Abstract
Objectives
Through recent genome-wide association studies (GWAS), several groups have reported significant association between variants in the alpha 1C subunit of the L-type voltage-gated calcium channel (CACNA1C) and bipolar disorder (BP) in European and European-American cohorts. We performed a family-based association study to determine whether CACNA1C is associated with BP in the Latino population.
Methods
This study consisted of 913 individuals from 215 Latino pedigrees recruited from the United States, Mexico, Guatemala, and Costa Rica. The Illumina GoldenGate Genotyping Assay was used to genotype 58 single-nucleotide polymorphisms (SNPs) that spanned a 602.9 kb region encompassing the CACNA1C gene including two SNPs (rs7297582 and rs1006737) previously shown to associate with BP. Individual SNP and haplotype association analyses were performed using Family-Based Association Test (version 2.0.3) and Haploview (version 4.2) software.
Results
An eight-locus haplotype block that included these two markers showed significant association with BP (global marker permuted p = 0.0018) in the Latino population. For individual SNPs, this sample had insufficient power (10%) to detect associations with SNPs with minor effect (odds ratio = 1.15).
Conclusions
Although we were not able to replicate findings of association between individual CACNA1C SNPs rs7297582 and rs1006737 and BP, we were able to replicate the GWAS signal reported for CACNA1C through a haplotype analysis that encompassed these previously reported significant SNPs. These results provide additional evidence that CACNA1C is associated with BP and provides the first evidence that variations in this gene might play a role in the pathogenesis of this disorder in the Latino population.
Keywords: bipolar disorder, calcium channels, genetic association studies, haplotypes, Hispanic Americans, L-type, pedigree, polymorphism, single nucleotide
Bipolar disorder (BP) is an often chronic and debilitating mental disorder affecting approximately 1–4% of the population (1, 2). Family, twin, and adoption studies on BP have shown that there is a substantial genetic component (3, 4), as BP has one of the highest heritability rates of all known psychiatric disorders (1). Over the past several years, genome-wide association studies (GWAS) have produced several successes in identifying genetic variants that contribute to genetically complex human disorders (5–16). Recently, a few research groups have performed independent GWAS of BP (11, 17–22), with little overlap amongst the single-nucleotide polymorphisms (SNPs) most strongly associated with BP (23). However, a meta-analysis of three GWAS of BP identified a SNP within the calcium channel, voltage-dependent, L-type, alpha 1C subunit (CACNA1C) gene, rs1006737, as being strongly associated with the risk of BP (21). A subsequent meta-analysis combining data from GWAS of BP and major depressive disorder (MDD) reported two SNPs in a 10.5-kb region within the CACNA1C gene which exceeded a genome-wide significance level (rs1006737 and rs7297582) (20).
There has been increasing evidence in recent years that alterations in calcium signaling are involved in the pathophysiology of BP (32). The CACNA1C gene located on chromosome 12p13.3 encodes the major constituent of the brain L-type voltage-gated calcium channels that are critical to dendritic calcium influx in response to synaptic activity (33, 34). Reports have associated SNPs in CACNA1C with an increase in grey matter density in the amygdala and hypothalamus (35), total grey matter (36), and brainstem volume (37). Neuroimaging studies of healthy individuals report that the rs1006737 risk allele modulates brain function during tasks of reward and emotional processing (38, 39), verbal fluency (40), attention networks (41), and episodic and prefrontal cortex (PFC) working memory (38, 42). The CACNA1C rs1006737 polymorphism also impacts ventrolateral PFC activation during fear processing in BP carriers of the risk allele but not their unaffected relatives (43). In addition, studies have shown that carriers of the rs1006737 variant show a pronounced effect on measures of psychosocial functioning, including significantly higher scores on tests for depression, anxiety, interpersonal sensitivity, obsessive-compulsive thoughts, and neuroticism, even in the absence of disease (44). These studies suggest that calcium channel dysfunction may contribute in part to the genetic etiology of BP through alterations in the functional activity of brain circuitries.
Although there is mounting evidence that CACNA1C is involved in the genetic etiology of BP, the genetic studies performed to date have been almost exclusively centered on populations with European ancestry. The replication of significant GWAS findings in diverse populations is needed in order to corroborate initial findings from European studies and characterize their genetic contribution to BP in other ethnicities. In order to address this issue, we have designed a family-based association study to try and validate the GWAS-significant CACNA1C SNPs in a large set of families with Latin American (Latino) ancestry.
Materials and methods
Study samples
The sample consisted of 215 Latino pedigrees in which BP probands and extended family members were recruited from the United States (Texas, New Mexico, California), Mexico (Mexico City, Monterrey), Costa Rica, and Guatemala. Inclusion criteria required a proband with a bipolar disorder type I (BP-I) diagnosis with at least one sibling with a clinical diagnosis of BP-I or schizoaffective BP (SABP), and a minimum of two additional first-degree relatives willing to participate. Extended pedigrees were collected when possible to include other family members with a history of affective or psychotic disorders. The sample consisted of 157 case-parent trios and 258 affected subjects with one parent genotyped. All subjects reported ancestry from Mexico or Central America. Family structure based on number of affected family members and country of origin is listed in Table 1. Previous genetic structure analysis has shown that these populations are closely related, with high levels of admixture consisting of three major ancestral populations (Caucasian, Native American, and African) (45).
Table 1.
Distribution of families per country of origin based on corresponding number of affected individualsa
| United States | Mexico | Costa Rica | Guatemala | |||||
|---|---|---|---|---|---|---|---|---|
| Affected individuals | Families | Mean (range) | Families | Mean (range) | Families | Mean (range) | Families | Mean (range) |
| 1 | 16 | 3.56 (2–6) | 22 | 3.05 (3–4) | 5 | 3.40 (3–5) | 2 | 3.00 (3) |
| 2 | 29 | 3.69 (2–7) | 45 | 3.84 (3–8) | 12 | 4.67 (3–9) | 9 | 3.56 (3–5) |
| 3 | 14 | 4.00 (3–8) | 22 | 5.05 (3–13) | 13 | 5.85 (4–10) | 3 | 4.00 (4) |
| 4 | 2 | 4.00 (4) | 7 | 5.86 (4–8) | 5 | 7.80 (5–14) | 2 | 6.00 (6) |
| 5 | 1 | 8.00 (8) | 2 | 8.50 (8–9) | 1 | 8.00 (8) | ||
| 7 | 1 | 10.00 (10) | ||||||
Affected individuals = number of family members with a diagnosis of bipolar disorder type I and/or schizoaffective bipolar disorder; families = number of families per country of origin; mean = average number of individuals per family; range = range of individuals per family.
There is no data presented for families with six affected individuals
Subjects signed Institutional Review Board (IRB)-approved written informed consent forms prior to enrolling in the study.
The procedures were approved by the IRB of Texas Tech University Health Science Center and respective IRBs in each participating site and country, and the study was performed in accordance with the Helsinki Declaration of 1975.
All study participants were diagnosed using DSM-IV criteria (46), by a best-estimation consensus procedure using the Diagnostic Interview for Genetic Studies (DIGS) (47), Family Interview for Genetic Studies (FIGS) (48), and available psychiatric records as previously described (49). The 215 Hispanic pedigrees under study contained 929 individuals: 466 of whom were diagnosed with BP-I and 16 of whom were diagnosed with SABP. Of the study participants with a diagnosis of BP-I or SABP, 200 were male and 282 were female.
Genotyping
DNA was isolated from lymphoblastoid cell lines established and stored for each study participant at the National Institute of Mental Health (NIMH) Center for Collaborative Genetic Studies. SNP selection covering the CACNA1C gene was based on a tagging SNP approach (r2 ≥ 0.9) using the SNP browser™ Software version 4.0.1 based on data from the CEU (Utah residents with ancestry from northern and western Europe) HapMap population (50). Known CACNA1C variants rs1006737 (20, 21) and rs7297582 (20), which reached genome-wide significance in previous meta-analysis GWAS studies, were included in the panel. Additional proximal SNPs to these known variants were chosen from the National Center for Biotechnology Information (NCBI) dbSNP (51) based on criteria that SNPs were validated and had heterozygosity > 0.4. A total of 58 SNPs were genotyped using a custom-designed Illumina GoldenGate SNP assay according to the manufacturer’s protocol (Illumina, San Diego, CA), blind to subject diagnosis and characteristics.
Genetic markers failing standard quality control criteria were excluded from further analysis: missing genotype > 0.1, minor allele frequency < 0.01, and exact Hardy–Weinberg equilibrium p < 0.01. Also excluded from the analysis were individuals with a missing genotype rate > 0.1, or Mendelian errors > 2. The discrepancy rate on 2% duplicate genotyping was < 0.002. A total of 57 SNPs for 213 pedigrees (913 individuals) were retained for further statistical analysis.
Statistical analyses
Analyses of individual SNPs were completed with the Family-Based Association Test (FBAT) version 2.0.3 (52), which was used for testing association under the null hypothesis of ‘no association and no linkage’ using an additive genetic model. To thoroughly test the locus identified by the two individual SNPs previously reported to be in association with BP, we also identified a haplotype block which included those SNPs and analyzed the haplotypes for association. Haploview version 4.2 (53) was used to visualize linkage disequilibrium (LD) relationships between all genotyped variants (57 SNPs) within and surrounding the CACNA1C region (54) and to construct LD blocks following the D′ method described by Gabriel et al. (55). Haplotype analyses were performed using the Haplotype-Based Association Testing (HBAT) assessment in the FBAT program. Haplotype-specific and global-permuted p-values were calculated for individual haplotype tests conducted under biallelic mode in haplotype FBAT, assuming an additive genetic model. In order to maximize power in the statistical analyses, permutation procedures were implemented to calculate the empirical p-values derived from 10,000 permutations. The significance level for all statistical tests was two-tailed (p < 0.05).
Power analyses were estimated using the transmission disequilibrium test (TDT) for discrete traits of the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) with α = 0.05 and D′ of 0.9. The current family study design (n = 215) yielded an estimate of 0.65 [odds ratio (OR) = 1.75] and 0.82 (OR = 2.00) for a gene of moderate effect and a power estimate of 0.40 (OR = 1.5) and 0.10 (OR =1.15) for a gene with minor effect. We calculated that our sample had reasonable power to detect a common risk variant, but would be insufficient to detect a susceptibility locus with a minor effect as seen in genetic studies of the CACNA1C gene for BP. To make optimal use of our data, we performed haplotype-based tests to increase the power of the association study (55).
Results
The primary aim of this study was to test to see if findings of association of SNPs rs1006737 and rs7297582 in the CACNA1C gene with BP, as reported in a European ancestry cohort by Liu et al. (20), were valid in other populations, specifically in the Latino population. When tested individually, we found no significant association between these two SNPs and BP (rs7297582, Z = 0.306, p = 0.760; rs1006737, Z = 0.725, p = 0.469) in the Latino population studied.
Given the negative findings of association for SNPs rs1006737 and rs7297582, we wished to test whether SNPs in LD with these variants would associate with BP in our Latino cohort. We used a tagged-SNP approach to cover the CACNA1C gene in addition to several SNPs in close proximity to rs1006737 and rs7297582, assuming that these SNPs were not functional SNPs but rather variants in LD with true causal mutation. Twelve LD blocks were identified in our sample (Fig. 1). Haploblock 4 encompasses both rs1006737 and rs7297582 SNPs, previously shown to associate with BP. Allele frequencies and the results of FBAT for single SNP analysis for the eight SNPs comprising Haploblock 4 are shown in Table 2. Univariate (single-marker) FBAT demonstrated no preferential transmission for any of the variant alleles tested (Table 2).
Fig. 1.
Calcium channel, voltage-dependent, L-type, alpha 1C subunit (CACNA1C) haplotype blocks in the Latino population. Linkage disequilibrium plot of the CACNA1C gene (shown as a black line) and adjacent regions with respect to the 57 single-nucleotide polymorphisms (SNPs) with minor allele frequency > 10%. The data visualized here using Haploview V4.2 (53) are based on the current genotyping data from the Latino population. Squares indicate pair-wise r2 values on a red-scale with D′ = 1 (red) through to D′ = 0 (white). Linkage disequilibrium blocks, as defined in Haploview, are represented by the triangular lines. Order of Blocks and SNPs (SNPs within Blocks in bold) from left to right: Block 1 (14 kb): rs765125, rs2299657, rs2283269; rs2238013; rs2238019; Block 2 (18 kb): rs2283281, rs758723, rs2299660, rs2299661, rs2239131; rs2238034; rs2238042; rs2239014; rs2238044; rs758171; Block 3 (16 kb): rs2239024, rs2238056; Block 4 (69 kb): rs769087, rs1006737, rs2159100, rs4765905, rs2370413, rs7297582, rs758170, rs1860002; Block 5 (20 kb): rs2239047, rs2283301, rs2238070; rs740418; Block 6 (31 kb): rs2238078, rs2283304, rs2283306, rs2239062, rs2238084; rs2239063; rs1009281; Block 7 (1 kb): rs2239066, rs2239068, rs2239071; rs2239074; rs2239087; Block 8 (7 kb): rs1860094, rs887001; rs1541452; rs4765951; Block 9 (14 kb): rs2239117, rs4765959; rs2370600; rs929492; Block 10 (7 kb): rs2238090, rs11832738; rs215976; Block 11 (16 kb): rs2283326, rs215993; rs758561; Block 12 (7 kb): rs1420725, rs740727.
Table 2.
Family-Based Association Test univariate results under additive model for eight SNPs with linkage disequilibrium in CACNA1C gene
| Marker | Allele | Allelic frequency | Families | Z | p-value |
|---|---|---|---|---|---|
| rs769087 | G | 0.715 | 98 | −0.372 | 0.710 |
| A | 0.285 | 98 | 0.372 | 0.710 | |
| rs1006737a | G | 0.712 | 100 | −0.725 | 0.469 |
| A | 0.288 | 100 | 0.725 | 0.469 | |
| rs2159100 | G | 0.711 | 99 | −0.588 | 0.557 |
| A | 0.289 | 99 | 0.588 | 0.557 | |
| rs4765905 | C | 0.710 | 101 | −0.585 | 0.559 |
| G | 0.290 | 101 | 0.585 | 0.559 | |
| rs2370413 | G | 0.520 | 130 | −0.323 | 0.746 |
| A | 0.480 | 130 | 0.323 | 0.746 | |
| rs7297582a | G | 0.697 | 108 | −0.306 | 0.760 |
| A | 0.303 | 108 | 0.306 | 0.760 | |
| rs758170 | G | 0.713 | 102 | −0.639 | 0.523 |
| A | 0.287 | 102 | 0.639 | 0.523 | |
| rs1860002 | A | 0.561 | 123 | −1.161 | 0.246 |
| G | 0.439 | 123 | 1.161 | 0.246 |
SNP = single-nucleotide polymorphism; CACNA1C = calcium channel, voltage-dependent, L type, alpha 1C subunit; families = number of informative families.
SNPs associated with bipolar disorder in previous studies.
Since the use of haplotypes may provide greater statistical power in association studies of common genetic variation than can be obtained with individual SNPs (55), we conducted haplotype analyses focusing on Haploblock 4 which contained SNPs previously associated with BP. To determine whether any specific haplotype would confer a higher risk for BP, the specific and global-haplotype FBAT tests of association were performed. One common and three rare haplotypes displayed significant association under an additive model (Table 3). The p-values of the whole haplotype permutation test were also statistically significant (χ2 sum, p = 0.0018) (Table 3). Of note, among these four disease-associated haplotypes, three (GGGCGGGA, GGGCGAGA, and AAAGAGGA) showed protective effect with negative Z-scores and one showed a susceptibility effect with a positive Z-score (GAAGGAAG). Interestingly, the specific alleles previously shown to be associated with BP on SNPs rs1006737 (A) and rs7297582 (A), were also seen on the GAAGGAAG haplotype showing increased risk (Z = 2.236, p = 0.031). The common haplotype significantly associated with BP was present across all countries in our cohort, while the other three haplotypes were very rare.
Table 3.
Haplotype analysis for eight SNPs with linkage disequilibrium in CACNA1C gene
| Haplotype | Frequency | Families | S-E(S) | Var(S) | Z | p-value | rank(S_obs) | P_2side |
|---|---|---|---|---|---|---|---|---|
| GGGCAGGA | 0.463 | 119 | 8.497 | 60.133 | 1.096 | 0.273 | 7432.5 | 0.283 |
| AAAGGAAG | 0.255 | 93 | 3.057 | 46.619 | 0.448 | 0.654 | 17063.5 | 0.651 |
| GGGCGGGG | 0.102 | 59 | 5.412 | 23.034 | 1.128 | 0.259 | 7647 | 0.290 |
| GGGCGGGA | 0.093 | 52 | −13.865 | 22.972 | −2.893 | 0.004a | 52692.5 | 0.004a |
| GGGCGAGG | 0.023 | 16 | 0.305 | 6.082 | 0.124 | 0.902 | 22174 | 0.837 |
| AAAGGGAG | 0.017 | 15 | 0.185 | 4.574 | 0.087 | 0.931 | 27104.5 | 0.977 |
| GGGCAGGG | 0.012 | 11 | −2.418 | 3.209 | −1.350 | 0.177 | 45600 | 0.269 |
| GGGCGAGA | 0.008 | 5 | −2.528 | 1.218 | −2.291 | 0.022a | 52591 | 0.009a |
| AAAGGGGG | 0.008 | 5 | 0.694 | 1.934 | 0.499 | 0.618 | 17009.5 | 0.644 |
| GGGCGGAG | 0.004 | 4 | 0.075 | 1.419 | 0.063 | 0.950 | 25296 | 0.952 |
| AAAGGAAA | 0.003 | 3 | −0.498 | 1.171 | −0.460 | 0.646 | 29224 | 0.895 |
| GGGCAAGA | 0.002 | 1 | 1 | 0.5 | 1.414 | 0.157 | 6667 | 0.248 |
| AAAGAGGA | 0.002 | 1 | −1.917 | 0.543 | −2.601 | 0.009a | 52076.5 | 0.028a |
| GAAGGAAGb | 0.002 | 2 | 2.5 | 1.25 | 2.236 | 0.025a | 847 | 0.031a |
| Whole marker permutation test (χ2sum, p = 0.0018) | ||||||||
Haplotypes comprised of single-nucleotide polymorphisms (SNPs): rs769087, rs1006737, rs2159100, rs4765905, rs2370413, rs7297582, rs758170, and rs1860002. Haplotypes with frequency ≤ 0.001 not shown. CACNA1C = calcium channel, voltage-dependent, L type, alpha 1C subunit; Frequency = frequency of the haplotype; S = test statistics for the observed number of transmitted alleles; E(S) = expected value of S under the null hypothesis (i.e., no linkage or association); P_2side = two-sided p-values using permutation test based on 10,000 permutations.
p-values < 0.05.
Bipolar risk alleles are bolded and underlined.
Discussion
In the present study, we attempted to validate, in nearly 1,000 Latinos, the significant CACNA1C variants associated with BP identified in populations of predominantly European ancestry (11, 18, 20, 21). When analyzed individually, we were unable to replicate association of SNPs rs7297582 and rs1006737 with BP in the Latino population. However, these two previously identified SNPs have not been shown to be functionally significant within the CACNA1C gene and may only be in LD with functional variants at this locus. Moreover, allele frequencies of genotyped markers in strong LD with the two previously associated SNPs are highly variable across ancestral populations (hapmap.ncbi.nlm.nih.gov), suggesting the likelihood of a differential profile of CACNA1C variants with ethnicity (Table 4). Given the negative findings of our single locus tests compared to previous associations found in the Caucasian population, combined with the finding that haplotypes at this locus are associated with BP in the Latino population, the possibility remains that there might be additional functional variants in both the European and Latino ancestry populations that might still be in strong LD with rs7297582 and rs1006737.
Table 4.
Allele frequencies among different populations
| Marker | Allele | Current study | CEU | JPT + CHB | YRI |
|---|---|---|---|---|---|
| rs769087 | G | 0.715 | 0.642 | 0.936 | 0.364 |
| A | 0.285 | 0.358 | 0.064 | 0.636 | |
| rs1006737a | G | 0.712 | 0.627 | 0.933 | 0.383 |
| A | 0.288 | 0.373 | 0.067 | 0.617 | |
| rs2159100 | G | 0.711 | 0.642 | 0.928 | 0.383 |
| A | 0.289 | 0.358 | 0.072 | 0.617 | |
| rs4765905 | C | 0.710 | 0.636 | 0.928 | 0.400 |
| G | 0.290 | 0.364 | 0.072 | 0.600 | |
| rs2370413 | G | 0.520 | 0.508 | 0.372 | 0.950 |
| A | 0.480 | 0.492 | 0.628 | 0.050 | |
| rs7297582a | G | 0.697 | 0.575 | 0.950 | 0.958 |
| A | 0.303 | 0.425 | 0.050 | 0.042 | |
| rs758170 | G | 0.713 | 0.642 | 0.930 | 0.527 |
| A | 0.287 | 0.358 | 0.070 | 0.473 | |
| rs1860002 | A | 0.561 | 0.517 | 0.652 | 0.058 |
| G | 0.439 | 0.483 | 0.348 | 0.942 |
CEU, CEPH = Utah residents with ancestry from northern and western Europe; JPT + CHB = Japanese in Tokyo and Han Chinese in Beijing; YRI = Yoruba in Ibadan, Nigeria.
Single-nucleotide polymorphisms associated with bipolar disorder in previous genome-wide association studies of European ancestry.
It is known that in the presence of multiple tightly linked markers a haplotype test may be more powerful to detect association than corresponding single-locus tests (56). Haplotype-based analysis of the haploblock encompassing the two previously reported SNPs associated with BP provided significant evidence for association with BP in the Latino population. Our complementary LD block-based analysis revealed that an eight-locus haploblock (consisting of rs769087, rs1006737, rs2159100, rs4765905, rs2370413, rs7297582, rs758170, and rs1860002) provided stronger evidence of association than single-marker analysis. In affected individuals, three distinct haplotypes (GGGCGGGA, GGGCGAGA, and AAAGAGGA) were under-represented and one haplotype (GAAGGAAG) was over-represented, giving rise to a significant permutated whole marker haplotypic association (χ2 sum, p = 0.0018). We performed a conditional analysis to see if the association was driven by the two previously reported risk variants. We removed the two variants and performed the family-based haplotype association test with the remaining six variants under identical parameters outlined in the methods section. These results were very similar to our initial findings, in which one common and three rare haplotypes were significantly associated (see Supplementary Table 1). The whole marker permutation test was also significant, indicating that the association is not driven by these two previously reported SNPs.
Since the fruition of this study, The Psychiatric GWAS Consortium (PGC) BP Working Group published their results of the most comprehensive GWAS of BP to date, including 16,731 samples and a replication sample of 46,918 individuals. They report a genome-wide significant SNP, rs4765913, in CACNA1C associated with BP (57). A combined analysis of PGC BP and PGC schizophrenia showed stronger association of this SNP compared to the BP GWAS alone (26, 57). The PGC significant variant falls between two haploblocks identified in our Latino population (Fig. 2). This SNP is approximately 6 kb downstream from the proximal marker of the associated haploblock and approximately 16 kb upstream of the next closest marker. Although we did not test SNP rs4765913 directly, we believe it is very likely, due to its proximity, to be in strong LD with the haploblock associated with BP in our Latino cohort, thus providing additional support for our findings.
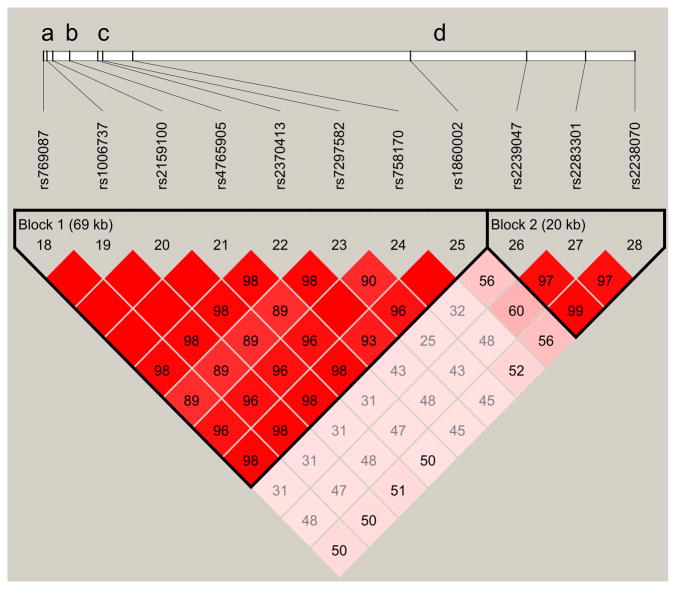
Fig. 2.
Calcium channel, voltage-dependent, L-type, alpha 1C subunit (CACNA1C) haploblocks in the Latino population in relation to single-nucleotide polymorphisms (SNPs) previously associated with bipolar disorder. Haploview V4.2 (53) was used to define linkage disequilibrium (LD) blocks represented by the triangular lines, based on the current genotyping data from the Latino population. The 111.5 kb CACNA1C gene region (shown as a black line) is depicted with respect to 11 genotyped SNPs forming the two haploblocks. Block 1 consists of 8 SNPs in strong LD including rs1006737a and rs7297582c, SNPs that have previously shown to associate with bipolar disorder (20, 21). Ripke et al. (27) reported rs4765905b associated with bipolar disorder and schizophrenia. Subsequent to this study, Sklar et al. (57) reported a bipolar disorder associated SNP (rs4765913d), which is positioned between Blocks 1 and 2. Squares indicate pair-wise r2 values on a red-scale with D′ = 1 (red) through to D′ = 0 (white).
Another recently published study reported a significant association of rs4765905 located within CACNA1C with BP and schizophrenic patients (n = 16,373) compared to control subjects (n = 14,044) (27). SNP rs4765905 was also genotyped in our current study and is nested in the LD block used in this association study. Taken as a whole, these results provide compelling evidence that this region of the CACNA1C gene plays a role in the pathogenesis of BP.
There are several limitations of the present study. First, compared to the previous studies in European ancestry populations, this was a smaller sample size. Larger sample sizes might be required in order to detect statistically significant association for single, common alleles in the Latino population. Secondly, this study only utilized published SNPs and did not use a sequencing approach to detect other potential SNPs or genetic variants within the gene which might be more directly causal for the disease. Finally, the combination of families from several countries may introduce heterogeneity into the analysis. However, if that were to be the case, this would diminish the chances of identifying association of gene variants. There are also some strengths to the present study: family-based association analyses were used, which prevent potential false positive results due to stratification in case-control analyses, and a narrow phenotype of carefully diagnosed BP-I and SABP cases were utilized as the affected phenotype of interest, drawn from families with multiplex BP cases.
In conclusion, these results provide the first evidence that genetic variations in CACNA1C might play a role in the pathogenesis of BP in the Latino population. We provide further support to previous case-control studies in Caucasian populations, strengthening the notion that genetic variation of CACNA1C influences the susceptibility to BP. Further investigations of the CACNA1C gene are thus warranted to increase our understanding of its role in the pathogenesis of BP. Careful replication with appropriate designs in independent sample sets from the Latino population is needed as advocated for stringent confirmation of genetic associations. Furthermore, comprehensive fine-mapping and resequencing will be required to characterize the disease gene architecture of this region in the Latino population.
Supplementary Material
Acknowledgments
We thank all the patients and their families for their support and participation. This study was funded in part by the National Institutes of Mental Health (RO1-MH0698567) and by the Center of Excellence in Neurosciences at the Paul L. Foster School of Medicine. We utilized the Illumina genotyping system of the Institutional Genomic Resource Core at the University of Texas Health Science Center at San Antonio under the Cancer Center Support Grant (P30 CA54174) through the gracious help of Teresa Johnson-Pais, Mandy Rolando, and Korri Weldon. We also acknowledge the help of Carolina Livi in preparing the illustrations.
Footnotes
Disclosures
The authors of this paper do not have any commercial associations that might pose a conflict of interest in connection with this manuscript.
References
- 1.McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Archiv Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 2.Merikangas KR, Akiskal HS, Angst J, et al. Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Archiv Gen Psychiatry. 2007;64:543–552. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edvardsen J, Torgersen S, Roysamb E, et al. Heritability of bipolar spectrum disorders. Unity or heterogeneity? J Affect Disord. 2008;106:229–240. doi: 10.1016/j.jad.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 5.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett JC, Lee JC, Lees CW, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki K, Tanaka T. Genome-wide association study to identify SNPs conferring risk of myocardial infarction and their functional analyses. Cell Mol Life Sci. 2005;62:1804–1813. doi: 10.1007/s00018-005-5098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 14.Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 17.Smith EN, Bloss CS, Badner JA, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sklar P, Smoller JW, Fan J, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott LJ, Muglia P, Kong XQ, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Blackwood DH, Caesar S, et al. Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Mol Psychiatry. 2011;16:2–4. doi: 10.1038/mp.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira MA, O’Donovan MC, Meng YA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baum AE, Akula N, Cabanero M, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keers R, Farmer AE, Aitchison KJ. Extracting a needle from a haystack: reanalysis of whole genome data reveals a readily translatable finding. Psychol Med. 2009;39:1231–1235. doi: 10.1017/S0033291708005084. [DOI] [PubMed] [Google Scholar]
- 24.Green EK, Grozeva D, Jones I, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casamassima F, Huang J, Fava M, et al. Phenotypic effects of a bipolar liability gene among individuals with major depressive disorder. Am J Med Genet B Neuropsychiat Genet. 2010;153B:303–309. doi: 10.1002/ajmg.b.30962. [DOI] [PubMed] [Google Scholar]
- 26.Hamshere ML, Walters JT, Smith R, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ripke S, Sanders AR, Kendler KS, et al. Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyegaard M, Demontis D, Foldager L, et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry. 2010;15:119–121. doi: 10.1038/mp.2009.69. [DOI] [PubMed] [Google Scholar]
- 29.Moskvina V, Craddock N, Holmans P, et al. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry. 2009;14:252–260. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1. 2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Shimada M, Miyagawa T, Kawashima M, et al. An approach based on a genome-wide association study reveals candidate loci for narcolepsy. Hum Genet. 2010;128:433–441. doi: 10.1007/s00439-010-0862-z. [DOI] [PubMed] [Google Scholar]
- 32.Manji HK, Moore GJ, Rajkowska G, Chen G. Neuroplasticity and cellular resilience in mood disorders. Mol Psychiatry. 2000;5:578–593. doi: 10.1038/sj.mp.4000811. [DOI] [PubMed] [Google Scholar]
- 33.Casamassima F, Hay AC, Benedetti A, Lattanzi L, Cassano GB, Perlis RH. L-type calcium channels and psychiatric disorders: a brief review. Am J Med Genet B Neuropsychiat Genet. 2010;153B:1373–1390. doi: 10.1002/ajmg.b.31122. [DOI] [PubMed] [Google Scholar]
- 34.Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrier E, Pompei F, Ruberto G, Vassos E, Collier D, Frangou S. Initial evidence for the role of CACNA1C on subcortical brain morphology in patients with bipolar disorder. Eur Psychiatry. 2011;26:135–137. doi: 10.1016/j.eurpsy.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Kempton MJ, Ruberto G, Vassos E, et al. Effects of the CACNA1C risk allele for bipolar disorder on cerebral gray matter volume in healthy individuals. Am J Psychiatry. 2009;166:1413–1414. doi: 10.1176/appi.ajp.2009.09050680. [DOI] [PubMed] [Google Scholar]
- 37.Franke B, Vasquez AA, Veltman JA, Brunner HG, Rijpkema M, Fernandez G. Genetic variation in CACNA1C, a gene associated with bipolar disorder, influences brainstem rather than gray matter volume in healthy individuals. Biol Psychiatry. 2010;68:586–588. doi: 10.1016/j.biopsych.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 38.Bigos KL, Mattay VS, Callicott JH, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Archiv Gen Psychiatry. 2010;67:939–945. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wessa M, Linke J, Witt SH, et al. The CACNA1C risk variant for bipolar disorder influences limbic activity. Mol Psychiatry. 2010;15:1126–1127. doi: 10.1038/mp.2009.103. [DOI] [PubMed] [Google Scholar]
- 40.Krug A, Nieratschker V, Markov V, et al. Effect of CACNA1C rs1006737 on neural correlates of verbal fluency in healthy individuals. Neuroimage. 2010;49:1831–1836. doi: 10.1016/j.neuroimage.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 41.Thimm M, Kircher T, Kellermann T, et al. Effects of a CACNA1C genotype on attention networks in healthy individuals. Psychol Med. 2011;41:1551–1561. doi: 10.1017/S0033291710002217. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar SN, Huang RQ, Logan SM, Yi KD, Dillon GH, Simpkins JW. Estrogens directly potentiate neuronal L-type Ca2+ channels. Proc Natl Acad Sci U S A. 2008;105:15148–15153. doi: 10.1073/pnas.0802379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jogia J, Ruberto G, Lelli-Chiesa G, et al. The impact of the CACNA1C gene polymorphism on frontolimbic function in bipolar disorder. Mol Psychiatry. 2011;16:1070–1071. doi: 10.1038/mp.2011.49. [DOI] [PubMed] [Google Scholar]
- 44.Erk S, Meyer-Lindenberg A, Schnell K, et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Archiv Gen Psychiatry. 2010;67:803–811. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- 45.Campos-Sanchez R, Barrantes R, Silva S, et al. Genetic structure analysis of three Hispanic populations from Costa Rica, Mexico, and the southwestern United States using Y-chromosome STR markers and mtDNA sequences. Hum Biol. 2006;78:551–563. doi: 10.1353/hub.2007.0002. [DOI] [PubMed] [Google Scholar]
- 46.American Psychiatric Association. Task Force on DSM-IV. Diagnostic and Statistical Manual of Mental Disorders: DSM-IV. 4. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 47.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Archiv Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 48.Maxwell ME. Family Interview for Genetic Studies (FIGS): Manual For FIGS. Bethesda: Clinical Neurogenetics Branch, Intramural Research Program, National Institute of Mental Health; 1992. [Google Scholar]
- 49.Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM. Best estimate of lifetime psychiatric diagnosis: a methodological study. Archiv Gen Psychiatry. 1982;39:879–883. doi: 10.1001/archpsyc.1982.04290080001001. [DOI] [PubMed] [Google Scholar]
- 50.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 51.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laird NM, Lange C. Family-based methods for linkage and association analysis. Adv Genet. 2008;60:219–252. doi: 10.1016/S0065-2660(07)00410-5. [DOI] [PubMed] [Google Scholar]
- 53.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 54.Barrett JC. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- 55.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 56.Clayton D, Chapman J, Cooper J. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27:415–428. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- 57.Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.