Significance
This is a unique report of receptor tyrosine kinase (RTK) “superacceptor” activity in which mutated EGFRs associated with lung cancer preferentially adopt the “acceptor” or “receiver” position in the presence of WT epidermal growth factor receptor (EGFR) or ErbB-2. The mechanism of superacceptor activity is defined by biochemical reconstitution data in combination with the first crystal structure of the L834R/T766M (L858R/T790M in alternate numbering) mutant EGFR kinase asymmetric dimer in an active conformation. The data imply that mutant/wild-type interactions play a key role in tumorigenesis as well as sensitivity of cells to various EGFR tyrosine kinase inhibitors, which could be therapeutically important. Notably, none of the previous studies involving mutated EGFR have studied the contribution of WT EGFRs in heterogeneous cell populations, although in nearly all instances wild-type EGFR alleles are preserved within EGFR mutant tumor cells.
Keywords: mutation, TKI, lapatinib, WZ-4002
Abstract
The initiation of epidermal growth factor receptor (EGFR) kinase activity proceeds via an asymmetric dimerization mechanism in which a “donor” tyrosine kinase domain (TKD) contacts an “acceptor” TKD, leading to its activation. In the context of a ligand-induced dimer, identical wild-type EGFR TKDs are thought to assume the donor or acceptor roles in a random manner. Here, we present biochemical reconstitution data demonstrating that activated EGFR mutants found in lung cancer preferentially assume the acceptor role when coexpressed with WT EGFR. Mutated EGFRs show enhanced association with WT EGFR, leading to hyperphosphorylation of the WT counterpart. Mutated EGFRs also hyperphosphorylate the related erythroblastic leukemia viral oncogene (ErbB) family member, ErbB-2, in a similar manner. This directional “superacceptor activity” is particularly pronounced in the drug-resistant L834R/T766M mutant. A 4-Å crystal structure of this mutant in the active conformation reveals an asymmetric dimer interface that is essentially the same as that in WT EGFR. Asymmetric dimer formation induces an allosteric conformational change in the acceptor subunit. Thus, superacceptor activity likely arises simply from a lower energetic cost associated with this conformational change in the mutant EGFR compared with WT, rather than from any structural alteration that impairs the donor role of the mutant. Collectively, these findings define a previously unrecognized mode of mutant-specific intermolecular regulation for ErbB receptors, knowledge of which could potentially be exploited for therapeutic benefit.
The gene encoding the epidermal growth factor receptor (EGFR) tyrosine kinase is somatically mutated in a substantial fraction of patients with lung cancer. The majority of primary activating EGFR mutations occur within the tyrosine kinase domain (TKD). The most frequent of these, which occur with a combined frequency of 90% (1), are exon 19 deletions that eliminate four amino acids (LREA) from the TKD and exon 21 missense mutations that substitute arginine for leucine at position 834 (L834R) (also identified as L858R in an alternative numbering of the human EGFR sequence that includes the 24 residue signal sequence) (2).
Exon 19 deletions and L834R substitutions are associated with increased sensitivity to EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib, translating to a 70% radiographic response rate in patients (3–5). Unfortunately, all individuals with metastatic disease eventually develop progressive disease after 10–16 mo of treatment with EGFR TKIs. The most common mechanism of acquired resistance is mutation at a second site in the EGFR TKD (the gatekeeper residue), T766M (T790M). This mutation confers resistance by increasing affinity for ATP, with which inhibitors must compete for binding, and also by modestly decreasing intrinsic affinity for TKIs (6).
Biochemical and crystallographic studies have shown that activation of the wild-type (WT) EGFR TKD involves formation of an asymmetric dimer in which one molecule allosterically activates its neighbor by promoting the reversal of intramolecular autoinhibitory interactions—acting as a “donor” or “activator” TKD that activates the “acceptor” or “receiver” TKD (7, 8). Crystal structures of individual L834R and T766M EGFR-TKD mutants show that these variants also form asymmetric dimers (6, 9), but whether the double mutant L834R/T766M adheres to the same configuration in the active state is unclear. Biochemical data indicate that the oligomerization potential of mutated EGFRs is enhanced relative to WT. For example, native gel and multiangle light scattering studies showed that the L834R substitution promotes formation of dimers and higher order oligomers of the EGFR TKD (10). Consistent with this observation, cell-based studies have demonstrated a reduced dependence on ligand stimulation for activation of mutated EGFRs. All mutated EGFR TKDs seen in lung cancer show an increase in catalytic efficiency over WT (6, 9, 11, 12). Interestingly, the doubly mutated L834R/T766M EGFR TKD has a two-to fivefold higher catalytic efficiency (kcat/Km) than either the singly mutated L834R or T766M mutant TKDs (6).
Here, using a biochemical and structural approach, we sought to determine whether mutated EGFRs most commonly associated with primary drug sensitivity and acquired resistance in lung cancer adhere to the well-established asymmetric dimer model. Our biochemical reconstitution data demonstrate that these EGFR mutations do in fact adhere to this model, but with the striking difference that the oncogenic mutants preferentially assume the receiver role when expressed together with WT EGFR. Our crystal structure of active EGFR-L834R/T766M reveals an asymmetric dimer interface essentially the same as that in WT EGFR. We further show that mutated EGFRs can hyperphosphorylate WT EGFR as well as the related family member erythroblastic leukemia viral oncogene (ErbB)2 as a result of their preferential adoption of the acceptor or receiver position in the asymmetric dimer. These findings have important implications for understanding mechanisms of drug sensitivity and resistance in EGFR mutant lung cancers and shed light on the distinct properties of different mutated forms of EGFR.
Results
Effects of Disrupting the N-Lobe Acceptor Site on Activity of Lung-Cancer–Associated EGFR Mutants.
Activation of the WT EGFR TKD involves formation of an asymmetric dimer, in which the C-lobe of one donor monomer contacts the N-lobe of an acceptor (Fig. 1), leading to allosteric activation of the acceptor through conformational changes in its N-lobe (8). Point mutations that disrupt this asymmetric dimer interface block EGFR activation (8). N-lobe substitutions, such as I682Q, block acceptor function, whereas C-lobe substitutions, such as V924R, abolish the capacity of EGFR TKDs to serve as donors (Fig. 1). Activity of these mutated variants is recovered, however, if TKDs with N-lobe and C-lobe mutations are mixed (8)—allowing the N-lobe–mutated TKD to serve as donor and the C-lobe–mutated TKD as acceptor (Fig. 1).
Fig. 1.
Allosteric activation of the WT EGFR kinase. Kinase activation occurs by asymmetric dimerization of tyrosine kinase domains (TKDs), creating the asymmetric dimerization interface (Far Left). Mutations at specific residues in this interface enforce TKDs to act solely as donors (I682Q) or acceptors (V924R), which are inactive when expressed alone due to an inability to dimerize (Center two panels). Coexpression of an enforced donor with an enforced acceptor restores dimerization capacity to 50% wild-type levels (Far Right).
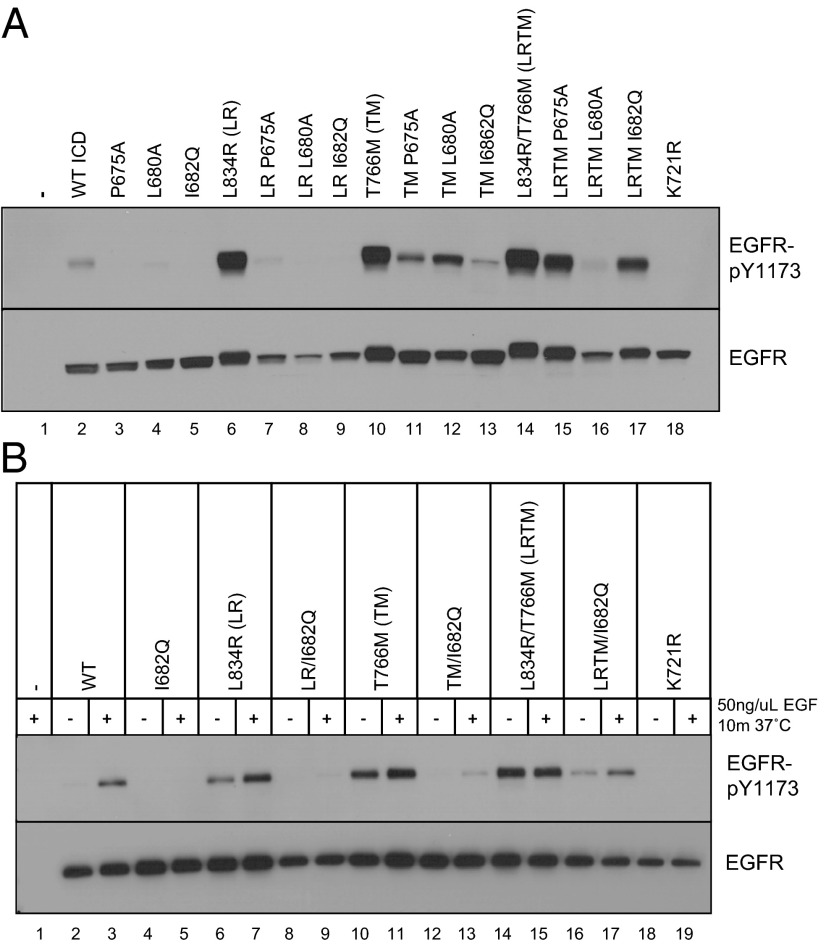
To ask how disrupting asymmetric dimer formation affects the activity of various oncogenic EGFRs found in lung cancer, we introduced individual N-lobe mutations that disrupt the asymmetric interface into WT, L834R, T766M, or L834R/T766M EGFR TKD backgrounds. Initially, we studied the effects of these mutations in the context of overexpressed epitope-tagged intracellular domains (ICDs; amino acids 645–1186), which do not require ligand binding for detectable activity (13). The mutated EGFR-ICDs were transiently expressed in HEK293 cells, and their autophosphorylation was measured by immunoblotting with antibodies specific for pY1173 in EGFR. A catalytically inactive EGFR-ICD containing the K721R mutation was studied in parallel to establish a baseline for inactivity. Consistent with previous studies (13), transiently expressed WT EGFR-ICD was constitutively autophosphorylated to some degree (Fig. 2A), and the lung-cancer–associated L834R (LR), T766M (TM), and L834R/T766M (LRTM) mutations all promoted large increases in this tyrosine phosphorylation (Fig. 2A).
Fig. 2.
N-lobe dimerization interface mutations have different effects in WT and lung-cancer–associated EGFRs. EGFR-ICD constructs (A) or full-length EGFR constructs (B) encoding either WT or kinase domain mutants (LR, L834R; TM, T766M; LRTM, L834R/T766M) with or without N-lobe mutations at the asymmetric dimer interface were transiently expressed in HEK293 cells. In B, cells were serum starved and treated with or without 50 ng/mL EGF for 10 min as indicated. Lysates were subjected to SDS/PAGE and immunoblotting for EGFR-pY1173 and total EGFR.
For each activated variant, we next analyzed the effects of three different N-lobe mutations that were designed to disrupt the acceptor site in the asymmetric dimer interface: P675A, L680A, and I682Q. All three mutations reduced WT EGFR-ICD activity, completely for the P675A and I682Q mutations, although some residual phosphorylation was seen for the L680A mutation. These same mutations also reduced autophosphorylation of EGFR-ICDs activated by lung-cancer–associated mutants, but the relative effects were surprisingly different. All of the N-lobe mutations greatly reduced activity of the L834R-mutated ICD (Fig. 2A), with only the P675A (as opposed to L680A) mutant retaining some residual activity. By contrast, the T766M-mutated EGFR-ICD retained significant autophosphorylation activity for all three N-lobe mutations, in the order L680A > P675A > I682Q. The same was true for L834R/T766M EGFR-ICDs, but intriguingly with a different order (P675A > I682Q > L680A) (Fig. 2A). These data suggest that T766M-containing lung-cancer–associated EGFR mutants have enhanced resistance to N-lobe mutations that disrupt the asymmetric dimer interface.
Activating mutations, such as L834R and T766M, may be said to exhibit a shifted conformational equilibrium in which the mutated TKD exists predominantly in the active structural state. The bulk of structural changes that take place upon EGFR TKD activation occur within the N-lobe. Thus, the activating mutations (L834R and T766M) will tend to stabilize asymmetric dimer formation, whereas L680A, P675A, and I682Q mutations have the opposite effect. The intriguing differences in the rank order of N-lobe mutations that mediate loss of tyrosine phosphorylation seen with T766M versus L834R/T766M lung cancer variants (versus WT) in Fig. 2A likely reflect subtle differences in the energetics of asymmetric dimer formation between lung cancer variants and altered distribution of binding energy across the residues at the interface.
To ensure that these results were not simply an artifact of using the ICD system for our studies, we also performed analogous experiments with full-length EGFR variants. As expected, autophosphorylation of intact WT EGFR was ligand dependent (Fig. 2B), whereas the EGFRs harboring lung-cancer–associated mutations showed significant constitutive activity (further enhanced by EGF in the cases of L834R and T766M). Consistent with the ICD results, the I682Q N-lobe mutation dramatically decreased both constitutive and ligand-dependent autophosphorylation of WT, L834R, and T766M-EGFRs, but had a smaller effect on L834R/T766M-EGFR activity. L834R/T766M-EGFRs retained ligand-independent activity even in the presence of the I682Q N-lobe mutation (Fig. 2B). Similar differences were also seen for the L680A N-lobe mutant (Fig. S1). Taken together, these observations argue that T766M- and L834R/T766M-mutated EGFRs differ from WT and L834R mutants in their energetic reliance on single residues at the asymmetric dimer interface.
Effects of Disrupting the C-Lobe Donor Site on Activity of Lung-Cancer–Associated EGFR Mutants.
We next used a similar approach to examine the effect of C-lobe mutations that disrupt the asymmetric dimer interface in WT EGFR. We introduced a V924R substitution into intact WT, L834R, T766M, and L834R/T766M EGFRs and assessed its effects on EGFR tyrosine phosphorylation. Consistent with published data, the V924R mutation rendered WT EGFR inactive in the presence of ligand (8). The V924R mutation also reduced (but did not completely abolish) EGF-induced autophosphorylation of the L834R- and T766M-EGFRs. However, L834R/T766M-EGFRs containing the V924R C-lobe mutation retained significant activity both in the absence and presence of ligand (Fig. 3A).
Fig. 3.
C-lobe dimerization interface mutations have different effects in WT and lung-cancer–associated EGFRs and ErbB-4. (A) Full-length EGFR constructs encoding either WT or kinase domain mutants (LR, L834R; TM, T766M; LRTM, L834R/T766M) with or without C-lobe mutations at the asymmetric dimer interface were transiently expressed in HEK293 cells. Cells were serum starved and treated with or without 50 ng/mL EGF for 10 min as indicated. Lysates were subjected to SDS/PAGE and immunoblotting for EGFR-pY1173 and total EGFR. (B) EGFR-ICD constructs encoding either WT or kinase domain mutants (LRTM, L834R/T766M) with or without single or double C-lobe mutations at the asymmetric dimer interface were transiently expressed in HEK293 cells. Lysates were subjected to SDS/PAGE and immunoblotting for EGFR-pY1173 and total EGFR. (C) ErbB-4–ICD constructs encoding either WT or kinase domain mutant (T796M) with or without C-lobe mutation at the asymmetric dimer interface were transiently expressed in HEK293 cells. Lysates were subjected to SDS/PAGE and immunoblotting for anti-pY and total ErbB-4. Two times total lysate was loaded in lanes labeled as 2×.
Another C-lobe mutation in the asymmetric dimer interface that disrupts WT EGFR activity is M928R (8). To determine the effect of V924R and M928R substitutions on the activity of the L834R/T766M variant, we introduced one or both substitutions into L834R/T766M-mutated EGFR-ICDs. As expected, autophosphorylation of WT EGFR-ICDs was completely abrogated by the V924R mutation, whether alone or combined with M928R. Surprisingly, when M928R was combined with a V924R mutation in the L834R/T766M EGFR-ICDs, there was no further reduction in tyrosine autophosphorylation (Fig. 3B). These data further suggest a different distribution of the energy of asymmetric dimer formation across specific residues in the interface for the lung-cancer–associated EGFR mutants compared with WT EGFR.
Activating Kinase Domain Mutations Also Confer Reduced Sensitivity to Asymmetric Dimer Interface Mutations in ErbB-4.
We also wondered if the properties observed for lung-cancer–associated EGFR TKD mutants would apply to other ErbB family members. ErbB-4 is the fourth member of the ErbB family of receptor tyrosine kinases and its TKD shares 89% identity with the EGFR TKD. Crystallographic and biochemical data suggest that ErbB-4 TKDs form an EGFR-like active asymmetric dimer (14). As previously reported, ErbB-4 ICDs are constitutively active when expressed in HEK293 cells (Fig. 3C) (13). Introduction of an artificial T796M “gatekeeper” mutation into ErbB-4 (analogous to EGFR T766M) led to increased activity. As with EGFR, a C-lobe mutation in the asymmetric dimer interface, V954R (analogous to EGFR V924R), essentially abolished WT ErbB-4–ICD autophosphorylation (Fig. 3C), but did not abrogate activity of ErbB-4–ICD harboring the activating gatekeeper T796M mutation. Thus, the effect of activating tyrosine kinase domain mutations on sensitivity to mutations in the asymmetric dimer interface applies to ErbB-4 as well as EGFR.
Lung-Cancer–Associated EGFR Mutants Show Altered Donor/Acceptor Complementation Properties.
As mentioned above, EGFR TKDs harboring N-lobe or C-lobe mutations can complement one another in the asymmetric dimer. Although neither the N-lobe mutated nor the C-lobe mutated TKD is active on its own, their coexpression results in reconstitution of EGFR activity to ∼50% (8) (Fig. 1). Given the reduced sensitivity of T766M-mutated EGFR to disruption of the asymmetric dimer interface, we asked whether lung-cancer–associated EGFR mutants retain this complementation effect, as a measure of the dependence of their activity on asymmetric dimer formation. In a first set of experiments using ICD constructs, we mixed N-lobe (I682Q)-mutated WT, L834R, or L834R/T766M-EGFRs with their C-lobe (V924R)-mutated counterparts. We initially anticipated that the complementation seen with WT EGFR would also be seen for lung-cancer–associated EGFR mutants, and that combining N- and C-lobe–mutated ICDs would increase tyrosine phosphorylation to levels greater than that seen for either N- or C-lobe mutant alone.
No such complementation was seen, however, with either L834R/T766M-mutated or L834R-mutated EGFR-ICDs (Fig. 4A and Fig. S2). For the L834R/T766M-mutated EGFR-ICD, both I682Q and V924R mutations reduced autophosphorylation as expected (Fig. 4A, lanes 7 and 8). However, mixing the N-lobe and C-lobe–mutated variants of L834R/T766M EGFR-ICD resulted in no complementation (Fig. 4A, lane 9), failing to increase autophosphorylation above the level seen for the V924R mutant alone. Analogous results were obtained for the L834R-mutated EGFR-ICD (Fig. S2, lanes 2–4). Given the model in Fig. 1, we would expect at a minimum that a “donor-competent” (I682Q-mutated/donor) L834R/T766M EGFR-ICD should be able to activate an “acceptor-competent” (V924R-mutated/acceptor) L834R/T766M EGFR-ICD. Comparison of lanes 8 and 9 in Fig. 4A, however, indicated that it cannot.
Fig. 4.

Lung-cancer–associated EGFRs demonstrate altered donor/acceptor biochemical complementation properties but still form asymmetric dimers. (A) ICD constructs containing mutations that enforce either donor-only (I682Q) or acceptor-only (V924R) behavior in the WT asymmetric dimer were used to assess the mechanism of activation of the lung-cancer–associated mutant L834R/T766M EGFR (LRTM). The indicated combinations of ICD constructs were coexpressed in HEK293 cells, and lysates were subjected to SDS/PAGE and immunoblotting. As predicted by the allosteric model of activation, autophosphorylation levels of coexpressed WT V924R and WT I682Q (lane 5) are half that of WT EGFR (lane 2). Autophosphorylation levels of coexpressed LRTM/I682Q and LRTM/V924R are not greater than either ICD expressed alone (lane 9 compared with lanes 7 and 8), showing that the mutated ICD behaves differently from WT. Coexpression of a WT acceptor mutant (V924R) with LRTM/I682Q dramatically diminishes activity (lane 10). By contrast, coexpression of LRTM/V924R with a WT donor mutant (I682Q) results in apparent superacceptor activity as described in the text (lane 11). (B) Crystal structure of L834R/T766M reveals an asymmetric dimer. The crystals contain two independent molecules that form a typical asymmetric dimer interaction. The L834R and T766M mutations are indicated, as are the locations of I682 and V924 in the asymmetric dimer interface. The irreversible inhibitor PD168393 was included to stabilize the protein and facilitate crystallization. See Fig. S5 for a comparison of the asymmetric dimer interface in this structure with that of WT EGFR.
To investigate this issue further, we asked whether the L834R/T766M EGFR-ICD functions as an effective donor or acceptor for WT EGFR-ICD. Coexpressing WT donor (I682Q) with L834R/T766M/V924R-EGFR acceptor activated the mutated acceptor to a level similar to that seen with no dimer interface mutations (Fig. 4A, lanes 6, 8, and 11), meaning that WT donors could achieve what L834R/T766M-mutated donors could not (Fig. 4A, lane 9). By contrast, coexpression of WT acceptors (V924R) with L834R/T766M-mutated donors (I682Q) surprisingly led to reduced activity compared with L834R/T766M-mutated donors expressed alone (Fig. 4A, lanes 7 and 10). Similar results were obtained with full-length receptors (Fig. S3). Supporting data indicate that donor-enforced (I682Q) lung cancer variant ICDs containing the catalytically inactivating point mutation K721R, also retain equivalent capacities to elicit tyrosine phosphorylation of an acceptor-enforced (V924R) WT ICD compared with donor-enforced (I682Q) WT ICDs with or without the K721R mutation (Fig. S4). Excepting broad structural differences, these data suggest that EGFRs harboring lung cancer mutations may prefer to act as acceptors rather than donors to WT EGFR TKDs.
The L834R/T766M-Mutated EGFR TKD Forms WT-Like Asymmetric Dimers.
To better understand these biochemical findings, we determined the crystal structure of the L834R/T766M mutant EGFR kinase. Although the structure is of relatively low resolution (4 Å) (Table S1), it clearly shows that the L834R/T766M mutant kinase forms an asymmetric dimer interaction that is closely similar to that seen for WT EGFR (Fig. 4B). The crystals contain two molecules in the asymmetric units, “A” and “B,” that are related by the asymmetric dimer interaction. In addition, crystal packing creates a chain of these interactions that extends throughout the crystal. Thus, both molecules A and B are in the active conformation. Although there is a small shift in the relative orientation of the interacting acceptor N-lobes and donor C-lobes compared with WT EGFR, specific asymmetric dimer interactions are preserved in the L834R/T766M mutant (Fig. S5). We expect that these differences arise from differences in the crystal lattice in the two structures, rather than from differences induced by the mutations per se. Comparison with other available EGFR kinase structures reveals that the present structure is most similar to the T766M mutant (Protein Data Bank, PDB ID 2JIU), which superimposes with an rmsd of 1.1 Å. However, the activation loop is mostly disordered in the present structure. The irreversible inhibitor PD168393 forms a covalent bond with Cys773 and binds essentially as previously observed in WT EGFR (15).
While we were preparing this manuscript, two groups reported structures of L834R/T766M mutant EGFR (16, 17). In all of these recently reported L834R/T766M structures, formation of an asymmetric dimer was precluded by experimental conditions including: (i) introduction of the V924R mutation, (ii) cocrystallization with a Mig6 peptide that binds the C-lobe and blocks the asymmetric dimer, or (iii) cocrystallization with a “type II” inhibitor that stabilizes the C-helix in the outward, inactive conformation. Thus, these structures exhibit inactive conformations, in contrast to the active state we observe in the present structure.
Lung-Cancer–Associated EGFR Mutant Activity Is Concomitant with Increased WT EGFR Monomer Association.
It is well established that EGFR activation normally involves an increase in dimeric and oligomeric receptor populations (18)—driven by ligand binding for the WT receptor. Chemical cross-linking, analytical ultracentrifugation (AUC), and other in vitro studies have shown a correlation between dimerization/oligomerization and activity (8, 19, 20). Although EGFR variants harboring lung-cancer–associated mutations have been widely studied, the impact of these mutations or their activating effects on oligomerization state has not been well characterized.
To determine if the increased tyrosine phosphorylation observed for L834R/T766M-mutated EGFR-ICD in the presence of WT EGFRs in Fig. 4A (lane 11) is due to increased interaction, we used coimmunoprecipitation assays (Fig. 5A). Flag-epitope–tagged EGFR-ICDs containing the donor-enforcing I682Q mutation were expressed either alone or in combination with myc-epitope–tagged WT or lung-cancer–associated EGFR-ICDs containing the acceptor-enforcing V924R mutation. We observed an increase in the association of WT I682Q (donor) monomers with lung-cancer–associated acceptor monomers. Moreover, the increase in association was concomitant with the reported catalytic activities of the L834R and L834R/T766M kinases (6, 11). Thus, both L834R and L834R/T766M substitutions in the acceptor TKD appear to enhance the strength of the donor/acceptor interaction—consistent with the idea that these mutations promote the active TKD conformation that is required for formation of the asymmetric dimer interface.
Fig. 5.
Lung-cancer–associated mutated EGFR acceptor activity is concomitant with increased WT EGFR monomer association and hyperphosphorylation. (A) Coexpression of ICD constructs containing the donor-enforcing I682Q in a WT background (green) or the acceptor-enforcing V924R (blue) in the lung-cancer–associated mutant backgrounds L834R (LR) or L834R/T766M EGFR (LRTM) was used to assess the level of monomer association. Lysates from the indicated combinations were subjected to immunoprecipitation with anti-Flag, followed by SDS/PAGE and immunoblotting for protein levels with anti-Myc and anti-Flag as indicated. Levels of coimmunoprecipitated acceptor-enforced mutant ICDs with donor I682Q are greater than levels of coimmunoprecipitated levels of V924R with I682Q (lanes 4 and 5 compared with lane 3), indicating that the lung-cancer–associated mutations increase the affinity of these interactions. (B) Coexpression of increasing amounts of Y992 truncated LRTM-ICD relative to a fixed amount of Flag-tagged WT ICD results in hyperphosphorylation of the WT population as assessed by Western blotting for anti-pY1173 and anti-EGFR. (C) Coexpression of increasing amounts (as indicated in micrograms) of myc-tagged lung-cancer–associated mutant ICDs relative to a fixed amount of Flag-tagged WT ICD results in dose-dependent hyperphosphorylation of the WT population as assessed by Flag immunoprecipitation and subsequent Western blotting for antiphosphotyrosine. Lysates were also immunoblotted with anti–EGFR-pY1173, anti-Myc, and anti-Flag, which show that the level of pY1173 signal cannot be explained solely by the level of the Myc-tagged ICDs harboring lung-cancer–associated mutations. Also, more L834R-ICD (LR) compared with L834R/T766M-ICD (LRTM) is required to hyperphosphorylate WT ICD to levels equivalent to that of the Flag-tagged TKD mutant ICD (compare lanes 4 with 6 and 9 with 11).
Lung-Cancer–Associated EGFRs Hyperphosphorylate Coexpressed WT EGFR.
In EGFR mutant lung cancer cells, the most common primary mutations (Exon 19 deletion or L834R) and acquired resistance mutations (T766M) usually co-occur alongside WT EGFR or other ErbB receptors (21, 22). It is therefore important to understand the effects of lung-cancer–associated EGFR mutants on the activity of coexpressed WT EGFR, which our results so far suggest are difficult to predict. Understanding the effects of mutated EGFR on coexpressed ErbB-2, which has been suggested to be the preferred heterodimeric partner of ErbB receptors (23), is also of great importance.
To assess these questions, we expressed the WT EGFR ICD alone (60 kDa) or in combination with increasing concentrations of a truncated form of the L834R/T766M-mutated EGFR-ICD that ends at Y992 (LRTMΔCT; 39 kDa). The truncation was used to allow separation of the coexpressed proteins by SDS/PAGE to discern individual protein expression levels and to assess the degree of phosphorylation of each protein independently. Coexpressing increasing concentrations of L834R/T766M ΔCT relative to WT resulted in significantly elevated phosphorylation of the WT EGFR-ICD—far above that seen when it is expressed alone (Fig. 5B). However, no dose dependence was observed as the amount of transfected mutated construct was increased beyond 25% of that used for WT. Note that the truncated L834R/T766M ΔCT construct lacks most of the EGFR autophosphorylation sites—underlining the ability of this protein to trans-hyperphosphorylate the WT ICD. We also used immunoprecipitation studies to separate WT and mutated ICDs in assessing WT EGFR hyperphosphorylation induced by coexpression with lung-cancer–associated EGFR mutants. Flag-tagged WT EGFR-ICDs were coexpressed with increasing concentrations of myc-tagged variants harboring lung-cancer–associated mutations [L834R (LR) or L834R/T766M (LRTM)] and immunoprecipitated using anti-Flag. Autophosphorylation of WT protein was then assessed using antiphosphotyrosine immunoblotting (Fig. 5C). Phosphotyrosine signals in the anti-Flag immunoprecipitates rose concomitantly with increasing concentrations of the coexpressed myc-tagged mutated EGFR-ICDs (Fig. 5C), for both LR-myc and LRTM-myc. Maximum WT autophosphorylation reached a level similar to that seen when Flag-tagged mutated EGFR-ICDs were assessed alone. Strikingly, coexpression of either LR- or LRTM-mutated EGFR-ICD with WT EGFR-ICD, at levels far below that of WT, led to strong hyperphosphorylation of the WT EGFR-ICD compared with that seen when WT EGFR-ICD was expressed alone (compare lane 1 in Fig. 5 to lanes 3–5 and lanes 8–10). Notably, lower amounts of the L834R/T766M–EGFR-ICD were required to hyperphosphorylate WT EGFR-ICD compared with the L834R-mutated EGFR-ICD.
To ensure that these results were not an artifact of coprecipitation of the lung-cancer–associated mutants in the anti-Flag immunoprecipitates, whole cell lysates were also probed with anti–EGFR-pY1173. The level of Y1173 phosphorylation seen upon coexpressing mutated and WT EGFR-ICDs greatly exceeded what can be explained by enhanced autophosphorylation of the mutant alone. This is best illustrated by comparing lanes 2 and 3 in Fig. 5C. These lanes display similar levels of Y1173 phosphorylation, yet the anti-myc blot shows that lane 3 contains >20-fold less LR-myc protein than lane 2. Because the WT-flag protein in lane 3 can generate only a weak pY1173 signal on its own (see lane 1), this observation establishes that EGFR Y1173 phosphorylation is greatly enhanced when WT-flag and LR-myc ICDs are coexpressed over what is seen for either protein expressed alone. The same argument applies for LRTM-myc when lanes 7 and 8 in Fig. 5C are compared. Unlike data shown in Fig. 5B in which L834R/T766M-ICDs harboring a truncation at Y992 were used, these data (using nontruncated mutant ICDs) also suggest dose-dependent hyperphosphorylation of WT EGFRs. Taken together, these data demonstrate that mutated EGFR variants can directly influence coexpressed WT EGFRs, leading to their hyperphosphorylation.
Collectively, these data establish that mutated EGFRs favor a position within dimers as acceptors in the presence of WT EGFRs. Through the increased catalytic activity of the mutated proteins (as previously reported) and an increased affinity for WT EGFR donors (Fig. 5A, lanes 3–5), we propose that mutated EGFRs seen in lung cancer have greatly enhanced acceptor (“superacceptor”) activity that allows them to hyperphosphorylate coexpressed WT protein in a directionally defined manner. We suggest that the capacity of these EGFR mutants to hyperphosphorylate WT proteins therefore be termed superacceptor activity.
To investigate this phenomenon in an orthogonal manner, we used EGFR-targeted TKIs that selectively block activity of WT or T766-mutated TKDs. We first used lapatinib, a dual EGFR/ErbB-2 TKI that selectively inhibits WT but not lung-cancer–associated mutant EGFRs (24, 25) to ask what would happen to EGFR autophosphorylation levels upon coexpression of mutated and WT EGFRs if only the WT receptor was selectively inhibited. We expressed Flag-tagged WT EGFR-ICD either alone (Fig. 6A, lanes 2–5) or in combination with approximately 1/10th the amount of myc-tagged L834R/T766M-mutated ICD (Fig. 6A, lanes 6–9). In parallel, myc-tagged L834R/T766M-mutated ICD was also expressed alone at this lower level (Fig. 6A, lanes 10–13). Each condition was subjected to treatment with increasing amounts of lapatinib, and lysates were both immunoblotted with anti–EGFR-pY1173 to assess total EGFR phosphorylation and immunoprecipitated with anti-Flag to isolate the WT ICD and assess its phosphorylation by antiphosphotyrosine immunoblotting (Fig. 6A). As expected, 100 nM lapatinib was sufficient to almost completely inhibit Y1173 autophosphorylation of WT ICDs expressed alone (Fig. 6A, lane 5), but had virtually no effect on activation of L834R/T766M-ICDs (26, 27) (Fig. 6A, lane 13). Crucially, 100 nM lapatinib had no effect on the hyperphosphorylation of WT ICD caused by coexpression of the mutated ICD, arguing that such hyperphosphorylation arises from trans-phosphorylation of WT ICD by a mutated ICD kinase that is not susceptible to lapatinib inhibition (Fig. 6A). Quantification of four replicate experiments to determine the relative change in pY1173 signal to total EGFR compared with untreated controls showed that for each group (WT, WT plus LRTM, or LRTM alone), there was no significant change in the ratios in the presence of up to 100 nM lapatinib, with the exception of cells expressing WT EGFR-ICD only. For the latter group, there was a substantial reduction to 0.18 ± 0.31% in pY1173 levels relative to EGFR (Fig. S6).
Fig. 6.
Hyperphosphorylation of WT-EGFR-ICD in the presence of lung-cancer–mutated EGFR-ICD is insensitive to lapatinib, whereas WZ-4002 restores normal WT EGFR-ICD tyrosine phosphorylation levels. (A) Coexpression of myc-tagged lung-cancer–associated mutant ICD with 1.0 μg of Flag-tagged WT ICD results in hyperphosphorylation of the WT population as assessed by anti-Flag immunoprecipitation and subsequent Western blotting for antiphosphotyrosine. Lapatinib treatment (100 nM) results in inhibition of kinase activity of singly expressed WT EGFR-ICD, but not inhibition of the hyperphosphorylation induced by coexpressing 1/10th the concentration of LRTM-myc-ICD, consistent with trans-phosphorylation by the lapatinib-insensitive mutated TKDs. (B) Coexpression of myc-tagged lung-cancer–associated mutant ICD with 1.0 μg of Flag-tagged WT ICD results in hyperphosphorylation of the WT population as assessed by anti-Flag immunoprecipitation and subsequent Western blotting for antiphosphotyrosine. Treatment of WT- and LRTM-ICD coexpressing cells with the T766M-specific TKI WZ-4002 (100 nM), reduces hyperphosphorylation of the WT ICD (lane 5). WZ-4002 treatment does not inhibit WT kinase activity of singly expressed WT-EGFR-ICD, but conversely, completely inhibits kinase activity of singly expressed LRTM-ICD-myc as assessed by Western blotting (lanes 3 and 7, respectively).
We also used WZ-4002, a TKI that specifically targets EGFR variants containing the T766M mutation (28). WZ-4002 has been shown to be a T766M-specific inhibitor by virtue of a substituted pyrimidine backbone (rather than a quinazoline backbone), so that the T766M mutation does not lead to steric inhibition of its binding to the nucleotide-binding pocket as seen with erlotinib and lapatinib (28). In the same assay used for Fig. 6A, we asked if targeted inhibition of the T766M-mutated EGFR-ICD population with WZ-4002 could reduce levels of WT phosphorylation to those seen for WT EGFR-ICD expressed alone (Fig. 6B). In agreement with previously published data (28), treatment with 100 nM WZ-4002 of the L834R/T766M-mutated EGFR kinase expressed alone in cells resulted in almost complete inhibition of Y1173 phosphorylation (Fig. 6B, lanes 6 and 7), but had little effect on autophosphorylation of the WT ICD (Fig. 6B, lanes 2 and 3). Importantly, inhibiting the L834R/T766M-mutated EGFR-ICD with WZ-4002 substantially reduced its ability to induce hyperphosphorylation of the coexpressed WT EGFR-ICD (Fig. 6B, lanes 4 and 5). To ensure these results were relevant to an existing EGFR mutant lung adenocarcinoma cell line, we expressed WT EGFR containing a C-terminal Myc epitope tag (EGFR-myc) in H1975 lung adenocarcinoma cells (Fig. S7). H1975 cells contain both WT and L834R/T766M-mutated EGFRs, in which the mutations occur in cis. In agreement with the data in Fig. 6B, treatment of H1975-EGFR-myc cells with WZ-4002 resulted in a substantial reduction of tyrosine phoshophorylation of the immunoprecipitated EGFR-myc population, indicating transphosphorylation of the ectopic EGFR-myc by the endogenous EGFR-L834R/T766M (Fig. S7). Collectively, these experiments with two different EGFR TKIs and two different cell types strongly support the hypothesis that mutated EGFRs (acting as acceptors) directionally hyperphosphorylate their coexpressed WT counterparts (which act as donors).
Lung-cancer–Associated EGFRs Also Promote Hyperphosphorylation of ErbB-2.
We next asked whether lung-cancer–mutated EGFRs can also promote hyperphosphorylation of coexpressed ErbB-2. As seen for WT EGFR, coexpression of mutated EGFR-ICDs with ErbB-2–ICDs led to ErbB-2 hyperphosphorylation (Fig. 7A). Coexpressing either L834R-mutated EGFR-ICD or L834R/T766M-mutated EGFR-ICD with the WT ErbB-2 ICD resulted in a substantially greater increase in tyrosine phosphorylation of ErbB-2 (assessed by immunoblotting with ErbB-2–specific anti-pY1221/2) compared with that seen with WT EGFR-ICD (Fig. 7A, compare lanes 8 and 12 with 4). Moreover, the EGFR-ICDs harboring lung-cancer–associated mutations were much more effective as acceptors than as donors: their counterparts containing V924R mutations had a much greater capacity to promote ErbB-2–ICDs hyperphosphorylation than the I682Q variants. Thus, the L834R- and L843R/T766M-mutated EGFR-ICDs also appear to function as superacceptors for ErbB-2. These data further suggest that there may be a conformational/directional preference for mutant EGFR association with ErbB-2 that is distinct from WT EGFR association with ErbB-2.
Fig. 7.
Lung-cancer–associated EGFRs induce hyperphosphorylation of coexpressed ErbB-2 by virtue of superacceptor activity. (A) Coexpression of lung-cancer–associated mutant ICDs with WT ErbB-2 ICDs in HEK293 cells results in hyperphosphorylation of the ErbB-2–ICD population as assessed by anti–ErbB-2 immunoprecipitation and subsequent Western blotting for anti–ErbB-2–pY1221/22. Coexpression of WT EGFR-ICD with ErbB-2–ICD results in a slight increase in phospho–ErbB-2, whereas coexpression of either lung-cancer–associated ICD (LR or LRTM) with ErbB-2 results in hyperphosphorylation of ErbB-2–ICD (lanes 4, 8, and 12, respectively). Unlike lung-cancer–associated EGFRs containing the donor-enforcing I682Q mutation, combination of lung-cancer–associated EGFR-ICDs containing acceptor-enforcing V924R mutation with ErbB-2–ICD results in hyperphosphorylation of ErbB-2–ICD at levels equivalent to the combination of ErbB-2–ICD with lung-cancer–associated parent EGFR-ICDs (lane 10 compared with lane 8 and lane 14 compared with lane 12). (B) Coexpression of intact WT EGFR with intact ErbB-2 results in an increase in tyrosine-phosphorylated ErbB-2 relative to ErbB-2 expressed alone, whereas ErbB-2 coexpression with lung-cancer–associated mutant EGFR (LRTM) results in a significant increase in phospho-ErbB-2 levels. Treatment of cells coexpressing ErbB-2 and EGFR-L834R/T766M with WZ-4002 in the presence of EGF abrogates the EGFR mutant-induced hyperphosphorylation of ErbB-2 (compare lane 16 to lane 11). A comparatively less dramatic effect on ErbB-2 tyrosine phosphorylation is observed for cells coexpressing ErbB-2 and WT EGFR treated with WZ-4002 in the presence of EGF (compare lane 15 to lane 9). (C) Coexpression of of 0.1 μg myc-tagged lung cancer-associated mutant ICD with 1.0 μg of Flag-tagged WT EGFR or ErbB-2–ICD results in additive increases in pAkt or pStat-3 levels compared with singly expressed ICDs, whereas a synergistic increase in pErk levels is observed upon coexpression (compare lanes 2–4 to lanes 6 and 7).
To ensure that these results were not an artifact of using the ICD system, we expressed either full-length WT or L834R/T766M-mutated EGFR alone or in combination with full-length ErbB-2 in HEK293 cells (Fig. 7B). ErbB-2 expressed alone was also included as a control. Lysates from serum-starved cells treated or not with EGF and 100 nM WZ-4002 were probed either by immunoblotting with antibodies to total EGFR, EGFR-pY1068, ErbB-2, and ErbB-2–pY1221/1222, or immunoprecipitation with anti–ErbB-2 followed by immunoblotting with antibodies against ErbB-2–pY1221/1222 or total ErbB-2. Consistent with the ICD data described above, coexpressing ErbB-2 with intact mutated EGFRs led to increased ErbB-2 activation compared with coexpression with WT-EGFR (Fig. 7B, compare lanes 8 and 9 with 10 and 11). As expected, no ligand-dependent increase in phospho–ErbB-2 levels was detected for singly expressed ErbB-2, whereas immunoblotting for phospho-EGFR indicated the anticipated EGF-dependent increase for EGFR populations. In addition, immunoblotting and immunoprecipitation of lysates from serum-starved cells treated with both EGF and WZ-4002 (the T766M-specific TKI) showed a dramatic decrease in ErbB-2 activation in samples coexpressing mutated EGFR and ErbB-2 compared with those coexpressing WT EGFR and ErbB-2 (Fig. 7B, compare lane 9 with 15 and 11 with 16). Collectively, these data show that in addition to directionally hyperactivating WT EGFRs, mutated EGFRs can also promote hyperphosphorylation of ErbB-2, apparently functioning as superacceptors.
Finally, we examined whether differences in canonical EGFR signaling pathways exist for coexpressed WT EGFR/ErbB-2 and mutant EGFR populations compared with either singly expressed protein (Fig. 7C). Augmented levels of activated Akt [also known as protein kinase B (PKB)], Erk, and Stat-3 are known to occur as a result of the constitutive activity of the most frequently mutated EGFRs in lung cancers compared with their WT counterparts. Reports that use either ectopic expression systems or patient-derived tumor cells indicate a significant change in the activity of these proteins that have well-defined roles in prosurvival and proliferative phenotypes (29, 30). WT EGFR, ErbB-2, or EGFR-L834R/T766M ICDs were expressed alone or in combination, and the effects on downstream signaling of coexpressing ErbB-2 or EGFR with lung cancer variants were examined (Fig. 7C). Coexpressing EGFR-L834/T766M-ICD with 10-fold more of either WT EGFR-ICD or ErbB-2–ICD resulted in substantial enhancement of Erk, Akt, and Stat-3 phosphorylation compared with that seen for any of the ICDs expressed alone (Fig. 7C, compare lanes 2–4 with 6 and 7). The levels of Akt and Stat-3 phosphorylation approximated those expected upon adding the effects of EGFR or ErbB-2 ICD with that of EGFR-L834/T766M-ICD. By contrast, the effect on Erk phosphorylation was dramatically greater than expected from simple addition, indicating that coexpression of either WT EGFR-ICD or ErbB-2–ICD with EGFR-L834/T766M-ICD has a synergistic effect on Erk activation. These data indicate that superacceptor activity of lung cancer mutants toward WT EGFR or ErbB-2 can result in heightened signaling output, which should in turn affect the phenotype of cells harboring both WT and mutated EGFR alleles.
Discussion
Several members of the eukaryotic protein receptor tyrosine kinase (RTK) superfamily are known oncogenic drivers due to overexpression or mutation. However, the impact of activating tyrosine kinase domain mutations on the mechanism of intermolecular protein–protein interaction and subsequent kinase activation is unknown. It is assumed that for most WT RTKs, ligand-binding drives dimerization of both the extracellular and intracellular regions (31). So far, the TKD dimerization mode of only a handful of these proteins has been resolved, and typically only for the WT RTKs. In recent years, a number of characteristics that help define the molecular consequences of oncogenic EGFRs, such as altered ATP affinity, signaling, and TKI resistance, have been described (6, 11, 22, 30, 32). However, the impact of EGFR TKD mutations on receptor dimerization, including both homodimerization of mutant receptors and heterodimerization with WT, remains poorly understood.
Our data indicate that EGFR variants harboring lung-cancer–associated mutations are greatly enhanced in their capacity to act as acceptors to activate WT ICDs but retain only WT-like donor activity. In fact, EGFR ICDs harboring lung-cancer–associated mutations are more potent than WT ICDs in hyperphosphorylating either WT EGFR or ErbB-2 when limited to acceptor function with a V924R mutation. This phenomenon is concomitant with an increased heterodimerization/oligomerization affinity of the mutant for the WT protein. To our knowledge, the description of superacceptor activity displayed by oncogenic EGFR mutants represents a unique observation.
In the current model of EGFR regulation, an inactive state is maintained by a delicate balance of factors that includes a relatively stable inactive conformation of the kinase domain that is incompatible with formation of the asymmetric dimer (33), as well as steric restraints on asymmetric dimer formation imposed by the extracellular portion of the receptor (34). Formation of the asymmetric dimer requires rearrangements of the N-lobe including an inward rotation of the C-helix as well as reorganization of the activation loop, as revealed by structural comparisons of active and inactive EGFR. In physiologic activation, ligand binding to the extracellular domain and accompanying rearrangements must provide the energy to induce this conformational change in the acceptor kinase domain. In the context of the L834R and L834R/T766M mutants, the balance is tipped by destabilization of the inactive conformation of the kinase by these mutations (9, 16). These mutations do not appear to “lock” the kinase in an active conformation, as we find that they still depend on asymmetric dimer formation for full activity (Figs. 2–4 and Figs. S1–S4) Furthermore, structures of the L834R/T766M mutant in which asymmetric dimerization is blocked reveal an inactive conformation (16).
The underlying physical basis for the superacceptor phenomenon can be readily explained in light of this model and the structural and biochemical findings described here. Because L834R and L8343R/T766M destabilize the inactive conformation of the kinase, the energetic cost of inducing the active conformation in the acceptor kinase is lower in the mutants than in WT EGFR. Consequently, in a mixed population of WT and oncogenic mutant EGFR, the lung cancer mutants preferentially assume the active, acceptor position. Because the mutants still depend on dimerization for full activity (Fig. 3A), it is reasonable to assume that, in a mutant homodimer, only the acceptor subunit is catalytically fully active. When a stoichiometric amount of WT EGFR is coexpressed with L834R or T766M EGFR, it will form heterodimers in which the mutated EGFR adopts the acceptor position. Thus, expression of WT together with mutant EGFR will increase the net activity of the mutant, by allowing more of the mutant to assume the active, acceptor position.
Consistent with our observation of superacceptor activity, we found that mutated EGFRs hyperphosphorylate WT EGFRs when coexpressed in cells. This is a critical finding due to the heterologous nature of patient-derived EGFR-mutant lung cancer tumor cells, which usually contain two copies of EGFR: the mutant allele and the wild-type allele (35). Even in EGFR mutant cell lines in which the mutant EGFR allele is amplified, the WT allele is usually never lost, implying a critical role for WT EGFRs alongside their mutated counterparts in tumorigenesis. Notably, ErbB-2, which we show is hyperphosphorylated by mutant EGFRs, is also coexpressed in EGFR mutant tumor cells (36). Interestingly, others have found that oncogenic K-Ras can activate the WT Ras isoforms H- or N-Ras via Sos, and that the K-Ras–dependent activation of WT Ras isoforms contributes to cellular signaling and tumorigenesis (37, 38). Thus, mutant-mediated activation of WT signaling counterparts may be a common theme in tumorigenesis.
Our results suggest that mutated EGFRs may unduly influence the activation state of coexpressed WT ErbB receptor neighbors within individual cells. Mutant EGFR superacceptor activity may thus affect cellular transformation as well as sensitivity of cells to specific TKIs. This phenomenon may aid in our understanding of the differences seen in preclinical versus clinical studies of TKIs, because preclinical in vitro studies usually use engineered cell populations expressing only mutated EGFRs and/or patient-derived cell lines in which the entire EGFR population (WT and mutant) activity level is surveyed. In support of this argument, we recently showed that amplification and overexpression of ErbB-2 in the context of mutated EGFRs can drive TKI resistance (36). Collectively, these findings pave the way for the identification of new modes of mutant-specific intermolecular regulation for ErbB receptors and potentially other RTK members. The knowledge of such properties could potentially be exploited for therapeutic benefit.
Materials and Methods
Reagents, Antibodies, Cell Lines, and Plasmids.
HEK293, NIH 3T3, and H1975 cells were obtained from the American Type Culture Collection and maintained in DMEM or RPMI-1640 (GIBCO) supplemented with 10% FBS (Atlanta Biologicals) at 37 °C at 5% CO2. EGF was purchased from R&D Systems. The following antibodies were used: monoclonal anti-Flag from Sigma; polyclonal anti–EGFR-pY1173, anti-EGFR (16F8), and monoclonal anti-Myc from Santa Cruz Biotechnologies; polyclonal anti-EGFR from Millipore; monoclonal anti–EGFR-pY1068 from R&D Systems; monoclonal anti–ErbB-2 from NeoMarkers; and polyclonal anti–ErbB-2-pY1221/1222 and anti–EGFR-L858R (L834R) from Cell Signaling. The respective HRP-conjugated secondary antibodies were purchased from Cell Signaling Technology. ECL reagent from Perkin-Elmer Life Sciences was used for immunoblotting. Stripping and reprobing of blots was performed according to manufacturer’s recommendations.
The plasmids encoding EGFR-ICD-Flag and ErbB-4–ICD–Flag, kind gifts of Graham Carpenter (Vanderbilt University), were cloned as previously described (13) and their sequences verified. The ErbB-2–ICD–Flag construct was created by subcloning the entire coding sequence of the ErbB-2 intracellular domain from pcDNA 3.1-ErbB2 using 5′ GCCTGAAGCTTCACCATGAAGCGACGGCAGCAGAAGATCCGGAAGTACACG-3′ and 5′-CATCTAGACACTGGCACGTCCAGACCCAGGTAC-3′ primers containing flanking HindIII and XbaI restriction sites as well as minimal Kozak sequence and start methionine. The ErbB-2 ICD insert was ligated into pFlag-CMV5.1 using the HindIII and XbaI sites. Site-directed mutagenesis was used to make all mutants with the use of PfuUltra polymerase (Stratagene) (Table S2 shows primer sequences). Site-directed mutagenesis of pMSCVpuro-EGFR was used to make all pMSCVpuro-EGFR mutants. All constructs were confirmed at the nucleotide level by direct sequencing. Phoenix-ampho cells, pMSCVpuro-EGFR, and lapatinib were kind gifts from C. L. Arteaga (Vanderbilt University). WZ-4002 was purchased from Selleck Chemicals.
Cell Culture and Transfections/Transduction.
Transient transfection for expression in mammalian cells was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. A total of 1–2 μg of each expression plasmid was used per 60-mm dish. To assess ligand-dependent EGFR activation, cells were serum starved overnight and treated with 25–50 ng/mL EGF.
Stable expression of full-length EGF receptor in H1975 cells was carried out via retroviral transduction with viral supernatants harvested from Phoenix cells transiently transfected with pMSCVpuro-EGFR plasmid. H1975 cells were selected and maintained with puromycin.
Cell Lysis, Immunoblotting, and Immunoprecipitation.
Cells at 80–90% confluence were either untreated or treated with EGF as indicated and then washed with PBS. Cells were scraped and lysed in ice-cold lysis buffer containing 1% Triton, 10% glycerol, 50 mM Hepes (pH 7.2), and 100 mM NaCl, 1 mM EDTA, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonylfluoride, 10 μg/mL aprotinin, and 25 μg/mL leupeptin. Lysates were cleared by centrifugation at 13,000 rpm in an eppendorf 5415D rotor for 10 min at 4 °C. SDS sample buffer was added to lysates, and samples were boiled for 5 min. Samples were then subjected to SDS/PAGE on either 4–20% or 8% polyacrylamide gels, transferred to nitrocellulose, and immunoblotted. For immunoprecipitation, cell lysates were prepared as described above, and 0.5–1.0 mg whole cell lysate was precleared with 15 μL recombinant protein G Sepharose (Life Technologies) for 30 min at 4 °C. Precleared lysates were incubated overnight with 1 μg of the indicated antibody at 4 °C. A total of 20 μL protein G Dynabeads was added to each sample and incubated up to 4 h at 4 °C. Immunoprecipitates were washed and eluted. Samples were subjected to SDS/PAGE as described above.
EGFR L834R/T766M+PD168393 complex structure determination.
A construct spanning residues 672–998 of the human EGFR and bearing both the L834R and T766M mutations in a GST-fusion format was generated using the Baculogold system (Pharmingen) and expressed in Sf9 insect cells. Purification of the EGFR mutant protein was done as described (9) except that the small molecule inhibitor PD168393 was added to the lysate at a final concentration of 2 μM and incubation of the lysate with glutathione beads was done at 4 °C overnight to ensure that the kinase molecules were fully labeled by PD168393. Crystals of L834R/T766M+PD168393 were prepared by hanging-drop vapor diffusion. The reservoir solution was 0.1 M Bicine pH 9, 0.5 M NaCl, 18% PEG4000, 5 mM Tris(2-carboxyethyl)-phosphine (TCEP). The crystals were flash frozen in liquid nitrogen after quick dumping in the reservoir solution supplemented with 25% ethylene glycol. Diffraction data were collected at Argonne National Laboratory ID24 beamline at 100K. Data were processed and merged with the program HKL2000 (39). The structure was determined by molecular replacement using the EGFR L834R kinase structure (PDB 2itv) (9) as the search model. The deformable elastic network (DEN) restrained refinement in CNS (40) was then used to refine the structure and the recently released high resolution L834R/T766M structure (PDB 3w2p) was used as the reference model in the DEN refinement. Repeated rounds of manual refitting and crystallographic refinement were performed using COOT (41) and CNS/DEN. Statistics of diffraction data processing and structure refinement are shown in Table S1.
Supplementary Material
Acknowledgments
We acknowledge Graham Carpenter and members of the W.P. laboratory for fruitful discussions and Jin H. Park for efforts in further analysis of the mutated EGFRs. This work was supported in part by grants from the V Foundation (to W.P.) and the National Cancer Institute [R01-CA121210, P01-CA129243, and U54-CA143798 (to W.P.); R01-CA116020 and P01-CA154303 (to M.J.E.); and R01-CA079992 (to M.A.L.)]. W.P. received additional support from Vanderbilt Unversity’s Specialized Program of Research Excellence in Lung Cancer, Grant CA90949 and the Vanderbilt-Ingram Cancer Center Core Grant, P30-CA68485.
Footnotes
Conflict of interest statement: Rights to testing of EGFR T790M mutations were licensed by Memorial Sloan-Kettering Cancer Center on behalf of W.P. and others to MolecularMD.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4LL0).
See Commentary on page 15169.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220050110/-/DCSupplemental.
References
- 1.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 3.Mitsudomi T, et al. West Japan Oncology Group Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Viteri S, Molina MA, Benlloch S, Taron M. Epidermal growth factor receptor tyrosine kinase inhibitors as first-line treatment in advanced nonsmall-cell lung cancer. Curr Opin Oncol. 2010;22(2):112–120. doi: 10.1097/CCO.0b013e32833500d2. [DOI] [PubMed] [Google Scholar]
- 6.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci USA. 2009;106(51):21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149(4):860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 11.Carey KD, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66(16):8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18(12):1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci USA. 2007;104(49):19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu C, et al. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16(3):460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blair JA, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3(4):229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 16.Gajiwala KS, et al. Insights into the aberrant activity of mutant EGFR kinase domain and drug recognition. Structure. 2013;21(2):209–219. doi: 10.1016/j.str.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Sogabe S, et al. Structure-based approach for the discovery of Pyrrolo[3,2d]pyrimidine-based EGFR T790M/L858R mutant inhibitors. ACS Medicinal Chemistry Letters. 2012;4:201–205. doi: 10.1021/ml300327z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorissen RN, et al. Epidermal growth factor receptor: Mechanisms of activation and signalling. Exp Cell Res. 2003;284(1):31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 19.Fanger BO, Stephens JE, Staros JV. High-yield trapping of EGF-induced receptor dimers by chemical cross-linking. FASEB J. 1989;3(1):71–75. doi: 10.1096/fasebj.3.1.2783412. [DOI] [PubMed] [Google Scholar]
- 20.Mohammadi M, et al. Aggregation-induced activation of the epidermal growth factor receptor protein tyrosine kinase. Biochemistry. 1993;32(34):8742–8748. doi: 10.1021/bi00085a004. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzahar E, et al. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16(10):5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusnak DW, et al. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther. 2001;1(2):85–94. [PubMed] [Google Scholar]
- 25.Wood ER, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64(18):6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 26.Li D, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27(34):4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilmer TM, et al. Impact of common epidermal growth factor receptor and HER2 variants on receptor activity and inhibition by lapatinib. Cancer Res. 2008;68(2):571–579. doi: 10.1158/0008-5472.CAN-07-2404. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305(5687):1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 30.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117(12):3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shtiegman K, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26(49):6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- 33.Endres NF, Engel K, Das R, Kovacs E, Kuriyan J. Regulation of the catalytic activity of the EGF receptor. Curr Opin Struct Biol. 2011;21(6):777–784. doi: 10.1016/j.sbi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endres NF, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152(3):543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladanyi M, Pao W. Lung adenocarcinoma: Guiding EGFR-targeted therapy and beyond. Mod Pathol. 2008;21(Suppl 2):S16–S22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- 36.Takezawa K, et al. HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2(10):922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeng HH, Taylor LJ, Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat Commun. 2012;3:1168. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3(1):112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, editor. Methods in Enzymology. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 40.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464(7292):1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.