Summary
RIP1 is a central mediator of cell death in cell stress, but can also mediate cell survival by activating NF-κB. Here, we show that RIP1 is a switch in EGFR signaling. EGFRvIII is an oncogenic mutant that does not bind ligand and is co-expressed with EGFRwt in glioblastoma (GBM). EGFRvIII recruits ubiquitin ligases to RIP1 resulting in K63-linked ubiquitination of RIP1. RIP1 binds to TAK1 and NEMO forming a EGFRvIII-RIP1 signalosome that activates NF-κB. RIP1 is essential for EGFRvIII-mediated oncogenicity and correlates with NF-κB activation in GBM. Surprisingly, activation of EGFRwt with EGF results in a novel negative regulation of EGFRvIII with dissociation of the EGFRvIII-RIP1 signalosome, loss of RIP1 ubiquitination, NF-κB activation, and association of RIP1 with FADD and Caspase-8. If EGFRwt is overexpressed with EGFRvIII, adding EGF leads to a RIP1 kinase dependent cell death. The EGFRwt-EGFRvIII-RIP1 interplay may regulate oncogenicity and vulnerability to targeted treatment in GBM.
Keywords: EGFRvIII, glioblastoma, EGFR wild type, NF-kappaB, RIP1, NEMO, TAK1, antagonism
Introduction
The receptor interacting protein (RIP, RIP1, RIPK1) is widely expressed and is a member of a kinase family that mediates cellular responses to inflammation and stress (Festjens et al., 2007; Meylan and Tschopp, 2005). RIP1 is also an important mediator of cell death (Weinlich et al., 2011). RIP1 plays a key role in apoptotic cell death induced by stimuli such as TNFα, DNA damage, or cIAP inhibitors (Bertrand et al., 2008; Petersen et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007). In addition, RIP1 plays a key role in necrotic forms of cell death triggered when Caspase activation is blocked (Feoktistova et al., 2011; Hitomi et al., 2008; Tenev et al., 2011). Necrotic cell death under these conditions requires the kinase activity of RIP1 and is inhibited by a specific inhibitor of RIP1 kinase, termed Necrostatin-1 (Degterev et al., 2008).
RIP1 is an essential component of signaling platforms involved in either NF-κB activation or cell death signaling (Bertrand and Vandenabeele, 2011). For example, RIP1 forms complexes with IKKγ (NEMO), TRADD and TAK1 to activate NF-κB in response to stressful stimuli. The core signaling complex involved in cell death signaling by RIP1 includes RIP1, FADD and Caspase-8 (Feoktistova et al., 2011; Tenev et al., 2011). K63-linked ubiquitination, may play a key role in determining the partners RIP1 associates with, and in determining whether RIP1 acts as a prosurvival adaptor protein or participates in cell death (Bertrand et al., 2008; Declercq et al., 2009; Ea et al., 2006; O'Donnell et al., 2012; O'Donnell et al., 2007; O'Donnell and Ting, 2012; Wang et al., 2008). The cIAPs and TRAF2 play an important role as ubiquitin ligases that mediate K63-linked ubiquitination of RIP1 (Bertrand et al., 2008). cIAP inhibitors/SMAC mimetics target cIAPs leading to a RIP1 dependent cell death. Thus, substantial evidence suggests that RIP1 plays a pivotal role in determining cell survival or death depending on the cellular context.
Aberrant activation of NF-κB is widespread in human cancer (Karin et al., 2002; Pacifico and Leonardi, 2006) including GBM (glioblastoma), and generally promotes survival of tumor cells. It has been suggested that two major pathways activate NF-κB in GBM (Bredel et al., 2011). Firstly, EGFR gene amplification and aberrant EGFR signaling are detected in 40-50% of GBMs. EGFR signaling is known to activate NF-κB (Habib et al., 2001; Sun and Carpenter, 1998; Yang et al., 2012). Secondly, NF-κB inhibitor-α (NFKBIA, which encodes lκBα) is commonly deleted in non-EGFR amplified GBMs, resulting in NF-κB activation (Bredel et al., 2011). A specific and oncogenic EGFR mutant (EGFR Type III, EGFRvIII, de2-7, ΔEGFR) can be detected in about one third of GBMs (Hatanpaa, 2010; Huang et al., 2009). EGFRvIII is generated from a deletion of exons 2-7 of the EGFR gene, which results in an in frame deletion of 267 amino acids from the receptor. EGFRvIII is unable to bind ligand and thought to signal constitutively.
EGFRvIII is usually co-expressed with EGFR wild type (wt) in GBM (Biernat et al., 2004; Ekstrand et al., 1991). The deletion of exons 2-7 in EGFRvIII occurs in EGFR gene amplified tumors (Biernat et al., 2004; Frederick et al., 2000), suggesting that the genetic abnormalities that lead to overexpression of EGFRwt and to expression of EGFRvIII co-exist in individual tumor cells. Immunohistochemical studies also suggest that the two receptors are expressed in the same tumor cells, although in some GBMs, EGFRvIII may be expressed in a more focal distribution compared to EGFRwt. However, even focally distributed EGFRvIII is expressed in cells with EGFRwt expression (Biernat et al., 2004; Nishikawa et al., 2004). A recent study has suggested that EGFRvIII activates NF-κB via mTORC2 (Tanaka et al., 2011). NF-κB plays an important role in EGFRvIII-mediated oncogenicity (Bonavia et al., 2012; Tanaka et al., 2011).
We previously showed that RIP1 is commonly expressed at high levels in GBM and confers a worse prognosis (Park et al., 2009). In this study, we propose an essential role for RIP1 in EGFRvIII mediated NF-κB activation. Silencing RIP1 inhibits EGFRvIII mediated NF-κB activation and oncogenicity in an orthotopic mouse model. Expression of EGFRvIII leads to recruitment of the ubiqitin ligases, TRAF2 and cIAP2, to RIP1 and to EGFRvIII resulting in K63-linked ubiquitination of RIP1 and NF-κB activation. EGFRvIII forms a signaling platform that incorporates RIP1, NEMO and TAK1. Both EGFRvIII and RIP1 are required for assembly of this signalosome. Importantly, we show that EGFRvIII mediated activation of NF-κB is negatively regulated by co-expressed EGFRwt. Thus, addition of EGF to cells which activates EGFRwt, results in a rapid loss of RIP1 ubiquitination and loss of its association with EGFRvIII, NEMO and TAK1 and a loss of EGFRvIII-mediated NF-κB activation. RIP1 now becomes associated with the death adaptor FADD in an EGF-dependent manner. When EGFRwt is overexpressed, as is common in GBM and other human cancers, EGF becomes a death signal and this cell death requires the kinase activity of RIP1. Thus, RIP1, previously implicated in cell stress and inflammatory signaling, is also a key life/death switch in a major RTK signaling system and turns a normally trophic signal into a death signal. These findings reveal a novel antagonistic interaction between EGFRwt and EGFRvIII and add a layer of complexity to our understanding of EGFRvIII and NF-κB activation in GBM.
Results
EGFRvIII activates NF-κB in glioma cell lines
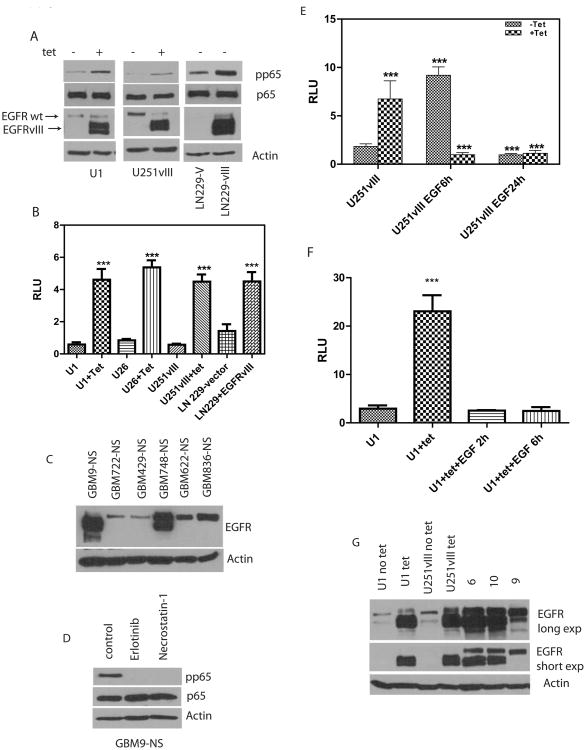
EGFRvIII is a tumor specific EGFR mutant not expressed in normal tissues or in established GBM cell lines. Stable expression of EGFRvIII in U87MG cells results in activation of NF-κB. (Bonavia et al., 2012; Tanaka et al., 2011). Since EGFRvIII does not bind ligand, we generated U87MG cell lines conditionally expressing EGFRvIII in response to tetracycline (Figure 1A) as a tool to examine regulated EGFRvIII signaling. Two tetracycline inducible clones were identified and named U1 and U26. EGFRvIII can be distinguished from EGFRwt on Western blots as a faster migrating band (Figure 1A). Consistent with previous studies, conditional expression of EGFRvIII in U1 and U26 cells leads to activation of NF-κB as determined by phosphorylation of the p65 subunit of NF-κB (Figure 1A and SFigure 1A). We also confirmed phosphorylation of p65 in U251-vIII cells (Figure 1A). U251vIII cells conditionally express EGFRvIII in response to tetracycline (Ramnarain et al., 2006). Additionally, constitutive EGFRvIII expression in LN229 cells also results in phosphorylation of phospho-p65 (Figure 1A) demonstrating that EGFRvIII activates NF-κB in multiple GBM cell lines. Additionally, we confirmed that EGFRvIII activated the transcriptional activity of NF-κB by reporter assays in the various cell lines using an NF-κB luciferase reporter as shown in Figure 1B. All the GBM cell lines used express endogenous EGFRwt.
Figure 1.
EGFRvIII activates NF-κB. A. Various cell lines with stable expression of EGFRvIII and endogenous EGFRwt. U1 cells are U87MG cells conditionally expressing EGFRvIII in response to tetracycline. U251vIII cells are U251MG cells conditionally expressing EGFRvIII in response to tetracycline. LN229-V cells are transfected with empty vector while LN229-vIII cells have stable and constitutive expression of EGFRvIII. Endogenous EGFRwt can be detected in cells as a slower migrating band above the EGFRvIII. Expression of EGFRvIII in various cell lines results in increased phosphorylation of the p65 subunit of NF-κB. B. Reporter assays show increased transcriptional activity of NF-κB using a luciferase assay. Conditional or constitutive expression of EGFRvIII results in increased NF-κB reporter activity (1way ANOVA, p<0.0001) (RLU: relative luciferase units). C. We screened a panel of early passage primary GBM cultures grown as neurospheres for presence of EGFRwt and EGFRvIII (NS: Neurosphere). D. GBM9-NS cells were treated with either Erlotinib (10μM) or Necrostatin (300nM) overnight followed by Western blot for phospho-p65. E. A reporter assay showing that the transcriptional activity of NF-κB in tetracycline treated U251vIII is abolished by exposure to EGF. Data were analyzed by 1way ANOVA followed by followed by Newman-Keuls multiple comparison test to validate the levels of significance in between groups and were statistically significant for all groups (p<0.05). F. Conditional expression of EGFRvIII in U1 cells results in activation of NF-κB reporter activity which is abolished by addition of EGF (50ng/ml) (P <0.00001, 1way ANOVA) G. EGFRvIII levels in U1 and U251vIII cells are comparable to EGFRvIII levels in resected GBM specimens (GBMs 6 and 10 have high levels of EGFRvIII).
In addition, we examined a panel of primary GBM stem like cultures grown as neurospheres for the presence of EGFRvIII (Figure 1C). We selected GBM9 neurosphere cultures (GBM9-NS) that retain substantial expression of EGFRvIII with a low level of EGFRwt and demonstrate phosphorylation of the p65 subunit of NF-κB. Importantly, inhibition of EGFR kinase activity by Erlotinib addition abolished phosphorylation of p65 in GBM9-NS, indicating that EGFRvIII is driving NF-κB activation in these cells (Fig 1D). Erlotinib also blocks NF-κB transcriptional activity in GBM9-NS (SFigure 1B). A dose response for the concentration of Erlotinib required to inhibit EGFR in U1 cells is shown in SFigure 1C-D. A second EGFR kinase inhibitor, AG1478, also blocked EGFR phosphorylation in U1 cells (SFigure 1E).
Next, we examined the effect of combined EGFRwt and EGFRvIII activation by exposing these cells to EGF. Unexpectedly, we found that exposure of cells to EGF leads to a complete loss of EGFRvIII-mediated NF-κB transcriptional activation in multiple cell lines (Figure 1E-F). Adding EGF to U251vIII cells not exposed to tetracycline (not expressing EGFRvIII) results in activation of NF-κB (Figure 1E). However, when EGFRvIII is expressed, ligand-mediated EGFRwt activation not only fails to activate NF-κB, but also abolishes EGFRvIII-mediated NF-κB activation. Similarly, Figure 1F and SFigure 1F-G show that addition of EGF abolishes EGFRvIII-mediated activation of NF-κB in U1 cells, U26 cells, and LN229-vIII cells. Electrophoretic mobility shift assays also demonstrate a loss of EGFRvIII-mediated NF-κB DNA binding in U1 cells in response to EGF (SFigure 2A). Thus, EGF-mediated activation of EGFRwt results in a complete reversal of EGFRvIII-mediated NF-κB activation in multiple cell lines. We examined the ability of EGF to abolish EGFRvIII-mediated NF-κB activation in U251vIII cells with silenced EGFRwt. We find that EGF did not abrogate EGFRvIII-mediated NF-κB activation in EGFRwt silenced cells (SFigure 2B-C). The levels of EGFRvIII expressed in our experimental system are similar to GBM tumors (Figure 1G).
Next, we examined whether co-expression of both EGFRwt and EGFRvIII can be detected in the same cells in primary GBM cultures. Primary neurosphere cultures from both GBM9-NS and 748-NS show co-expression of both EGFRwt and EGFRvIII (Figure 1C). We generated single cell suspensions from these tumors by limiting dilution and expanded single cell clonal populations. Multiple colonies derived from single cells were examined for expression of EGFRwt and EGFRvIII by Western blot. In all clonal populations from both tumors both EGFRvIII and EGFRwt are co-expressed (SFigure 3). Our data thus supports the evidence from previous studies that EGFRvIII is usually co-expressed with EGFRwt in the same tumor cells.
A role for RIP1 in EGFRvIII mediated NF-κB activation
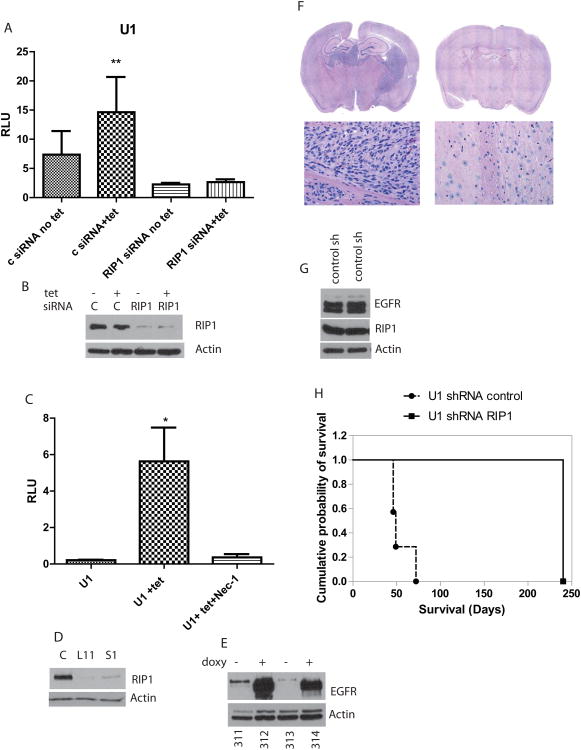
While it is well known that EGFRvIII activates NF-κB, the underlying mechanisms are not well understood. On the other hand, NF-κB activation by cytokines such as TNFα has been extensively characterized and involves RIP1. Furthermore, we previously reported that RIP1 forms a complex with EGFRwt when both are expressed at high levels although the significance or mechanism of this interaction was not investigated (Habib et al., 2001). We examined whether EGFRvIII uses a mechanism similar to TNFα to activate NF-κB. To examine a possible role for RIP1 in EGFRvIII signaling, we silenced RIP1 in U1 cells and U251-vIII cells. Silencing RIP1 inhibits EGFRvIII-induced NF-κB reporter activity (Figure 2A-B and SFigure 4A-B). EGFRvIII-mediated NF-κB activation is abolished by Necrostatin-1, a specific inhibitor of RIP1 kinase (Figure 2C, 1D and SFigure 4C). Thus, both Necrostatin-1 and Erlotinib inhibit EGFRvIII-mediated NF-κB activation. These findings suggest that RIP1 is required for EGFRvIII-mediated NF-κB activation. A necrostatin-1 dose response for the concentration of necrostatin-1 required to inhibit the RIP1 activity in U1 cells was determined by EGFRvIII-driven phosphorylation of phospho-p65 (SFigure 4D-E). RIP1 knockdown by siRNA also inhibits phosphorylation of p65 (SFigure 4F).
Figure 2.
A. RIP1 is essential for EGFRvIII mediated activation of NF-κB transcriptional activity in U1 cells. siRNA mediated knockdown of RIP1 blocks the ability of EGFRvIII to activate NF-κB. Addition of tetracycline results in expression of EGFRvIII in both RIP1 silenced and control (scrambled) siRNA control but NF-κB is activated only in control siRNA cells (1way ANOVA, p=0.0013). B. Silencing of RIP1 is shown by Western blot. C. EGFRvIII mediated transcriptional activity of NF-κB is blocked by Necrostatin-1 in U1 cells (p=0.017). D. Stable silencing of RIP1 in U1 cells using RIP1 shRNA. Two independent clones with efficient silencing of RIP1 L11 and S1 are shown along with RIP1 levels in control scrambled shRNA (C) expressing cells. E. Demonstrates that doxycycline penetrates into the intracranial tumors to induce expression of EGFRvIII. Protein lysates were made from U1 derived intracranial tumors in mice followed by Western blot with EGFR. Mice 312 and 314 were exposed to doxycycline in drinking water and food, and show expression of both EGFRvIII and EGFRwt while 311 and 313 were not exposed to doxycycline and demonstrate expression of endogenous EGFRwt only. F. H&E sections from a representative brain section from control shRNA group showing presence of brain tumor while the brain section from the L11 mouse is free of tumor. G. Expression of EGFRvIII and RIP1 in tumors from two mice from control shRNA group. Lysates were prepared directly from tumor tissue followed by Western blot. H. Kaplan-Meier survival analysis of RIP1 silenced cells (L11) compared to control shRNA expression implanted intracranially in mice (n=8). The log rank test was significant (X2(1) =15.47, p <.0001). All mice were exposed to doxycycline in food and water.
Silencing RIP1 inhibits EGFRvIII mediated oncogenicity
Next, we examined the biological consequences of silencing RIP1 in EGFRvIII expressing cells. We used shRNA to stably silence RIP1 in U1 cells using lentiviral shRNA directed against RIP1 or control shRNA followed by puromcyin selection. Stable clones with silenced RIP1 were identified (Figure 2D). We found that silencing RIP1 resulted in significantly decreased numbers of cells in culture compared to controls (SFigure 5A-C). Next, we tested U1 RIP silenced cells for growth in an orthotopic xenograft model of GBM in athymic nude mice. We first confirmed that EGFRvIII was conditionally expressed in U1 (control shRNA) cells in intracranial tumors in animals exposed to doxycycline in drinking water and feed (Figure 2E). Next, we injected cells with silenced RIP1 or control shRNA intracranially into groups of nude mice. Animals were sacrificed at the onset of neurological symptoms or after eight months. Kaplan-Meier survival analyses were computed to compare the two groups on time until the appearance of neurological symptoms (the event), where time was defined as number of days elapsed from injection to symptom appearance. The median number of days until the event for the shRIP1-L11 group was 240 days (100% censored data), as opposed to 57 days for control shRNA group (Figure 2H). The log rank test was significant (χ²(1) =15.47, p <.0001), indicating that shRIP-1-L11 mice remained symptom-free for a significantly longer period of time than the control mice, Surprisingly, no animals injected with the shRIP1 silenced cells formed tumors, while all of the animals with shControl cells formed tumors as shown in Figure 2F-H. Both RIP1 and EGFRvIII can be detected in tumors from U1control shRNA mice (Figure 2G). This experiment suggests that RIP1 is required for EGFRvIII mediated oncogenicity.
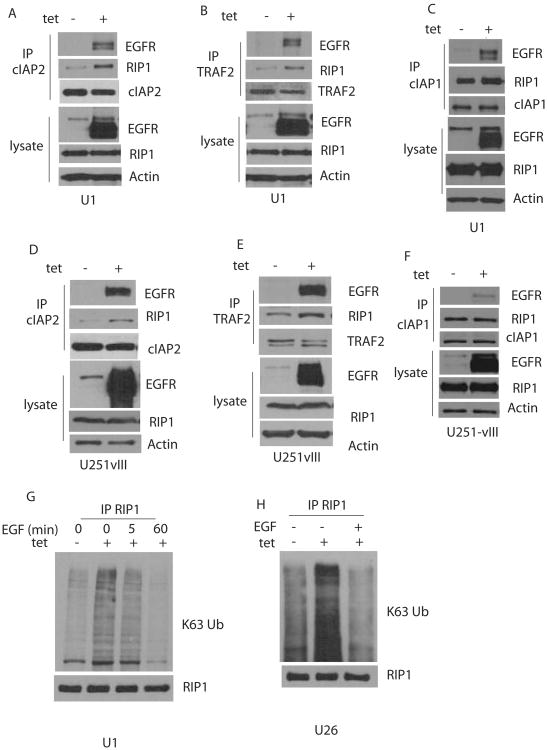
EGFRvIII recruits the ubiquitin ligases c-IAP1, c-IAP2 and TRAF2 to RIP1, resulting in K-63 linked polyubiquitination of RIP1
The ubiquitin ligases involved in K63 linked ubiquitination of RIP1 have been identified as c-IAP1, c-IAP2 and TRAF2. Thus, we tested whether conditional EGFRvIII expression leads to increased recruitment of ubiquitin ligases to RIP1. Figure 3A-C shows that expression of EGFRvIII in U1 cells results in increased associated of cIAP2 and TRAF2 with RIP1. cIAP1 is associated with RIP1 in these cells, but the cIAP1-RIP1 association does not increase with EGFRvIII expression. We also detected an increased recruitment of c-IAP1, cIAP2 and TRAF2 to EGFRvIII. As is discussed below, RIP1 is also recruited to EGFRvIII suggesting that RIP1 ubiquitination may occur in EGFRvIII associated complexes. A similar result is detected in U251vIII cells (Figure 3D-F). Expression of EGFRvIII results in increased K63-linked ubiquitination of RIP1 in U1 cells (Figure 3G). Importantly, the K63-linked ubiquitination of RIP1 is lost when EGF is added to cells, consistent with the loss of EGFRvIII-mediated NF-κB activaton. Similar results were found in U26 cells and U251vIII cells (Fig. 3H and SFigure 5D). In U251 cells, RIP1 appears to be constitutively K63-linked ubiquitinated as has also been reported for other cancer cell types (Feoktistova et al., 2011). Addition of EGF results in a loss of K63 linked RIP1 ubiquitination although this is delayed compared to U1 cells (SFigure 5D).
Figure 3.
EGFRvIII recruits ubiquitin ligases to RIP1 resulting in K63-linked ubiquitination of RIP1. A. U1 cells were exposed to tetracycline to express EGFRvIII followed by immunoprecipitation with cIAP2 antibodies followed by Western blot with EGFR and RIP1. B-C U1 cells were exposed to tetracycline to express EGFRvIII followed by immunoprecipitation with TRAF2 or cIAP-1 antibodies followed by Western blot with EGFR and RIP1. D-F show the same result in U251vIII cells. G. U1 cells were exposed to tetracycline, with or without EGF (50ng/ml) as indicated followed by IP with RIP1 and Western blot with K63 specific ubiquitin antibodies or RIP1 antibodies. An increased K63-linked ubiquitination of RIP1 is detected in tetracycline treated cells which decreases following exposure to EGF for 1h. H. The same results in a second U87MG clone conditionally expressing EGFRvIII (U26).
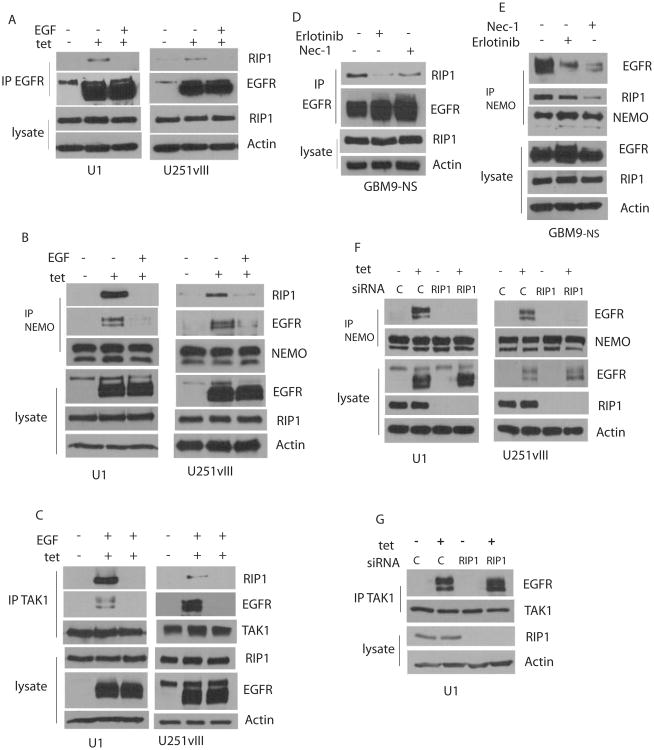
Formation of a EGFRvIII-RIP1 signaling platform
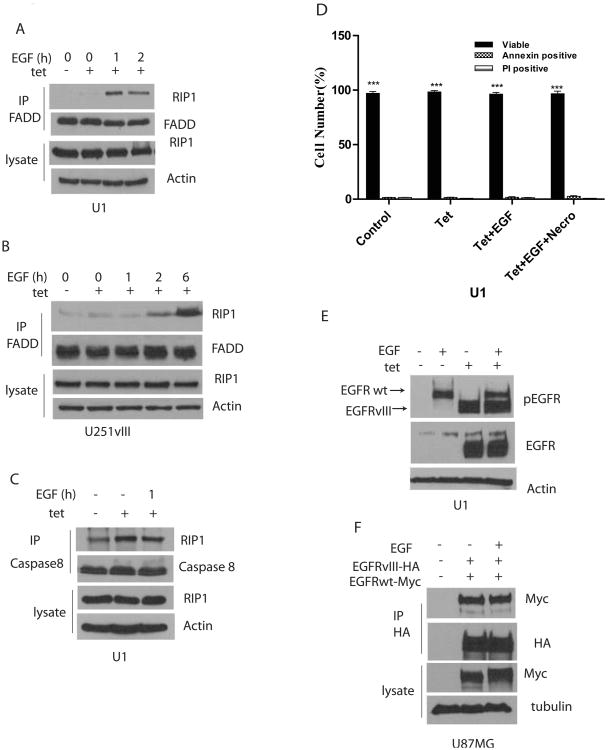
RIP1 associates with EGFRvIII in U1 and U251vIII cells. Thus, when we immunoprecipite with EGFR antibodies and perform Western blot analysis with RIP1, the RIP1 -EGFRvIII association is detectable only in cells exposed to tetracycline. Interestingly, the RIP1-EGFRvIII association is rapidly lost when EGF is added (Figure 4A). No significant association of RIP1 with EGFRwt is detected with or without ligand. IKKy (NEMO) is an important partner for RIP1 in NF-κB activation. We find that NEMO forms a complex with RIP1 that is dependent on EGFRvIII, as demonstrated by immunoprecipitation with NEMO antibodies followed by Western blotting with RIP1. These data suggest that EGFRvIII induces formation of an NF-κB signaling complex. Remarkably, the RIP1-NEMO complex dissociates when EGF is added (Figure 4B). Furthermore, NEMO also binds to the EGFRvIII, suggesting the formation of a RIP1-NEMO containing signaling complex associated with EGFRvIII. TAK1 is another important component of the NF-κB activation network and phosphorylates the IKK complex, an event that culminates in NF-κB activation. TAK1 also forms a complex with RIP1 when EGFRvIII is expressed and dissociates from RIP1 and EGFRvIII when EGF is added to cells (Figure 4C). Exposure of GBM9-NS cells to an EGFR kinase inhibitor, Erlotinib, blocks the EGFRvIII-RIP1 association, as does necrostatin-1, albeit to a lesser extent (Figure 4D). In these cells, the NEMO-RIP1 association is not inhibited by Erlotinib while it is inhibited by Necrostatin-1 suggesting that RIP1 kinase activity is required (Figure 4E). Association of NEMO with the EGFRvIII, however, is decreased by both Erlotinib and Necrostatin-1 (Figure 4E). Importantly, RIP1 siRNA knockdown abolishes recruitment of NEMO but not TAK1 to EGFRvIII in U1 and U251-vIII cells (Figure 4F-G). RIP1 does not influence EGFR levels in these cells. These experiments suggest that expression of EGFRvIII leads to recruitment of RIP1, NEMO and TAK1 to EGFRvIII and that EGFRvIII promotes association of RIP1 with NEMO and TAK1 in this complex.
Figure 4.
Formation of an EGFRvIII associated signaling platform. A. Expression of EGFRvIII in tetracycline treated U1 cells (left panel) or U251vIII cells (right panel) results in association with RIP1. The EGFRvIII-RIP1 association is abolished by EGF. The experiment was conducted by treating cells to EGF followed by immunoprecipitation with EGFR antibody and Western blot with RIP1 antibody. No association with EGFRwt is detected. B. Expression of EGFRvIII in tetracycline treated cells induces association of RIP1 and NEMO and NEMO and EGFRvIII in U1 cells (left panel) and U251vIII cells (right panel). EGF treatment abolishes the RIP1-NEMO association and the NEMO-EGFRvIII association. Immunoprecipitation was conducted with NEMO antibodies followed by Western blot with RIP1 antibodies. C. Expression of EGFRvIII in tetracycline treated cells induces association of RIP1 and TAK1 and TAK1 and EGFRvIII in U1 cells (left panel) and U251vIII cells (right panel). EGF treatment abolishes the RIP1-TAK1 association and the TAK1-EGFR association. Immunoprecipitation was conducted with TAK1 antibodies followed by Western blot with RIP1 antibodies. EGF treatment was for 15 min in AC. D. In GBM9-NS cells, the RIP1-EGFRvIII association is diminished by Necrostatin-1 and to a lesser degree by Erlotinib. Immunoprecipitation was conducted with EGFR antibodies followed by Western blot with RIP1 antibody. E. In GBM9-NS cells, the NEMO-EGFR association is decreased by both Necrostatin-1 and Erlotinib, while the NEMO-RIP1 association is decreased by Erlotinib, but not Necrostatin-1. Immunoprecipitation was conducted with NEMO antibodies followed by Western blot with EGFR or RIP1. F. RIP1 was silenced using RIP1 siRNA in U1 (left panel) and U251vIII cells (right panel), followed by immunoprecipitation with NEMO and Western blot with EGFR in the presence or absence of tetracycline. The NEMO-EGFRvIII association is lost if RIP1 is silenced. G. The EGFRvIII-TAK1 association is unaffected by RIP1 silencing in U1 cells.
Formation of a RIP1-FADD complex in response to EGF
While EGFRvIII expression leads to increased association of NEMO and TAK1 with RIP1, it does not induce the association of FADD with RIP1. Interestingly, when EGF is added to EGFRvIII expressing cells, RIP1 loses its association with NEMO and TAK1 and forms a second complex with FADD. The FADD-RIP1 association is EGF-dependent and can be detected in U1 as well as U251vIII cells (Fig. 5 A-B). Caspase 8 appears to be complexed with RIP1 in U1 cells (Figure 5C). Thus, a death complex with RIP, FADD and Caspase 8 forms in the presence of EGF. However, we do not detect cell death when EGF is added to U1 cells expressing EGFRvIII (Figure 5D).
Figure 5.
Association of RIP1 with FADD and Caspase 8. A. Association of RIP1 with FADD following EGF exposure in EGFRvIII expressing U1 cells. FADD becomes associated with RIP1 after EGF exposure for 1h in tetracycline treated cells. Immunoprecipitation was done with FADD antibody followed by Western blot with RIP1. B. Association of RIP1 with FADD following EGF exposure in U251-vIII cells. C. Association of RIP1 and Caspase 8 in U1 cells. RIP1 is associated with Caspase 8 even in the absence of EGFRvIII in these cells. D. In U1 cells there is no increase in the number of Annexin positive or PI positive cells with either tetracycline or EGF exposure (50ng/ml for 24h). E. EGFRvIII becomes constitutively tyrosine phosphorylated in U1 cells following expression. Addition of EGF results in increased tyrosine phosphorylation of EGFRwt but has no effect on EGFRvIII tyrosine phosphorylation. F. EGFRvIII and EGFRwt form a complex with each other that does not appear to be influenced by EGF. EGFRwt-Myc or EGFRvIII-HA were transfected in U87MG cells followed by immunoprecipitation with Myc and Western blot with HA antibody.
EGFRvIII becomes constitutively tyrosine phosphorylated and activated upon expression in cells. Next, we examined whether EGF influenced EGFRvIII tyrosine phosphorylation in U1 cells. If EGF is added to EGFRvIII expressing cells, tyrosine phosphorylation of EGFRvIII in unaltered (Figure 5E). As has been reported previously (Luwor et al., 2004), we find that EGFR coimmunoprecipitates with EGFRvIII, but the association does not appear to be altered by EGF (Figure 5F).
Increased expression of EGFRwt leads to ligand and RIP1 dependent cell death
In the experiments described above, we observed that RIP1 becomes associated with FADD in response to EGF, but this does not result in cell death. These experiments were conducted in cells with a relatively low level of endogenous EGFRwt. We examined whether overexpression of EGFRwt would result in a RIP1 dependent cell death. It should be noted that the EGFR gene amplification is detected in 40-50% of GBMs and results in overexpression of both EGFRwt as well as mutant forms of EGFR. Thus, EGFRwt is usually expressed at high levels in tumors cells expressing EGFRvIII. We stably overexpressed EGFRwt in U1 that conditionally express EGFRvIII in response to tetracycline (U1wt cells). The level of EGFRwt in U1wt cells is similar to levels in actual GBM tumors (SFigure 5E).
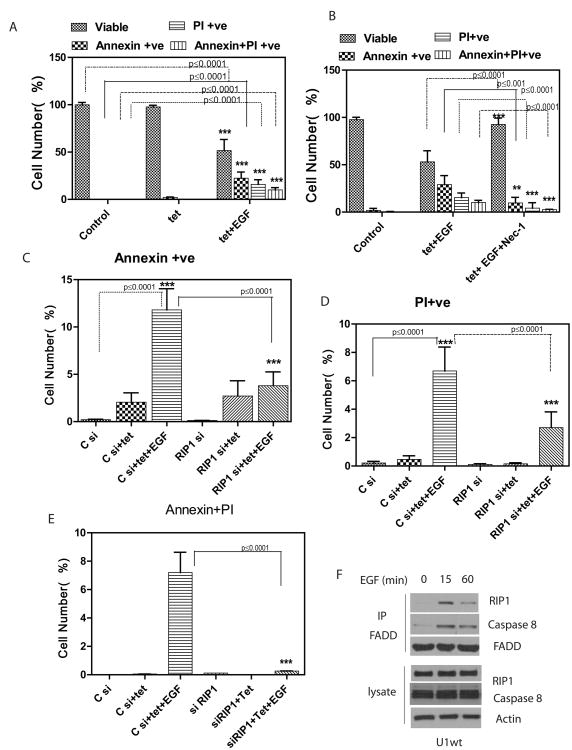
Next, we examined whether exposure of U1wt cells to EGF results in cell death. Expression of EGFRvIII by exposure of cells to tetracycline induces no cell death. However, if cells are exposed to EGF for 24h, there is a robust cell death that has elements of both apoptosis and necrosis (Figure 6A and SFigure 6A). Importantly, this cell death is blocked by the use of necrostatin-1, indicating a requirement for RIP1 kinase activity (Figure 6B). A FACS plot of original data for Figure 6A-B is shown in SFigure 7A. If EGF exposure is continued for 72h, the cell death detected is primarily necrotic and again blocked by Necrostatin-1 (SFigure 6A). siRNA knockdown of RIP1 also blocks EGF-induced cell death in U1wt cells, confirming the essential requirement for RIP1 in EGF-mediated cell death (Figure 6C-E and SFigure 6B). EGF induced and RIP1 dependent cell death is also detected in a second cell line expressing both high levels of EGFRwt and EGFRvIII (U251vIIIwt cells, SFigure 6C-D).
Figure 6.
A. U1wt cells were exposed to EGF (50ng/ml) for 24 hours and an Annexin-FACS assay was performed. Unstained cells represent viable cells. Annexin positive cells are undergoing apoptosis. PI (Propidium iodide) positive cells are undergoing necrosis. In “Control” cells are treated with control vehicle (PBS). In the absence of EGF there is no cell death. When EGF is added to tetracycline treated cells, the number of viable cells is decreased (p<0.0009) and there is evidence of both apoptotic (p<0.005) and necrotic cell death (p<0.005). Double stained cells (Annexin+PI positive) are also shown. B. Necrostatin-1 (300nM) inhibits the EGF induced cell death demonstrating the requirement for RIP1 in EGF-induced cell death. There is an increase in viable cells (p<0.009) and a decrease in apoptotic (p<0.001) and necrotic cells (p<0.001). As a control for Necrostatin-1 cells were treated with DMSO in EGF+tet cells. Data were analyzed by 1way ANOVA followed by Newman-Keuls Multiple Comparison test. C. siRNA knockdown of RIP1 in U1wt cells inhibits EGF induced apoptotic cell death. Cells were transfected with control (scrambled) or RIP1 siRNA followed by EGF exposure for 24h and FACS analysis to compare Annexin positive cells. Exposure to EGF leads to substantial increase in apoptotic (Annexin positive) cells in control siRNA cells. This apoptotic cell death is inhibited in RIP1 siRNA cells (p<0.0001). D. siRNA knockdown of RIP1 in U1wt cells inhibits EGF induced necrotic cell death. Exposure to EGF (24h) leads to substantial increase in necrotic (PI positive) cells in control siRNA cells. This necrotic cell death is inhibited in RIP1 siRNA cells (p<0.0001). E. shows double stained cells (positive for Annexin and PI) for the same experiment. F. RIP1 and Caspase 8 form a complex with FADD in response to EGF in U1wt cells. Upon addition of EGF, there is a rapid increased association of RIP1 and Caspase 8 with FADD. Cells were exposed to tetracycline in all lanes to induce expression of EGFRvIII.
We also confirmed a loss of EGFRvIII-induced NF-κB activation in U1wt cells in response to EGF (SFigure 6E). This loss of NF-κB activation is consistent with the EGF-induced switch to cell death, since NF-κB activation generally promotes cell survival. Furthermore, treatment of U1wt cells with EGF results in a rapid formation of a death signaling complex with increased EGF-dependent association of FADD with RIP1 and Caspase-8 (Figure 6F). Moreover, the complex persists for up to 24 hours (SFigure 6F-G).
Expression of EGFR, RIP1 and phosphorylation of p65 in GBMs
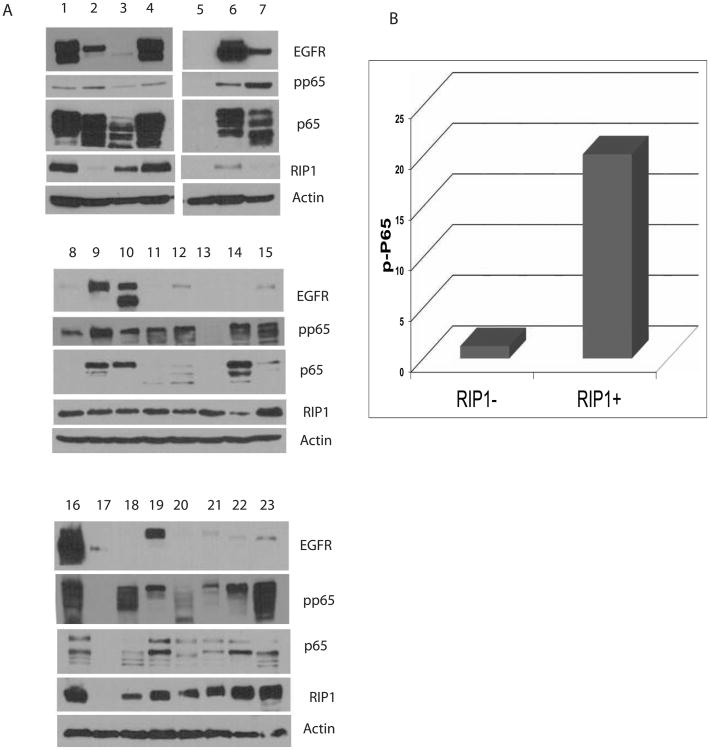
Next, we examined expression of EGFR, RIP1 and phospho-p65 in tumor lysates made directlyfrom GBM tissue (Figure 7A). EGFRvIII mRNA levels were also detected by quantitative realtime PCR (SFigure 7B). In this series of 23 GBMs, 30% of tumors express EGFRvIII. In 6/7GBMs, EGFRvIII is expressed in the presence of EGFRwt, while EGFRvIII appears to be expressed without EGFRwt in one tumor. While phosphorylation of p65 can be detected inevery tumor that expresses EGFRvIII, p65 is also phosphorylated in tumors that are not EGFR amplified, consistent with an additional mechanism(s) of NF-κB activation in GBM. Aspreviously noted, a major mechanism of NF-κB activation in GBMs is deletion of NFKBIA, detected usually in GBMs that do not have amplified EGFR. Importantly, RIP1 is expressed widely in GBMs and the presence of RIP1 correlates with phosphorylation of p65 in GBM asshown in Figure 7A-B (p<0.05, Pearson correlation). Thus, the major components of the EGFRvIII-RIP1-pp65 network are detectable in GBM.
Figure 7.
A. Western blot showing expression of EGFRwt, EGFRvIII, and phospho-p65 in a panel of 23 GBMs. Lysates were made directly from resected tumors followed by Western blot. EGFRvIII is expressed in only a subset of GBMs (usually with EGFRwt) while phosphorylation of p65 can be detected in the majority of GBMs as can RIP1. B. RIP1 correlates with phosphorylation of p65 in GBMs. There was a significant positive correlation between the expression of RIP1 and pp65 using Pearson correlation (r=.49, p<.05).
Discussion
RIP1 has emerged as a key mediator of survival and cell death in the context of cellular stress and inflammatory signaling. RIP1 has been reported to play an essential role in activation of NF-κB in response to TNF-alpha, TLR3 and DNA damage. Recent studies have elucidated a key role for RIP1 in both apoptotic and necrotic forms of cell death. In general, the prosurvival actions of RIP1 are likely to be mediated largely by activation of NF-κB. Post-translational modifications of RIP1, such as K63-linked polyubiquitination, may determine the nature of RIP1 signaling partners and the biological outcome. Receptor tyrosine kinase (RTK) signaling pathways are of central importance in cancer. Our study suggests that RIP1 play a role as a cell life/death switch in a classical oncogenic RTK signaling pathway in an interaction between EGFRwt and the oncogenic EGFR mutant EGFRvIII.
We propose a model for EGFRvIII-mediated activation of NF-κB that involves recruitment of components of the network that mediates TNFα induced NF-κB activation. Firstly, EGFRvIII recruits the ubiquitin ligases c-IAP1, c-IAP2 and TRAF2 to RIP1 and to EGFRvIII resulting in K63-linked ubiquitination of RIP1. EGFRvIII forms a signaling complex by recruiting RIP1, NEMO and TAK1 to EGFRvIII and also increases the association of RIP1 with NEMO and TAK1. Importantly, RIP1 is required for the recruitment of NEMO to EGFRvIII. An unusual feature of EGFRvIII mediated NF-κB activation is the requirement for the kinase activity of RIP1. Previous studies suggested that RIP1 kinase activity is dispensable for NF-κB activation. However, in a recent study it was shown that Etoposide-induced NF-κB activation occurs in two phases and that RIP1 kinase activity is required for the second phase (Biton and Ashkenazi, 2011).
Our study demonstrates that RIP1 is required for EGFRvIII-mediated oncogenicity. Stable silencing of RIP1 in U87MG cells expressing EGFRvIII results in slowed proliferation of cells in culture and importantly, abrogates EGFRvIII mediated tumorigenicity in an in vivo orthotopic model. In our experiment, none of the eight animals with inoculated with RIP1-silenced EGFRvIII cells formed intracranial tumors over an observation period of eight months while all of the animals with control shRNA formed intracranial tumors in about 57 days. Presumably, the lack of EGFRvIII-mediated tumorigenicity results from a loss of the ability of EGFRvIII to activate NF-κB.
An intriguing observation is the unexpected finding that ligand mediated activation of EGFRwt abolishes EGFRvIII-mediated NF-κB activation using RIP1 as a switch. This is unexpected since both EGFRwt and EGFRvIII are known to activate NF-κB, and one would predict a synergistic effect. Synergistic interactions reported previously include EGFRvIII mediated induction of the EGFRwt ligand HB-EGF in an autocrine loop and paracrine effects of EGFRvIII on EGFRwt via an IL-6 mediated pathway (Inda et al., 2010; Ramnarain et al., 2006). The antagonistic effect of EGFRwt on EGFRvIII is highly novel and to our knowledge, antagonistic interactions between RTKs in the EGFR family have not been reported previously. EGFRvIII is expressed almost exclusively in tumors with EGFR gene amplification and EGFRwt overexpression. As discussed, immunohistochemical studies examining expression of EGFRwt and EGFRvIII and our data support the co-expression of EGFRwt and EGFRvIII in individual tumor cells. Our current studies show that ligand mediated activation of EGFRwt results in a dissociation of the EGFRvIII associated signaling complex composed of RIP1, NEMO and TAK1 as well as a separation of RIP1 from NEMO and TAK1. This is associated with loss of K63-linked RIP1 ubiquitination and a complete loss of NF-κB activation. We propose that activation of EGFRwt induces conformational changes in EGFRvIII leading to a loss of EGFRvIII signalosome, a hypothesis that will be examined in future studies.
Our study provides evidence that a RIP1 switch operates in EGFR signaling. Firstly, EGFRvIII forms a physical complex with RIP1 and associated signaling proteins, recruits ubiquitin ligases to RIP1 resulting in K63-linked ubiquitination of RIP1 leading to NF-κB activation via a RIP1 dependent pathway. Ligand-mediated activation of EGFRwt results in a dissolution of the EGFRvIII signalosome, leading to a loss of RIP1 association with key signaling proteins, such as NEMO, TAK1, EGFRvIII, deubiquitination of RIP1, and a complete loss of NF-κB activation. Addition of EGF now results in an association of RIP1 with FADD and Caspase-8. Thus, addition of EGF in the presence of EGFRvIII shifts RIP1 towards a cell death role. While exposure to EGF does not result in cell death when cells express a low level of EGFRwt, when EGFRwt is overexpressed to levels similar to those detected in actual GBM tumors, EGF becomes a death signal. This is remarkable since EGF is normally a trophic factor. This cell death has features of both apoptosis and necrosis and it requires the kinase activity of RIP1. Thus, the addition of EGF results in unleashing the RIP1 switch all the way from an EGFRvIII-RIP1-NF-κB oncogenic signal to a RIP1-FADD-Caspase 8 cell death signal. Although it is somewhat counter-intuitive that a growth factor receptor would induce cell death, there are precedents for life/death signaling switches. For example, Caspase-8, traditionally viewed as an apoptotic protein also has a role in survival (Dillon et al., 2012). Similarly, increased expression of EGFRwt has previously been implicated in growth suppression and apoptosis (Armstrong et al., 1994; Gill and Lazar, 1981).
RIP1 is expressed in most GBMs and is co-expressed with EGFRvIII in tumors showing evidence of NF-κB activation, suggesting that the mechanisms detected in our experimental system may be operational in GBM. Thus, EGFRvIII mediated activation of NF-κB may be a major target for treatment. Since RIP1 mediates EGFRvIII mediated activation of NF-κB, inhibition of RIP1 using either chemical inhibition of silencing approaches could be a potentially important treatment in GBM. In addition, our data suggest that EGFRwt activation could be a mechanism for activating the cell death function of RIP1 in GBM. This is a very novel approach since so far all effort has been focused on inhibiting the EGFR in GBM with limited success (Karpel-Massler et al., 2009). Furthermore, the availability of endogenous EGFRwt ligand(s) in the vicinity of EGFRvIII expressing GBM cells may influence the oncogenic effects of EGFRvIII by abrogating EGFRvIII mediated NF-κB activation and by diverting RIP1 into a cell death pathway that renders tumor cell populations vulnerable to targeted or conventional therapy.
Experimental Procedures
Plasmids, transfection and generation of Cell lines
The glioblastoma cell lines U251MG and U87MG were used to generate cell lines conditionally expressing EGFRvIII using the T-Rex Tet-on system from Invitrogen as we have described previously (Ramnarain et al., 2006). EGFRwt with an HA tag and EGFRvIII with an HA tag was cloned into PcDNA3.1 (Neo) vector using standard molecular techniques. Lipofectamine 2000 (Invitrogen) was used for all transfections. To generate cell lines that express high levels of EGFRwt constitutively along with conditional expression of EGFRvIII, we stably transfected EGFRwt into U1 and U251vIII cells followed by selection in G418. Primary GBM cultures were generated directly from human GBMs. A papain dissociation system was used to dissociate tumors and cells were then cultured in Neurobasal medium supplemented with B27 without Vitamin A, and with EGF (10ng/ml) and bFGF (10ng/ml). LN229-EGFRvIII cells have been described previously (Li et al., 2004). At least 3 independent experiments were done unless otherwise indicated.
Luciferase assays
Cells were plated in 48 well dishes followed by transfection with either along with NF-κB-LUC plasmid using lipofectamine. A dual-luciferase reporter assay system was used according to the instructions of the manufacturer (Promega, Madison WI). Firefly luciferase activity was measured in a luminometer and normalized on the basis of Renilla luciferase activity.
RNA interference
For transient silencing of EGFR, we used a pool of siRNA sequences directed against human RIP1 or control (scrambled) siRNA obtained from Dharmacon or Qiagen. siRNA was performed according to the manufacterer's protocol using Lipofectamine 2000 reagent (Invitrogen). For stable silencing of RIP1, ready to use RIP1 lentiviral particles or control shRNA particles were obtained from Santa Cruz Biotechnology followed by puromycin selection according to the manufacterer's protocol. For stable silencing of EGFRwt we used two shRNA sequences directed against exon 3 of EGFRwt (Smith et al., 2005) or control vector followed by selection in G418.
Antibodies, Reagents and Western blotting
Western blot and immunoprecipitation was performed according to standard protocols. For analysis of RIP1 ubiquitination we followed a protocol described previously (Biton and Ashkenazi, 2011) and as described in supplemental methods. Details of antibodies used in this study are outlined in supplemental methods. Necrostatin-1 was obtained from Tocris Bioscience and used at a concentration of 300nM (overnight exposure). Erlotinib and AG-1478 were obtained from Selleck Chemicals and used at a concentration of 10uM (overnight). EGF was purchased from Peprotech and used at a concentration of 50ng/ml.
Primary Tumors
Frozen tissue specimens of human GBMs were received from the Research tumor bank at UT Southwestern Medical Center according to IRB approved protocols. Resected tumors were initially frozen at −80°C.
Cell Death/Annexin Assay
Annexin assay was done by using Annexin −V-FLUOS Staining kit (Roche applied Science). Cells were cultured in DMEM (10% FBS and Pen/Strep). Cells (1×106) were plated in 6 well plates and treated with EGF for 24h or 72h if indicated. The cells were trypsinized and washed 2 times with 1X PBS. The cells were incubated for 25 minutes at Room temperature with Propidium Iodide and Annexin −V-FLUOS labeling solution in incubation buffer (supplied by the manufacturer). Annexin and or PI positive cells were detected by Flow Cytometry.
Orthotopic implants
For intracerebral stereotactic inoculation, 2 × 105 cells were suspended in PBS and matrigel (5 μL) and injected into the right corpus striatum of the brains of 6-8 week old nude mice using a stereotactic frame (BALB/c nu/nu).Animals were monitored for neurological signs and sacrificed when neurological signs appeared or after 240 days. All animal studies were done under IACUC approved protocols.
Statistical Anaysis
Error bars represent the means ± standard deviations of three independent experiments. All data were analyzed for significance using GraphPad Prism 5.0 software, where P < 0.05 was considered statistically significant. One-way ANOVA and two-tail t-test were used to compare groups. Survival analysis for mice is described in the results.
Additional information about experimental procedures is provided in Supplemental methods.
Supplementary Material
Highlights.
EGFRvIII expression activates NF-κB by promoting ubiquitination of RIP1
RIP1 complexes with EGFRvIII, NEMO and TAK1 to activate NF-κB and promotes oncogenicity
EGFRwt activation abolishes the EGFRvIII-RIP1 signalosome and NF-κB activation
EGFRwt activation results in formation of a RIP1-FADD-Caspase-8 complex and cell death
Acknowledgments
We thank Dr. Mien-Chie Hung (MD Anderson Cancer Center) for the EGFRwt-Myc plasmid and Dr. James Van Brocklyn (Ohio State University) for GBM9-NS cells. This work was supported in part by NIH grants RO1 NS062080 to AH, by RO1 grants CA139217 and CA102792 to DAB, and RO1 CA149461 to SB and by grants from National Aeronautics and Space Administration (NNX13AI13G) and the Cancer Prevention and Research Institute of Texas (RP100644) to SB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong DK, Kaufmann SH, Ottaviano YL, Furuya Y, Buckley JA, Isaacs JT, Davidson NE. Epidermal growth factor-mediated apoptosis of MDA-MB-468 human breast cancer cells. Cancer Res. 1994;54:5280–5283. [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Vandenabeele P. The Ripoptosome: death decision in the cytosol. Mol Cell. 2011;43:323–325. doi: 10.1016/j.molcel.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 2004;14:131–136. doi: 10.1111/j.1750-3639.2004.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton S, Ashkenazi A. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell. 2011;145:92–103. doi: 10.1016/j.cell.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Bonavia R, Inda MM, Vandenberg S, Cheng SY, Nagane M, Hadwiger P, Tan P, Sah DW, Cavenee WK, Furnari FB. EGFRvIII promotes glioma angiogenesis and growth through the NF-kappaB, interleukin-8 pathway. Oncogene. 2012;31:4054–4066. doi: 10.1038/onc.2011.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–232. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T, Mak TW, Wallach D, Green DR. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep. 2012;1:401–407. doi: 10.1016/j.celrep.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires sitespecific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- Gill GN, Lazar CS. Increased phosphotyrosine content and inhibition of proliferation in EGF-treated A431 cells. Nature. 1981;293:305–307. doi: 10.1038/293305a0. [DOI] [PubMed] [Google Scholar]
- Habib AA, Chatterjee S, Park SK, Ratan RR, Lefebvre S, Vartanian T. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-kappa B (NF-kappa B)-inducing kinase to activate NF-kappa B. Identification of a novel receptor-tyrosine kinase signalosome. J Biol Chem. 2001;276:8865–8874. doi: 10.1074/jbc.M008458200. [DOI] [PubMed] [Google Scholar]
- Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor (EGFR) in glioma: Signal transduction, neuropathology, imaging and radioresistance. Neoplasia. 2010;12:675–684. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- Inda MD, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Karpel-Massler G, Schmidt U, Unterberg A, Halatsch ME. Therapeutic inhibition of the epidermal growth factor receptor in high-grade gliomas: where do we stand? Mol Cancer Res. 2009;7:1000–1012. doi: 10.1158/1541-7786.MCR-08-0479. [DOI] [PubMed] [Google Scholar]
- Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HK. Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Oncogene. 2004;23:4594–4602. doi: 10.1038/sj.onc.1207602. [DOI] [PubMed] [Google Scholar]
- Luwor RB, Zhu HJ, Walker F, Vitali AA, Perera RM, Burgess AW, Scott AM, Johns TG. The tumor-specific de2-7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene. 2004;23:6095–6104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21:53–56. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- O'Donnell MA, Hase H, Legarda D, Ting AT. NEMO inhibits programmed necrosis in an NFkappaB-independent manner by restraining RIP1. PLoS One. 2012;7:e41238. doi: 10.1371/journal.pone.0041238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MA, Ting AT. NFkappaB and ubiquitination: partners in disarming RIPK1-mediated cell death. Immunol Res. 2012;54:214–226. doi: 10.1007/s12026-012-8321-7. [DOI] [PubMed] [Google Scholar]
- Pacifico F, Leonardi A. NF-kappaB in solid tumors. Biochem Pharmacol. 2006;72:1142–1152. doi: 10.1016/j.bcp.2006.07.032. [DOI] [PubMed] [Google Scholar]
- Park S, Hatanpaa KJ, Xie Y, Mickey BE, Madden CJ, Raisanen JM, Ramnarain DB, Xiao G, Saha D, Boothman DA, et al. The receptor interacting protein 1 inhibits p53 induction through NF-kappaB activation and confers a worse prognosis in glioblastoma. Cancer Res. 2009;69:2809–2816. doi: 10.1158/0008-5472.CAN-08-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- Smith K, Gunaratnam L, Morley M, Franovic A, Mekhail K, Lee S. Silencing of epidermal growth factor receptor suppresses hypoxia-inducible factor-2-driven VHL-/- renal cancer. Cancer Res. 2005;65:5221–5230. doi: 10.1158/0008-5472.CAN-05-0169. [DOI] [PubMed] [Google Scholar]
- Sun L, Carpenter G. Epidermal growth factor activation of NF-kappaB is mediated through IkappaBalpha degradation and intracellular free calcium. Oncogene. 1998;16:2095–2102. doi: 10.1038/sj.onc.1201731. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, Dang J, Zhu S, Yang H, De Jesus J, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Weinlich R, Dillon CP, Green DR. Ripped to death. Trends Cell Biol. 2011;21:630–637. doi: 10.1016/j.tcb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, You MJ, Koh MY, Cote G, Aldape K, et al. EGFR-Induced and PKCepsilon Monoubiquitylation-Dependent NF-kappaB Activation Upregulates PKM2 Expression and Promotes Tumorigenesis. Mol Cell. 2012;48:771–784. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.