Abstract
IFN-γ engenders strong anti-proliferative responses, in part through activation of p53. However, the long-known IFN-γ-dependent upregulation of human Trp-tRNA synthetase (TrpRS), a cytoplasmic enzyme that activates tryptophan to form Trp-AMP in the first step of protein synthesis, is unexplained. Here we report a nuclear complex of TrpRS with the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) and with poly (ADP-ribose) polymerase 1 (PARP-1), the major PARP in human cells. The IFN-γ-dependent poly (ADP-ribosyl)ation of DNA-PKcs (which activates its kinase function) and concomitant activation of p53 were specifically prevented by Trp-SA, an analog of Trp-AMP that disrupted the TrpRS/DNA-PKcs/PARP-1 complex. The connection of TrpRS to p53 signaling in vivo was confirmed in a vertebrate system. These and further results suggest a surprising evolutionary expansion of the protein synthesis apparatus to a nuclear role that links major signaling pathways.
Aminoacyl-tRNA synthetases (aaRSs) are universal ancient proteins that establish the rules of the genetic code through specific aminoacylation of tRNAs, which constitutes the first step of translation1. The 20 enzymes (one for each amino acid) are divided into two classes (I and II) according to the architecture of the active site for synthesis of aminoacyl-AMP (AA-AMP), which is transferred to the 3’-end of tRNA 2,3. We were especially interested in the long-standing observation that the class I human TrpRS is highly induced by IFN-γ 4–6. IFN-γ is associated with strong anti-proliferative7 and anti-angiogenic effects8 and the activation of p53 through phosphorylation of Ser159. In addition to TrpRS, IFN-γ stimulates production of other angiostatic factors like MIG10 and IP1011. While the synthetase is mainly located in the cytoplasm, under IFN-γ stimulation TrpRS is secreted and its embedded anti-angiogenic function is activated by removal of an appended N-terminal domain (by proteolytic processing or alternative splicing)12,13. The activated form of TrpRS binds to VE-cadherin on the surface of endothelial cells and inhibits the formation of cadherin-mediated endothelial cell-cell junctions that are critical for vasculature development14.
In addition to these considerations, we were interested by the observation that eukaryotic tRNA synthetases have progressively (in evolution) added domains that are often not associated with the canonical aminoacylation function15. The WHEP domain is a common appended motif found in a number of human tRNA synthetases including TrpRS, HisRS, GlyRS, MetRS and the bifunctional Glu-ProRS15. The three WHEP domains in Glu-ProRS were found to mediate protein-protein and protein-RNA interactions that regulate translation of genes associated with the inflammatory response16. Separately, recent work showed that human LysRS is phosphorylated in stimulated mast cells17. Phosphorylation releases LysRS from the multi-tRNA-synthetase complex in the cytoplasm and redirects the protein to the nucleus, where it interacts with the microphthalmia-associated transcription factor (MITF) to activate transcription of genes that control the immune response. These observations show that at least some tRNA synthetases, long thought to only play a role in translation in cytoplasm, have distinct ex-tranational nuclear functions. Hence, it remained a subject of investigations to determine whether these functions originated from the need to coordinate these nuclear signaling pathways with translation in the cytoplasm. In this aspect, we were interested in the previously reported nuclear presence of TrpRS18,19 which was suggestive of a possible novel nuclear function for TrpRS.
Here we found that, upon IFN-γ stimulation, nuclear TrpRS increased in concentration and appeared in a complex where it served as a bridge between the catalytic subunit of DNA-PKcs and PARP-1. The subsequent cascade of biochemical events that emanated from this ternary complex led to activation of p53. Importantly, a novel domain, which is dispensable for aminoacylation, was added to the N-terminus of TrpRS at the time of vertebrates and was found to be essential for enabling TrpRS to link IFN-γ and p53 signaling.
RESULTS
Nuclear partners of TrpRS
As IFN-γ is known to upregulate TrpRS expression4–6, we started our investigation by analyzing the effect of IFN-γ stimulation on TrpRS nuclear localization. Under normal conditions, little TrpRS could be detected in the nuclear fraction of 3B11 cells (Supplementary Fig. 1a). However, upon IFN-γ treatment, along with its overexpression in the cytoplasm, TrpRS was easily detected in the nucleus of variety of cell lines (Supplementary Fig. 1b). This observation demonstrated that IFN-γ-mediated upregulation and translocation of TrpRS into the nucleus was not specific to a particular cell line. Interestingly, plasmid-based transient overexpression of TrpRS alone also led to its nuclear localization (Supplementary Fig.1c). Confocal microscopic study also demonstrated the nuclear localization of TrpRS during IFN-γ stimulation (Supplementary Fig. 1d) or by ectopic overexpression (Supplementary Fig. 1e). In addition to other possible effects from IFN-γ stimulation, this observation suggested that detection of nuclear localization occurred because of increased expression of TrpRS and the possibility of a nuclear localization signal (NLS) in the primary sequence. A multiple sequence alignment revealed four potential bipartite NLS elements near the C-terminus, sequentially designated as M1 to M4, respectively (Supplementary Fig. 1f). Subsequent mutational analysis confirmed that human TrpRS contains a bipartite NLS 448RRKEVTDEIVKEFMTPRK465 (M3 and M4) that was responsible for its nuclear import (Supplementary Fig. 1g).
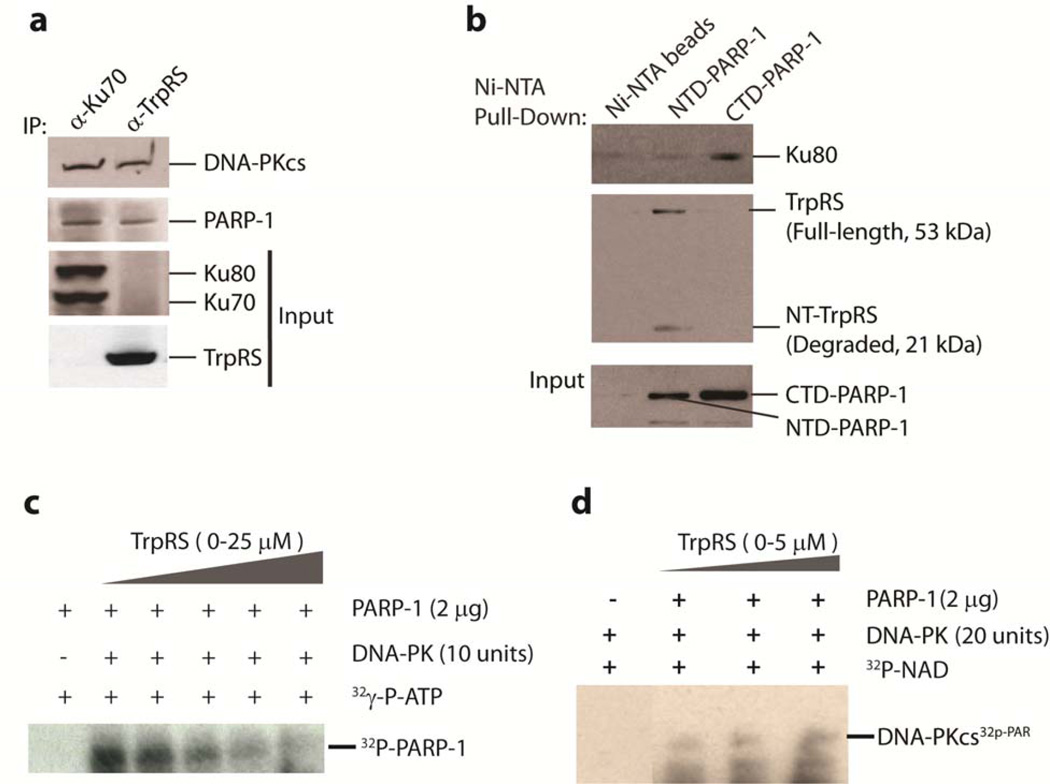
Having identified the NLS of TrpRS, we attempted to identify potential nuclear partners for TrpRS. The nuclear fraction from HeLa cells that were treated with IFN-γ was isolated and a co-immunoprecipitation was performed using polyclonal antibodies directed against TrpRS (α-TrpRS). A mass spectrometric identification of the immunoprecipitated proteins yielded DNA-PKcs (the catalytic subunit of DNA-PK (465 kD)) and PARP-1 (110 kDa) as major potential interacting partners (Fig. 1a). The identities of DNA-PKcs and PARP-1 and their interaction with TrpRS were also confirmed by reverse co-immunoprecipitation using a-DNA-PKcs and α-PARP-1 (Supplementary Fig. 2b).
Figure 1.
Identification of DNA-PKcs and PARP-1 as nuclear interacting partners of TrpRS. a. SDS-PAGE gel showing protein bands that were co-immunoprecipitated with α-TrpRS antibodies from HeLa cells treated with IFN-γ (lysates (WCL) or nuclear extracts (lane 2 to 7, duplicates)), or with IgG control (lane 1) (Full-length gels are presented in Supplementary Fig. 2a). b and c. Mapping of the interaction domain on TrpRS and on DNA-PK. b. A schematic representation of human TrpRS domain constructs is depicted at the top. (ES = Eukaryote-Specific, AD = Aminoacylation Domain, ABD = Anticodon Binding Domain, NLS = Nuclear Localization Signal) Ni-NTA pull-down experiment showing WHEP domain of TrpRS was responsible for the interaction with DNA-PKcs and PARP-1. DNA-PKcs (and PARP-1) was found to interact with full-length- and NT-TrpRS, and with the WHEP domain alone, but not with mini- and T2-TrpRS. c. Immunoprecipitation experiment showing the interaction of kinase domain of DNA-PKcs (KD-DNA-PKcs) with TrpRS. A schematic representation of DNA-PKcs domain constructs is depicted at the top. N-terminal (NT-DNA-PKcs) and the middle domain (MD-DNA-PKcs) regions of DNA-PKcs do not interact with TrpRS.
WHEP domain needed for TrpRS/DNA-PK/PARP-1 interaction
As a class I tRNA synthetase, TrpRSs from all organisms contain the conserved Rossmann-fold catalytic and anticodon binding domains20,21. In addition, human TrpRS has a vertebrate-specific helix-turn-helix WHEP domain (∼60 aa) appended at the N-terminus15. The WHEP domain is completely disordered in the co-crystal structure of the human TrpRS/tRNA complex21, consistent with its dispensable nature for aminoacylation20. The flexible conformation of the WHEP domain suggested that it may be important for interactions with other partners. To test if the WHEP domain was involved with the interaction with PARP-1 and DNA-PKcs, we used two natural variants lacking the WHEP domain. One was mini-TrpRS (aa 48–471) and the other was T2-TrpRS (aa 94–471). The former is produced by alternative splicing22 and the latter by natural proteolysis12. Two additional constructs were the N-terminal fragment NT-TrpRS (aa 1–186), which covers the entire eukaryote-specific region of TrpRS20,21, and the WHEP domain by itself (aa 1–60). Ni-NTA pull-down experiments with these His-tagged proteins showed that neither mini- nor T2-TrpRS interacted with PARP-1 or DNA-PKcs. In contrast, both NT-TrpRS and the WHEP domain interacted with PARP-1 and DNA-PKcs (Fig. 1b). Thus, the WHEP domain of TrpRS was responsible and sufficient for mediating the interaction with DNA-PKcs and PARP-1.
To understand the determinants on DNA-PKcs that interact with TrpRS, three different constructs of the 4128 amino acid DNA-PKcs were also created. These constructs were based largely on predicted domain boundaries in the sequence23. Only the fragment containing the C-terminal kinase domain of DNA-PKcs (V5-KD-DNA-PKcs) interacted with TrpRS (Fig. 1c). Neither the N-terminal (aa 1–1025, NT-DNA-PKcs) nor the middle (aa 1000–2184, MD-DNA-PKcs) portions of DNA-PKcs interacted with TrpRS (Fig. 1c), thus suggesting that the C-terminal kinase region of DNA-PKcs was responsible for the interaction.
Although DNA-PKcs and PARP-1 are substrates for each other24, interestingly, purified DNA-PKcs and PARP-1 do not interact in vitro25. Hence, their interaction is believed to be mediated through Ku70/80, the co-factor of DNA-PKcs in DNA repair which specifically binds to damaged DNA26. However Ku70/Ku80 was not seen in our mass spectrometric analysis. These observations led us to consider TrpRS as an alternative functional bridge such that a DNA-PKcs/TrpRS/PARP-1 ternary complex may form independent of DNA damage. (In experiments described later, we established that DNA-PKcs and PARP-1 formed a ternary complex, and not just separate binary complexes, with TrpRS.) To explore this idea, we prepared HeLa cell lysates and first confirmed that endogenous DNA-PKcs, PARP-1, and Ku70/Ku80 could be co-immunoprecipitated with antibodies (α-Ku70) directed against Ku70 (Fig. 2a). While the DNA-PKcs/Ku70/80/PARP-1 complex was readily detected, it contained no endogenous TrpRS. Conversely, immunoprecipitation with α-TrpRS antibodies pulled down DNA-PKcs and PARP-1, as expected, but revealed no detectable Ku70/80 (Fig. 2a). These experiments confirmed our mass spectrometric analysis showing the absence of Ku70 and Ku80 from the TrpRS/DNA-PKcs/PARP-1 complex and suggested that the binding of TrpRS and Ku70/80 to DNA-PKcs and PARP-1 are mutually exclusive.
Figure 2.
TrpRS facilitates the poly(ADP-ribosy)lation of DNA-PKcs. a. Mutually exclusive binding of TrpRS and Ku70/80 to DNA-PKcs and PARP-1. Co-IP experiments were performed with α-TrpRS or α-Ku70 antibodies incubated with HeLa cell lysates. The proteins that were bound with the antibodies were separated by SDS-PAGE and immunoblotted with α-DNA-PKcs, α-PARP-1, α-Ku70/80 and α-TrpRS. TrpRS did not co-immunoprecipitate with Ku70/80 and vice versab. Ni-NTA pull-down experiment showing domain-specific interaction of PARP-1 with TrpRS and with Ku70/80. TrpRS interacted with the N-terminal (NTD-PARP-1) and Ku70/80 interacted with the C-terminal (CTD-PARP-1) domains of PARP-1. c. TrpRS downregulated DNA-PK-dependent phosphorylation of PARP-1 in vitro in a concentration-dependent manner. DNA-PK (10 units) was incubated with a series of TrpRS concentrations for 30 min in protein kinase assay. The phosphorylated [32P]PARP-1 was separated from free γ-[32P]ATP by SDS-PAGE and visualized by exposure to x-ray film. (Full-length blots are presented in Supplementary Fig. 2d).dIn vitro demonstration of the ability of TrpRS to facilitate the PARylation of DNA-PKcs. Purified DNA-PK (purchased from Promega corporation) and purified recombinant PARP-1 were mixed with 32p-NAD and incubated with an increasing concentration of TrpRS (upto 5 µM) in the reaction mixture. 32p-PARylated DNA-PKcs was detected after running an SDS-PAGE and using x-ray film (Full-length blots are presented in Supplementary Figure 2e).
Distinct Domains of PARP-1 Interact with TrpRS and Ku70/80
The mutual exclusive nature of the two complexes, along with the observation that both TrpRS (Fig. 1c) and Ku70/8027 interact with the C-terminal region harboring the kinase domain of DNA-PKcs, prompted us to further characterize the domain-specific interactions of PARP-1 with TrpRS and Ku70/80. N-terminal (aa 1–372, NTD-PARP-1) and C-terminal (catalytic) domains of PARP-1 (aa 661–1014, CTD-PARP-1) were each cloned with a His-tag and expressed in E. coli. Mixing the bacterial extracts with HeLa cell lysates, Ni-NTA pull-down experiments showed distinct interactions of Ku70/80 and of TrpRS with PARP-1--Ku70/80 interacted with the catalytic domain of PARP-1 (CTD-PARP-1) and TrpRS interacted with the N-terminal region (NTD-PARP-1) (Fig. 2b). Interestingly, an N-terminal WHEP-domain-containing degradation fragment of TrpRS was also found to be co-eluted with NTD-PARP-1 (Fig. 2b). This observation is consistent with the conclusion that the N-terminal WHEP domain was responsible for mediating the interaction with DNA-PKcs and PARP-1. Therefore, the potential ternary complex of DNA-PKcs/TrpRS/PARP-1 was formed through the WHEP domain of TrpRS bridging the kinase domain of DNA-PKcs to the N-terminal domain of PARP-1..
Although purified DNA-PKcs and PARP-1 do not interact in vitro25, DNA-PK is known to phosphorylate PARP-124, in a reaction possibly facilitated by Ku70/80 bridging PARP-1 to the kinase domain of DNA-PKcs27. Because we found that Ku70/80 interacts with the C-terminal catalytic domain of PARP-1, we wondered if CTD-PARP-1 contained the phosphorylation site for DNA-PK. To test this idea, we carried out an in vitro DNA-PK (containing Ku70/80 and ds DNA) kinase assay using the different domains PARP-1. As expected, only CTD-PARP-1 (not NTD-PARP-1) was phosphorylated (Supplementary Fig. 2c). Therefore, through its interaction with Ku70/80 in the presence of dsDNA , DNA-PK phosphorylated PARP-1 at its catalytic CTD. This result is consistent with the earlier report that the major phosphorylation sites for DNA-PK are at the C-terminal domain of PARP-124. On the other hand, TrpRS appeared to bridge the N-terminal region of PARP-1 to the kinase domain of DNA-PKcs. Thus, Ku70/80 and TrpRS differ in the way they orient PARP-1 with respect to DNA-PKcs.
Effect of TrpRS on modifications of PARP-1 and DNA-PKcs
Once phosphorylated by DNA-PKcs, PARP-1 cannot poly(ADP-ribosyl)ate (PARylate) DNA-PKcs24. Because of the inverse relationship of the phosphorylation status of PARP-1 and its ability to PARylate DNA-PKcs, we were interested to see if TrpRS can influence the DNA-PKcs-dependent phosphorylation of PARP-1 and, conversely, the PARP-1-dependent PARylation of DNA-PKcs. For this purpose, we performed in vitro kinase and PARylation assays. In the kinase assay, PARP-1 was the substrate for DNA-PKcs and, in the PARylation assay, DNA-PKcs was the substrate for PARP-1. As expected, TrpRS inhibited the DNA-PK mediated phosphorylation of PARP-1 (Fig. 2c) and facilitated the PARylation of DNA-PKcs (Fig. 2d).
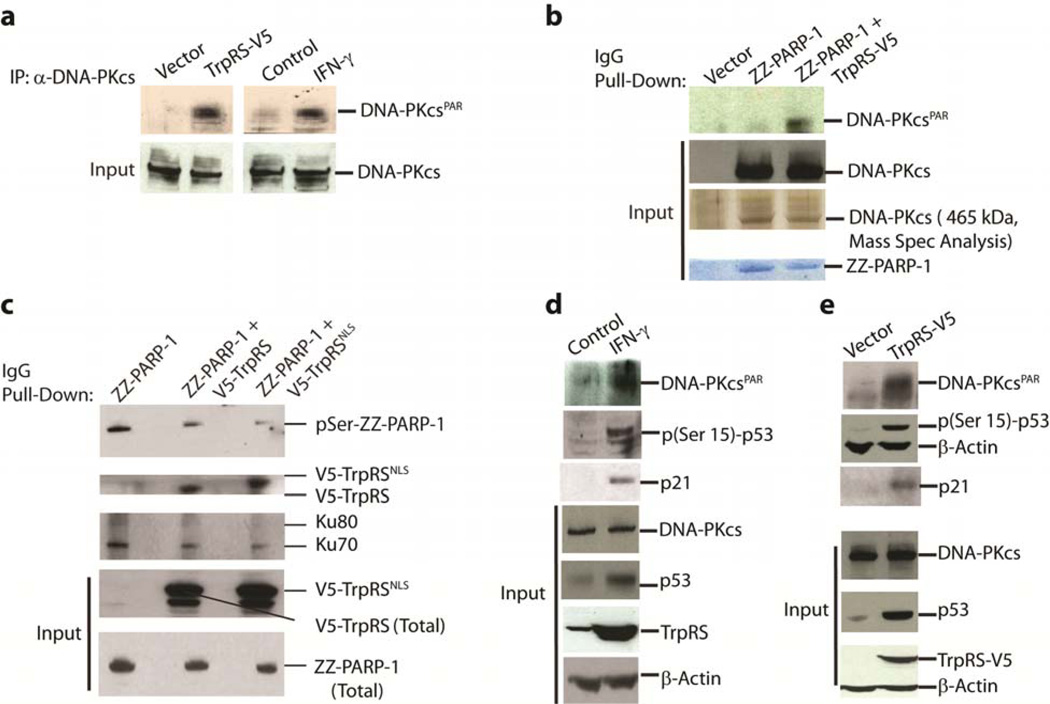
Having demonstrated in vitro that TrpRS facilitates the PARylation of DNA-PKcs (DNA-PKcsPAR) and inhibits the phosphorylation of PARP-1, we wanted to see if similar effects occurred in vivo when TrpRS was overexpressed in the absence of IFN-γ stimulation. For this purpose, TrpRS-V5 was ectopically overexpressed in HeLa cells and this overexpression resulted in substantial PARylation of DNA-PKcs (Fig. 3a, left panel). Consistently, IFN-γ stimulation to upregulate endogenous TrpRS also resulted in a strong PARylation of DNA-PKcs (Fig. 3a, right panel). Furthermore, in an IgG pull-down experiment using ZZ-PARP-1 (a fusion gene of PARP-1 having an IgG binding domain of Protein A (ZZ) at the N-terminus), with TrpRS-V5 ectopically expressed, PARP-1-associated-DNA-PKcs was also PARylated (DNA-PKcsPAR). In contrast, without overexpression of TrpRS-V5, PARP-1 associated DNA-PKcs (through Ku70/80) was mostly unmodified (Fig. 3b). (The identity of DNA-PKcs on these gels was confirmed by mass spectrometry.)
Figure 3.
TrpRS overexpression via transfection or IFN-γ stimulation activated DNA-PKcs and p53 by promoting DNA-PKcs PARylation. a,. Overexpression of TrpRS by transient transfection (left panel) or IFN-γ stimulation (right panel) led to the PARylation of DNA-PKcs (immunoblotting). b. Bridging of DNA-PKcs with PARP-1 by TrpRS led to the PARylation of DNA-PKcs. Only the ZZ-PARP-1 pulled down from samples co-transfected with TrpRS-V5 contained the PARylated DNA-PKcs (upper 2 panels). The identity of the specific band as DNA-PKcs was confirmed by mass spectrometry. (Full-length blots/gels are presented in Supplementary Fig. 3). c. TrpRS downregulated the auto-phosphorylation of PARP-1. Upper panel shows the phosphorylation status (pSer) of the immunoprecipitated ZZ-PARP-1 from HeLa cells transfected with plasmids encoding either ZZ-PARP-1 alone or co-transfected with V5-TrpRS and V5-TrpRSNLS (an NLS (KKKRKV) was inserted into the C-terminus of V5-TrpRS). Displacement of Ku70/80 (2nd panel from top) by overexpressed V5-TrpRS was demonstrated by the increased association of V5-TrpRSNLS with the immunoprecipitated ZZ-PARP-1 (3rd panel). d. IFN-γ stimulated over-production of TrpRS also led to the activation of p53 and production of the downstream marker p21. Levels of total and poly(ADP)ribosylated DNA-PKcs (DNA-PKcsPAR), of phosphorylated p53, and of p21 were probed by immunoblotting with specific antibodies. eIn vivo overexpression of TrpRS in HeLa cells led to the activation of p53. Levels of total and poly(ADP)ribosylated DNA-PKcs (DNA-PKcsPAR), of total and phosphorylated p53, and of p21 were probed by immunoblotting.
Similarly, to demonstrate the influence of TrpRS overexpression on the in vivo phosphorylation of PARP-1, ZZ-PARP-1 was co-transfected into HeLa cells with genes encoding either V5-tagged TrpRS (TrpRS- V5) or V5-tagged TrpRS with an additional nuclear localization sequence appended to the C-terminus (designated as TrpRSNLS-V5). (The rationale for using TrpRSNLS-V5 was that the enhanced nuclear localization of TrpRS, in the absence of IFN-γ stimulation, should result in a greater response than that achieved with wild-type TrpRS alone.) ZZ-PARP-1 from all three samples was pulled down using IgG and blotted with anti-phospho-serine/threonine antibodies (DNA-PKcs is a serine/threonine protein kinase). Diminished phosphorylation of ZZ-PARP-1 was seen when V5-TrpRS was co-expressed, and this diminution was even greater with TrpRSNLS-V5 (Fig. 3c). Concomitantly, co-expression of TrpRS-V5 also diminished the binding of Ku70/80 to ZZ-PARP-1. This diminution was even more efficient with TrpRSNLS-V5 (Fig 3c). These data support the idea that, when TrpRS displaces Ku70/80 and functionally bridges PARP-1 to DNA-PKcs, it facilitates PARylation of DNA-PKcs and down-regulates the phosphorylation of PARP-1.
TrpRS facilitates phosphorylation of p53
As PARylation of DNA-PKcs (DNA-PKcsPAR) is known to stimulate its kinase activity on p53 (pSer15-p53)25, we tested for p53 phosphorylation (Ser15) in vivo under conditions where TrpRS was overexpresssed. p53 is a well-known substrate for DNA-PK both in vitro28 and in vivo29 and phosphorylation of p53 at Ser15 by DNA-PK (or other kinases (e.g., ATM) with similar specificity) is required for its stabilization and activation30. As expected, IFN-γ stimulation to upregulate endogenous TrpRS resulted in strong PARylation of DNA-PKcs with concomitant phosphorylation (Ser15) of p53 (Fig. 3d). Similarly, we found that ectopic overexpression of TrpRS, in the absence of IFN-γ stimulation, also resulted in activation of DNA-PKcs by PARP-1-mediated PARylation and the concomitant phosphorylation of p53 at Ser15 (Fig. 3e). In both instances (Fig. 3d and e), the upregulation of expression of p21 from a downstream target gene of pSer15-p53 was also seen. These results are consistent with the previous work25 where in vitro that PARylation of DNA-PKcs stimulates its kinase activity on p53. As the phosphorylated form of PARP-1 is known to be pro-angiogenic31 and activated p53 is known to be anti-angiogenic32, these results are also consistent with the anti-angiogenic role of TrpRS when secreted.
Importantly, while the WHEP domain was sufficient and necessary for the bridging of homodimeric TrpRS to PARP-1 and DNA-PKcs (Fig. 1b), overexpression of the WHEP domain alone did not result in PARylation of DNA-PKcs, even though a complex formed between the WHEP domain and DNA-PKcs (Supplementary. Fig. 4a), and even though some of the WHEP domain could enter the nucleus (presumably due to its small size (Supplementary Fig. 4b)). These results suggested that the rest of TrpRS (which encodes the active site for Trp-AMP synthesis and harbors the NLS) has a role in making a functional complex. To show more clearly that DNA-PKcs and PARP-1 form a ternary complex with TrpRS (as opposed to separate binary complexes), experiments were designed to establish that a ‘pulldown’ of KD-DNA-PKcs simultaneously captured NTD-PARP-1 and dimeric TrpRS and, conversely, a ‘pulldown’ of NTD-PARP-1 simultaneously captured KD-DNA-PKcs and dimeric TrpRS (Supplementary Fig. 4c). In contrast, we could not detect a ternary complex with the WHEP domain or with a rationally designed TrpRS monomer (Supplementary Fig. 4d). These results are consistent with a model where DNA-PKcs binds to one and PARP-1 to the other WHEP domain of dimeric TrpRS.
To understand better the role of the WHEP domain in conjunction with the active site, we re-examined our previously obtained crystal structure of human TrpRS20. The structure captured the homodimeric TrpRS in a state of half-site reactivity where the reaction intermediate Trp-AMP occupied one active site, while the other active site was empty. Interestingly, when the active site is bound with Trp-AMP, the WHEP domain (blue) (Fig. 4a) is resolved in the crystal structure and folds back towards the active site as a cap while, at the same time, the WHEP domain of the other subunit, which lacks bound Trp-AMP, is disordered along with other residues with which it interacts. As the disposition of the WHEP domain may affect its availability for protein-protein interactions and is determined by the occupancy of the active site, we hypothesized that a bound ligand such as Trp-AMP could negatively affect the interaction of TrpRS with DNA-PKcs and PARP-1.
Figure 4.
Occupancy of TrpRS active site determined the WHEP domain conformation and dictated the DNA-PK/TrpRS/PARP-1 complex formation and the associated activities. a. Open (left) and closed (right) conformations of human TrpRS controlled by the binding of Trp-AMP. The human TrpRS structure was solved as a homodimer in the asymmetric unit (PDB 1R6T) . One subunit in the asymmetric unit has a bound Trp-AMP in the active site (closed conformation) while the active site of the other subunit is empty (open conformation). The WHEP domain (blue), part of the eukaryote-specific patch (purple, Y83-Y100), and the loop harboring the ‘KMSAS’ motif (green) are resolved only when Trp-AMP is bound. b and c. Dose-dependent inhibition of Trp-SA on PARylation of DNA-PKcs and phosphorylation of p53 that was induced by TrpRS overexpression by IFN-γ stimulation (b) or by transient transfection (c). d. Specificity of Trp-SA in inhibiting PARylation of DNA-PKcs and phosphorylation of p53 under IFN-γ stimulation. (Full-length blots are presented in Supplementary Fig. 5a and b) e. Effect of Trp-SA (a Trp-AMP analog) on the interaction of TrpRS with DNA-PKcs and PARP-1. Co-immunoprecipitation experiments with α-TrpRS and α-Ku70, in the presence and absence of Trp-SA (50 µM), revealed that Trp-SA abolished the interaction of TrpRS with DNA-PKcs and PARP-1 and promoted association of DNA-PKcs and PARP-1 with Ku70/80. The observation is consistent with the idea that binding of Ku70/80 and TrpRS are mutually exclusive.
To test this idea, we used a non-hydrolyzable analog of Trp-AMP designated as Trp-SA (5’-O-[N-(9L-tryptophanyl) sulfamoyl] adenosine), and investigated the effect of Trp-SA on DNA-PK and p53 activation. We added various amount of Trp-SA into media containing cultured cells that were overexpressing either endogenous TrpRS (by IFN-γ stimulation) (Fig. 4b) or exogenous TrpRS-V5 (Fig. 4c). Strikingly, Trp-SA suppressed the PARylation of DNA-PKcs and the activation of p53 in a dose-dependent manner (Fig. 4 b and c). The treatment with Trp-SA (for 2 hours) did not affect expression of TrpRS, DNA-PKcs or β-actin (Fig. 4 b and c), or the nuclear localization of TrpRS. Thus, while the active site of TrpRS is essential for protein synthesis, the partial inhibition of TrpRS activity by Trp-SA was not sufficient to reduce overall protein synthesis within a short period of time. (This result is consistent with other work showing that cell and even animal survival can be supported by reduced levels of tRNA synthetase activity33.) Nevertheless, to rule out the possibility that the effects of Trp-SA on DNA-PK PARylation and p53 were caused by inhibition of protein synthesis, a non-hydrolyzable adenylate analog of glycyl-AMP (Gly-SA) was used as a control. Gly-SA (25 uM) did not suppress DNA-PK PARylation and did not diminish p53 phosphorylation, thus suggesting the effect of Trp-SA was specific to TrpRS and independent of any general effect on protein synthesis (Fig. 4d). To further confirm this result, we used puromycin at a concentration (25 uM) known to inhibit protein synthesis in HeLa cells34. Puromycin, like Gly-SA, did not diminish the phosphorylation of p53, which occurred on overexpression of TrpRS (Supplementary Fig. 5c). Addition of Trp-SA to whole cells, followed by preparation of lysates and immunoprecipitations, showed that Trp-SA acted by disrupting the interaction of TrpRS with DNA-PKcs and PARP-1 (Fig. 4e).
In further experiments, and in an attempt to link the production of p(Ser15)-p53 to an activity dependent on DNA-PKcs, we used the DNA-PKcs-specific inhibitor KU57788 (Axon MedChem, Netherlands) that is reported to have a >1000-fold specificity for DNA-PKcs over other kinases including ATM35. Overexpression of TrpRS in the presence of KU57788 sharply suppressed the phosphorylation of p53 (Supplementary Fig. 5c). While we cannot exclude the possibility that, in addition to DNA-PKcs, KU57788 inhibits an undefined kinase, the most straightforward interpretation is that DNA-PKcs has a role in the production of p(Ser15)-p53, when TrpRS was overexpressed.
Moreover, the rise in total p53 that accompanies overexpression of TrpRS was associated with a similar rise in p(Ser15)-p53. Because the rise in total p53 can be blocked by inhibitors directed at TrpRS or DNA-PKcs, which in turn block production of p(Ser15)-p53 (Supplementary Fig. 5c), we suggest that stabilization of p53 by phosphorylation explains at least in part the rise in total p53 that occurs upon overexpression of TrpRS. Finally, we compared the time course of PARylation of DNA-PKcs and of appearance of p(Ser15)-p53 upon overexpression of recombinant or endogenous TrpRS (using IFN-γ induction) (Supplementary Fig. 6). These data showed that, whether by overexpression of exogenous (Supplementary Fig. 6a) or of endogenous TrpRS (IFN-γ stimulation (Supplementary Fig. 6b)), DNA-PKcsPAR rises in a systematic way for the duration of the time course (28 hours), and in a way that followed the increase of TrpRS. In the case of total p53 and p(Ser15)-p53, their rises were more or less systematic and correlated to the levels of TrpRS for the first 20 hours.
Effect of TrpRS on p53 signaling in vivo in a vertebrate
The in vitro biochemical analysis and the cell-based assays showed the essential role of the WHEP domain for TrpRS-dependent p53 signaling. The WHEP domain was added to TrpRS in vertebrates at the time of fish15. The appearance of the new domains in tRNA synthetases correlates with their involvement in ex-translational biology in higher eukaryotes15. To assess the importance of this novel pathway at the animal level, we established an in vivo system in developing zebrafish (which is known to exhibit IFN signaling pathways36,37). In these experiments, we replaced IFN-γ upregulation of TrpRS by directly injecting synthetic mRNAs encoding TrpRS, mini-TrpRS, T2-TrpRS, and TrpRSNLS into wild-type and p53 mutant (p53M214K/M214K , designated as p53m/m) zebrafish embryos 38. These 4 constructs tested the effects of TrpRS and, if any, their dependence on the presence of the WHEP domain that is lacking in mini-TrpRS and T2-TrpRS (Fig. 5 and Supplementary Fig. 7). The phenotypic consequences and specific expressions of p53-dependent downstream genes (to monitor p53 activation) were then observed, within a few days, in the context of the whole organism.
Figure 5.
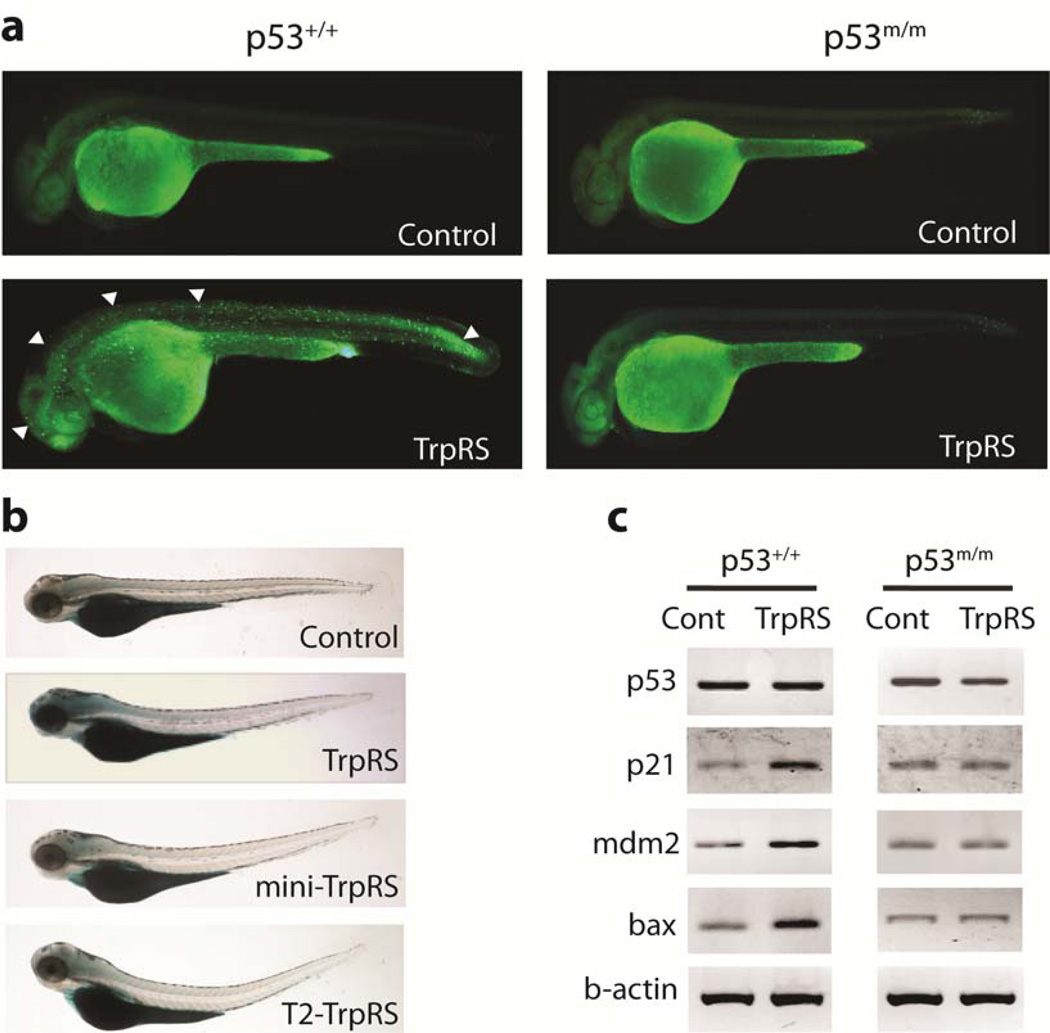
TrpRS mediated activation of p53 in zebrafish. a. Acridine orange (AO) staining of live zebrafish embryos at 30 hours post-fertilization (hpf). AO-positive fluorescent staining was observed in the brain, spinal cord, trunk, and tail of p53+/+embryos injected with TrpRS-encoding mRNA, but was not observed in mutant p53 (p53m/m) embryos. Representative regions of AO-positive cells are indicated by white arrowheads. b. Senescence-associated β galactosidase (SA-β-gal) staining of 3.5 days post-fertilization (dpf) embryos. Those embryos injected with TrpRS-encoding mRNA showed significant SA-β-gal induction in the brain and throughout the head, whereas mini-TrpRS and T2-TrpRS mRNAs as well as the control did not show any detectable changes. c. Genes involved in p53 downstream signaling pathway were analyzed by semi-quantitative RT-PCR in control embryos and in embryos injected with TrpRS-encoding mRNA.
In addition to uninjected control embryos, synthetic mRNA encoding GFP was introduced as an injection control. TrpRS-encoding-mRNA injected embryos displayed cell death in the developing brain and neural tube at 30 hours post-fertilization (hpf) (Fig. 5a). Out of 30 embryos injected with TrpRS-encoding mRNA, 26 gave positive results, i.e, ∼80% of the total embryos responded positively to the injection. Some of the fish embryos had severe abnormal phenotypes which were not counted as positive. In contrast, no abnormalities were observed in embryos injected with GFP-encoding mRNA. In addition, injection of mRNA encoding either mini- or T2-TrpRS did not induce detectable cell death. Significantly, TrpRS-induced cell death was suppressed by the p53 mutant (p53m/m) background (Fig. 5a) as well as by coinjection of a p53-directed morpholino. These results in the vertebrate model supported the cell-based observations that WHEP domain-containing TrpRS regulated p53-mediated cell death signaling.
To further study the activation of p53, we applied zebrafish embryos for staining with senescence-associated β galactosidase (SA-β gal), a marker that is upregulated by p53-induced cellular senescence in vivo39,40. As shown in Figure 5b, the staining was especially prominent in the head region of the developing embryos that were injected with TrpRS mRNA. In contrast, the intensity was greatly reduced in control embryos and in those injected with mini- or T2- TrpRS mRNA. These effects of injection of TrpRS RNA were significantly reduced in p53m/m zebrafish (Supplementary Fig. 7a).
We then determined the expression levels of zebrafish p53, p21WAF1/CIP1, mdm2, and bax, relative to β-actin as a control. Semi quantitative reverse transcriptase PCR (RT-PCR) was performed on RNA extracted from individual mRNA-injected embryos. When compared to wild-type and control-injected embryos, no obvious change of p53 mRNA levels was detected in embryos injected with TrpRS-encoding RNA (Fig. 5c). However, consistent with p53 activation, downstream targets of the p53 pathway, such as mdm2, p21WAF1/CIP1 and bax, were correspondingly upregulated in wild-type (normal) p53 embryos injected with TrpRS-encoding mRNA(Fig. 5c, left panels). More importantly, the upregulation of mdm2, p21WAF1/CIP, and bax were counteracted in the p53 mutant fish embryos (Fig. 5c, right panels). In further support of the role of TrpRS as a component of p53 signaling, TrpRSNLS (with an additional, appended NLS at the C-terminus to further enhance nuclear localization) was even more effective than TrpRS in inducing p53-dependent phenotypes (Supplementary Fig. 7b–d) as well as in upregulation of the target genes (Supplementary Fig. 7e). These results with single embryos were representative of similar analyses on additional embryos (wild-type, n = 4; TrpRS, n = 10; mini-TrpRS, n = 6; and T2-TrpRS, n = 6; TrpRSNLS, n = 10) and suggested that, in the presence of overexpressed TrpRS, the p53-driven apoptotic and senescence pathway is activated. This activation was apparent in the observed increases in acridine orange-positive or senescence-associated β galactosidase-positive cells in embryonic phenotypes 40,41.
Based on the above findings, a mutually exclusive binding model for TrpRS and Ku70/80 with DNA-PK and PARP-1 in the nucleus is proposed in Figure 6. In the presence of Trp-AMP, the WHEP domain caps the active site and Ku70/80 bridges between DNA-PK and PARP-1. However, in the presence of nuclear TrpRS and in the absence of Trp-AMP (as a result of IFN-γ stimulation), Ku70/80 is displaced as the ‘flipped out’ WHEP domain bridges the C-terminal kinase domain of DNA-PKcs to the N-domain of PARP-1. Consequently, PARylation of DNA-PKcs is stimulated, which in turn promotes the DNA-PKcs kinase activity for p53 phosphorylation and activation. Simultaneously, binding of TrpRS prevents Ku70/80 from binding to DNA-PKcs and PARP-1 and subsequent phosphorylation of PARP-1(Fig. 6).
Figure 6.
Cartoon illustration of the nuclear function of TrpRS. In the absence of nuclear TrpRS, Ku70/80 bridges between DNA-PK and PARP-1 and orients the C-domain of PARP-1 to be phosphorylated by DNA-PKcs. In the presence of nuclear TrpRS, Ku70/80 is displaced through the WHEP domain that bridges the C-terminal kinase domain of DNA-PKcs to the N-domain of PARP-1. Consequently, nuclear TrpRS stimulates PARsylation of DNA-PKcs and, in turn, the DNA-PKcs kinase activity that leads to p53 phosphorylation and activation. Because TrpRS excluded Ku70/80 from binding to DNA-PKcs and PARP-1, it also inhibited the Ku70/80-mediated PARP-1 phosphorylation by DNA-PKcs. The ability of TrpRS to bind to DNA-PKcs and PARP-1 was controlled by the occupancy of the active site. Only when the active site was non-occupied could the WHEP domain be opened and available for interaction with DNA-PKcs and PARP-1.
Discussion
In our work, we detected a complex of DNA-PKcs/Ku70/80/PARP-1, which formed in the presence of damaged DNA, and of DNA-PKcs/TrpRS/PARP-1. Further work showed that the two complexes were mutually exclusive (Fig 2a and b). Using different domains of PARP-1, we found that the CTD of PARP-1 interacts with the Ku70/80 bridging proteins and is phosphorylated by DNA-PKcs. In contrast, TrpRS bridged the N-terminal part of PARP-1 to the kinase domain of DNA-PKcs, so that Ku70/80 and TrpRS differed in the way they oriented PARP-1 with respect to DNA-PKcs. In this orientation, DNA-PKcs was PARylated by PARP-1. Thus, the two orientations of DNA-PKcs and PARP-1 had different functional consequences.
The novel orientation facilitated by TrpRS as a bridging protein is through the WHEP domains that are joined to the N-termini of the homodimeric TrpRS subunits, with one binding partner, e.g. DNA-PKcs, binding to one WHEP domain and the other partner, e.g. PARP-1, to the other WHEP domain. TrpRS is one of ten a class I tRNA synthetases, of which some (e.g., CysRS, IleRS, LeuRS, ValRS, ArgRS GlnRS, GluRS) have monomeric structures while others are dimers2. Although a complete active site is contained within each subunit of TrpRS, tRNATrp binds across the dimer interface, so that the dimer is required for aminoacylation activity21. Interestingly, the function of TrpRS as a bridging protein for DNA-PKcs and PARP-1 provides a second rationale for the homodimeric structure of the synthetase. This homodimeric quaternary structure is conserved for TrpRSs through evolution15. Thus, the addition of the WHEP domain to TrpRS at the stage of vertebrates exploited a pre-existing dimeric state that could facilitate a bridging function for the synthetase.
The phosphorylation and activation of p53 described here appeared to be implemented, at least in part, by the nuclear TrpRS interaction with DNA-PKcs and PARP-1. This activation— through phosphorylation of Ser15—is linked to the robust anti-angiogenic32 and antiproliferative functions of p5340–42. Thus, the role of nuclear TrpRS may cooperate with its extracellular role as an angiostatic procytokine12. In addition, the possible activation of a major tumor suppressor by nuclear TrpRS is suggestive of a broader role that goes beyond angiostasis and is consistent with well-documented functional integration of IFN signaling and p53 activation7,9,42.
Because overexpression of TrpRS, in the absence of IFN-γ stimulation, was associated with p53 activation (Fig. 3d), and because addition of the TrpRS-specific inhibitor Trp-SA, and not of another tRNA synthetase or protein synthesis inhibitor, prevented IFN-γ-stimulated activation of p53 (Fig. 4b), TrpRS appears to have a role in IFN-γ-induced p53 activation. Its apparent role as a component of this functional integration may be part of the reason that TrpRS is strongly upregulated upon IFN-γ stimulation. Also of possible relevance is the long-standing and largely unexplained observation that IFN-γ stimulation upregulates tryptophan-degrading indoleamine 2,3-dioxygenase (IDO)43,44. Our results suggest that nuclear TrpRS may link IFN-γ stimulation to p53 activation though emptying of the Trp-AMP binding pocket to thereby promote a ‘flipped out’ position of the novel WHEP domain that was added to TrpRSs at the stage of vertebrates (Fig. 4a–e). Consequently, simultaneous upregulation of IDO would promote activation of the nuclear role of TrpRS.
Interestingly, the work on LysRS 17 and the new results presented here on TrpRS show that at least some tRNA synthetases, long thought to only play a role in translation in cytoplasm, have distinct ex-translational nuclear functions. It remains a subject of future investigations to determine whether these functions originated from the need to coordinate these nuclear signaling pathways with translation in the cytoplasm.
METHODS
Cell culture, transfection and IFN-γ treatment
HeLa cells were cultured in a humidified incubator with 5% CO2 in DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Invitrogen) and 1x penicillin/streptomycin. The cells were transfected with pcDNA6-TrpRS-V5, pcDNA6-V5 or pcDNA3.1-ZZ-PARP-1 using Lipofectamine LTX (Invitrogen). To induce TrpRS expression by IFN-γ, the cells were incubated with human IFN-γ (R & D system, Minneapolis, MN) at a series of concentrations from 0 to 1000 units/ml for 8–28 hours prior to being used in experiments described.
Recombinant protein purification
DNA encoding either full-length TrpRS (aa 1–471), mini-TrpRS (aa 48–471), T2-TrpRS (aa 94–471), N-terminal fragment (aa 1–186) or WHEP domain (aa 1–60) was cloned into Nde-I/Hind-III sites of pET-20b vector (Novagen, Gibstown, NJ). The expressed proteins include a 6-His tag from the vector sequence. The full-length PARP-1 and its variants (N-terminal domain (NTD) and C-terminal domain (CTD)) as well as the kinase domain (KD) of DNA-PKcs were cloned into pET 20b vector. All constructs were sequenced (Next Generation Sequencing Core Facility, The Scripps Research Institute, La Jolla) to confirm the perfection of cloning. All proteins having a C-terminal His-tag were expressed in E. coli strain BL21 (DE3) by induction for 4 hr with 1 mM isopropyl β-D-thiogalactopyranoside. Proteins were purified from the supernatants of lysed cells using Ni-NTA agarose (Qiagen, Chatsworth, CA) column chromatography according to the manufacturer’s instruction. ZZ-PARP-1, with an appended IgG binding domain of Protein A at the N-terminus, was purified from the HeLa cells after a 48 hrs transfection by binding to IgG agarose beads. Immunoprecipitate was washed three times, subjected to SDS-PAGE and immunoblotted with PARP-1 antibody to confirm the identity of the protein purified.
In vitro DNA-PK protein kinase activity assay
DNA-PK protein kinase activity was measured as per the manufacturer’s assay protocol (Promega Corporation, Madison, WI). In short, 5 µl recombinant TrpRS (0–25 µM) and 5 µl PARP-1 (1.25mg/ml ) were incubated with 5 µl DNA-PK (10 units) at 4oC for 15 min, then incubated with 10 µl DNA-PK protein kinase assay cocktail (50 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 2 µg/ml calf thymus DNA, 0.2 mM cold ATP, 40 µg/ml BSA, and 1 µCi [32P]-γ-ATP (Perkin-Elmer, Boston, MA)) at 30°C for 30 min. DNA-PK dependent phosphorylation of NTD-PARP-1 and CTD-PARP-1 (2mg/ml) was conducted similarly as per the protocol where they were used as substrate (10 µl each) for kinase activity. The reaction was stopped by addition of 25µl 2x SDS sample buffer (5% SDS, 10 mM DTT) and heated at 95°C for 3 min. The [32P]-PARP-1 or [32P]-CTD-PARP-1 were separated from free γ-[32P]ATP on SDS-PAGE, dried and visualized by exposure to x-ray film.
In vitro PARylation assay
20 units of DNA-PK (Promega Corporation, Madison, WI) was mixed with 5 µl recombinant TrpRS (0–5 µM) and 5 µl recombinant PARP-1 (2µg ) at 4°C for 15 min, then incubated with 10 µl PAR assay cocktail (50 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 200 nM NAD, 40 µg/ml BSA, and 1 µCi [32P]-NAD (Perkin-Elmer, Boston, MA)) at 30°C for 30 min. The reaction was stopped by addition of 25µl 2x SDS sample buffer (5% SDS, 10 mM DTT) and heated at 95°C for 3 min. The [32P]-PARylated DNA-PKcs was separated from free [32P]-NAD on SDS-PAGE, dried and visualized by exposure to x-ray film.
Acridine orange staining and detection of cell death in zebrafish embryos
Live zebra-fish embryos were dechorionated in pronase (2.0 mg/ml in egg water, 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) for 3 to 5 min and rinsed five times in egg water at 28 hpf At 30 hpf, embryos were incubated in 10 µg/ml acridine orange (AO; Sigma A-6014) in egg water for 30 min at 28.5°C, followed by 3-time quick rinses embryos were anesthetized in 160 µg/ml tricaine (3-aminobenzoic acid ethyl ester, Sigma A-5040), and mounted laterally on the glass slide 41. All animal experiments were approved by and conducted in accordance with the guidelines established by the Institutional Animal Care and Use Committee (IACUC) at The Scripps Research Institute, IACUC approval number 09-0009.
Senescence-associated β-galactosidase (SA-β-gal) activity assay in zebrafish embryos
Zebrafish embryos at 3.5 dpf were washed in phosphate buffer saline (PBS) and fixed overnight in 4% paraformaldehyde in PBS. After fixation, samples were washed twice in PBS, pH 7.5, twice again in PBS, pH 6.0, and then incubated at 37 °C (in the absence of CO2) for 12–16 h with SA-β-gal staining solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2 in PBS at pH 6.0) 40.
Statistical Analysis
Data processing and statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) version 14.0. This software was used to perform statistical tests where appropriate.
More detailed description of the experiments is provided in Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Prof. David Chen, UT Southwestern Medical School, Dallas, TX for the DNA-PKcs clone and Prof. Paul Chang, Department of Biology, MIT, Cambridge, MA for the ZZ-PARP-1 clone. This work was supported by grant GM15539 from the National Institutes of Health and by a fellowship from the National Foundation for Cancer Research.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 2.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 3.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 4.Fleckner J, Rasmussen HH, Justesen J. Human interferon gamma potently induces the synthesis of a 55-kDa protein (gamma 2) highly homologous to rabbit peptide chain release factor and bovine tryptophanyl-tRNA synthetase. Proc. Natl. Acad. Sci. U S A. 1991;88:11520–11524. doi: 10.1073/pnas.88.24.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin BY, Anderson SL, Xing L, Powell RJ, Tate WP. Interferon induces tryptophanyl-tRNA synthetase expression in human fibroblasts. J. Biol. Chem. 1991;266:24245–24248. [PubMed] [Google Scholar]
- 6.Bange FC, Flohr T, Buwitt U, Bottger EC. An interferon-induced protein with release factor activity is a tryptophanyl-tRNA synthetase. FEBS Lett. 1992;300:162–166. doi: 10.1016/0014-5793(92)80187-l. [DOI] [PubMed] [Google Scholar]
- 7.Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 8.Lindner DJ. Interferons as antiangiogenic agents. Curr. Oncol. Rep. 2002;4:510–514. doi: 10.1007/s11912-002-0065-4. [DOI] [PubMed] [Google Scholar]
- 9.Kim KS, Kang KW, Seu YB, Baek SH, Kim JR. Interferon-gamma induces cellular senescence through p53-dependent DNA damage signaling in human endothelial cells. Mech. Ageing. Dev. 2009;130:179–188. doi: 10.1016/j.mad.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Sgadari C, et al. Mig, the monokine induced by interferon-gamma, promotes tumor necrosis in vivo. Blood. 1997;89:2635–2643. [PubMed] [Google Scholar]
- 11.Angiolillo AL, et al. Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J. Exp. Med. 1995;182:155–162. doi: 10.1084/jem.182.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakasugi K, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U S A. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor M, et al. Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J. Biol. Chem. 2008;283:2070–2077. doi: 10.1074/jbc.M706028200. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Q, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampath P, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 17.Yannay-Cohen N, et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell. 2009;34:603–611. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Popenko VI, et al. Immunoelectron microscopic location of tryptophanyl-tRNA synthetase in mammalian, prokaryotic and archaebacterial cells. Eur. J. Cell. Biol. 1993;62:248–258. [PubMed] [Google Scholar]
- 19.Popenko VI, Cherni NE, Beresten SF, Zargarova TA, Favorova OO. Immune electron microscope determination of the localization of tryptophanyl-tRNA-synthetase in bacteria and higher eukaryotes. Mol. Biol. (Mosk) 1989;23:1669–1681. [PubMed] [Google Scholar]
- 20.Yang XL, et al. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XL, et al. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO. J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Shue E, Ewalt KL, Schimmel P. A new gamma-interferon-inducible promoter and splice variants of an anti-angiogenic human tRNA synthetase. Nucleic Acids Res. 2004;32:719–727. doi: 10.1093/nar/gkh240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ariumi Y, et al. Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene. 1999;18:4616–4625. doi: 10.1038/sj.onc.1202823. [DOI] [PubMed] [Google Scholar]
- 25.Ruscetti T, et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 26.Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J. Cell Biol. 2007;197:183–186. doi: 10.1083/jcb.200705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J. Biol. Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 28.Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol. Cell. Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo RA, McLure KG, Lees-Miller SP, Rancourt DE, Lee PW. DNA-dependent protein kinase acts upstream of p53 in response to DNA damage. Nature. 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 30.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 31.Beckert S, et al. IGF-I-induced VEGF expression in HUVEC involves phosphorylation and inhibition of poly(ADP-ribose)polymerase. Biochem. Biophys. Res. Commun. 2006;341:67–72. doi: 10.1016/j.bbrc.2005.12.148. [DOI] [PubMed] [Google Scholar]
- 32.Teodoro JG, Parker AE, Zhu X, Green MR. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science. 2006;313:968–971. doi: 10.1126/science.1126391. [DOI] [PubMed] [Google Scholar]
- 33.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 34.Grueneberg DA, et al. Kinase requirements in human cells: IV. Differential kinase requirements in cervical and renal human tumor cell lines. Proc. Natl. Acad. Sci. U S A. 2008;105:16490–16495. doi: 10.1073/pnas.0806578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 36.Aggad D, et al. In vivo analysis of Ifn-gamma1 and Ifn-gamma2 signaling in zebrafish. J. Immunol. 2010;185:6774–6782. doi: 10.4049/jimmunol.1000549. [DOI] [PubMed] [Google Scholar]
- 37.Sieger D, Stein C, Neifer D, van der Sar AM, Leptin M. The role of gamma interferon in innate immunity in the zebrafish embryo. Dis. Model Mech. 2009;2:571–581. doi: 10.1242/dmm.003509. [DOI] [PubMed] [Google Scholar]
- 38.Berghmans S, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. U S A. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishi S, et al. The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet. 2008;4:e1000152. doi: 10.1371/journal.pgen.1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidi S, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.