Abstract
Background Human ageing is a complex, multifactorial process and early developmental factors affect health outcomes in old age.
Methods Metabolomic profiling on fasting blood was carried out in 6055 individuals from the UK. Stepwise regression was performed to identify a panel of independent metabolites which could be used as a surrogate for age. We also investigated the association with birthweight overall and within identical discordant twins and with genome-wide methylation levels.
Results We identified a panel of 22 metabolites which combined are strongly correlated with age (R2 = 59%) and with age-related clinical traits independently of age. One particular metabolite, C-glycosyl tryptophan (C-glyTrp), correlated strongly with age (beta = 0.03, SE = 0.001, P = 7.0 × 10−157) and lung function (FEV1 beta = −0.04, SE = 0.008, P = 1.8 × 10−8 adjusted for age and confounders) and was replicated in an independent population (n = 887). C-glyTrp was also associated with bone mineral density (beta = −0.01, SE = 0.002, P = 1.9 × 10−6) and birthweight (beta = −0.06, SE = 0.01, P = 2.5 × 10−9). The difference in C-glyTrp levels explained 9.4% of the variance in the difference in birthweight between monozygotic twins. An epigenome-wide association study in 172 individuals identified three CpG-sites, associated with levels of C-glyTrp (P < 2 × 10−6). We replicated one CpG site in the promoter of the WDR85 gene in an independent sample of 350 individuals (beta = −0.20, SE = 0.04, P = 2.9 × 10−8). WDR85 is a regulator of translation elongation factor 2, essential for protein synthesis in eukaryotes.
Conclusions Our data illustrate how metabolomic profiling linked with epigenetic studies can identify some key molecular mechanisms potentially determined in early development that produce long-term physiological changes influencing human health and ageing.
Keywords: Ageing, metabolomics, epigenetics, twin studies, developmental origins of health and disease, birthweight
Introduction
Human ageing is a multifactorial process influenced by genetic, lifestyle and environmental factors. Genetic variation explains only ∼20–25% of the variability of human survival to the mid 80s,1,2 implying the existence of molecular changes over time which must relate to environmental, epigenetic and lifestyle factors.
Several markers for the ageing process, such as telomere length and circulating levels of dehydroepiandrosterone sulphate (DHEAS), have been identified previously.3,4 Peripheral blood leukocyte telomere length has been shown to be a systemic marker for biological ageing.5 DHEAS is a major but poorly understood circulating steroid in human blood, whose levels are known to strongly correlate with age.
The biochemistry of ageing is complex, with biologically significant changes occurring in many different types of molecules.6 Metabolomics is a novel technology which aims to profile all low-molecular-weight metabolites that are present in biological samples which can investigate the molecular changes seen with ageing. Recently, a study on ageing in 2162 normal individuals with a wide age range (32–81 years) using a panel of 163 metabolites showed that metabolic profiles are strongly correlated with age.7 A small study of 269 individuals using a broader metabolomic platform also reported correlations with age for 51 metabolites (P < 0.05 and false discovery rate Q < 0.15).8
In this study we assess, using a non-targeted metabolomic platform, the extent to which metabolomic profiles are correlated with chronological age and ageing-related traits in a large twin population (n = 6055). We further examine the role of specific metabolites on a likely developmental determinant of healthy ageing (i.e. birthweight) and investigate the potential pathways involved by using epigenome-wide association study (EWAS) data.
Methods
A flow chart of the study rationale is presented in Supplementary Figure 1, available as Supplementary data at IJE online.
Discovery cohort
Study subjects were twins enrolled in the TwinsUK registry, a national register of adult twins.9 In this study we analysed data from 6055 twins with metabolomic profiling available.
The replication cohort
consisted of individuals from the follow-up study KORA F4 (Cooperative Health Research in the Region of Augsburg) drawn from the general population of the region of Augsburg, Germany.10 In total, 887 individuals with fasting serum metabolomic profiles available using the Metabolon platform and measures of FEV1 were analysed.
Phenotype definition
Data were included on body mass index (BMI, body weight in kilograms divided by height in square metres), forced expiratory volume in 1 s (FEV1), forced expiratory vital capacity (FVC), hip bone mineral density (BMD), systolic and diastolic blood pressure (SDP and DBP, total and HDL cholesterol, albumin, leukocyte telomere length and dehydroepiandrosterone sulfate (DHEAS). For details on phenotype measurements see Supplementary Appendix 1, available as Supplementary data at IJE online.
Metabolomics measurements
Non-targeted mass spec-based metabolomic profiling was performed on 1052 fasting serum samples and 5003 fasting plasma samples from participants in the TwinsUK study, using the Metabolon platform, as described previously.11,12 Full details and quality control are included in the supplementary data as Supplementary Texts 2 and 3, available at IJE online. In all, 280 known metabolites were measured in the 1052 serum samples, and 281 known and 175 unknown metabolites were measured in the 5003 plasma samples.
Statistical analysis
Statistical analysis was carried out using Stata version 11.
Step 1. Identify metabolites associated with chronological age
For each metabolite, we calculated the residuals by running linear regression adjusting for sex, BMI and batch effect. Linear regressions for each metabolite residuals accounting for familial relatedness using random intercept linear regression were carried out:
| (1) |
where Xij is the metabolite residual of twin j from pair i; ζj is the family-specific error component which represents the omitted family characteristics or unobserved heterogeneity. We accounted for multiple testing using Bonferroni correction (P = 0.05/280 metabolite residuals).
Step 2. Identify a metabolite panel of independent metabolites associated with chronological age
Although the hypothesis underlying our work is that metabolite levels change with age, in order to investigate which compounds are correlated independently of each other with age we carried out a stepwise linear regression, with age as the dependent variable. This was performed on all the metabolite residuals identified in Step 1, to look for a panel of independent metabolites which could be used as a surrogate for age. We use as cutoff being removed from the model 0.0000001. Adjusting for serum/plasma did not change the results.
Step 3. Test for association with mortality
A Cox regression model was used to estimate the proportional hazards ratio for mortality as a function of the linear combination of 22 metabolites. The variable derived in Figure 1 was used as the independent variable and death was the outcome variable (mean follow-up time 7.33 years, SD 4.46).
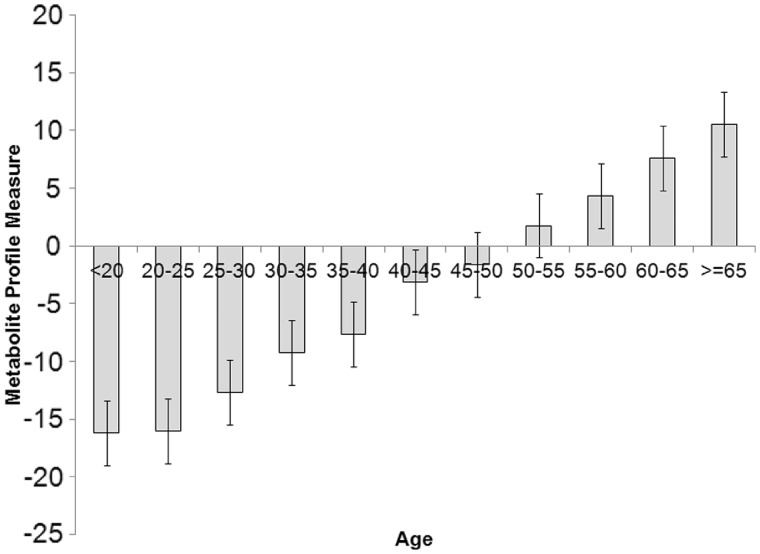
Figure 1.
Metabolite profile measures and age. The metabolite profile measure was calculated for each study participant using the coefficients from the stepwise regression on age of the 22 metabolites in Table 1. The mean and standard error of this variable was then computed for 5 years of age intervals in the study sample
Step 4. Assess whether the metabolite panel associated with chronological age is also associated with known age-related traits
We tested for association with age-related traits: telomere length, SBP, DBP, FEV1, FVC, BMD, DHEAS, total and HDL cholesterol and albumin, comparing the model:
| (2) |
with the model:
 |
(3) |
where ζj is the family-specific error component.
We calculated the coefficient of determination R2 and the Akaike information criterion (AIC) to compare the proportion of variance explained by models (2) and (3) and to assess which of the models better fitted the data.
Step 5. Replication in the KORA sample of C-glyTrp
Linear regressions between C-glyTrp and age and FEV1 were performed. Both analyses were adjusted for sex and BMI. Linear regression for FEV1 was also adjusted for age and height. Meta-analyses of the combined discovery and validation results were conducted using inverse-variance fixed-effects meta-analysis.
Step 6. Heritability of C-glyTrp in TwinsUK
We estimated heritability using structural equation modelling to separate the observed phenotypic variance into three latent sources of variation: additive genetic variance (A), shared/common environmental variance (C), and non-shared/unique environmental variance (E).
Step 7. Discordant twin analysis
We selected 86 monozygotic twins with a minimum difference in birthweight of 750 g (which corresponds to 2SD of the average difference between twins-mean difference at 1060 g, SD difference 320 g). A linear regression was performed on the difference in circulating levels of C-glyTrp and the difference in birthweight.
Step 8. Epigenetic study of C-glyTrp in TwinsUK
DNA methylation levels were obtained using the 27k Illumina CpG methylation probe array in 172 female twins aged 32 to 80 years, randomly selected from the discovery cohort. QC measures were applied, as previously described13 and 24 641 autosomal probes passed quality control. We tested for association between whole-blood DNA methylation patterns and C-glyTrp, adjusting for age, sex, BMI, metabolomic batch, methylation chip, sample position on methylation chip and family relatedness. Adjusting for zygosity did not change the results.
We followed up the association between C-glyTrp metabolite levels with DNA methylation levels at three probes in an independent sample of 350 individuals from the TwinsUK cohort. DNA methylation levels in the follow-up sample were obtained using the Illumina Infinium 450k array.14 We meta-analysed results across the discovery (n = 172) and replication (n = 350) samples using inverse-variance fixed-effect meta-analysis. Probes in both the discovery and replication samples were standardized to have mean zero and variance 1.
Results
A total of 6055 individuals aged 17–85 years from the TwinsUK cohort were included in the analysis of 280 blood metabolites. The demographic characteristics of the study population are presented in Supplementary Table S1 (available as Supplementary data at IJE online).
Age was found to correlate with 165 metabolites after accounting for multiple testing (P < 1.8 × 10−4), family relatedness, sex and body mass index (BMI) (see Supplementary Table S2, available as Supplementary data at IJE online). The majority of these metabolites were related to lipid pathways (73) and amino acid pathways (49). There were also 11 carbohydrates, 9 xenobiotics, 7 nucleotides, 6 intermediates in the energy pathway, 6 cofactors and vitamins and 4 peptides.
After a stepwise linear regression including these 165 metabolites, we identified a panel of 22 independent metabolites associated with age (Table 1), achieving an R2 = 59%. The majority of metabolites were lipids (nine) and amino acids (seven) plus two intermediates in the energy pathway, two xenobiotics, one carbohydrate and one nucleotide. Many of the identified correlations between age and compounds have been previously described in the literature such as that with steroids,15 creatinine16 and citrulline.17 This serves as a proof of principle for the other metabolites identified here.
Table 1.
List of metabolitesa associated with age, and their association with FEV1, and hip BMD
| Age |
FEV1 |
Hip BMD |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Type | Sub-pathway | beta | SE | P | beta | SE | P | beta | SE | P |
| aspartate | a-a | Alanine and aspartate metabolism | −1.05 | 0.17 | 9.2 × 10−10 | 0.001 | 0.01 | 9.0 × 10−1 | 0.003 | 0.002 | 2.3 × 10−1 |
| creatine | a-a | Creatine metabolism | 1.43 | 0.18 | 7.1 × 10−15 | −0.004 | 0.01 | 6.5 × 10−1 | −0.01 | 0.002 | 6.9 × 10−4 |
| creatinine | a-a | Creatine metabolism | −3.11 | 0.22 | 2.1 × 10−45 | 0.001 | 0.01 | 9.5 × 10−1 | 0.01 | 0.003 | 7.3 × 10−7 |
| glutamate | a-a | Glutamate metabolism | 1.6 | 0.22 | 4.4 × 10−13 | −0.03 | 0.01 | 6.3 × 10−4 | −0.002 | 0.003 | 4.2 × 10−1 |
| serine | a-a | Glycine, serine threonine metabolism | −2.17 | 0.23 | 9.7 × 10−21 | 0.03 | 0.01 | 7.9 × 10−3 | −0.002 | 0.003 | 4.3 × 10−1 |
| C-glycosyltryptophanb | a-a | Tryptophan metabolism | 2.47 | 0.24 | 1.3 × 10−23 | −0.05 | 0.01 | 2.9 × 10−5 | −0.02 | 0.003 | 4.8 × 10−9 |
| citrulline | a-a | Urea cycle; arginine, proline, metabolism | 2.04 | 0.2 | 1.4 × 10−23 | 0.01 | 0.01 | 1.4 × 10−1 | 0.0003 | 0.003 | 9.0 × 10−1 |
| threitol | ch | Pentose metabolism | 1.3 | 0.22 | 2.2 × 10−9 | 0.01 | 0.01 | 5.0 × 10−1 | 0.001 | 0.003 | 6.1 × 10−1 |
| citrate | e | Krebs cycle | 3.03 | 0.34 | 5.7 × 10−19 | 0.04 | 0.02 | 1.9 × 10−2 | 0.0003 | 0.005 | 9.4 × 10−1 |
| phosphate | e | Oxidative phosphorylation | −2.15 | 0.34 | 2.6 × 10−10 | −0.004 | 0.02 | 8.2 × 10−1 | 0.01 | 0.004 | 2.6 × 10−2 |
| octanoylcarnitine | l | Carnitine metabolism | 1.31 | 0.19 | 2.5 × 10−12 | −0.02 | 0.01 | 1.0 × 10−1 | −0.003 | 0.003 | 2.7 × 10−1 |
| 4-androsten-3beta, 17beta-dioldmonosulfateb | l | Sterol/steroid | −1.79 | 0.25 | 9.3 × 10−13 | 0.01 | 0.01 | 6.3 × 10−1 | 0.01 | 0.003 | 2.0 × 10−5 |
| 4-androsten-3beta, 17beta-diol disulfate 2b | l | Sterol/steroid | −1.79 | 0.24 | 1.7 × 10−13 | 0.01 | 0.01 | 2.5 × 10−1 | 0.01 | 0.003 | 1.4 × 10−1 |
| eicosapentaenoate (EPA; 20:5n3) | l | Essential fatty acid | 1.87 | 0.26 | 1.1 × 10−12 | 0.02 | 0.01 | 1.6 × 10−1 | −0.01 | 0.003 | 1.1 × 10−1 |
| 3-carboxy-4-methyl-5-propyl-2- furanpropanoate (CMPF) | l | Fatty acid, dicarboxylate | 1.27 | 0.22 | 1.4 × 10−8 | 0.02 | 0.01 | 4.9 × 10−2 | 0.01 | 0.003 | 1.8 × 10−3 |
| 10-heptadecenoate (17:1n7) | l | Long-chain fatty acid | 3.52 | 0.39 | 3.8 × 10−19 | −0.04 | 0.02 | 3.9 × 10−2 | 0.001 | 0.01 | 8.2 × 10−1 |
| dihomo-linoleate (20:2n6) | l | Long-chain fatty acid | −2.29 | 0.28 | 3.2 × 10−16 | 0.02 | 0.01 | 2.0 × 10−1 | −0.002 | 0.004 | 6.7 × 10−1 |
| myristoleate (14:1n5) | l | Long-chain fatty acid | −1.79 | 0.31 | 8.1 × 10−9 | 0.01 | 0.01 | 6.9 × 10−1 | −0.003 | 0.004 | 4.0 × 10−1 |
| palmitoyl sphingomyelin | l | Sphingolipid | 1.9 | 0.17 | 1.1 × 10−28 | 0.005 | 0.01 | 6.0 × 10−1 | −0.01 | 0.002 | 8.1 × 10−4 |
| urate | n | Purine and urate metabolism, | −1.55 | 0.22 | 7.0 × 10−12 | −0.03 | 0.01 | 2.2 × 10−2 | 0.003 | 0.003 | 3.0 × 10−1 |
| erythritol | x | Sugar, sugar substitute, starch | 2.35 | 0.29 | 1.5 × 10−15 | 0.01 | 0.01 | 5.2 × 10−1 | −0.002 | 0.004 | 6.3 × 10−1 |
| 1,7-dimethylurate | x | Xanthine metabolism | 1.69 | 0.17 | 1.0 × 10−23 | 0.01 | 0.01 | 1.4 × 10−1 | 0.0002 | 0.002 | 9.3 × 10−1 |
a-a, amino acid; ch, carbohydrate; e, energy; l, lipid; n, nucleotide; x, xenobiotic.
aMetabolite concentrations were median normalized using the day median and inverse normalized. For each metabolite, residuals were calculated by running a linear regression and adjusting for sex, BMI, batch effect and family relatedness.
bHere age is the dependent variable as we searched for a panel of independent metabolites which could be used as a surrogate for age.
For each study participant we then added the 22 metabolites using the regression coefficient, and created a metabolite-derived age variable. Plotting this variable in 5-year age groups shows a clear linear relationship with age (Figure 1). The value of this combination of 22 metabolites as surrogate measure of chronological age is confirmed by testing their relationship to mortality using data on 188 death events over an average follow-up time of 7.4 years. The variable created in Figure 1 results in a Cox proportional hazards ratio for death HR = 1.08 (95% CI 1.06-1.09, P < 1 × 10−26) per ‘metabolic age’ year. An important question is whether these metabolites contribute additional information to existing age-related traits. We ran linear regressions on age and the 22 independently associated metabolite residuals, adjusting for each of them in turn (see Supplementary Tables S3 and S4, available as Supplementary data at IJE online). All metabolites remained associated (P < 1 × 10−7) with age when adjusting for telomere length and all but two (both steroids) when adjusting for DHEAS.
Given the strong association with age, we hypothesized that these age-correlated 22 metabolites may be predictors of age-related traits after adjustment for chronological age. We considered the following traits: telomere length, systolic and diastolic blood pressure, two measures of lung function (FEV1 and FVC, BMD, DHEAS, total and HDL cholesterol and albumin, all of which have been previously reported to be strongly associated with age.18–21
These 22 metabolites are not, after adjusting for age, all equally associated with the various ageing related traits (Supplementary Tables 5–10, available as Supplementary data at IJE online), but adding all 22 metabolites to the model E [(ageing-related traitij) = β0 + βichronological ageij + ζj + εij] increased the proportion of explained variance for each ageing-related trait. In particular, it added 2% to the total variance explained for FEV1 and FVC, 2.5% for BMD, 4% for both SBP and DBP and for albumin, 5% for telomere length, 26.6% for DHEAS, 23.7% for HDL cholesterol and 46.33% for total cholesterol. The model including the 22 metabolites had in all cases the best fit (a lower AIC), indicating that the panel reflects the process of biological ageing after adjustment for chronological age.
From the main analysis (Table 1), the association with the level of C-glyTrp is of particular interest as it has not previously been reported to be an age-related metabolite and may point to novel molecular pathways involved in ageing. On its own C-glyTrp is correlated with age (beta = 0.03, SE = 0.001, P = 7.0 × 10−157), and age-related traits such as lung function (FEV1 beta = −0.04, SE = 0.008, P = 1.8 × 10−8 adjusted for age and confounders) and BMD (beta = −0.01, SE = 0.002, P = 1.9 × 10−6). Moreover it is strongly and independently correlated with chronological age after adjusting for the presence of the other 21 metabolites (beta = 2.47, SE = 0.24, P = 1.3 × 10−23) and with FEV1 (beta = −0.05, SE = 0.01, P = 2.9 × 10−5), BMD (beta = −0.02, SE = 0.003, P = 4.8 × 10−9) (Table 1), albumin (beta = −0.39, SE = 0.09, P = 1.6 × 10−5) (Supplementary Table S5, available as Supplementary data at IJE online), total cholesterol (beta = -0.11, SE = 0.03, P = 2.7 × 10−5) and HDL cholesterol (beta = −0.06, SE = 0.01, P = 7.8 × 10−5) (Supplementary Table S6, available as Supplementary data at IJE online), FVC (beta = −0.04, SE = 0.01, P = 6.5 × 10−3) (Supplementary Table S7, available as Supplementary data at IJE online), SBP (beta = −0.74, SE = 0.39, P = 5.50 × 10−2) and DBP (beta = −0.86, SE = 0.27, P = 1.4 × 10−3) (Supplementary Table S8, available as Supplementary data at IJE online).
As this metabolite is potentially a novel marker of ageing, we replicated the association in an independent population (KORA n = 887, Germany). Supplementary Table S11 (available as Supplementary data at IJE online) shows the demographic characteristics of the replication cohort. C-glyTrp is associated with chronological age in KORA (beta = 0.004, SE = 0.001, P = 4.14 × 10−8) and it is associated on meta-analysis (beta = 0.012, SE = 0.001, P = 2.29 × 10−101). It is also associated with FEV1 in the KORA cohort (beta = −0.037, SE = 0.019, P = 4.87 × 10−2) and associated on meta-analysis (fixed effects: beta = −0.04, SE = 0.01, P = 2.43 × 10−9) (see Table 2).
Table 2.
Meta-analysis results for C-glyTrp with chronological age and FEV1; beta, SE and P-values are reported also for the discovery and replication cohorts. All analyses are adjusted for sex, BMI, batch effect and family relatedness. FEV1 is also adjusted for chronological age and heighta
| Phenotype | Discovery |
Replication |
Fixed effect |
||||||
|---|---|---|---|---|---|---|---|---|---|
| beta | SE | P | beta | SE | P | beta | SE | P | |
| Chronological age | 0.03 | 0.001 | 7.0 × 10−157 | 0.004 | 0.001 | 4.14 × 10−8 | 0.012 | 0.001 | 2.29 × 10−101 |
| FEV1 | −0.04 | 0.008 | 1.80 × 10−8 | −0.037 | 0.019 | 4.87 × 10−2 | −0.04 | 0.01 | 2.43 × 10−9 |
aThe betas, SEs and P-values are not adjusted for the presence of other metabolites.
Given its consistent association in two independent cohorts, we further explored its biology.
Serum levels of 2-(a-mannopyranosyl)-L-tryptophan have been shown to be a more accurate measure of renal function than serum creatinine concentration, and renal function can be compared in subjects independently of age and muscle mass when 2-(a-mannopyranosyl)-L-tryptophan concentration is measured.22 In the discovery cohort we find that levels of C-glyTrp are correlated with those of serum creatinine (squared correlation coefficient R2 = 0.17, P < 0.0001).
Tryptophan metabolism has already been indicated as a pathway affected by ageing in a metabolomic study of murine livers,23 but the association we see is strongest with glycosylated tryptophan, C-glyTrp.
We assessed possible genetic variants associated with levels of this metabolite by looking at previously published genome-wide association scan data from both the KORA and the TwinsUK cohorts.24 However, we failed to find any genome-wide significant genetic associations with this metabolite24 and, despite reasonable statistical power, we did not find any association with chaperone proteins or glycosylation-related protein encoding genes which would be the obvious genetic determinants.
Taking advantage of the twin nature of our data we ran a heritability analysis25 (1319 MZ pairs, 1256 DZ pairs) and found that the levels of this metabolite have a heritability (h2) of 28%, meaning that 72% of the variance in its levels is not defined by a common genetic component. Thus, levels of this marker could be to a large extent environmentally or epigenetically mediated.
It has been postulated that the health outcomes during ageing can be determined during early development.26 The epidemiological observations that low birthweight is associated with increased rates of coronary heart disease, stroke, type 2 diabetes adiposity, the metabolic syndrome and osteoporosis in adult life26 have been extensively replicated.
We hypothesized that C-glyTrp might be related also to early development. This metabolite is also strongly associated with lower weight at birth in our twin population (beta = −0.06, SE = 0.01, P = 2.5 × 10−9), and birthweight explained 0.88% (R2 = 0.0088) of the variance in adult levels of C-glyTrp, suggesting that levels of this metabolite may reflect reduced growth and/or accelerated ageing in adult life.
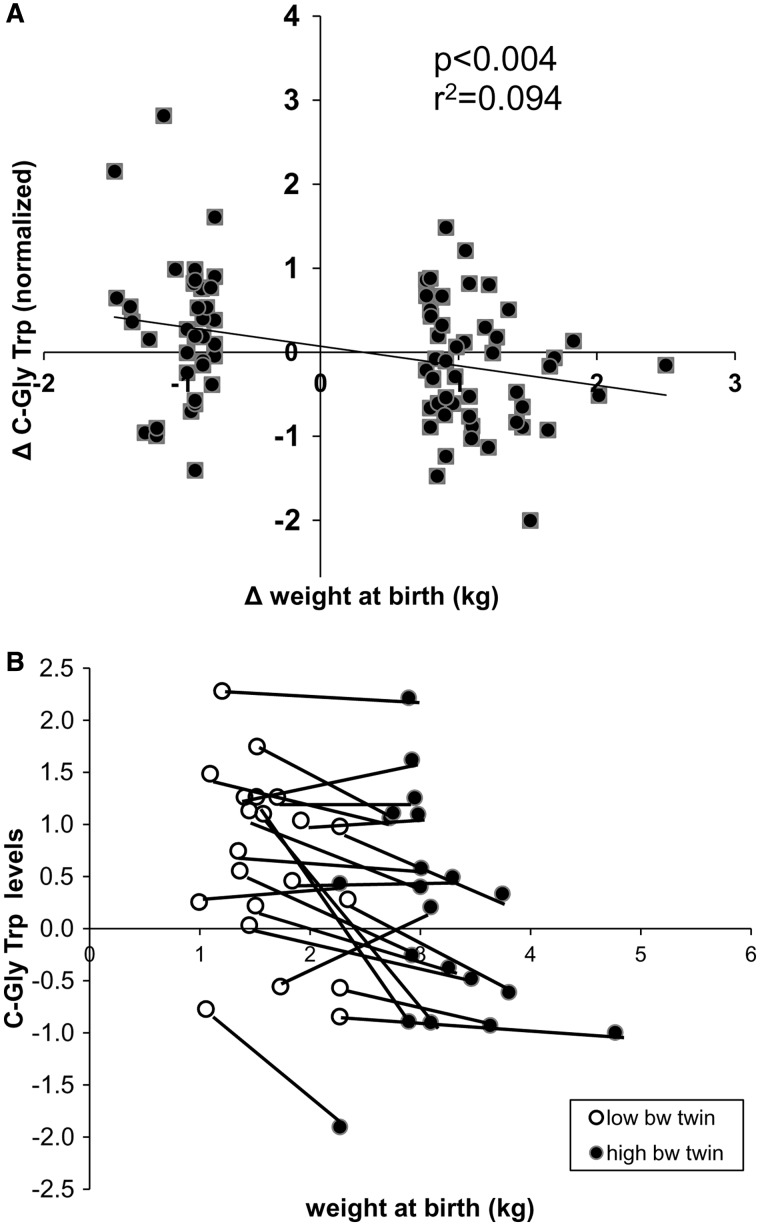
In twins there is considerable variation and on average a birthweight difference of ∼290 g27 within a pair. Interestingly, having selected 85 monozygotic twins (i.e. genetically identical) highly discordant for weight at birth (750 g, 2SD of the average difference between twins), we find a correlation (R2 = 0.094, P < 0.004) between the difference in the levels of C-glyTrp (adjusted for confounders) and the difference in the weight at birth between twins (Figure 2). These data give further support to a non-genetic contribution to the levels of C-glyTrp which appear to be influenced by early development.
Figure 2.
(A) Correlation between the difference in levels of C-glyTrp and the difference in birthweight in 85 monozygotic twins discordant for birthweight (minimum difference 750 g) (B) Relationship between C-glyTrp levels and weight at birth in the 20 most discordant monozygotic twin pairs. The twin in the pair with the highest birthweight is shown in dark circles, the one with lower birthweigh in white circles. Each twin pair is connected by a line
Given the low heritability with this marker and the association of C-glyTrp with birthweight differences, we hypothesized that it may be epigenetically mediated. We compared metabolite levels at C-glyTrp with genome-wide DNA methylation profiles from the Illumina HumanMethylation27 DNA Analysis BeadChip assay in 172 individuals from the discovery cohort.13 The analyses were adjusted for age, sex, BMI, metabolomic batch, methylation chip, sample position on methylation chip and family relatedness. We found 3CpG-sites (cg12757143, cg20367961, cg25999867) at which DNA methylation levels were associated with levels of C-glyTrp with P < 2 × 10−6 (see Table 3).
Table 3.
Association between three methylation probes and C-glyTrp levels in combined discovery and replication samples (total n = 522)
| Discovery |
Replication |
Meta-analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probe | Chr | Positiona | Nearest gene | beta | SE | beta | SE | beta | SE | P |
| cg12757143 | 9 | 140473704 | WDR85 | −0.25 | 0.06 | −0.18 | 0.04 | −0.20 | 0.04 | 2.9 × 10−8 |
| cg20367961 | 1 | 41950237 | EDN2 | −0.23 | 0.06 | 0.04 | 0.05 | −0.08 | 0.04 | 4.7 × 10−2 |
| cg25999867 | 11 | 134145940 | GLB1L3 | −0.21 | 0.05 | −0.02 | 0.03 | −0.06 | 0.03 | 3.0 × 10−2 |
aBase pair position on genome build 37.
We followed up the association between DNA methylation levels at the three probes with C-glyTrp metabolite levels in a replication sample of 350 additional individuals from the TwinsUK cohort. In two of the three probes the direction of association in both the discovery and the replication sets was the same, and for one of the probes (cg12757143) the significance level after adjustment for potential confounders achieved Bonferroni significance (P < 1.3 × 10−7) (Table 3).
Probe cg12757143 maps to the promoter of the WDR85 gene in chromosome 9. WDR85 is a WD repeat-containing protein that plays a role in the first step of diphthamide biosynthesis.28 The translation elongation factor 2 in eukaryotes (eEF-2) contains a unique post-translationally modified histidine residue, termed diphthamide, which serves as the only target for diphtheria toxin and Pseudomonas aeruginosa exotoxin A.
Diphtheria toxin and exotoxin A inhibit host translation through ADP ribosylation of eEF2 and cause cell death by inducing apoptosis.29 ADP ribosylation occurs on diphthamide, a post-translationally modified histidine uniquely present in EF2 and conserved among all eukaryotes.28 Importantly the diphthamide modification on eEF2 has recently been shown to be essential for mRNA translation and embryonic development in mice.30
Therefore we find that epigenetic regulation at a gene implicated in the regulation of eEF2 which is key for cell cycle and embryonic development is associated with circulating levels of C-glyTrp. This is consistent with the strong association between C-glyTrp and birthweight and suggests that the relationship between this metabolite and ageing may be related to apoptotic pathways. One of the other two probes whose association was not replicated mapped to the EDN2 gene which encodes for endothelin. Plasma levels of endothelin are increased in end-stage renal disease and have been implicated in renal inflammation and hypertension.31 The second probe mapped to GLB1L3 encoding β-galactosidase-1-like protein 3 which has been implicated in age-related retinal degeneration.32
Discussion
In this study we report a panel of 22 biochemical metabolites which combined are strongly correlated with chronological age in humans (R2 = 59%). Importantly this panel can also account for some of the variation in ageing-related clinical traits measured as lung function and bone mineral density even after adjusting for chronological age.
Because of its novelty, we focused on one specific ageing-related metabolite, C-glyTrp. This metabolite has already been proposed as a particularly useful marker for normal renal function regardless of the age and muscle mass of the subjects.22
In our twin population, we found a strong relationship between C-glyTrp and birthweight, a well-known developmental determinant of health status in mid life and old age, suggesting that its levels may be influenced by early human development. However, other factors related to birthweight could also influence levels of C-glyTrp and the data shown here do not demonstrate causality. The difference in birthweight explained 9.4% of the variance in the difference of C-glyTrp between genetically identical twins in a pair. The correlation between this metabolite and methylation at the promoter of WDR85, involved in regulating diphthamide synthesis, a key process for RNA translation, cell cycle and embryonic development, further suggests a role for this compound in both ageing and early development.
We note some study limitations. First, there is a female predominance in our study sample (93% of the study sample are women). Second, birthweight is the only early life measure collected in the TwinsUK cohort and we could not test other early life measures which may have strengthened our data. Finally, we have been unable to access an independent population with combined birthweight and metabolomic data on which to replicate this result. Nonetheless, this result appears to be robust as the metabolite levels are not only strongly associated in independent individuals from the cohort, but are also replicated in within-pair differences in discordant identical twin pairs.
Conclusions
Observational and experimental evidence increasingly supports a relation between growth and development during foetal and infant life and health in later years, termed the developmental origins of health and disease (DOHaD).26 The data from the present study provide specific molecular insights for this hypothesis. The results illustrate how metabolomic profiling joined by epigenetic studies may help to identify novel molecular mechanisms implicated in subtle early life influences which produce long-term physiological changes that influence human health.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This work was supported by: European Community’s Seventh Framework programme (FP7/2007-2013) EurHEALTH Ageing HEALTH-F2-2011-277849; Pfizer; the Wellcome Trust European Community’s Seventh Framework Programme (FP7/2007-2013 to TwinsUK); ENGAGE project grant agreement (HEALTH-F4-2007-201413 to TwinsUK); the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas NHS Foundation Trust in partnership with King's College London (to TwinsUK); ERC Advanced Principal Investigator award to T.D.S.; the Wellcome Trust (Grant 098051 to P.D. and L.T.); the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria to the Helmholtz Zentrum München-German Research Center for Environmental Health and Munich Center of Health Sciences (MC Health), Ludwig-Maximilians-Universität, as part of LMU innovative and KORA; the Competence Network ASCONET, specifically the subnetwork COSYCONET, and the KORA-Age project funded by the German Federal Ministry of Education and Research to KORA. K.S. is supported by ‘Biomedical Research Program' funds at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation. The statements made herein are solely the responsibility of the authors.
Supplementary Material
Acknowledgements
We thank James Nisbet for technical assistance and the Wellcome Trust Genotyping Facility for DNA methylation typing of the TwinsUK samples. We thank Roche Diagnostics Australia Pty Ltd, Castle Hill, NSW, Australia, who provided support for the analysis of DHEAS.
Conflict of interest: R.PM. is an employee of Metabolon Inc. S. J. and M.J.B are full-time employees and shareholders of Pfizer.
KEY MESSAGES.
Using metabolomic profiling we have identified a specific metabolite, highly correlated with age and ageing traits, such as lung function and bone mineral density.
This metabolite, a glycosylated amino acid, is strongly associated with birthweight, a developmental determinant of healthy ageing.
This glycosylated amino acid is also associated with methylation levels at a probe that maps to the promoter of a regulator of translation elongation factor 2.
The findings provide molecular mechanisms for the developmental origins of adult disease hypothesis and highlight the importance of epigenetic factors in this process.
References
- 1.Fraser GE, Shavlik DJ. Ten years of life: Is it a matter of choice? Arch Intern Med. 2001;161:1645–52. doi: 10.1001/archinte.161.13.1645. [DOI] [PubMed] [Google Scholar]
- 2.Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet. 1996;97:319–23. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 3.Gerashchenko BI. At a crossroads of cancer risk and aging: the role of telomeres. Exp Oncol. 2010;32:224–27. [PubMed] [Google Scholar]
- 4.Feldman HA, Longcope C, Derby CA, et al. Age trends in the level of serum testosterone and other hormones in middle-aged men: longitudinal results from the Massachusetts male aging study. J Clin Endocrinol Metab. 2002;87:589–98. doi: 10.1210/jcem.87.2.8201. [DOI] [PubMed] [Google Scholar]
- 5.Valdes AM, Andrew T, Gardner JP, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366:662–64. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 6.Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6:4716–23. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- 7.Singman PZ, Yu Z, Prehn C, et al. Human serum metabolic profiles are age-dependent. Aging Cell. 2012;11:960–7. doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawton KA, Berger A, Mitchell M, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–97. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 9.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and Healthy Ageing Twin Study. Int J Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wichmann HE, Gieger C, Illig T. KORA-gen — resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 11.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 12.Reitman ZJ, Jin G, Karoly ED, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Nat Acad Sci U S A. 2011;108:3270–75. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell JT, Tsai PC, Yang TP, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450 000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 15.Baylis D, Bartlett DB, Syddall HE, et al. Immune-endocrine biomarkers as predictors of frailty and mortality: a 10-year longitudinal study in community-dwelling older people. Age (Dordr) 2013;35:963–71. doi: 10.1007/s11357-012-9396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candow DG. Sarcopenia: current theories and the potential beneficial effect of creatine application strategies. Biogerontology. 2011;12:273–81. doi: 10.1007/s10522-011-9327-6. [DOI] [PubMed] [Google Scholar]
- 17.De Ceuleneer M, Van Steendam K, Dhaenens M, Deforce D. In vivo relevance of citrullinated proteins and the challenges in their detection. Proteomics. 2012;12:752–60. doi: 10.1002/pmic.201100478. [DOI] [PubMed] [Google Scholar]
- 18.Knudson RJ, Lebowitz MD, Holberg CJ, Burrows B. Changes in the normal maximal expiratory flow-volume curve with growth and aging. Am Rev Respir Dis. 1983;127:725–34. doi: 10.1164/arrd.1983.127.6.725. [DOI] [PubMed] [Google Scholar]
- 19.Kanis JA. Diagnosis of osteoporosis and assessment of fracture risk. Lancet. 2002;359:1929–36. doi: 10.1016/S0140-6736(02)08761-5. [DOI] [PubMed] [Google Scholar]
- 20.Baker GT, 3rd, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23:223–39. doi: 10.1016/0531-5565(88)90025-3. [DOI] [PubMed] [Google Scholar]
- 21.Johnson TE. Recent results: biomarkers of aging. Exp Gerontol. 2006;41:1243–46. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Yonemura K, Takahira R, Yonekawa O, Wada N, Hishida A. The diagnostic value of serum concentrations of 2-(alpha-mannopyranosyl)-L-tryptophan for normal renal function. Kidney Int. 2004;65:1395–99. doi: 10.1111/j.1523-1755.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 23.Houtkooper RH, Argmann C, Houten SM, et al. The metabolic footprint of aging in mice. Scientific Rep. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neale M, Cardon L. Methodology for Genetic Studies of Twins and Families. Dordrecht: Kluwer Academic Publishers; 1992. [Google Scholar]
- 26.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conley D, Strully KW, Bennett NG. Twin differences in birth weight: the effects of genotype and prenatal environment on neonatal and post-neonatal mortality. Econ Hum Biol. 2006;4:151–83. doi: 10.1016/j.ehb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Carette JE, Guimaraes CP, Varadarajan M, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–35. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 29.Sharma AK, FitzGerald D. Pseudomonas exotoxin kills Drosophila S2 cells via apoptosis. Toxicon. 2010;56:1025–34. doi: 10.1016/j.toxicon.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Bachran C, Gupta P, et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Nat Acad Sci U S A. 2012;109:13817–22. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyauchi Y, Sakai S, Maeda S, et al. Increased plasma levels of big-endothelin-2 and big-endothelin-3 in patients with end-stage renal disease. Life Sci. 2012;91:729–32. doi: 10.1016/j.lfs.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Le Carre J, Schorderet DF, Cottet S. Altered expression of beta-galactosidase-1-like protein 3 (Glb1l3) in the retinal pigment epithelium (RPE)-specific 65-kDa protein knock-out mouse model of Leber's congenital amaurosis. Mol Vis. 2011;17:1287–97. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.