Abstract
We report that K5.Smad7 mice, which express Smad7 transgene by a keratin-5 promoter, were resistant to radiation-induced oral mucositis, a painful oral ulceration. In addition to NF-κB activation known to contribute to oral mucositis, we found activated TGF-β signaling in oral mucositis. Smad7 dampened both pathways to attenuate inflammation, growth inhibition and apoptosis. Additionally, Smad7 promoted oral epithelial migration to close the wound. Further analyses revealed that TGF-β signaling Smads and their co-repressor CtBP1 transcriptionally repressed Rac1, and Smad7 abrogated this repression. Knocking down Rac1 in mouse keratinocytes abrogated Smad7-induced migration. Topically applying Smad7 protein with a cell permeable Tat-tag (Tat-Smad7) to oral mucosa showed preventive and therapeutic effects on radiation-induced oral mucositis in mice. Thus, we have identified novel molecular mechanisms involved in oral mucositis pathogenesis and our data suggest an alternative therapeutic strategy to block multiple pathological processes of oral mucositis.
Oral mucositis, a severe oral ulceration, is a common adverse effect of a large dose of radiation for bone marrow transplant or craniofacial radiotherapy for cancer1,2. Severe oral mucositis could require feeding tubes, management of severe pain, and prematurely halting radiotherapy1. Excessive inflammation and epithelial ablation are key features of oral mucositis3,4. Palifermin, a KGF (human keratinocyte growth factor) recombinant protein, is approved for preventing oral mucositis in bone-marrow transplant patients1. Two Palifermin clinical trials in head and neck cancer patients showed that Palifermin reduced severe oral mucositis incidence from 67% and 69% to 51% and 54%, respectively5,6. Other oral mucositis drugs in clinical trials or pre-clinical studies include growth factors7–9, agents for radioprotection10–12, anti-inflammatory agents or immune modulators13–16.
The modest effects of Palifermin and drugs being developed in the above mentioned categories highlight the need for identification of biomarkers for novel therapies. However, the lack of routine diagnostic biopsies or discarded tissues from oral mucositis patients has hindered this effort. To overcome this limitation, we utilized a keratinocyte-specific Smad7 transgenic mouse model as a tool for biomarker discovery and experimental therapy in oral mucositis. Smad7 was initially identified as a signaling antagonist of the TGF-β superfamily, which blocks TGF-β-induced growth inhibition and apoptosis in keratinocytes17. Smad7 also reduces inflammation by antagonizing NF-κB activation18. These facts compelled us to examine if Smad7 can alleviate oral mucositis.
RESULTS
K5.Smad7 mice were resistant to oral mucositis
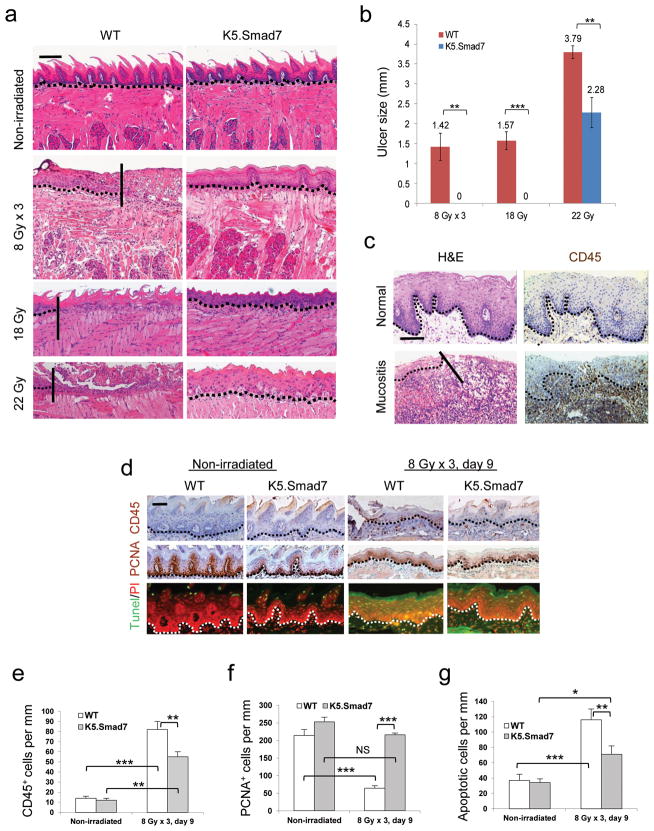
We generated K5.Smad7 transgenic mice, which express Smad7 in keratinocytes as previously described19, and confirmed transgene expression in oral epithelia (Supplementary Fig. 1a,b). We exposed K5.Smad7 mice and wildtype littermates to cranial radiation to determine the biological equivalent dose (BED)20 required to induce oral mucositis in mice. We found that 8 Gy x 3 (BED = 43.2), a regimen relevant to hypo-fractionated radiotherapy in clinic, was the minimal dose needed to induce oral mucositis (Fig. 1a,b). To evaluate the potency of Smad7 effects, we also tested single doses of cranial radiation and found that oral mucositis severity correlated with BED values between 18 Gy (BED = 50.4) and 22 Gy (BED = 70.4) (Fig. 1a,b, Supplementary Fig. 1c ). By day 9 after initiation of radiation, wildtype mice developed oral ulcers (Fig. 1a,b). K5.Smad7 oral mucosa prior to irradiation had morphology similar to wildtype mice, but exhibited resistance to radiation-induced oral mucositis (Fig. 1a,b). Histological analyses revealed that wildtype mice developed oral mucositis (Fig. 1a) similar to that in humans (Fig. 1c). K5.Smad7 oral epithelia typically showed radiation dose-dependent damage, i.e. thinning epithelium and flattened tongue papillae after 8 Gy x 3 radiation, and more damaged (hypo- or hypertrophic) epithelial cells after 18 Gy and 22 Gy radiation (Fig. 1a). Consistent with increased leukocyte infiltration in human oral mucositis lesions (Fig. 1c), lesions in wildtype mice harbored numerous infiltrated leukocytes (Fig. 1d,e) consisting of neutrophils, macrophages, and lymphocytes (Supplementary Fig. 1d); all were substantially reduced in K5.Smad7 oral mucosa (Fig. 1d,e and Supplementary Fig. 1d). Because it is difficult to capture human oral mucositis pathology at the acute phase, we utilized a mouse model to assess proliferation and apoptosis when ulcers are just formed. Similar to previous reports7,12, proliferative cells were sparse in irradiated wildtype oral epithelium, but were seen more in irradiated K5.Smad7 oral epithelium (Fig. 1d,f). Conversely, apoptotic cells were significantly reduced in irradiated K5.Smad7 oral mucosa compared to wildtype mice (Fig. 1d,g).
Figure 1. K5.Smad7 mice are resistant to radiation-induced oral mucositis.
(a) H&E staining of tongues from nonirradiated and irradiated (day 9 after initiation of radiation) wild-type (WT) and K5.Smad7 tongues. The vertical lines in the images of tongues from WT mice highlight the ulcer boundary. Scale bar, 50 μm. (b) Sizes (mean ± s.e.m) of tongue ulcers. n = 8 for WT, n= 7 for K5.Smad7 in 8 Gy x 3 radiation; n = 5 for WT, n = 4 for K5.Smad7 in 18 Gy radiation; n = 5 per group for WT and K5.Smad7 in 22 Gy radiation. (c) Human ventricular posterior of the tongue (top) and radiation-induced tongue mucositis (bottom) visualized using H&E (left) and CD45 (right) staining. The solid diagonal line indicates the ulcer boundary, and dotted lines indicate the basement membrane. Scale bar, 50 μm. (d) Immunostaining of CD45, proliferating cell nuclear antigen (PCNA) and TUNEL assay in irradiated sections adjacent to an ulcer from WT mice and in damaged areas from K5.Smad7 mice. PI: propidium iodide. Dotted lines indicate the basement membrane. Scale bar, 25 μm. (e–g) Quantification (mean ± s.d.) of staining in d. n =3 or 4 per group. *P < 0.05, **P < 0.01, ***P < 0.001, NS: no significance determined by two-tailed Student’s t test.
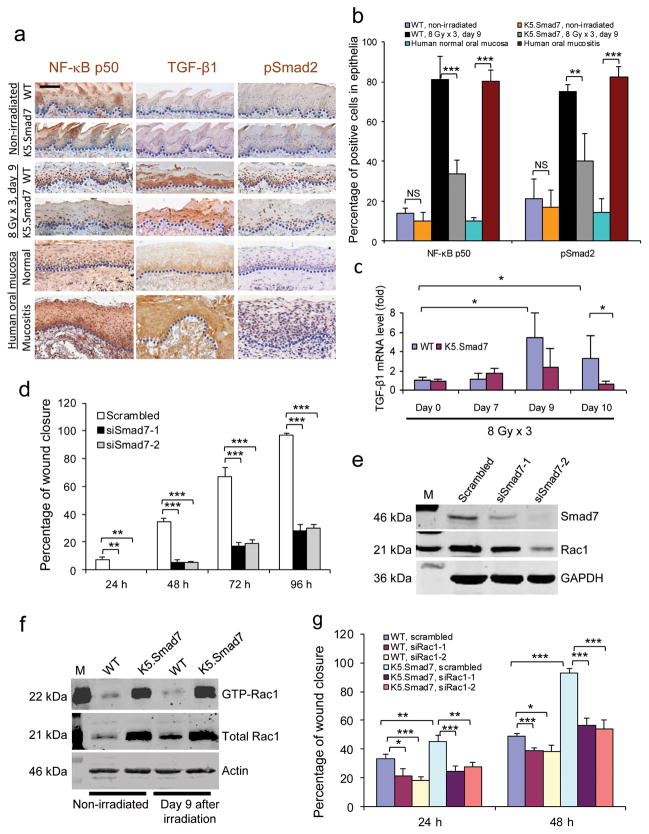
As expected, cells with nuclear NF-κB p50 subunit were significantly increased in oral mucositis compared to non-irradiated wildtype oral mucosa (Fig. 2a,b). Interestingly, TGF-β1, an immune suppressant in internal organs21,22 but pro-inflammatory in oral mucosa23, together with its activated signaling mediator, phosphorylated (p) Smad2, were also increased in oral mucositis compared to non-irradiated oral mucosa in wildtype mice (Fig. 2a,b). We also detected similar changes in human oral mucositis lesions (Fig. 2a,b). Irradiated K5.Smad7 oral epithelia significantly reduced cells positive for nuclear NF-κB p50 and pSmad2, even though they still had abundant TGF-β1 protein (Fig. 2a,b). TGF-β1 mRNA in irradiated wildtype oral mucosa was significantly increased on day 9 and day 10 (Fig. 2c). TGF-β1 mRNA level in K5.Smad7 mucosa was similar to wildtype mucosa at earlier time points but was back to normal by day 10 (Fig. 2c). These data suggest that TGF-β1 transcription is not inhibited by Smad7, but its more rapid decline in K5.Smad7 mucosa could be a consequence of accelerated healing. Phospho-Smad1/5/8, markers for activated BMP signaling24, were not affected by Smad7 before and after radiation (Supplementary Fig. 1e). This result is consistent with the ability of Smad7 to preferentially inhibit TGF-β signaling.
Figure 2. Molecular alterations attenuated by Smad7.
(a) Immunostaining of NF-κB subunit p50, TGF-β1 and pSmad2 in irradiated tongue sections of WT mice adjacent to an ulcer and sections from the damaged area of K5.Smad7 mice, as well as in human samples from nonirradiated oral mucosa and radiation-induced mucositis. Dotted lines delineate epithelial-stromal boundary. Scale bar, 25 μm. (b) Quantification (mean ± s.d.) of nuclear NF-κB subunit p50 and pSmad2 in a. n = 3 or 4 per group. (c) Quantitative RT-PCR (mean ± s.d.) of TGF-β1 (normalized to keratin 5; n = 6 per group for day 0, n = 4 for day 7 and day 9, and n = 7 for day 10). (d) Quantification (mean ± s.d.) of human oral keratinocyte migration (see images in Supplementary Fig. 2). Scrambled, scrambled siRNA; siSmad7-1 and siSmad7-2, two different siRNAs specific to Smad7. n = 3 per group. (e) Western blot analysis of Rac1 72 h after Smad7 knockdown. The knockdown efficiency of siSmad7-1 and siSmad7-2 can be estimated from the blot. M: molecular markers; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (f) Western blot analysis of total and activated (GTP-bound) Rac1 (GTP-Rac1) protein. (g) Effect of Rac1 knockdown on Smad7-mediated keratinocyte migration (see knockdown efficiency in Supplementary Fig. 3a and images in Supplementary Fig. 3d). n = 3 per group. Data are presented as mean ± s.d. siRac-1 and siRac1-2 are two siRNAs specific for Rac1. *P <0.05, **P < 0.01, ***P < 0.001, NS: no significance determined by two-tailed Student’s t test.
Rac1 contributed to Smad7-mediated keratinocyte migration
To determine if Smad7 contributes to healing in human oral keratinocytes, we knocked down Smad7 in spontaneously immortalized human oral keratinocytes (NOK-SI)25, which harbor a p53 (R283Q) mutation and p16INKa promoter methylation but no mutations in any of the TGF-β signaling components (unpublished data from Gutkind laboratory). Smad7 knockdown blunted keratinocyte migration after wounding (Fig. 2d and Supplementary Fig. 2a). Conversely, knocking down TGF-β1 accelerated keratinocyte migration (Supplementary Fig. 2b–d), consistent with accelerated wound healing seen in mice null for TGF-β1 or Smad326,27. To search for molecular mechanisms associated with Smad7-mediated keratinocyte migration, we examined Rac1, a protein indispensable for oral wound healing25. Rac1 was reduced after Smad7 knockdown (Fig. 2e). We expected that TGF-β1 overexpression in oral mucositis would activate Rac1 through a Smad-independent mechanism28. However, although total Rac1 protein increased by 2-fold after irradiation, activated Rac1 protein did not change considerably in wildtype tongues (Fig. 2f). In K5.Smad7 oral mucosa, both total and activated Rac1 were significantly increased by 4-fold and 8-fold, respectively, compared to wildtype oral mucosa (Fig. 2f). To determine the functional significance of Smad7-induced Rac1 activation, we knocked down Rac1 in primary keratinocytes isolated from wildtype and Smad7 transgenic neonatal skin and assayed for cell proliferation and migration. Rac1 knockdown showed modestly reduced proliferation in wildtype and Smad7 keratinocytes (Supplementary Fig. 3a–c) but almost completely abrogated Smad7-induced migration (Fig. 2g and Supplementary Fig. 3d), suggesting that increased Rac1 contributes to Smad7-mediated cell migration.
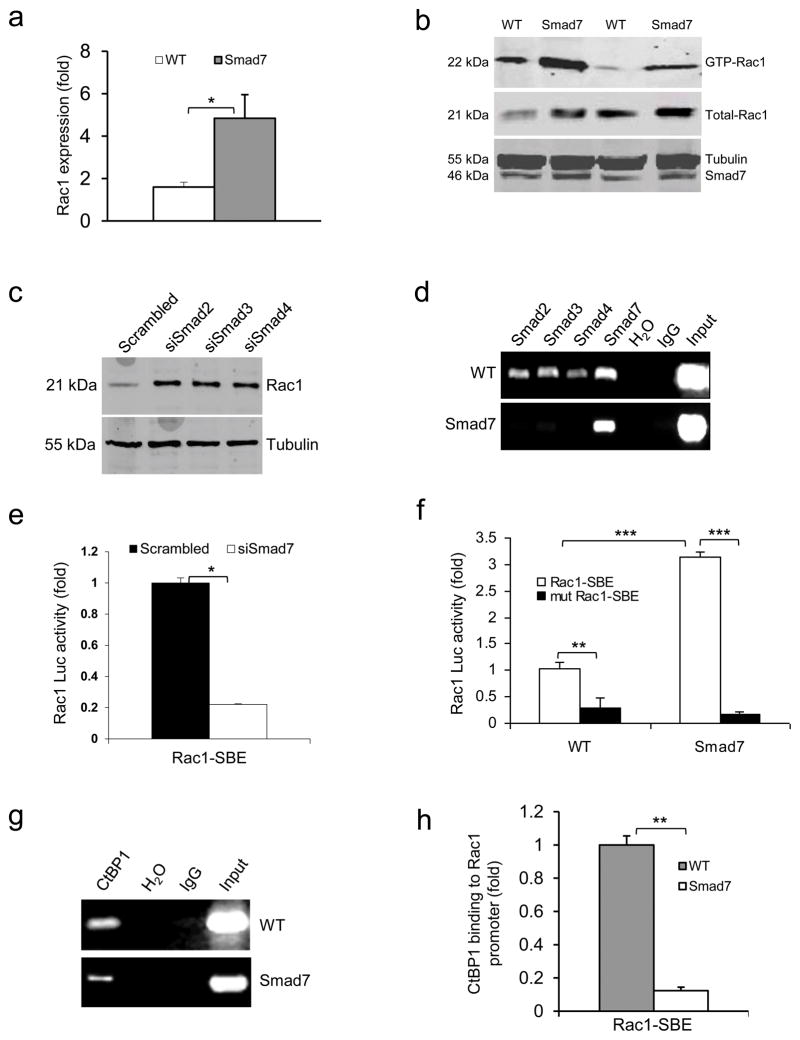
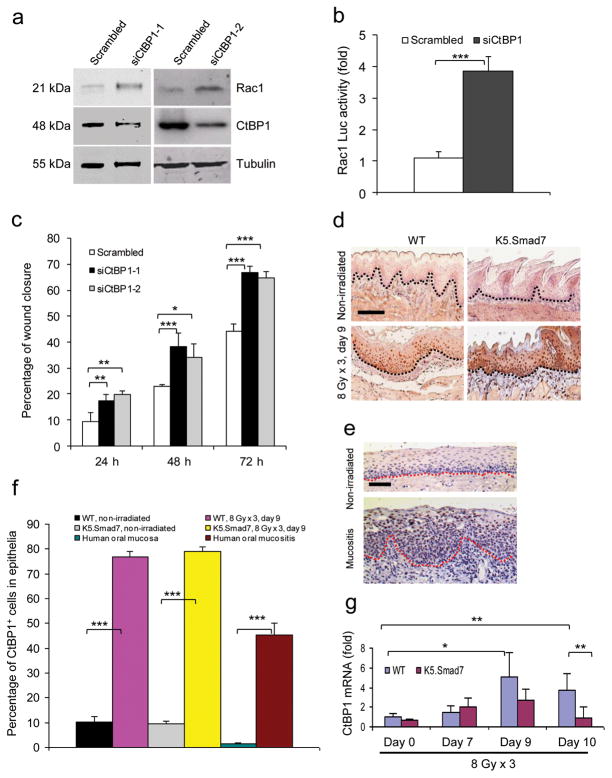
We observed that increased Rac1 mRNA levels in Smad7 transgenic keratinocytes correlated with total and active Rac1 protein levels (Fig. 3a,b and Supplementary Fig. 4a,b), suggesting that increased Rac1 activation in Smad7 keratinocytes is, at least in part, a consequence of increased Rac1 transcripts. Further, Rac1 protein increased by ~3 fold (Fig. 3c) after knockdown of individual Smads in NOK-SI cells (Supplementary Fig. 4c–e). These data suggest that normal Smad signaling represses Rac1 transcription. Among the two putative Smad binding elements (SBEs) in the mouse Rac1 promoter (−2.1 Kb and −1.5 Kb upstream of the coding sequence), which are in similar regions of the human Rac1 promoter, chromatin immunoprecipitation (ChIP) identified Smad-2, -3, -4, and -7 binding to the −1.5 Kb site (Fig. 3d) but not the −2.1 Kb site (not shown) in wildtype keratinocytes; binding of Smad-2, -3 and -4 was significantly reduced in Smad7 transgenic keratinocytes (Fig. 3d). Luciferase reporter assays using a SBE-containing Rac1-Luc construct show that knockdown of Smad7 in wildtype keratinocytes significantly reduced luciferase activity (Fig. 3e). Conversely, Smad7 transgenic cells had increased luciferase activity compared to wildtype cells, and mutating the SBE attenuated this increase (Fig. 3f). Thus, Smad7 binding to SBE appears necessary to expel signaling Smads to abrogate Rac1 repression. Among known Smad transcriptional co-repressors29–31, we found that CtBP1 bound to the Rac1 promoter SBE-1.5 Kb site in wildtype keratinocytes (Fig. 3g), and Smad7 transgene expression significantly reduced CtBP1 binding to the SBE (Fig. 3g,h). When CtBP1 was knocked down in NOK-SI cells, Rac1 protein and Rac1-Luc activity were increased compared to keratinocytes transfected with scrambled siRNA (Fig. 4a,b), suggesting that CtBP1 binding to SBE-1.5 Kb represses Rac1 expression. Further, knocking down CtBP1 in NOK-SI cells increased their migration (Fig. 4c and Supplementary Fig. 4f). Upon examination of CtBP1 protein in radiation-induced oral mucositis, we found that CtBP1 is barely detectable in non-irradiated mouse and human oral mucosa (Fig. 4d–f); however, CtBP1 positive cells were significantly increased in irradiated oral mucosa of wildtype and K5.Smad7 mice as well as in human oral mucositis (Fig. 4d–f). Additionally, CtBP1 mRNA in irradiated wildtype oral mucosa was significantly increased on day 9 and day 10 (Fig. 4g). CtBP1 mRNA level in K5.Smad7 mucosa was similar to wildtype mucosa at earlier time points but declined to normal by day 10 (Fig. 4g). These results indicate that Smad7 does not reduce CtBP1 mRNA but instead inhibits CtBP1 binding to the Rac1 promoter by repelling the Smad/CtBP1 complex from the SBE binding site; further, more rapid CtBP1 reduction in K5.Smad7 mucosa serves as a marker of healing.
Figure 3. Smad7 leads to higher Rac1 expression by repressing individual Smad proteins and CtBP1 binding to the SBE of the Rac1 promoter.
(a) Rac1 mRNA levels (mean ± s.d.) in keratinocytes from WT and Smad7 transgenic mice. n = 4 per group. (b) Western blot analysis of GTP-bound Rac1 (GTP-Rac1) and total Rac1 in WT and Smad7 transgenic keratinocytes. Smad7 protein levels were determined by re-probing the tubulin western blot with an antibody to Smad7 (see an additional western blot and quantification in Supplementary Fig. 4a, b). (c) The amount of Rac1 protein after knocking down Smad2, Smad3 or Smad4 individually in human keratinocytes (See Supplementary Fig. 4c–e for Smad knockdown efficiencies). siSamd2-4, siRNAs specific for Smad2-4. (d) ChIP assay for Smad2, Smad3, Smad4, and Smad7 binding to the SBE -1.5 kb site of the Rac1 promoter in keratinocytes from WT and Smad7 transgenic mice. (e) Rac1 luciferase reporter assay in mouse keratinocytes. n = 6 per group. siSmad7, siRNA specific for Smad7; Rac1-SBE, the SBE-1.5 kb site of the Rac1 promoter. Data are the mean ± s.d. (f) Activities (mean ± s.d.) of Rac1-luc reporters containing SBE (Rac1-SBE) or mutant SBE (Mut Rac1-SBE) in keratinocytes from WT and Smad7 transgenic mice. n = 6 per group. (g) Images of ChIP assays of CtBP1 binding to the SBE-1.5 kb site of the Rac1 promoter in keratinocytes from WT or K5.Smad7 mice. (h) ChIP-quantitative PCR (mean ± s.d.) of CtBP1 binding to the SBE in g in keratinocytes from WT and Smad7 transgenic mice. n = 4 per group. *P < 0.05, **P < 0.01, ***P < 0.001 determined by two-tailed Student’s t test.
Figure 4. CtBP1-associated Rac1 repression contributes to the inhibition of keratinocyte migration.
(a) Western blot analysis of Rac1 protein after knockdown of CtBP1 in human oral keratinocytes. siCtBP-1 and siCtBP1-2 are two different siRNAs specific for CtBP1. (b) SBE-containing Rac1-luc reporter activity (mean ± s.d.). n = 6 per group. (c) Effect of CtBP1 knockdown on human oral keratinocyte migration (mean ± s.d.). n = 3 per group. (d) Immunostaining of CtBP1 in irradiated sections adjacent to the ulcer (WT) or from the damaged area (K5.Smad7). Dotted lines denote the basement membrane. Scale bar, 50 μm. (e) Immunostaining of CtBP1 in nonirradiated oral mucosa and radiation-induced oral mucositis in human specimens. Dotted lines denote the basement membrane. Scale bar, 50 μm. (f) Quantification of nuclear CtBP1-positive cells (mean ± s.d.) in d and e. n = 3 or 4 per group. (g) Quantitative RT-PCR (mean ± s.d.) for CtBP1 (normalized to keratin 5). n = 6 per group for day 0, n = 4 for day 7 and day 9, and n = 7 for day 10. *P < 0.05, **P < 0.01, ***P < 0.001 determined by two-tailed Student’s t test.
Tat-Smad7 alleviated radiation-induced oral mucositis
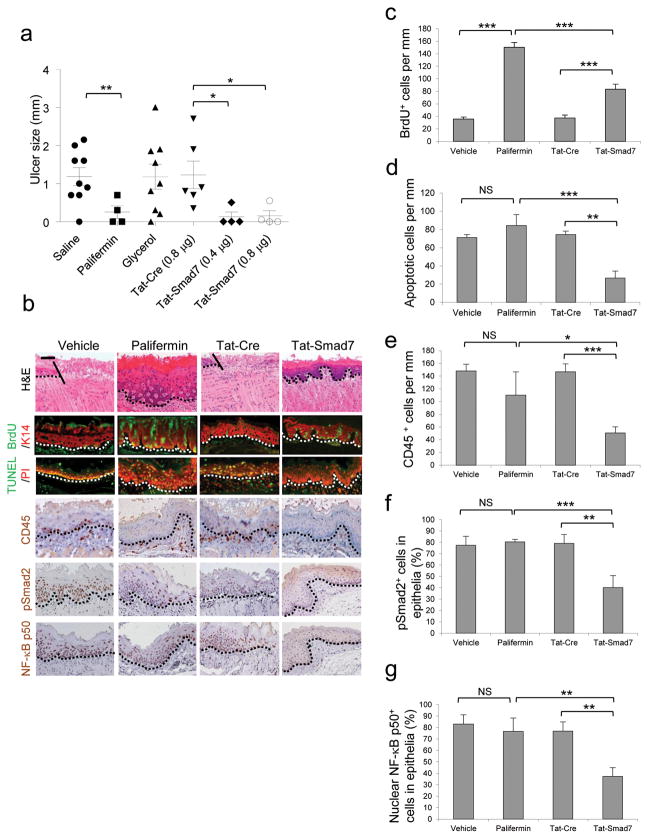
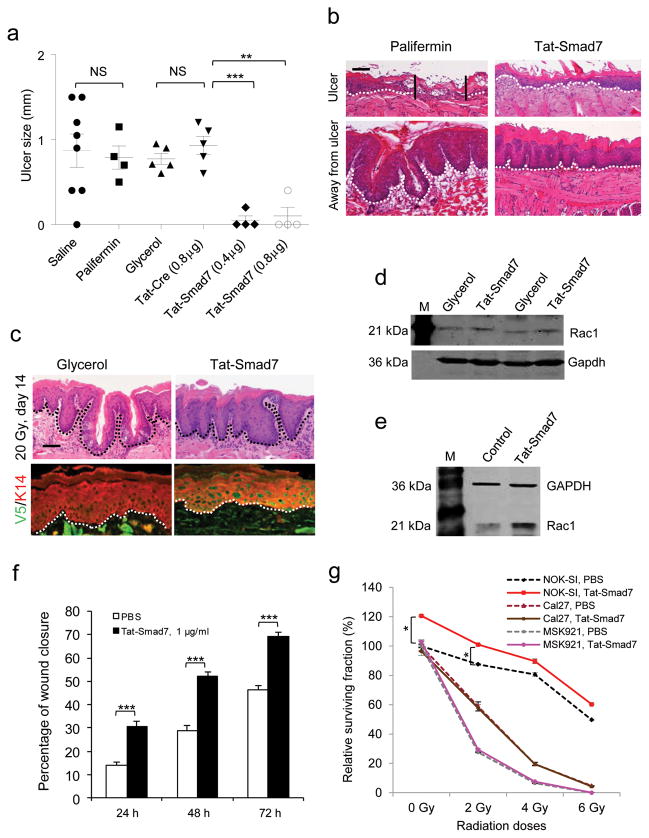
Smad7 transgene’s ability to block multiple pathological processes of oral mucositis prompted us to explore if localized Smad7 delivery can be used to prevent and treat oral mucositis. Because Smad7 is a nuclear protein, local Smad7 delivery needs to allow Smad7 rapidly entering into cells before saliva washes off the protein. Thus, we produced a recombinant human Smad7 with an N-terminal Tat-tag allowing proteins to rapidly permeate the cell membrane and enter the nucleus32–34. A V5 epitope was added to the C-terminal end of the Tat-Smad7 protein to track Tat-Smad7 cell penetration (Supplementary Fig. 5a-d). We tested Tat-Smad7 bioactivity using its ability to block Smad2 phosphorylation (Supplementary Fig. 5c). We also produced Tat-Cre recombinant protein with the same tags as a control (Supplementary Fig. 5e,f). For oral mucositis prevention, both Tat-Smad7 and Tat-Cre (in 50% glycerol/PBS) were topically applied to the oral cavity daily, starting 24 hours prior to radiation through day 8 after initiation of radiation, and treated tissues were examined on day 9. An additional group received Palifermin treatment with a clinical regimen, i.e., 6.25 mg kg−1 (i.p.) daily for 3 days prior to irradiation and daily for 3 days 24 hours after the last dose of radiation. Tat-Cre showed no effect compared to vehicle controls (Fig. 5a,b). Tat-Smad7 treatments showed preventive effects on ulcer formation similar to Palifermin (Fig. 5a). The dose-dependent effect of Tat-Smad7 was more obvious when used on animals given a 20 Gy (BED = 60) single dose of radiation that induced larger oral ulcers than fractionated radiation (Supplementary Fig. 5g). Microscopically, both Palifermin and Tat-Smad7 treated oral mucosa prevented open ulceration in the majority of cases (Fig. 5b). Palifermin treated mucosa exhibited more keratinocyte down-growth but also more damaged keratinocytes (condensed or charcoal-like nuclei, swelled mono- or multi-nucleated cells and shattered nuclear fragments in conified layers) than Tat-Smad7 treated mucosa (Fig. 5b). Immunostaining revealed that Palifermin increased proliferation more significantly than Tat-Smad7. Tat-Smad7 reduced apoptosis, leukocyte infiltration, nuclear pSmad2 and NF-κB p50 but Palifermin did not (Fig. 5b–g).
Figure 5. Oral Tat-Smad7 application prevents radiation-induced oral mucositis in mice.
(a) Dot blot graph (mean ± s.e.m.) of oral mucositis ulcer sizes on day 9 after initiation of 8 Gy x 3 radiation. Vehicle is saline or 50% glycerol/PBS. (b) Pathological alterations on day 9 after initiation of 8 Gy x 3 radiation. The scale bar represents 50 μm for the top row and 25 μm for the rest of rows. Dotted lines delineate epithelial-stromal boundary; the solid lines highlight the ulcer boundary. (c–g) Quantification (mean ± s.d.) of immunostaining shown in b. n = 3 or 4 per group. *P < 0.05, **P < 0.01, ***P < 0.001, NS, no significance determined by two-tailed Student’s t test.
To test if Tat-Smad7 can be used to treat existing oral mucositis, we exposed mice to fractionated (8 Gy x 3) cranial radiation and applied Tat-Smad7 (topically) or Palifermin (6.25 mg kg−1, i.p.) daily from day 6 after initiation of radiation (when mucosal damage was obvious) till day 9, and treated tissues were examined on day 10. Although beginning post-radiation administration of Palifermin at earlier time points than the current protocol reduced oral mucositis in mice35,36, Palifermin administration with the current protocol did not accelerate ulcer closure (Fig. 6a) regardless of its hyperproliferative effect on the entire oral mucosa (Fig. 6b). This is not surprising, as Palifermin is approved to prevent but not treat oral mucositis. Tat-Smad7 treated oral mucositis reduced ulcer sizes and pathological alterations after both fractionated and single dose radiation (Fig. 6a,b and Supplementary Fig. 6a–g). Away from ulcers, Tat-Smad7 treated oral mucosa exhibited less hyperplasia and more differentiated epithelia than Palifermin-treated oral mucosa (Fig. 6b). With a 20 Gy single dose radiation that caused slower healing than fractionated radiation, the effect of Tat-Smad7 on recovery after wound closure was more obvious. When vehicle treated ulcer was just re-epithelialized, Tat-Smad7 treated mucosa had almost recovered to normal morphology (Fig. 6c). Consistent with observations in K5.Smad7 mice, Tat-Smad7 increased Rac1 promoter activity and reduced CtBP1 binding to the SBE of the mouse Rac1 promoter (Supplementary Fig. 6h,i), and increased Rac1 protein in mouse oral mucositis and human oral keratinocytes (Fig. 6d,e). Tat-Smad7 treated human oral keratinocytes after wound scratch had accelerated wound closure (Fig. 6f and Supplementary Fig. 7a). Further, irradiated human oral keratinocytes increased nuclear pSmad2 and NF-κB p50, which were attenuated by Tat-Smad7 treatment (Supplementary Fig. 7b). In contrast, although Tat-Smad7 penetrated oral cancer cells efficiently (Supplementary Fig. 7c), it did not further elevated Rac1 protein level that is already abundant in cancer cells (Supplementary Fig. 7d). This result could account for faster migration of cancer cells than normal keratinocytes (Fig. 6f and Supplementary Fig. 7a, e–h) but the lack of promotion effects of Tat-Smad7 on migration in two oral cancer cell lines, MSK921 that does not contain genetic loss of TGF-β signaling components37 and Cal27 that has mutated Smad438 (Supplementary Fig. 7e–h). Colony assays show that survival of human oral keratinocytes was slightly increased by Tat-Smad7 treatment with or without radiation (Fig. 6g). Consistent with the notion that reduced survival after irradiation is more prominent in cancer cells than in normal cells, SCC cells showed substantial reductions in cell survival after radiation. Treatment with Tat-Smad7 did not affect survival in SCC cells with or without radiation (Fig. 6g).
Figure 6. Tat-Smad7 treatment on oral mucositis.
(a) Dot blot graph (mean ± s.e.m.) of ulcer sizes measured on day 10 after initiation of 8 Gy x 3 radiation. Glycerol is 50% glycerol/PBS. (b) H&E staining of an open ulcer in palifermin-treated but not Tat-Smad7-treated mucosa (top) and a comparison of epithelial thickness between palifermin-treated and Tat-Smad7-treated mucosa (bottom). Dotted lines delineate the basement membrane, and the vertical lines highlight the ulcer boundary. Scale bar, 50 μm. (c) Tat-Smad7 treatment in 20 Gy-induced oral mucositis after ulcers healed. V5 immunostaining visualizes Tat-Smad7 in oral epithelia (sections are away from the damaged regions); K14 immunostaining was used as counterstain. Green in muscle cells represents autofluorescence. Dotted lines delineate the basement membrane. Scale bar, 25 μm. (d) Rac1 western blot analysis of Tat-Smad7-treated mouse tongues on day 10 after initiation of 8 Gy x 3 radiation. M, molecular markers. (e) Rac1 western blot analysis of Tat-Smad7-treated normal human oral keratinocytes 48 h after treatment. Control, PBS. (f) Effect of Tat-Smad7 treatment on oral human keratinocyte migration (NOK-SI, see images in Supplementary Fig. 7a). n = 4 per group. Data are the mean ± s.d. (g) Survival curves (mean ± s.d.) of NOK-SI keratinocytes and SCC lines (Cal27 and MSK921) with or without Tat-Smad7 treatment. n = 4 per group for each radiation dose. *P < 0.05, **P < 0.01, ***P < 0.001, NS, no significance determined by two-tailed Student’s t test.
DISCUSSION
It is critical for oral mucositis prevention and treatment to overcome epithelial ablation due to massive apoptosis and blunted keratinocyte proliferation. Because Palifermin is a mitogen, it has stronger proliferative effects than Smad7. On the other hand, the anti-apoptotic effect of Smad7 is more potent than Palifermin. The proliferative and anti-apoptotic effects of Smad7 are more obvious in oral mucositis than in normal oral mucosa, when TGF-β1, a potent growth inhibitor and apoptosis inducer for epithelial cells39,40, was increased. Additionally, our data suggest that increased Rac1 activation is largely responsible for Smad7-mediated keratinocyte migration in wound closure. This finding was initially unexpected, given the documented role of TGF-β signaling in Rho/Rac activation in cancer cells via a Smad-independent mechanism41. Our study suggests that during oral mucositis, Smad-dependent Rac1 repression could overcome Smad-independent Rac1 activation (if any) due to increased Smad signaling (evidenced by increased pSmad2) and Smad transcriptional co-repressor CtBP1. When this repression is abrogated by Smad7, it permits Rac1 activation-mediated keratinocyte migration. However, in oral cancer cells, signaling Smads are lost or inactivated, or other mechanisms independently activate Rac1; hence Smad7-mediated abrogation of Rac1 repression would no longer occur. Although Rac1 activation also contributed to keratinocyte proliferation, knocking down Rac1 only partially attenuated the proliferative effect of Smad7. Therefore, Rac1’s contribution to proliferation appears to be limited, and blocking TGF-β1-induced growth arrest is also needed to overcome radiation-induced growth inhibitory effects.
Dampening excessive inflammation creates a microenvironment for oral mucositis healing13–16. The antagonistic effect of Smad7 on both TGF-β and NF-κB signaling18 could make Smad7 a more efficient anti-inflammatory molecule than other agents targeting only NF-κB. Because inflammatory cells produce cytokines that further activate TGF-β and NF-κB, reduced TGF-β and NF-κB signaling, found in K5.Smad7 or Tat-Smad7 treated oral mucosa after radiation, reflects the direct antagonistic effect of Smad7 on these two pathways and the consequence of reduced inflammatory cytokines from infiltrated leukocytes. However, Smad7 did not reduce NF-κB or TGF-β signaling below their normal physiological conditions. This incomplete blockade of NF-κB or TGF-β signaling may be beneficial to oral mucositis healing, as a complete loss of either pathway could induce excessive inflammation21,22,42.
The primary obstacle to using growth factors to treat oral mucositis in cancer patients is the potential risk of promoting cancer cell growth1,8,43. The majority of human oral cancers lose TGF-β signaling in tumor epithelial cells44,45; thus, anti-Smad-associated cell proliferation and migration by Smad7 would not be effective in cancer cells. In tumors with intact TGF-β signaling, activation of other oncogenic pathways could override TGF-β-induced tumor suppressive effects. These two scenarios could explain why we did not observe Smad7 increasing proliferation and migration in oral cancer cells with mutant or intact TGF-β signaling components. Additionally, TGF-β signaling promotes tumor invasion mainly through Smad-independent mechanisms after loss of TGF-β-induced tumor suppression. Thus, blocking TGF-β signaling by Smad7 in cancer cells could abrogate TGF-β-mediated tumor promotion effects, which behaves similarly to TGF-β inhibitors currently being used in clinical trials for advanced cancers46. Further, the potent anti-inflammatory effect of Smad7 may reduce the risk of tumor progression. Therefore, the long-term effects of Smad7 application on cancer need to be evaluated in vivo. We have not observed spontaneous tumor formation in K5.Smad7 mice19. Because Smad7 is not a secreted protein, local and short-term Smad7 protein delivery in oral mucositis treatment should have few systemic effects. Of note, several cell permeable proteins have moved from molecular mechanism studies into clinical trials47. In bone marrow transplant patients, whose oral epithelia do not contain cancer cells, Smad7 topical application may be suitable for both prevention and treatment of oral mucositis. For oral cancer patients, future studies should assess if Smad7 application compromises radiotherapy-mediated killing of oral cancer cells.
In summary, we describe a mouse model resistant to oral mucositis that suggests molecular targets for oral mucositis prevention and treatment. We provide evidence that Smad7-mediated oral mucositis healing is a result of targeting multiple pathogenic processes mediated by these molecules (Supplementary Fig. 8). Our data instigate future studies to determine the functional contributions of these molecular targets (e.g., TGF-β, NF-κB, CtBP1, Rac1) to oral mucositis pathogenesis and whether they can be used as predictive and therapeutic responsive markers of oral mucositis in patients.
Materials and Methods
Radiation-induced oral mucositis
The Institutional Animal Care and Use Committee at the University of Colorado Denver Anschutz Medical Campus and at the US NIH approved all animal experiments. We generated K5.Smad7 mice in as previously reported17,19, bred them into in the C57BL/6 background and used 8–10 weeks old male and female transgenic mice and wildtype littermates in radiation induced oral mucositis studies. The Results section specifies the number of mice used in each experiment; no gender differences were observed (not shown). We used 8–10 weeks old C3H females (Jackson Laboratory) or C57BL/6 mice for experiments with Tat-Samd7 treatment (no strain differences), and each figure specifies the number of mice used in each experiment. We induced oral mucositis by single-dose or fractionated cranial irradiation (specified in the main text) as previously reported48. Briefly, for 8 Gy x 3 fractionated cranial irradiation, we guided each animal into a customized mouse jig with standardized snout positioning and closed the end of the jig. For single-dose (18 Gy to 22 Gy, specified in the main text) cranial irradiation, we anesthetized each mouse with 30–50 μl of ketamine cocktail (32 mg ml−1 ketamine, 4.8 mg ml−1 xylazine, and 0.6 mg ml−1 acepromazine) during irradiation. We used lead to shield their bodies (except heads) in the RS2000 biological irradiator (Rad Source) with 1.126 Gy min−1, and monitored irradiated mice daily. We used the biological equivalent dose (BED) to compare different radiation fractionation regimens as previously described20. Briefly, BED = nd[1 + d/(α/β)], where n = number of fractions; d = radiation dose/fraction; α/β = 10. We harvested mouse tongues at time points specified in the main text, fixed tissues in 10% formalin, embedded them in paraffin, and cut 5 μm sections. We analyzed histological changes and measure ulcers using H&E stained slides.
Human samples
The First Affiliated Hospital of Kunming Medical University, China provided de-identified archived human tissue paraffin sections and approved the study as an exempt for human subjects. Oral mucositis lesions were from the tongue, buccal or oropharyngeal mucosa adjacent to recurrent oral cancers that had undergone radiotherapy. Non-irradiated oral mucosa sections were from surgically removed sleep apnea oral tissues and a tongue biopsy adjacent to a cyst (mucocele).
Immunofluorescence (IF), immunohistochemistry (IHC), and TUNEL assay for apoptosis
We performed IF and IHC as previously described49. Primary antibodies used were guinea pig antibody to K14 (1:400, Fitzgerald, 20R-CP200), rat antibody to CD4 (1:20, BD Bioscience, 550278), Ly-6G (1:20, BD Bioscience, 550291), BM8 (antibody to F4/80, 1:20, Invitrogen, MF48000), FITC-labeled antibody to BrdU (BD Bioscience, 347583), rat antibody to CD45 (1:50, BD Bioscience, 550539) for mouse samples, mouse antibody to CD45 (1:50, Abcam, Ab781) for human samples, chicken antibody to TGF-β1 (1:50, R&D, AF-101-NA), rabbit antibody to CtBP1 (1:100, Millipore, 07-306), rabbit antibody to NF-κB p50 (1:200, Santa Cruz Biotechnology, SC-7178), rabbit antibody to PCNA (1:200, Santa Cruz Biotechnology, SC-7907), rabbit antibody to pSmad2 (1:100, Cell Signaling Technology, 3101), and mouse antibody to V5 (1:500, Invitrogen, 460705). For IF, secondary antibodies to different species IgG were Alexa Fluor® 594 (red) or 488 (green) conjugated (1:200 for all, Invitrogen). For IHC, we used secondary biotinylated antibodies to different species IgG (1:300, Vector Labs) and developed using Vectastain ABC kit (Vector Labs). We used a Terminal deoxynucleotidyl transferase uridine nick end-labeling (TUNEL, G3250) kit (Promega) on formalin fixed tissue sections to detect apoptotic cells. We performed in vivo BrdU labeling by i.p. injection of 0.125 mg g−1 BrdU 1 hour prior to euthanization. We quantified PCNA or BrdU as cells mm−1 epithelial length including all epithelial cells, TUNEL or CD45-positive cells as cells mm−1 epithelial length including all epithelial layers and stroma above the muscle layer, nuclear pSmad2 or NF-κB p50 positive cells as the number of positive cells/existing total remaining epithelial cells (i.e., excluding sloughed epithelial cells induced by irradiation). Consecutive fields of slides were used to count BrdU-labeled cells using MetaMorph software.
Cell culture
We prepared Smad7 transgenic and wildtype primary keratinocytes from neonatal mouse skin as previously described49, and cultured them in PCT medium (CELLnTEC). We cultured spontaneously immortalized normal oral keratinocytes (NOK-SI) derived from gingival tissues of healthy volunteers and maintained in defined keratinocyte medium25. We also cultured oral cancer cells Cal27 (ATCC) and MSK921 (D. Raben’s lab, finger printed by University of Colorado Cancer Center Tissue Culture Core) in Dulbecco Modified Eagle Medium supplemented with 10% fetal bovine serum (GIBCO; Invitrogen). To assess the effect of Tat-Smad7 in irradiated cells, we cultured the above human cell lines in chamber slides (BD Bioscience, 354108), irradiated them with 3 Gy and added Tat-Smad7 (1μg ml−1) to the culture medium immediately after irradiation. Cells were fixed in 100% cold methanol 4 hours after Tat-Smad7 treatment for immunostaining of pSmad2, NF-κB p50 and V5.
Transfection with siRNA
When cultured keratinocytes reached 70% confluency, 100nM of target siRNA or scrambled siRNA (Dharmacon) was transfected using Lipofectamine® 2000 (Invitrogen). Cells were harvested 48~72 hours after transfection and subjected to western analyses to determine knockdown efficiency. For migration assays, siRNA was transfected when cells were plated. Target siRNAs included in this study are: mouse siRac1-1 (Invitrogen, MSS237708) and siRac1-2 (IDT, MMC.RNAI.N009007.12.3); human siSmad2 (Dharmacon, L-003561-00-0005), siSmad3 (Invitrogen, HSS106252), and siSmad4 (Invitrogen, HSS118066); human siCtBP1-150 and siCtBP1-2 51; human siSmad7-1 and siSmad7-252; human TGF-β1 (Dharmacon, J-012562-08-0005); mouse siSmad719.
Generation and characterization of Tat-Smad7 and Tat-Cre proteins
We constructed human Smad7 cDNA with a 5′-Tat-tag sequence encoding RKKRRQRRR and a 3′ V5-tag sequence, and cloned the construct into the pGEX-6p-1 protein expression vector (New England Biolabs) to make a GST-Tat-Smad7 fusion protein. We transformed Tat-Smad7 into BL-21 Star™ E. coli (Invitrogen) to produce Tat-Smad7 protein, followed by glutathione column purification and elution using enzymatic cleavage from the GST fusion (Precision enzyme, GE Life Sciences). Tat-Smad7 protein solution (in PBS) was diluted using culture medium to final concentrations of 0.1–2 μg ml−1 to assess protein transduction and data from 1 μg ml−1 Tat-Smad7 is presented. The same volume of PBS used in Tat-Smad7 stock solution was added to culture medium as vehicle control. We constructed Tat-Cre with the same 5′ Tat-tag and 3′ V5-tag as in Tat-Smad7 and cloned it into the pET101-Topo protein expression vector (Invitrogen) that contains a sequence encoding C-terminal 6XHis. We transformed Tat-Cre into BL-21 Star™ E. coli (Invitrogen) to produce Tat-Cre protein and purified it with Ni-NTA column. We verified the purity and size of both proteins using SDS-PAGE electrophoresis. To evaluate transduction and activity of Tat-Smad7 protein in vitro, we added Tat-Smad7 to primary mouse keratinocytes. We fixed slides in cold methanol for 5 minutes and stained for V5 and pSmad2. We verified Tat-Cre activity by digesting a 1,460 bp floxed fragment from the 7,650 bp vector pLL3.7 (Addgene). For in vivo treatments, we used 30 μl 50% glycerol/PBS as a vehicle control and Tat-Cre as a non-irrelevant protein control. We topically applied Tat-Smad7 or Tat-Cre (in 30 μl 50% glycerol/PBS, doses and regimens are specified in each figure) to mouse oral cavity and restricted mice from oral intake for 1 hour. We included i.p. injection of Palifermin in our studies with doses and regimens specified in the Results section. We i.p. injected normal saline as a vehicle control for Palifermin. We harvested samples on day 9 for prevention studies and on day 10 or day 14 for treatment studies. For oral treatments in non-irradiated mice, we used the same amount of Tat-Smad7 or vehicle control, and oral tissues were excised 12 hours after treatment.
In vitro keratinocyte proliferation assay
We determined in vitro keratinocyte proliferation by BrdU incorporation in wildtype and Smad7 transgenic keratinocytes. We transfected 70% confluent cells with Rac1 siRNAs, and changed to regular culture medium 24 hours later. We used an in situ cell proliferation kit (Roche Applied Science) to perform in vitro BrdU labeling and detection, and used MetaMorph software to count BrdU-labeled cells.
In vitro cell migration assays
When cells reached 100% confluency, we treated them with mitomycin C (Sigma) at 10 μg ml−1 for 2 hours to inhibit cell proliferation and introduced scratch wound with Fisherbrand pipet tip. We photographed cell migration daily. We performed migration assays when cells reached confluency after 24 to 36 hours of siRNA transfection, and used Image-J software to document cell migration as the wound area occupied with migrating cells. For Tat-Smad7 treatment, we exposed cells to Tat-Smad7 protein at 1 μg ml−1 or vehicle control (PBS) in medium after wound scratch, and changed medium every other day with freshly added Tat-Smad7 until migrating cells fully covered scratched wound.
Cell survival assay
We performed cell survival assays as previously described53 with slight modifications. Briefly, we plated cells in 12-well plates at 500 cells well−1 for non-irradiated wells, and increased up to 1,500 cells well−1 along with increased radiation doses. We irradiated cells 24 hours after they were plated. We added Tat-Smad7 at 1 μg ml−1 or the same volume of PBS used to dissolve Tat-Smad7 (control) to culture medium of irradiated and non-irradiated cells. We changed the medium every other day with freshly added Tat-Smad7 or PBS for 10 to14 days. We fixed colonies in methanol, stained them in 0.5% crystal violet solution (containing 25% methanol), counted and averaged them from 4 wells in each experiment. We performed two to three separate experiments for each cell line. We calculated the relative surviving fraction as previously described53, i.e., the absolute surviving fraction (colony numbers/total plated cells) under each radiation dose divided by the absolute surviving fraction of non-irradiated cells.
RNA extraction and analyses
We isolated total RNA using Trizol (Invitrogen) and performed quantitative (q) RT-PCR using Taqman® Assays-on-Demand™ probes (Applied Biosystems) as previously described54.
Western analysis
We performed protein extraction and western analyses as previously described54. The antibodies used in this study included rabbit antibody to Smad755 (1:500), rabbit antibodies to Smad2 (1:300, Zymed, 51-1300) and Smad4 (1:300, Epitomics, 1676-1), rabbit antibody to Smad3 (1:300, Cell Signaling Technology, 9513), mouse antibody to Rac1 (1:500, BD Biosciences, 610651), rabbit antibody to CtBP1 (1:500, Millipore, 07-306), mouse antibody to tubulin (1:3000, Sigma, T5168), mouse antibody to GAPDH (1:5000, Abcam, Ab8245) and goat antibody to actin (1:1000, Santa Cruz Biotechnology, SC1616). Gray-scale images were obtained using the Odyssey® v.1.2 software (LI-COR Biosciences).
Rac1 activation assay
We examined active GTP-bound Rac1 using a Biochem™ Kit for Rac1 activation (Cytoskeleton Inc, BK035). We cultured wildtype and Smad7 transgenic keratinocytes in 15 cm diameter tissue culture plates and prepared protein lysates using the provided lysis buffer. We used 1 mg of cell lysate to assay Rac1 activity and 50 μg of lysate to examine total Rac1 and Smad7 proteins. To measure GTP-bound Rac1 in mouse tongues, we ground half of the tongue to a powder in liquid nitrogen and lysed it with lysis buffer to extract protein, assayed GTP-bound Rac1 in 2 mg of protein lysate per sample and loaded 50 μg of protein lysate for total Rac1 protein western blot.
ChIP assays
We performed ChIP assays using the ChIP-IT express kit (Active Motive, 53009) as previously described31,56,57. We isolated DNA-protein complex from primary mouse keratinocytes. For ChIP, we incubated 6.3 μg sheared chromatin with protein-G magnetic beads and 2 μg each of rabbit antibodies to Smad2 (Cell Signaling Technology, 3122), Smad3 (Cell Signaling Technology, 9523), Smad4 (Cell Signaling Technology, 9515), Smad7 antibody (Santa Cruz Biotechnology, SC-11392), CtBP1 (Millipore) or a negative control rabbit IgG (Santa Cruz Biotechnology, SC-2027). We used eluted DNA from the protein-DNA complex for PCR analyses, and compared CtBP1 binding to the Rac1 promoter in wildtype and Smad7 transgenic keratinocytes by ChIP band intensities on gel images or by quantitative PCR using Power SYBR Green Master Mix (Applied Biosystems). Primers used to amplify the Rac1 SBE-1.5 Kb promoter regions:
5′-TGGAATTCCTGGTCTGGTTT-3′ (sense)
5′-GCCAAGCTGCTCTTCCAGTA-3′ (antisense)
5′-TCTCAGGGGGCCAAAGGTGTT-3′ (sense)
5′-TCCCAGCACCTGAATCACATGG-3′ (antisense)
Rac1 promoter luciferase reporter construct, site-directed mutagenesis and luciferase assay
We amplified the 883 bp fragment of −1671 bp to −789 bp of the Rac1 promoter, encompassing the SBE-1.5 Kb site, from wildtype mouse DNA using 5′ XhoI and 3′ HindIII tagged primers, and cloned this Rac1 promoter fragment into pGL4.26 vector (Promega) to make the Rac1 promoter-pGL4.26 luciferase reporter (Rac1-Luc) construct. For site-directed mutagenesis, we mutated the SBE sequence 5′-TGTCTGTGCT-3′ to 5′-TGATAGAGCT-3′. We co-transfected Rac1-Luc and pGL4.74 (1:20) with Smad7 siRNA19, CtBP1 siRNA or scrambled siRNA using Lipofectamine 2000 (Invitrogen) to primary mouse keratinocytes, or treat primary mouse keratinocytes with Tat-Smad7 treatment (1μg ml−1). We collected cell lysates and performed luciferase assays 48 hours after transfection or Tat-Smad7 treatment, using the Dual-Luciferase® Reporter Assay kit (Promega) following manufacturer’s instructions. We measured Rac1-luciferase activity with the Glomax machine (Promega) and expressed by the ratio of firefly activity to Renilla activity. Primers used for amplification of Rac1 promoter sequence were:
5′-ATCCTCGAG-TATCCTCCAGGTCTGGG-3′
5′-GCCAAGCTT-AGCGTCCAGCGTTAACCTG-3′
Statistical analysis
We analyzed statistical differences in molecular analyses and oral mucositis ulcer size using the Student’s t test and presented all data by mean ± s.d. except ulcer size, which were presented by mean ± s.e.m. We analyzed oral mucositis incidences by Fisher’s exact test.
Supplementary Material
Acknowledgments
This work was supported by US NIH grants AR061792 and DE015953 to XJW, R01CA115468 to QZ, R03DA033982 to RZ and QZ, Colorado State/University of Colorado Bioscience Discovery Evaluation Grant to XJW, QHZ and YR, and partially supported by P30CA046934 to University of Colorado Cancer Center. LB was supported by a grant (No. 81060189) from The National Natural Science Foundation of China (NSFC). FL was supported by grants (No. 81102596) from the National Science Foundation of China (NSFC), The Shanghai Rising-Star Program (NO.12QA1403300), and The Innovation Program of Shanghai Municipal Education Commission (NO.12YZ067). The authors thank S. Said, C. Marshall and C. Liu for searching human oral mucositis clinical samples, C.Y. Li and G. Gang for their input on radiotherapy in oral cancer patients. The authors also thank P. Garl for proof reading the manuscript.
Footnotes
Conflicts of Interest Statement: The authors have declared that no conflict of interest exists.
Author contributions
GH, QZ, XJW designed experiments and wrote the manuscript. DR provided input in radiotherapy dosing and regimens in oral cancer treatment and clinical care of radiation-induced oral mucositis and the MSK921 cell line. GH, LB, FL and DW performed the radiation-induced oral mucositis animal experiments and other histopathological/molecular analyses at the University of Colorado Denver, AMC. AC, AS, and JBM performed radiation-induced oral mucositis animal experiments at the NIH. YD, GB, and QZ performed protein purification. RZ, YR, and QZ contributed to optimization of Tat-Smad7 protein production. SG provided NOK-SI cells. PtD provided Smad7 antibody. LB and JBL provided human samples and performed immunostaining on those samples. DR, JBM, SG provided suggestions for manuscript revisions.
References
- 1.Sonis ST. Efficacy of palifermin (keratinocyte growth factor-1) in the amelioration of oral mucositis. Core evidence. 2010;4:199–205. doi: 10.2147/ce.s5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vagliano L, et al. Incidence and severity of oral mucositis in patients undergoing haematopoietic SCT--results of a multicentre study. Bone marrow transplantation. 2011;46:727–732. doi: 10.1038/bmt.2010.184. [DOI] [PubMed] [Google Scholar]
- 3.Scully C, Sonis S, Diz PD. Oral mucositis. Oral diseases. 2006;12:229–241. doi: 10.1111/j.1601-0825.2006.01258.x. [DOI] [PubMed] [Google Scholar]
- 4.Wu JC, Beale KK, Ma JD. Evaluation of current and upcoming therapies in oral mucositis prevention. Future Oncol. 2010;6:1751–1770. doi: 10.2217/fon.10.133. [DOI] [PubMed] [Google Scholar]
- 5.Le QT, et al. Palifermin reduces severe mucositis in definitive chemoradiotherapy of locally advanced head and neck cancer: a randomized, placebo-controlled study. J Clin Oncol. 2011;29:2808–2814. doi: 10.1200/JCO.2010.32.4095. [DOI] [PubMed] [Google Scholar]
- 6.Henke M, et al. Palifermin decreases severe oral mucositis of patients undergoing postoperative radiochemotherapy for head and neck cancer: a randomized, placebo-controlled trial. J Clin Oncol. 2011;29:2815–2820. doi: 10.1200/JCO.2010.32.4103. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, et al. R-Spondin1 protects mice from chemotherapy or radiation-induced oral mucositis through the canonical Wnt/beta-catenin pathway. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2331–2336. doi: 10.1073/pnas.0805159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu HG, et al. Therapeutic effect of recombinant human epidermal growth factor (RhEGF) on mucositis in patients undergoing radiotherapy, with or without chemotherapy, for head and neck cancer: a double-blind placebo-controlled prospective phase 2 multi-institutional clinical trial. Cancer. 2009;115:3699–3708. doi: 10.1002/cncr.24414. [DOI] [PubMed] [Google Scholar]
- 9.Weigelt C, Haas R, Kobbe G. Pharmacokinetic evaluation of palifermin for mucosal protection from chemotherapy and radiation. Expert opinion on drug metabolism & toxicology. 2011;7:505–515. doi: 10.1517/17425255.2011.566556. [DOI] [PubMed] [Google Scholar]
- 10.Haddad R, et al. Randomized phase 2 study of concomitant chemoradiotherapy using weekly carboplatin/paclitaxel with or without daily subcutaneous amifostine in patients with locally advanced head and neck cancer. Cancer. 2009;115:4514–4523. doi: 10.1002/cncr.24525. [DOI] [PubMed] [Google Scholar]
- 11.Cotrim AP, et al. Pharmacological protection from radiation +/− cisplatin-induced oral mucositis. International journal of radiation oncology, biology, physics. 2012;83:1284–1290. doi: 10.1016/j.ijrobp.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iglesias-Bartolome R, et al. mTOR Inhibition Prevents Epithelial Stem Cell Senescence and Protects from Radiation-Induced Mucositis. Cell stem cell. 2012;11:401–414. doi: 10.1016/j.stem.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fistarol SK, Itin PH. Anti-inflammatory treatment. Curr Probl Dermatol. 2011;40:58–70. doi: 10.1159/000321051. [DOI] [PubMed] [Google Scholar]
- 14.Elad S, et al. Topical immunomodulators for management of oral mucosal conditions, a systematic review; part I: calcineurin inhibitors. Expert Opin Emerg Drugs. 2010;15:713–726. doi: 10.1517/14728214.2010.528389. [DOI] [PubMed] [Google Scholar]
- 15.Elad S, et al. Topical immunomodulators for management of oral mucosal conditions, a systematic review; Part II: miscellaneous agents. Expert Opin Emerg Drugs. 2011;16:183–202. doi: 10.1517/14728214.2011.528390. [DOI] [PubMed] [Google Scholar]
- 16.O’Neill ID. Off-label use of biologicals in the management of inflammatory oral mucosal disease. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2008;37:575–581. doi: 10.1111/j.1600-0714.2008.00693.x. [DOI] [PubMed] [Google Scholar]
- 17.He W, et al. Overexpression of Smad7 results in severe pathological alterations in multiple epithelial tissues. EMBO J. 2002;21:2580–2590. doi: 10.1093/emboj/21.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong S, et al. Smad7 binds to the adaptors TAB2 and TAB3 to block recruitment of the kinase TAK1 to the adaptor TRAF2. Nat Immunol. 2007;8:504–513. doi: 10.1038/ni1451. [DOI] [PubMed] [Google Scholar]
- 19.Han G, et al. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Fowler JF, Harari PM, Leborgne F, Leborgne JH. Acute radiation reactions in oral and pharyngeal mucosa: tolerable levels in altered fractionation schedules. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2003;69:161–168. doi: 10.1016/s0167-8140(03)00231-7. [DOI] [PubMed] [Google Scholar]
- 21.Shull MM, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkarni AB, et al. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci U S A. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu SL, et al. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Research. 2004;64:4405–4410. doi: 10.1158/0008-5472.CAN-04-1032. [DOI] [PubMed] [Google Scholar]
- 24.Yan X, Chen YG. Smad7: not only a regulator, but also a cross-talk mediator of TGF-beta signalling. Biochem J. 2011;434:1–10. doi: 10.1042/BJ20101827. [DOI] [PubMed] [Google Scholar]
- 25.Castilho RM, et al. Rac1 is required for epithelial stem cell function during dermal and oral mucosal wound healing but not for tissue homeostasis in mice. PloS one. 2010;5:e10503. doi: 10.1371/journal.pone.0010503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch RM, et al. Incisional wound healing in transforming growth factor-beta1 null mice. Wound Repair Regen. 2000;8:179–191. doi: 10.1046/j.1524-475x.2000.00179.x. [DOI] [PubMed] [Google Scholar]
- 27.Ashcroft GS, Roberts AB. Loss of Smad3 modulates wound healing. Cytokine Growth Factor Rev. 2000;11:125–131. doi: 10.1016/s1359-6101(99)00036-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 30.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes & Development. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 31.Hoot KE, et al. HGF upregulation contributes to angiogenesis in mice with keratinocyte-specific Smad2 deletion. J Clin Invest. 2010;120:3606–3616. doi: 10.1172/JCI43304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks H, Lebleu B, Vives E. Tat peptide-mediated cellular delivery: back to basics. Advanced drug delivery reviews. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Cardarelli F, Serresi M, Bizzarri R, Beltram F. Tuning the transport properties of HIV-1 Tat arginine-rich motif in living cells. Traffic. 2008;9:528–539. doi: 10.1111/j.1600-0854.2007.00696.x. [DOI] [PubMed] [Google Scholar]
- 34.Kalvala A, et al. Enhancement of gene targeting in human cells by intranuclear permeation of the Saccharomyces cerevisiae Rad52 protein. Nucleic acids research. 2010;38:e149. doi: 10.1093/nar/gkq486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorr W, Reichel S, Spekl K. Effects of keratinocyte growth factor (palifermin) administration protocols on oral mucositis (mouse) induced by fractionated irradiation. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2005;75:99–105. doi: 10.1016/j.radonc.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Dorr W, Heider K, Spekl K. Reduction of oral mucositis by palifermin (rHuKGF): dose-effect of rHuKGF. International journal of radiation biology. 2005;81:557–565. doi: 10.1080/09553000500196136. [DOI] [PubMed] [Google Scholar]
- 37.Singh B, et al. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001;61:4506–4513. [PubMed] [Google Scholar]
- 38.Qiu W, Schonleben F, Li X, Su GH. Disruption of transforming growth factor beta-Smad signaling pathway in head and neck squamous cell carcinoma as evidenced by mutations of SMAD2 and SMAD4. Cancer Lett. 2007;245:163–170. doi: 10.1016/j.canlet.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ten Dijke P, Goumans MJ, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. J Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- 41.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 42.Pasparakis M, et al. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature. 2002;417:861–866. doi: 10.1038/nature00820. [DOI] [PubMed] [Google Scholar]
- 43.Maier P, Veldwijk MR, Wenz F. Radioprotective gene therapy. Expert opinion on biological therapy. 2011;11:1135–1151. doi: 10.1517/14712598.2011.580271. [DOI] [PubMed] [Google Scholar]
- 44.Bornstein S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu SL, et al. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes & Development. 2006;20:1331–1342. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nature reviews Drug discovery. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. British journal of pharmacology. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng C, et al. Prevention of radiation-induced oral mucositis after adenoviral vector-mediated transfer of the keratinocyte growth factor cDNA to mouse submandibular glands. Clin Cancer Res. 2009;15:4641–4648. doi: 10.1158/1078-0432.CCR-09-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han G, Li F, Ten Dijke P, Wang XJ. Temporal smad7 transgene induction in mouse epidermis accelerates skin wound healing. Am J Pathol. 2011;179:1768–1779. doi: 10.1016/j.ajpath.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 51.Bergman LM, Birts CN, Darley M, Gabrielli B, Blaydes JP. CtBPs promote cell survival through the maintenance of mitotic fidelity. Mol Cell Biol. 2009;29:4539–4551. doi: 10.1128/MCB.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong S, Lee C, Kim SJ. Smad7 sensitizes tumor necrosis factor induced apoptosis through the inhibition of antiapoptotic gene expression by suppressing activation of the nuclear factor-kappaB pathway. Cancer Res. 2007;67:9577–9583. doi: 10.1158/0008-5472.CAN-07-1179. [DOI] [PubMed] [Google Scholar]
- 53.Munshi A, Hobbs M, Meyn RE. Clonogenic cell survival assay. Methods in molecular medicine. 2005;110:21–28. doi: 10.1385/1-59259-869-2:021. [DOI] [PubMed] [Google Scholar]
- 54.Li AG, Lu SL, Zhang MX, Deng C, Wang XJ. Smad3 knockout mice exhibit a resistance to skin chemical carcinogenesis. Cancer Res. 2004;64:7836–7845. doi: 10.1158/0008-5472.CAN-04-1331. [DOI] [PubMed] [Google Scholar]
- 55.He W, et al. Overexpression of Smad7 results in severe pathological alterations in multiple epithelial tissues. Embo J. 2002;21:2580–2590. doi: 10.1093/emboj/21.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoot KE, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118:2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owens P, et al. Smad4-dependent desmoglein-4 expression contributes to hair follicle integrity. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.