–16). Although the pH is high (pH 9–11), given abundant hydrogen and methane for energy and sulfate as an electron acceptor, it is not surprising to see a taxonomically rich microbial community consisting of sulfur-oxidizing, sulfate-reducing, and methane-oxidizing bacteria, as well as methanogenic and anaerobic methane-oxidizing archaea (14). In contrast, terrestrial serpentinizing sites are, as expected, quite different from the Lost City site: a study of a highly alkaline spring system in Maqarin, Jordan (19) revealed microorganisms in these springs belonged mainly to the Proteobacteria. Microbial studies of two terrestrial serpentinizing sites, Cabeço de Vide Aquifer (CVA) in Portugal and Tablelands in Newfoundland, Canada, revealed that microbes belonging to classes Clostridia and Betaproteobacteria, especially related to genus Hydrogenophaga within Betaproteobacteria, were abundantly observed in the both sites (9, 10). The Hydrogenophaga strains are hypothesized to oxidize hydrogen in the serpentinizing system; however, it still remains unknown. In no case yet reported has a terrestrial site been shown to have a microbial population similar to that seen at the sulfur-driven deep-sea sites.
Here we report findings from a 3-y, multiple-season study of three different springs from The Cedars, using a culture-independent approach that targeted all domains of life, with the goal of characterizing the microbial diversity of these three sites. The three springs on which our work is focused differ with regard to their water input: being fed by two different (i.e., geochemically distinct) groundwater sources (shallow and deep) (7). Thus, although all of The Cedars springs share many extreme properties, they differ in terms of their geochemistry and morphology of the springs. In one of the springs we examined three subenvironments with different redox conditions. Each of the three springs is different from the others with regard to geochemistry as well as microbial diversity. In the springs that are fed by shallow groundwater, the microorganisms are “familiar,” being easily placed into known phylogenetic phyla and also being similar to microorganisms from other terrestrial sites. In contrast, in the spring fed by deep groundwater, many of the phylotypes are members of, or most closely related to, candidate divisions or classes that are defined only by the 16S rRNA clones recovered from various environments, and where no cultivated microorganisms are clustered.
Results and Discussion
Sampling Site Descriptions and Geochemical Characterizations.
Our work was focused on three distinct springs within The Cedars peridotite complex: Grotto pool spring 1 (GPS1) (Fig. S1 A and B), Nipple spring 1 (NS1) (Fig. S1 C and D), and Barns spring 5 (BS5) (Fig. S1 E and F). These springs are: (i) spatially isolated from one another, (ii) geochemically discernible, (iii) accessible for easy sample collection, and (iv) fed by different mixtures of deep and shallow serpentinizing waters (Table 1) (7). GPS1 possesses the highest pH and lowest redox potential of any spring we have studied at The Cedars. This spring also has a well-defined surface interface, allowing for direct sampling of the spring water from the subsurface (Fig. S1 A and B). NS1 is located at a higher elevation and has the lowest salinity of any ultrabasic spring we have measured at The Cedars (Fig. S1 C and D). As with GPS1, the emergent water of NS1 can be directly sampled before any mixing with the external environment, which is an unusual feature at most serpentinization sites, marine or terrestrial. In contrast, at BS5 the spring water that enters a small pool (approximately 80-cm wide and 40-cm deep) from the subsurface is exposed to sunlight, and mixed with air and external contaminants (i.e., leaf litter, insects, and so forth) (Fig. S1 E and F). Despite these factors, the waters of BS5 remain highly reducing and the slow flow rate of its spring at the bottom of the pool results in the formation of a stratified system [e.g., relatively oxic surface waters (Eh = −40 mV) and anoxic bottom waters (Eh = −560 mV)]. Although all three springs are inundated with meteoric water during the rainy season (November to April), the chemical and physical properties of each spring in the dry season were stable over the 3-y study period (Table 1), suggesting that the input of serpentinization water was the major factor determining the geochemistry of each site (7).
Table 1.
Chemical composition and environmental parameters of The Cedars springs
| Composition and parameters | GPS1 |
NS1 |
BS5 |
Creek |
||||||
| 2009 September |
2010 April |
2011 October |
||||||||
| 2009 September | 2010 April | 2011 October | 2010 April | 2011 October | Bottom | Surface | Bottom | Bottom | 2010 April | |
| pH | 11.9 | 11.9 | 11.9 | 11.5 | 11.5 | 11.6 | 11.5 | 11.5 | 11.5 | 8.36 |
| Temperature (°C) | 16–18 | 16–18 | 16–18 | 16–18 | 16–18 | 16–18 | 16–18 | 16–18 | 16–18 | 16–18 |
| Eh (mV) (vs. H2) | −650 | −650 | −700 | −635 | −600 | −550 | −40 | −590 | −560 | 65 |
| DO* | <dl | <dl | <dl | <dl | <dl | <dl | <dl | <dl | <dl | ND |
| Conductivity (µS/cm) | 3,000 | 3,000 | 3,000 | 740 | ND | 942 | 942 | 800 | ND | 444 |
| Na+ (mM) | 14.40 | 14.98 | ND | 0.96 | ND | 2.02 | 1.94 | 1.94 | ND | 0.06 |
| K+ (mM) | 0.13 | 0.12 | ND | 0.01 | ND | 0.03 | 0.03 | 0.02 | ND | <dl |
| Ca2+ (mM) | 1 | 1 | 0.93 | 1.43 | 1.3 | 1.20 | 1.14 | 1.26 | 1.15 | 0.06 |
| Mg2+ (mM) | <dl | 0.007 | <dl | 0.014 | 0.008 | <dl | <dl | 0.064 | 0.026 | 2.136 |
| Cl− (mM) | 8.81 | 8.76 | 8.63 | 0.97 | 0.92 | 1.56 | 1.42 | 1.45 | 1.35 | 0.12 |
| SO42− (mM) | <dl | <dl | <dl | 0.001 | 0.001 | 0.001 | <dl | 0.001 | <dl | 0.01 |
| PO43−, NO3−, NO22−, NH4+ (mM) | <dl | <dl | <dl | <dl | <dl | <dl | <dl | <dl | <dl | <dl |
| Flow rate (L/min) | ∼1.5 | ∼0.9 | 0.1–0.25 | NA | ||||||
| Air–water interface (cm2) | ∼1 | 2–4 | 2,500–4,000 | NA | ||||||
| TIC* | ND | ND | 0.035 | ND | 0.006 | ND | ND | ND | 0.070 | 2.940¶ |
| DOC* | ND | ND | 0.17 | ND | 0.14 | ND | ND | ND | 0.02 | 0.05¶ |
| N2 (% by vol)*† | 36.6 ± 0.4‡ | 63.1 ± 3.3 | 53.6 | NA | ||||||
| H2 (% by vol)*† | 50.9 ± 1.1‡ | 15.7 ± 1.0 | 34 | NA | ||||||
| CH4 (% by vol)*† | 7.4 ± 3.1‡ | 15.8 ± 0.0 | 5.3 ± 1.2 | NA | ||||||
| O2 (% by vol)*† | <dl‡ | <dl | <dl | NA | ||||||
| fdeep*§ | 1 | 0.08 ± 0.02 | 0.14 ± 0.02 | 0 | ||||||
| Suspended cell density (cells/mL)* | ND | ND | <10 | ND | 531 ± 376 | ND | 3,684 ± 593¶ | ND | 3,187 ± 1,126 | 18,986 ± 414¶| |
<dl, below our detection limit; DO, dissolved oxygen; DOC, dissolved organic carbon; NA, not available; ND, not determined; TIC, total inorganic carbon. Detection limits of ions, dissolved oxygen, and oxygen gas were 1–2 µM, 2 ppm, or 5 ppm respectively.
These data were published in our previous report (7).
The values are the average values over multiple sampling periods (June 2005, August 2005, August 2006, and September 2011).
Gas samples were not able to be collected at GPS1 because of the spring structure. All gases data from CS1, immediately below GPS, were assumed to be a good representation of the gases from the deep FSC groundwater of GPS1 (7).
The fraction of deep groundwater (fdeep) in each spring was calculated using a two-component mixing model.
Cell density, TIC, and DOC were analyzed with the samples collected in October 2011.
Comparisons of Community Structures Between the Springs in The Cedars.
Community diversity analyses were assessed by multiple clone-libraries of small subunit (16S and 18S) rRNA gene sequences in three different springs (Fig. 1 and Table S1). To understand the effect of the rainy seasons and the yearly community changes, the diversity analyses at GPS1 and BS5 were conducted twice in 2009, at the early (May) and late (September) dry season, and in October of 2011. In addition, microbial community structure was also determined at three subenvironments in BS5, including the spring water at the bottom of the pool where groundwater is continuously emerging from the subsurface, the surface water of the pool near the air/water interface, and biofilms on the surface of rocks in the bottom of the pool (Fig. 2). These data were used to address the geochemical factors affecting the microbial community.
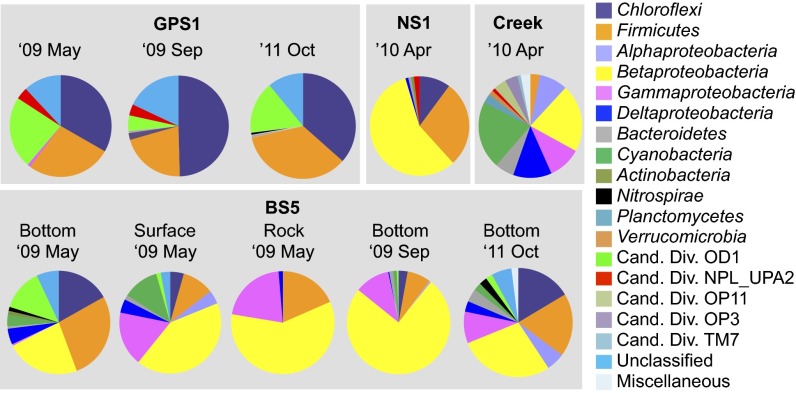
Fig. 1.
Phylum-level taxonomic distribution of the bacterial 16S rRNA gene community profile in The Cedars springs. Sampling times (year and month) and locations (spring and sublocation) are shown at the top of each pie chart. Phylum Proteobacteria is divided into class-level taxonomies.
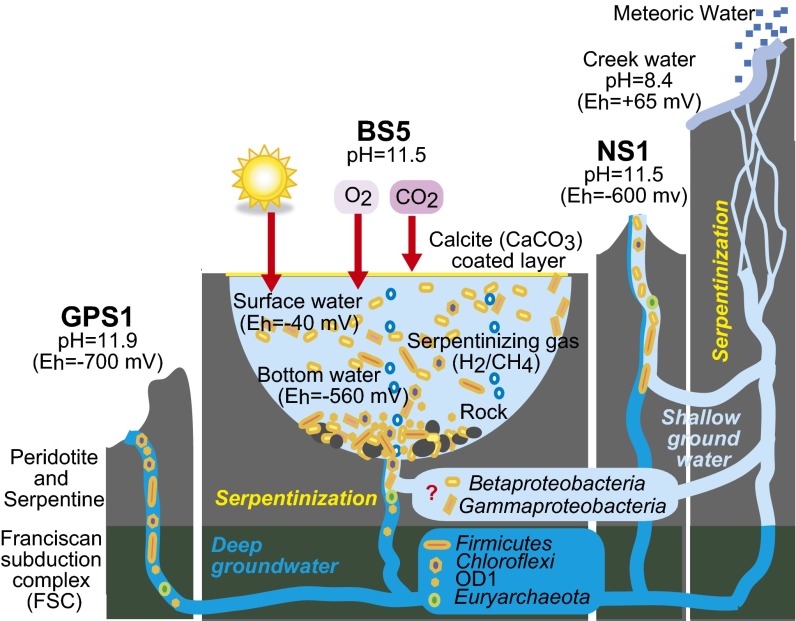
Fig. 2.
A schematic diagram showing the geological setting of The Cedars springs studied and their microbial communities. Dark and light blue colors indicate the deep or shallow groundwaters, respectively. Rods and circles in various colors indicate microbes.
Domain-specific PCR of the ultrabasic springs revealed the presence of both Bacteria and Archaea, and the absence or very low density of Eukarya, with the exception of the surface of BS5, where potential external contamination was unavoidable (Table S1). Rarefaction curve and α-diversity analysis (Chao1 richness and Shannon’s index) -targeted bacterial operational taxonomic units (OTU) (99% cutoff) revealed that the taxonomic diversities of the ultrabasic springs are low in comparison with that of the adjacent creek water, which is fed by meteoric water and snow melt (Fig. S2 and Table S1) (7). The lowest species richness was seen in GPS1, where the community was dominated by phyla Chloroflexi and Firmicutes, a candidate division OD1, and two undefined phyla in the SILVA database (20) (Fig. 1 and Table S1). NS1 and BS5 also contained phyla of Chloroflexi and Firmicutes, but were dominated by a class Betaproteobacteria at NS1 and classes of Beta- and Gammaproteobacteria at BS5. Only one archaeal phylotype was detected, and it was present in all of the ultrabasic springs at any year or season (Table S1). Sørensen’s similarity coefficients and multidimensional scale plots created on the basis of OTU composition indicated that the spring communities differ from each other, and from the creek, but are similar within the same springs at different seasons and in different years (Figs. S3 and S4).
Phylotype-Level Comparison in the Shallow and Deep Serpentinizing Groundwater Sources Community.
Detailed phylotype-level comparison showed that between 84% and 98% of bacterial sequences from each spring’s community were affiliated to 16 phylotypes (Fig. 3). As reported previously (7), principal component analyses of the three springs’ water chemistries indicated that there are two sources of groundwater: one that contacts only The Cedars’ peridotite body (shallow groundwater), and a deeper groundwater that flows through the peridotite body and contacts the marine sediments of the FSC below the peridotite body (deep groundwater). These groundwaters mix and discharge at NS1 and BS5, but only the deeper FSC contacting groundwater discharges from the GPS1 spring (Fig. 2).
Fig. 3.
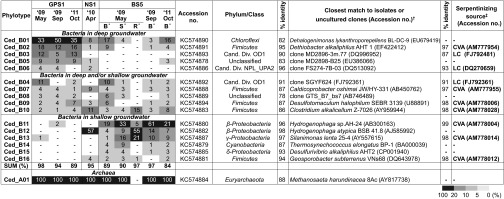
Heat-map chart showing the relative abundance of the microbial phylotypes in each community and their closest relatives. A dash (-) indicates not detected; Cand. Div., candidate division. *The letters (B, S, R) indicate the bottom (B) and surface (S) of the pool fed by the BS5 spring water and biofilm on rocks (R) at the bottom of the pool. †If the 16SrRNA identity of the closest isolated strain is less than 80%, the closest uncultured clone from a given environment is shown. ‡If the close sequence is detected in the other serpentinizing sites, the closest uncultured clone at the sites is described: CVA, Cabeço de Vide; LC, Lost City.
The microbial diversity dataset is consistent with this two-source model: 8 of 10 of the abundant phylotypes in GPS1, where only deep groundwater discharges, were detected at BS5 and NS1, both of which are fed by mixtures of shallow and deep groundwater as the water source, but some of the abundant phylotypes at BS5 and NS1 were not detected in GPS1 (Fig. 3). The presence of taxa specific to BS5 and NS1 indicated that different communities were established at each water source (Fig. 3). Identical 16S rRNA gene sequences of most of the major members in GPS1 were detected at NS1 and BS5, suggesting that all of the springs may share the deep groundwater source.
The deep groundwater source is likely anoxic because all of uncultured clone sequences that are closely related to those seen in GPS1 were recovered from anoxic environments (i.e., a deep-sea sediment, an anaerobic methane-oxidizing environment, and a hydrothermal vent, including Lost City) (Fig. 3) (16, 21). In contrast, although dissolved oxygen or oxygen in bubbling gas was below the detection limit (2 and 5 ppm, respectively) in all three springs (7), the community diversity in shallow groundwater may be affected by the oxygen from meteoric water or other sources, as the most abundant taxa detected at NS1 and BS5 are phylogenetically close to well-known aerobic hydrogen-oxidizing Hydrogenophaga strains in class Betaproteobacteria (22).
In addition, we also examined the differences between three locations in the BS5 pool (Fig. 3). Here we sampled from three sublocations: (i) bottom (i.e., close to the spring source) (Eh = −550 mV) (BS5/B), (ii) surface layers of the spring (Eh = −40 mV) (BS5/S), and (iii) rock from the bottom of the anoxic zone (BS5/R) (Fig. 2). The planktonic cell densities and water chemistries of the bottom and surface water were similar with the exception of Eh, which was much lower in the bottom waters (−550 mV vs. −40 mV) (Table 1). The Eh at the depth of 1, 3, 10, 20, and 40 cm below the surface of BS5 spring was −40, −93, −182, −251, and −550 mV, respectively, suggesting that exposure to the air was clearly affecting the environment, and oxygen from the air reacted with reduced molecules in the spring water biotically or abiotically. As aerobic bacteria have a very high affinity for oxygen, immediate consumption of this limiting electron acceptor likely causes its concentration to remain below our limit of detection.
Comparing the microbial community structure at the bottom and surface of the spring showed that the relative abundance of most of the phylotypes from the deep groundwater source (Ced_B01, B02, and B03) decreased significantly at the surface, but that of shallow groundwater-specific phylotypes belonging to Betaproteobacteria (Ced_B11and B12), Gammaproteobacteria (Ced_B13), and Cyanobacteria (Ced_B14) increased, suggesting a significant role for oxygen in regulating the abundance of both groups in the spring (Fig. 3). As for the biofilm on the rocks, the diversity richness was significantly lower than any of the water samples, suggesting a specialized microbial community inhabiting the water/mineral interfaces of the pool. Interestingly, the dominant Betaproteobacterial phylotype in the biofilm was different from the dominant one in the BS5 water and similar to that in NS1, suggesting a different ecophysiological strategy (ecological niche) for these environments.
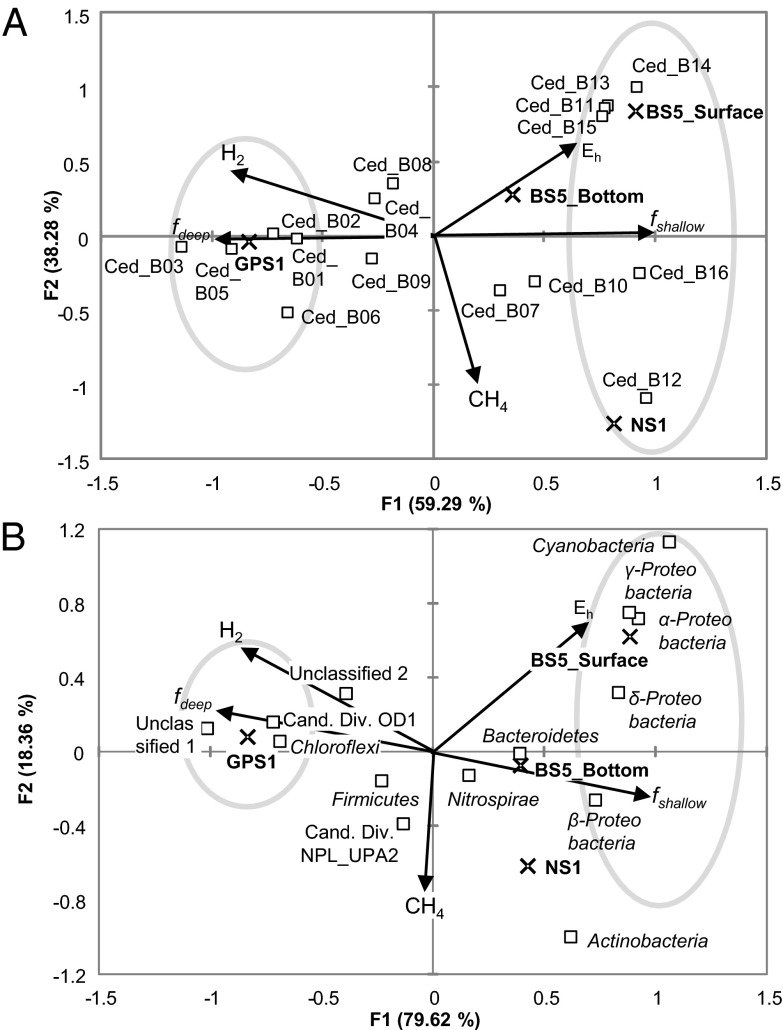
In searching for answers to the community profiles documented here, the most harmonious hypothesis was that the source groundwater (deep and shallow) provided different inocula (defined here as the source groundwater plus the resident microbial community) for the springs. Thus, the GPS1 community reflects an inoculum from the deep groundwater, whereas the NS1 community reflects a mixture of deep and shallow inocula. BS5 was further subdivided, with three different subenvironments, reflective of deep and shallow inocula plus strong selective pressure from the altered redox conditions. We began to test this hypothesis by canonical correspondence analysis (CCA) (Fig. 4) (23). Environmental factors examined in CCA were the fraction of deep and shallow groundwaters (fdeep, fshallow), Eh, and gas composition of hydrogen and methane. The fraction of deep or shallow groundwater was estimated based on Cl− concentration as described previously (7). Concentrations of Br−, I−, Li+, Rb+, Na+, K+, and Rb+, and conductivity were correlated with the fraction of deep groundwater with r2 ≥ 0.98 (7). The result of CCA for major phylotypes of The Cedars revealed that most of the phylotypes were closely related to the fraction of deep (Ced_B01, B02, B03, B05, and B06) or shallow (Ced_11, 12, 13, 14, 15, and 16) groundwater. Some of the phylotypes correlated with the fraction of shallow groundwater were associated with Eh as well (Ced_B11, 13, 14, and 15). In addition, CCA in phylum-level diversity indicated that the phylum Chloroflexi, an undescribed phylum, and a candidate division OD1 were tightly associated with the deep groundwater, the phylum Cyanobacteria, and classes of Alpha-, Beta-, Gamma-, and Deltaproteobacteria were associated with the shallow groundwater, and the phylum Cyanobacteria and classes of Alpha- and Gammaproteobacteria were correlated with both the fraction of shallow groundwater and Eh. Several of the phylotypes (Ced_B04, B07, B08, B09, and B10) and phyla (Firmicutes, Nitrospirae, Candidate Division NPL_UPA2, Unclassified 2, and Actinobacteria) were not correlated with sources, suggesting that the members belonging to these groups could adapt both types of groundwater or that their distributions are partially governed by factors not measured here. To this point in time, the data we have gathered and analyzed remain consistent with our hypothesis that the major driver for community diversity is the inoculum supplied by the groundwater. Additional springs should be analyzed to further test and refine the current hypothesis.
Fig. 4.
Canonical correspondence analysis diagram showing the relationships between five environmental variables and phylotypes (A) or phyla (B). Symbols indicate that phylotypes or phyla/classes (square), sampling sites (cross), and environmental settings (arrows). Areas surrounded by gray line designate phylotypes or phylum/class that are highly correlated with each fraction of water (fdeep or fshallow).
Taxonomic Comparison of Dominant Phylotypes in The Cedars with Those from Other Environments.
Understanding the accurate taxonomic position of the dominant phylotypes is important for describing the microbial members of the site. In addition, given that serpentinization is a global phenomenon, it was of interest to compare the phylotypes we have characterized with others from alkaline and serpentinizing environments. We thus constructed phylogenetic trees of the dominant phylotypes (Fig. S5).
Chloroflexi.
The most dominant phylotype (Ced_B01) in the GPS1 belongs to the clade of candidate order MSBL5 in the phylum Chloroflexi (Fig. S5A). No similar phylotype of Ced_B01 has been detected at any other alkaline or serpentinizing environments. The closest isolate, with 82% of 16S rRNA gene identity, is Dehalogenimonas lykanthroporepellens BL-DC-9, which is capable of reductive dehalogenation, a variety of polychlorinated alkanes (24). Although the phylotype Ced_B01 is most closely related to the family Dehalococcoidetes, it does not appear to be a member of this family.
Anaerobic Firmicutes.
Five of the dominant phylotypes from the three springs were clustered in the class Clostridia within the phylum Firmicutes (Fig. S5B). Two of the closest isolates are well-known alkaliphilic strains, such as Dethiobacter alkaliphilus AHT 1 (25) and Clostridium alkalicellum Z-7026 (26) (Fig. 3). Of note is the fact that four of the five phylotypes are closely related to the clones recovered from the other terrestrial serpentinizing sites [e.g., CVA in Portugal (9)] (Fig. S5B). The results may suggest that the detected phylotypes in the class Clostridia are specifically adapted to the terrestrial serpentinizing ecosystem. Interestingly, the most dominant phylotype (OTU25) at CVA, which is close to Candidatus Desulforudis audaxviator (9, 27) within the family Peptococcaceae, was not detected at The Cedars. This absence may be because this Clostridia strain is capable of reducing sulfate, which was detected at CVA but not at The Cedars (9) (Table 1). The result may further suggest that the composition of the microbial community in electron donor-rich and acceptor-poor environments is strongly affected by (or selected for) the presence or absence of available electron acceptors.
Betaproteobacteria (genus Hydrogenophaga).
Two abundant phylotypes (Ced_B11 and Ced_B12) at BS5 and NS1 were closely related with the genus Hydrogenophaga in the family Comamonadaceae (Fig. S5C). Hydrogenophaga strains are well-known hydrogen oxidizers and all isolated strains in this genus are neutrophilic (22). However, a sequence that was nearly identical (99% identity) to that of Ced_B11 was detected at CVA terrestrial serpentinizing sites as a highly abundant phylotype (9). Furthermore, several uncultured clones recovered from extremely alkaline groundwater (28) and alkaline soils (direct submission to the National Center for Biotechnology Information database) were also shown to be closely related to these clones from serpentinizing sites. In addition, a recent metagenomic analysis of the Tablelands, a continental serpentinite-hosted alkaline seep in Canada, confirmed the presence of hydrogenase genes from Hydrogenophaga spp. (10). All these lines of evidence indicate that strains closely associated with phylotype Ced_B11 have the ability to thrive in highly alkaline environments.
Gammaproteobacteria, Deltaproteobacteria, and Cyanobacteria.
Phylotype Ced_B13 in the class Gammaproteobacteria that was detected in BS5 is closely related to the alkaliphilic Silanimonas lenta strain 254 (29) and also to 16S rRNA gene clones recovered from CVA (Fig. S5D) (9). These three make a clade, suggesting that these strains live in a similar manner in the environment. Phylotype Ced_B15 in Deltaproteobacteria was detected only in BS5 and is closely related to the D. alkaliphilus strain AHT2 (25) isolated from a soda lake sediment, as well as to uncultured clones from alkaline soda lakes, alkaline springs, and soil (Fig. S5E). One phylotype in the phylum Cyanobacteria was detected only in BS5 (Fig. S5F), and its niche is likely the sunlit portion of this pond.
Candidate Divisions and Unclassified Phyla.
Phylotypes Ced_B03 and B04 are assigned to a candidate division OD1, whereas phylotype Ced_B06 is assigned to a candidate division NPL-UPA2 (Fig. S5G). Phylotypes Ced_B05 and B08 are unassigned to any proposed divisions or phylum classified by 16S rRNA genes from cultured strains or uncultured clones (Fig. S5G). All these phylotypes were detected at GPS1 repeatedly over all 3 y of the study. Although none of these phylotypes were closely related to taxa observed at CVA, phylotypes Ced_B03, B04, and B06 made a clade with several 16S rRNA clones from the oceanic serpentinizing site, Lost City, in the phylogenetic tree (13, 16). Phylotype Ced_B05 stands out as a highly distinct taxon at The Cedars, with no closely related sequences in current databases, although it is an out-group of candidate division OP3. These phylotypes, clustered in candidate divisions or undescribed phyla, may have a special interest with regard to ancient life because they are mainly present in the strictly anaerobic serpentinizing source. Such groundwater sources are viewed as potential analogs for early ecosystems on both Earth and Mars, where there was no oxygen and where a highly reducing mineralogy was likely widespread (2, 30, 31).
Euryarchaeota.
Only one identical (i.e., 100% identity) archaeal sequence (Phylotype Ced_A01) was detected in the entire study. Although its position in the phylogenetic tree places it most closely to the order Methanosarcinales, this sequence doesn’t fit with any of the well-known methanogen or anaerobic methanotroph clades (Fig. S5H). Abiotic methane is possibly produced during the serpentinizing reaction, but previous isotopic analyses suggested that the methane at The Cedars springs is primarily produced by acetate-using methanogens (7). Thus, it is possible that this archaeal group plays an important role in methane production at The Cedars.
Conclusions
The geomorphology of The Cedars springs has provided an opportunity to collect and study pristine, actively serpentinizing water samples from the subsurface, which is unique with respect to other presently known serpentinizing marine or terrestrial sites. According to current metabolic paradigms, the extreme environmental and minimal nutritional conditions at The Cedars peridotite present a major challenge for microbial survival and growth as we know it: a challenge that has been met, as evidenced by the microbial populations at The Cedars springs. This study revealed that stable but different microbial communities were established at each spring site.
The hypothesis we propose, based on the currently available correlation analyses and sequence comparisons, is that the major driver of diversity for these three springs is the inoculum provided by the source groundwaters, and that this is then strongly impacted by the exposure to the oxic interface. It is notable that many of the sequences we have seen might have been regarded as sequencing errors or as microbes just temporally present, but because they appeared multiple times over a 3-y period, this leaves little room for doubt. The use of multiple visits to this site allowed us to concentrate on a number of key organisms, the characterization of which is now a major focus of our efforts.
The microbial community established in sites mixed with shallow groundwater consisted mainly of phylotypes in classes of Betaproteobacteria and Clostridia, similar to other terrestrial serpentinizing sites, such as CVA and the Tablelands (9, 10). In contrast, the community structure in the deep groundwater-fed spring is clearly distinct from any of the other terrestrial serpentinization sites. Repeated sampling over a 3-y period confirmed the identity of permanent members of the deep groundwater community. Most of these candidate division members could be clustered into a clade with phylotypes reported from the oceanic serpentinizing site, Lost City (13, 14, 16), but not at the terrestrial sites. Considering that serpentinizing systems are globally distributed, and regarded by some as early (anoxic) earth analogs (30, 31), this work may lead us to truly interesting members of the “early life” serpentinization community.
Materials and Methods
Sampling Sites, Sample Collections, and Geochemical Analysis.
Samples for geochemical and microbiological analysis were collected from three different ultrabasic springs: GPS1, BS5, and NS1, and also the main creek within The Cedars, the host water of the Austin creek (Fig. S1) (7). Details about the water sample collections and geochemical analysis are described in SI Materials and Methods.
Gas sampling and analysis was conducted as described previously (7).
Small Subunit rRNA Gene Clone Libraries Analyses.
DNA were extracted by an UltraClean soil DNA extraction kit (MO Bio) with some modifications. A detailed description is available in SI Materials and Methods. In brief, universal bacterial (U27F and U1492R) (32), archaeal [Arc4F (33) and U1492R], and eukaryal [EukF (34) and EukR (35)] PCR primers were used to amplify the region of small-subunit (16S and 18S) rRNA genes. Approximately 600 bp of partial small subunit rRNA gene sequences were determined. The sequences were aligned and assigned to OTUs (classified as an OTU, >99% cutoff). Nearly full-length of 16S rRNA gene sequences (between the U27F and U1492R primer region) were determined for the phylotypes that occupy more than 1–2% of the spring communities. All sequences generated by this study have been deposited in the National Center for Biotechnology Information GenBank database under accession numbers KC574878–KC575114.
Phylogenetic affiliations were determined by the combination of SILVA (20), BLAST (36), Greengenes (37), and RDP (38). However, most of the 16S rRNA sequences from GPS1 were unassigned to taxa or assigned different taxa by programs. Therefore, the taxonomic assignment of each phylotype was made by the phylogenetic trees created with the closest relatives (SI Materials and Methods).
Correlation Between Community Composition and Environmental Factors.
Canonical correspondence analysis (XLSTAT) was performed for describing correlation between community composition and environmental factors (23). All of the bacterial communities in The Cedars springs in different years were applied to this analysis. Fraction of deep and shallow groundwater was estimated as described previously (7) (details are in SI Materials and Methods). Average values of water fractions, gas compositions, and Eh were used for this analysis because the values are stable among the seasons and years.
Supplementary Material
Acknowledgments
We thank Roger Raiche and David McCrory for kindly providing their private land for our research; Penny Morrill for the helpful discussions; and Orion Johnson for support in the field. This work was funded by National Science Foundation–Earth Sciences Grant 1024872 and by National Aeronautics and Space Administration Cooperative Agreement NNA13AA92A.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC574878–KC575114).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302426110/-/DCSupplemental.
References
- 1.Frost BR, Beard JS. On silica activity and serpentinization. J Petrol. 2007;48(7):1351–1368. [Google Scholar]
- 2.Sleep NH, Meibom A, Fridriksson T, Coleman RG, Bird DK. H2-rich fluids from serpentinization: Geochemical and biotic implications. Proc Natl Acad Sci USA. 2004;101(35):12818–12823. doi: 10.1073/pnas.0405289101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley DS, et al. AT3-60 Shipboard Party An off-axis hydrothermal vent field near the Mid-Atlantic Ridge at 30 degrees N. Nature. 2001;412(6843):145–149. doi: 10.1038/35084000. [DOI] [PubMed] [Google Scholar]
- 4.Kelley DS, et al. A serpentinite-hosted ecosystem: The Lost City hydrothermal field. Science. 2005;307(5714):1428–1434. doi: 10.1126/science.1102556. [DOI] [PubMed] [Google Scholar]
- 5.Barnes I, Lamarche VC, Jr, Himmelberg G. Geochemical evidence of present-day serpentinization. Science. 1967;156(3776):830–832. doi: 10.1126/science.156.3776.830. [DOI] [PubMed] [Google Scholar]
- 6.Barnes I, O'Neil JR. Presentday serpentinization in New Caledonia, Oman and Yugoslavia. Geochim Cosmochim Acta. 1978;42(1):144–145. [Google Scholar]
- 7.Morrill PL, et al. Geochemistry and geobiology of a present day serpentinization site in California: The Cedars. Geochim Cosmochim Acta. 2013;109:222–240. [Google Scholar]
- 8.Marques JM, et al. Origins of high pH mineral waters from ultramafic rocks, Central Portugal. Appl Geochem. 2008;23(12):3278–3289. [Google Scholar]
- 9.Tiago I, Veríssimo A. Microbial and functional diversity of a subterrestrial high pH groundwater associated to serpentinization. Environ Microbiol. 2012;15(6):1687–1706. doi: 10.1111/1462-2920.12034. [DOI] [PubMed] [Google Scholar]
- 10.Brazelton WJ, Nelson B, Schrenk MO. Metagenomic evidence for h(2) oxidation and h(2) production by serpentinite-hosted subsurface microbial communities. Front Microbiol. 2012;2:268. doi: 10.3389/fmicb.2011.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szponar N, et al. Geochemistry of a continental site of serpentinization in the Tablelands Ophiolite, Gros Morne National Park: A Mars analogue. Icarus. 2013;224(2):286–296. [Google Scholar]
- 12.Hosgormez H. Origin of the natural gas seep of Cirali (Chimera), Turkey: Site of the first Olympic fire. J Asian Earth Sci. 2007;30(1):131–141. [Google Scholar]
- 13.Schrenk MO, Kelley DS, Bolton SA, Baross JA. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ Microbiol. 2004;6(10):1086–1095. doi: 10.1111/j.1462-2920.2004.00650.x. [DOI] [PubMed] [Google Scholar]
- 14.Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol. 2006;72(9):6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brazelton WJ, Baross JA. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 2009;3(12):1420–1424. doi: 10.1038/ismej.2009.79. [DOI] [PubMed] [Google Scholar]
- 16.Brazelton WJ, et al. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci USA. 2010;107(4):1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989;21(6):705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 18.Horikoshi K. Alkaliphiles: Some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63(4):735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen K, Nilsson E, Arlinger J, Hallbeck L, O’Neill A. Distribution, diversity and activity of microorganisms in the hyper-alkaline spring waters of Maqarin in Jordan. Extremophiles. 2004;8(2):151–164. doi: 10.1007/s00792-004-0374-7. [DOI] [PubMed] [Google Scholar]
- 20.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue) D1:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber JA, Johnson HP, Butterfield DA, Baross JA. Microbial life in ridge flank crustal fluids. Environ Microbiol. 2006;8(1):88–99. doi: 10.1111/j.1462-2920.2005.00872.x. [DOI] [PubMed] [Google Scholar]
- 22.Willems A, et al. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (formerly Pseudomonas pseudoflava and “Pseudomonas carboxydoflava”), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis) Int J Syst Bacteriol. 1989;39(3):319–333. [Google Scholar]
- 23.ter Braak CJF. Canonical correspondence analysis: A new eigenvector technique for multivariate direct gradient analysis. Ecology. 1986;67(5):1167–1179. [Google Scholar]
- 24.Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA. Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int J Syst Evol Microbiol. 2009;59(Pt 11):2692–2697. doi: 10.1099/ijs.0.011502-0. [DOI] [PubMed] [Google Scholar]
- 25.Sorokin DY, Tourova TP, Mussmann M, Muyzer G. Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: Two novel representatives of reductive sulfur cycle from soda lakes. Extremophiles. 2008;12(3):431–439. doi: 10.1007/s00792-008-0148-8. [DOI] [PubMed] [Google Scholar]
- 26.Zhilina TN, et al. [Clostridium alkalicellum sp. nov., an obligately alkaliphilic cellulolytic bacterium from a soda lake in the Baikal region] Mikrobiologiia. 2005;74(5):642–653. Russian. [PubMed] [Google Scholar]
- 27.Chivian D, et al. Environmental genomics reveals a single-species ecosystem deep within Earth. Science. 2008;322(5899):275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- 28.Roadcap GS, Kelly WR, Bethke CM. Geochemistry of extremely alkaline (pH>12) ground water in slag-fill aquifers. Ground Water. 2005;43(6):806–816. doi: 10.1111/j.1745-6584.2005.00060.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee EM, Jeon CO, Choi I, Chang KS, Kim CJ. Silanimonas lenta gen. nov., sp. nov., a slightly thermophilic and alkaliphilic gammaproteobacterium isolated from a hot spring. Int J Syst Evol Microbiol. 2005;55(Pt 1):385–389. doi: 10.1099/ijs.0.63328-0. [DOI] [PubMed] [Google Scholar]
- 30.Schulte M, Blake D, Hoehler T, McCollom T. Serpentinization and its implications for life on the early Earth and Mars. Astrobiology. 2006;6(2):364–376. doi: 10.1089/ast.2006.6.364. [DOI] [PubMed] [Google Scholar]
- 31.Sleep NH, Bird DK, Pope EC. Serpentinite and the dawn of life. Philos Trans R Soc Lond B Biol Sci. 2011;366(1580):2857–2869. doi: 10.1098/rstb.2011.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishii S, et al. Functionally stable and phylogenetically diverse microbial enrichments from microbial fuel cells during wastewater treatment. PLoS ONE. 2012;7(2):e30495. doi: 10.1371/journal.pone.0030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giovannoni SJ, DeLong EF, Olsen GJ, Pace NR. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71(2):491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 35.Moon-van der Staay SY, et al. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol Oceanogr. 2000;45(1):98–109. [Google Scholar]
- 36.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.