Abstract
A central feature of the continuum of life in sexually reproducing metazoans is the cycle of the germline from one generation to the next. This volume describes the cycle of the germline for Caenorhabditis elegans, through chapters that are focused on distinct aspects or processes in germ cell development. Topics include sequential and dependent processes such as specification of germ cells as distinct from somatic cells, sex determination, stem cell proliferative fate versus meiotic development decision, recombination/ progression through meiotic prophase, contemporaneous processes such as gametogenesis, meiotic development and apoptosis, and continuing the cycle into the next generation through fertilization and the oocyte-to-embryo-transition. Throughout germ cell development, translational control and epigenetic mechanisms play prominent roles. These different aspects of germ cell development are seamlessly integrated under optimal conditions and are modified in the different reproductive strategies that are employed by C. elegans under harsh environmental conditions. In this chapter we set the stage by providing a brief background on the C. elegans system and germ cell development, indicating processes in the cycle of the germline that are covered in each chapter.
Keywords: C. elegans, germ cell, gametogenesis, meiosis, reproduction, somatic gonad, apoptosis, meiotic recombination, sex determination, fertilization, germline stem cell
1. Introduction
The ability of a metazoan to create new organisms via sexual reproduction is the basis for all animal life. This volume represents the current state of knowledge in the quest for understanding the reproductive system in C. elegans, an organism that exists primarily as a self-fertilizing hermaphrodite. The chapters are organized from the earliest inkling of germline development in the P lineage of the embryo (Wang and Seydoux, 2012, Chapter 2) to evolutionary comparisons between C. elegans reproduction and their closest relatives (Haag and Liu, 2012, Chapter 14). Each chapter focuses upon different aspects or processes in germ cell development and differentiation, the genetic and molecular events that are known to be involved with each process, and comparisons with other organisms.
2. Why C. elegans?
In 1897, Emile Maupas, a French zoologist and botanist, described C. elegans as a species of nematode that lives in rich humus in which “[he] came twice across.. in the surroundings of Algiers” (Maupas 1900). His sketches show remarkable detail for both forms of C. elegans: the hermaphrodite producing both sperm and oocytes, and the male producing sperm only. His sketches also reveal that the somatic cells are clearly divided in function. He described the hermaphrodite soma as having a female form with a vulva that is behind the middle point of the body, and the male form with a specialized tail structure with a flattened somewhat “heart-shaped bursa” containing “nine pairs of papillae”. As for the germline, he described the hermaphrodite gonad as a simple S-shaped tube that is divided into “vitellogen” containing the oocytes and “germigen” containing a well-developed central rachis surrounded by a layer of germ cells (Figure 1). He described the male gonad as filled with large numbers of spermatozoa with the exact shape and structure as seen in the hermaphrodites (Maupas 1900). For many subsequent years scientists largely ignored C. elegans. But in the 1960s, Sydney Brenner initiated the modern study of C. elegans as a model organism. Brenner had previously validated the hypothesis that mRNAs are read as triplet codons, and an addition or subtraction of one or two nucleotides created frameshift mutations (Brenner et al. 1965). He attributed the success of determining such basic biology to the use of bacteriophage, which he was able to grow in sufficient quantities to identify the rare mutations that led to the discoveries. He extrapolated that a simple multicellular organism would further the understanding of how cells interact with each other, and the biological basis of behavior, two key biological questions. He chose C. elegans for three reasons. First, like bacteriophage, the worm is small, easy to propagate in large numbers, and therefore amenable to genetic and biochemical analysis. Second, the worm is transparent, meaning the cells were visible in situ, and the organism would not need to be dissected into its parts before analysis. Finally, he was able to induce mutations, and as hermaphrodites are the dominant form, he could create homozygous mutations by selfing. Since a small portion of the population was male, Brenner could also recombine genotypes through crosses. By using standard complementation and linkage analyses of different mutations he isolated, Brenner created the first linkage map, confirming that the genome consists of five pairs of autosomes and a pair of X chromosomes in the hermaphrodite and five pairs of autosomes and one X chromosome in the male (Brenner 1974).
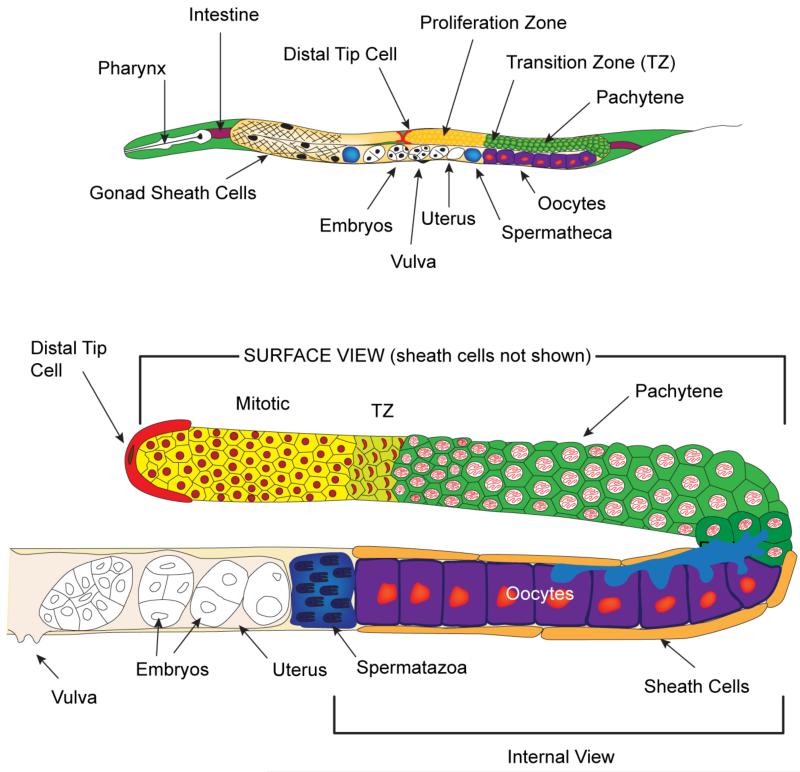
Figure 1. Schematic of adult C. elegans hermaphrodite and gonad.
(Top) Adult C. elegans hermaphrodite, highlighting the reproductive system, which contains two U-shaped gonad arms connected by a common uterus. The distal end of each gonad arm is capped by a somatic distal tip cell (DTC) that covers the distal end of the germline, containing the proliferative zone (yellow). A surface view of the left side U-shaped gonad arm shows the five pairs of somatic gonadal sheath cells covering the area from the transition zone to the spermatheca. On the right side, the gonad arm is shown without the sheath cells. The green cells represent the germ cells in meiotic prophase I, the purple cells represent the developing oocytes, the proximal darker blue area is the spermatheca, and the clear embryos are found within the uterus. (Bottom) A detailed view of one adult hermaphrodite gonad arm is shown. The upper part of the arm is shown as a surface view without the covering sheath cells. The transition zone is visible as a light green color. The lower part of the gonad is an internal view of the proximal region including the sheath cells. The oocytes closest to the loop are connected to the central rachis (light blue, also see Figure 4), which allow cytoplasmic material to enter, while the proximal 4 – 5 oocytes closest to the spermatheca are fully cellularized. See text and wormatlas.org < http://www.wormatlas.org/ > for details.
Further research in the 1970s cemented C. elegans as a premier model organism for reproductive studies. Hirsh et al. (1976) used Nomarski differential interference contrast microscopy to elucidate the different structures of the hermaphrodite gonad, focusing on oogenesis (Figure 2). Ward and Carrel (1979) examined spermatogenesis in both the hermaphrodite and the male, describing the cellular morphology of sperm and the developmental steps to create sperm. Kimble and Hirsh (1979) then used Nomarski microscopy based lineage analysis to trace the development of the hermaphrodite gonad from the early primordium of four cells at hatching to the adult with two U-shaped gonad arms, each containing ~1000 germ cells and a total of 143 somatic gonad cells (Figure 3). The male gonad also begins with a four cell primordium but follows a different pathway of development. Instead of two symmetrical arms, males have a single gonad that contains over 1000 germ cells and 56 somatic cells (Kimble and Hirsh 1979). These studies determined that the cells in the somatic gonad follow an essentially invariant lineage as is seen in the rest of the somatic tissues, whereas germline cell fates depend upon germ cell position within the gonad as well as time (Kimble and Hirsh 1979; Sulston and Horvitz 1977). Since Maupas’ early description of C. elegans and Sydney Brenner’s choice to use C. elegans as a contemporary model, research on this organism has ballooned into a large group of scientists studying almost every aspect of biology from the view of a nematode.
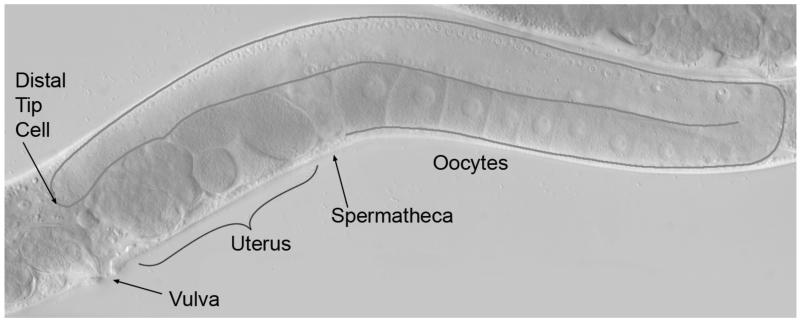
Figure 2. Morphology from Nomarski differential interference contrast (DIC) microscopy.
Live C. elegans adult hermaphrodite showing an interior view of the U-shaped gonad arm in the posterior half of the worm. The gonad is outlined in dark gray where the distal tip cell caps the distal end and developing oocytes populate the proximal end. In the distal half of the gonad, on the dorsal surface, germ cell nuclei are situated along the outer surface of the gonadal tube whereas the central rachis is devoid of germ cells/nuclei. Germ cells move from the very distal end to the proximal end via bulk flow. Overt oocyte development occurs from the loop region into the proximal half of the gonad on the ventral side. The spermatheca lies adjacent to the final oocyte. As an oocyte is ovulated into the spermatheca, a sperm fertilizes it and early development initiates in the uterus. The developing embryos are then released through the vulva to the external environment, where the remainder of embryogenesis occurs. Also see Figure 4.
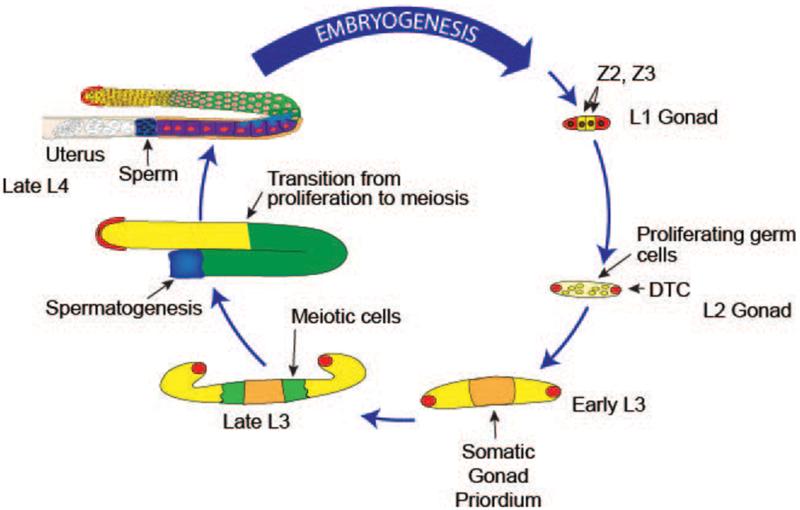
Figure. 3. Cycle of the germline: hermaphrodite gonadogenesis.
Gonadogenesis begins at L1 stage with four gonad precursor cells. Z2 and Z3, the germline progenitors, are sandwiched between the somatic precursors Z1 and Z4 (red), which will divide and form the DTCs and the remaining somatic gonad cells. Prior to the L3 stage, all germ cells proliferate mitotically (yellow). Beginning in early L3, germ cells that are farthest from the DTC leave the proliferative zone, enter meiotic development (green) and progress through meiotic prophase and gametogenesis in an assembly-line fashion. Germ cells that switch from the proliferative fate to meiotic development at this point will become sperm, by the end of the L4 stage in the proximal end of the gonad arm, while those that switch in L4 and adulthood will become oocytes. Migration of the two DTCs generates the U-shape of the gonad: starting in the L3 stage, the DTCs each migrate away from the centrally located gonad primordium along the ventral surface; at the L3/L4 molt, the DTCs each migrate centripetally to the dorsal surface; and in the L4 stage, each DTC migrates back toward the center of the animal. Germ cell proliferation and gametogenesis mediated germ cell volume expansion fills the gonadal tube as the DTCs migrate. Gonads in the schema are for illustrative purpose and are not to scale.
3. Experimental Approaches
Underpinning the entire body of research in C. elegans is the ability to identify the molecular events that control developmental and homeostatic processes via genetic and molecular analyses. Essential genetic approaches include forward genetics, reverse genetics, and creation of transgenic strains. For forward genetics or phenotype based studies, incubating C. elegans with chemical mutagens produces worms with various types of reproductive phenotypes (e.g. sterility) that can be dominant, recessive, or maternal effect, which can be isolated by screening different generation of animals. Secondary screens for enhancers, suppressors or synthetic phenotypes have further expanded the collection of mutations that affect germ cell development. Molecular identification of the genes containing the phenotype-causing lesions has moved from laborious positional cloning to the use of single nucleotide polymorphisms (SNPs), as many natural isolates of C. elegans, for example Hawaiian isolate CB4856, are highly polymorphic containing a SNP every ~1000 base pairs (Jakubowski and Kornfeld 1999; Wicks et al. 2001). From forward genetic screens, mutations in more than 500 genes have been identified that affect fertility (see WormBase at <http://www.wormbase.org/>). In fact, the advent of whole genome sequencing has revitalized the field of forward genetic screens (Sarin et al. 2008; Hobert 2010). Discovering the exact alterations that create the phenotypic change can be done quickly and precisely although as with any gene identification strategy, orthogonal approaches like transgene rescue and phenocopying by RNA interference (RNAi) are required to confirm the identification. With expense rapidly decreasing, whole genome sequencing of mutants is expanding in use.
At the other end of the spectrum are genome sequence driven reverse genetic approaches, which include deletion/ disruption of a gene of interest and RNAi mediated mRNA knockdown, to determine gene function from the resulting phenotype. Gene deletion by random chemical mutagenesis followed by PCR detection has been the workhorse approach (Barstead and Moerman 2006), largely facilitated by the Oklahoma-Vancouver and the NBP-Japanese consortia. More recently site-specific double strand break induction, either with the Mos1 transposase and a large collection of mapped Mos1 transposon insertions (Frokjaer-Jensen et al. 2008) or by designer Zn-finger or TALEN sequence specific nucleases (Wood et al. 2011), has generated gene deletions through imprecise repair by the non-homologous end-joining pathway, or gene replacements through homologous recombination via repair from a transgene template. Genetic epistasis analysis has been an important strategy for ordering genes into pathways/networks, employing null and gain-of-function mutations isolated from forward and reverse genetic approaches (e.g. Hodgkin, 1986).
RNAi, another reverse genetic approach whose mechanism was uncovered in C. elegans, reduces the mRNA level for the gene being tested (Fire et al. 1998). Gene inactivation occurs through an evolutionary conserved pathway of defense against invading viruses, which is triggered by double-stranded RNA (dsRNA). When C. elegans is fed bacteria expressing dsRNA, soaked in a solution of dsRNA, or injected with dsRNA, the organism protects itself by eliminating the incoming dsRNA and any complementary endogenous mRNA (Ahringer 2006). The system self-amplifies, and therefore, is a potent method of gene suppression. Furthermore, the use of a null mutation in the gene rrf-1, which encodes a somatically functioning RNA-dependent RNA polymerase, allows RNAi be largely restricted to the germline (Sijen et al. 2001). When the dsRNA is complementary to an endogenous mRNA, the resulting phenotypic change provides information on the in vivo function of the gene. RNAi is very amenable to high throughput screens, as well as high germline phenotype content screens, and has proven particularly useful in the analysis of the role of essential genes (embryonic or larval lethal) in adult germline development. RNAi can be applied after critical embryonic or larval events have occurred and/or the rrf-1 null mutation can be incorporated into the screening strain. If RNAi is restricted to late larval stages or young adults, the role the gene plays in adult gonad development/ function can be assessed. Green et al. (2011) used a short-term RNAi treatment to examine the function of 554 essential C. elegans genes in the anatomy and function of the adult germline, where 116 genes were completely uncharacterized. High-content phenotypic analysis, scoring 94 phenotypic features, parsed the genes into 102 functional classes allowing the uncharacterized genes to be placed into groups with known function, such as membrane trafficking, glycosylation, fatty acid synthesis, mitochondrial function, transcription, and MAP kinase signaling, to name a few.
Another key method for genetic analysis is the expression of wild type or altered gene products through the generation of transgenic lines. C. elegans germ cells have a very potent mechanism for silencing transgenes that are present as multiple tandem copies, which are generated in the course of constructing standard transgenic lines (Kelly and Fire 1998). Therefore, two methods have been employed to generate single/ low copy integrated transgenic lines that are not silenced, ballistic microparticle bombardment (Merritt and Seydoux 2010) and Mos1 transposase mediated integration into specific Mos1 transposable element landing sites in the genome (Frokjaer-Jensen et al. 2008). To distinguish whether gene function is required in the germline or somatic cells, genetic mosaic analysis using mitotically unstable extra-chromosomal transgene arrays can be employed (Yochem and Herman 2003). Important for genetic screens and phenotypic analysis is transgene tagging of genes and gene products to allow in vivo analysis of expression and localization as well as to mark cell types and various subcellular structures. Central to the in vivo analysis is the use of fluorescently tagged proteins like GFP, which was first employed in C. elegans (Chalfie et al. 1994). The combination of powerful genetic approaches, with visualization of germ cell development in live worms at the level of subcellular structures, for example GFP tagged plasma-membrane and mCherry tagged histones-chromosomes (Figure 4), has and will continue to rapidly advance our understanding of germ cell development.
Figure 4. Fluorescence image of C. elegans adult hermaphrodite.
Live C. elegans hermaphrodite highlighting the U-shaped gonad arm in the posterior half of the worm. The gonad is outlined in yellow where the distal tip cell caps the distal end and developing oocytes populate the proximal end. The strain, OD95 (Green et al. 2011), allows visualization of plasma membranes (green), with a GFP tag fused to the PH-domain from PLC delta, expressed in the germline and early embryo from the pie-1 promoter, and chromatin (red), with an mCherry tag fused to histone H2B, also expressed from the pie-1 promoter. Interior view shows, in the distal half of the gonad, germ cells that are situated along the outer surface of the gonadal tube with openings to the central rachis that is largely devoid of germ cells/nuclei. A single file row of growing oocytes in diakinesis is shown in the proximal half of the gonad. Asterisks indicate highly condensed nuclei that are indicative of apoptosis.
4. Development and Anatomy
In the C. elegans adult hermaphrodite, the germline resides within two U-shaped arms of the gonad, joined at a common uterus (Figure 1). The vulva lies just past the midpoint of the worm on the ventral side, and is a flat slit against the external cuticle. The tube shaped gonad contains germ cells in different stages of differentiation, sequentially developing in an assembly-line fashion from the proliferative germ cells near the somatic distal tip cell (DTC), through meiotic prophase I in the distal gonad and into the loop, and culminating with full-formed oocytes in the proximal gonad. The hermaphrodite specifies male germ cells in the L3 stage, which differentiate into sperm in the L3/L4 stage, and then specifies female germ cells from L4 through adulthood, which differentiate into oocytes. By contrast in the male, spermatogenesis occurs continuously. The pathway that controls germ cell sexual fate is discussed by Zanetti and Puoti (2012) in Chapter 3. Hermaphrodite self-sperm, as well as sperm from a male mating, reside in the spermatheca and await ovulation of the most proximal oocyte, at which point fertilization occurs followed by initiation of embryogenesis (Figures 1, 2 & 4). On the side of the spermatheca towards the vulva resides the uterus, which contains early embryos that eventually pass through the vulva and continue to develop externally. In contrast, the male has one gonad arm and only makes sperm, which passes through the seminal vesicle and vas deferens to be inserted into the vulva of the hermaphrodite via its specialized tail structure during mating. An unmated hermaphrodite is able to produce around 300 embryos, whereas, a mated hermaphrodite can produce up to 1000, demonstrating that the limiting factor for self-fertility is not oocyte production, but rather the amount of self-sperm formed by the hermaphrodite (Hodgkin and Barnes 1991). C. elegans propagate primarily as hermaphrodites, with two X chromosomes and a diploid set of autosomes (2X, 2A), but will produce males (genetically XO) by nondisjunction in about 0.1% of the progeny. If mating occurs, 50% of cross progeny will be male.
Wild-type C. elegans progress through their life cycle in about 3.5 days at 20° C, under optimal laboratory conditions, including constant temperature and unlimited food (equivalent to “feasting at the Hilton”). Under these conditions hermaphrodites are self-fertile for about 4 days with a typical life-span of about two weeks (Figure 5). Early during embryogenesis, the germline is set aside from the somatic cells, as described by Wang and Seydoux (2012) in Chapter 2. After the hermaphrodite lays an embryo, it continues to develop from an oval mass of cells into a small larva, progressing through 2-fold and 3-fold stages of development. Following hatching from the chitin eggshell that protects the embryo, the hatchling develops through four sequential larval stages (L1 to L4), molting its external cuticle with each successive stage. After the final molt emerges a sexually mature adult capable of laying new embryos, and the cycle begins again.
Fig. 5. Life Cycle of C. elegans and Potential Diapauses.
C. elegans adults lay embryos that pass through gastrulation, comma stage, 2-fold and 3-fold embryos before hatching in the L1 larval form. The larvae develop through L2, L3, and L4 molts before becoming adults. The cycle takes about 3.5 days at 20°C under rich nutritional conditions. When the food source becomes scarce, worms at different stages of development can enter diapause where reproductive development is halted and their metabolism is slowed (red arrows). If L1 larvae hatch in the absence of food, they enter L1 diapause, blocking initiation of further development. When food becomes available, L1 larvae reinitiate development. If L2 worms encounter environments with reduced/no food, overcrowding and/or high temperatures, they enter an alternate developmental form called dauer, where development of the reproductive system is arrested, which is resistant to stress and desiccation but is motile and active in searching for food. If the dauer finds food, then the worm enters the developmental program in the L3 stage and proceeds to adulthood. The third diapause occurs when L4/adults find themselves without food. The adult reproductive diapause halts reproduction by degrading most of the germline, but leaving proliferative zone cells that are apparently cell cycle arrested. Upon refeeding, worms in adult reproductive diapause resume germ cell proliferation, meiotic development and oogenesis and can become fertile. Bagging is another strategy to overcome starvation conditions. Here the adult worm stops laying embryos, and any embryos within the mother continue to develop. At hatching, these embryos eat the mother from the inside out. The mother does not survive, but ensures that the embryos have enough food to reach larval diapauses. See Hubbard et al (2012)(Chapter 5) for further discussions of the response of the reproductive system to alternative environments.
The reproductive tract is generated during post-embryonic development, with distinct aspects of gonadogenesis occurring during individual larval stages (Figure 3). L1 larvae have two primordial germ cells (Z2 and Z3) sandwiched between two somatic gonad precursors (Z1 and Z4), surrounded by a basal lamina. Z2 and Z3 begin proliferating in mid-L1 to populate the gonad with germ cells. The somatic precursors also begin to proliferate and constitute 12 cells before the second molt, including two DTCs, with one capping each of the two gonad arms, and 10 other cells that form the hermaphrodite somatic gonad primordium (Kimble and Hirsh 1979). The hermaphrodite DTC has two functions; a migratory/ morphogenetic leader function that gives rise to the U-shape of each gonad arm and a signaling role to promote the proliferative germ cell fate (the male DTCs only have the later function) (Hedgecock et al. 1987; Kimble and White 1981). The 10 somatic gonad primordium cells will give rise to the somatic gonadal sheath cells, the spermatheca and the uterus (Figure 1). The sheath cells are multi functional, including being necessary for oocyte maturation and ovulation, promoting meiotic prophase progression, and robust germ cell proliferation (Govindan et al. 2009; McCarter et al. 1997; Killian and Hubbard 2005).
From the L4 stage the hermaphrodite germline begins to look like its adult counterpart, with the gonad arm on the dorsal side of each U-shaped tube capped by the DTC. Germ cells adjacent to the DTC proliferate mitotically (collectively referred to as the proliferative or mitotic zone), while those some distance away from the DTC enter and progress through meiotic prophase (Figure 1). Pioneering laser ablation experiments by Kimble and White (1981), in which the DTC was killed or its position altered, demonstrated that the DTC promotes the proliferative fate and/or inhibits the meiotic fate, and that it establishes the mitotic–meiotic prophase polarity of the germline. Cells within the proliferative zone include the germline stem cells, with the DTC providing the niche. Subsequent studies found that the DTC signals the distal germ cells via a Notch receptor, such that loss of the signal results in all proliferative cells prematurely switching to meiotic development, while hyperactivation of the Notch pathway results in a germline tumor (Austin and Kimble 1987; Berry et al. 1997)(also see Hansen and Schedl, 2012, Chapter 4). In fact, the description and discovery of the C. elegans DTC controlled proliferative zone helped define the concept of a niche, which is describe by Morrison and Spradling (2008) as a specialized local microenvironment where stem cells reside and that directly promote the maintenance of stem cells.
As germ cells move by bulk flow away from the DTC, escaping its influence, they switch to the meiotic fate, entering and progressing through meiotic prophase and forming gametes. The meiotic and gametogenic events of oogenesis and spermatogenesis are significantly dimorphic, with some of the differences conserved between species. Kim et al (2012) describe the control of oogenesis and meiotic maturation in Chapter 10, Lui and Calaiacovo (2012) describe meiotic development during oogenesis in Chapter 6, while Chu and Shakes (2012) discuss both meiotic development and gamete formation during spermatogenesis in Chapter 7.
Spermatogenesis in both the hermaphrodite and male is rapid with meiotic prophase I, leptotene & zygotene (cells in this stage are in a region of the gonad arm called the transition zone, TZ), pachytene, diplotene and diakinesis, lasting 20-24 hours (Jaramillo-Lambert et al. 2007). Homologous chromosome pairing/ synapsis and initiation of meiotic recombination occurs in leptotene-zygotene, with the resolution of recombination occurring in the transition from pachytene to diplotene. In spermatogenesis all meiotic germ cells undergo two sequential meiotic divisions, without a prophase arrest, to generate four spermatids which, following spermiogenesis, form motile amoeboid sperm (Ward et al. 1981).
Meiotic prophase of oogenesis occurs over a considerably longer period, 54 – 60 hrs, presumably because of the requirement to make the very large, nutrient- and macromolecule rich oocyte and because of controls in the production and release of the oocyte for fertilization. In oogenesis, many more germ cells enter meiotic prophase than actually become oocytes, with the excess apparently functioning as nurse cells that produce macromolecules for oogenesis, with these cells being eliminated by apoptosis during late pachytene (Figure 4). In Chapter 9, Bailly and Gartner (2012) describe how germline cells undergo apoptosis as part of this developmental process, called physiological apoptosis, as well as apoptosis that occurs in response to DNA damage/ unrepaired meiotic recombination.
Oogenesis has an extended pachytene stage, in which germ cells synthesize RNAs and proteins that are donated to the oocyte (Gibert et al. 1984; Schisa et al. 2001). Much of the germline is a syncytium, where the plasma membrane does not fully surround the nucleus, leaving an opening that connects each nucleus and its surrounding cytoplasm to a common cytoplasm. By convention, each nucleus, surrounding cytoplasm and membranes is called a germ cell. While partially syncytial, trafficking of molecules is highly controlled and thus, unlike the syncytial Drosophila embryo, adjacent cells can display distinct behaviors such as being at different mitotic or meiotic cell cycle stage and display differences in molecular marker phenotypes. In pachytene, the germ cells are on the surface of the gonadal tube (Figure 1) with an interior nucleus/ cell free cytoplasmic region called the rachis or core (Figures 2 and 4). As germ cells progress from pachytene to diplotene, oocyte differentiation begins in the loop region with a single file row of growing oocytes found on the external surface of the gonadal tube and the rachis found on the internal surface. RNAs and proteins made in pachytene nuclei are deposited into the rachis and delivered to the growing oocytes via cytoplasmic streaming (Nadarajan et al. 2009; Wolke et al. 2007). The proximal 4 or 5 oocytes are in diakinesis, are fully cellularized (no longer connected to the rachis) and actively uptake yolk produced by the intestine (Grant and Hirsh 1999; Hall et al. 1999; Maddox et al. 2005). The most proximal oocyte (called -1), adjacent to the spermatheca, undergoes meiotic maturation (nuclear envelope breakdown, progression to metaphase of meiosis I and rearrangement of the oocyte cortex and cytoplasm), and is ovulated into the spermatheca and fertilized (McCarter et al. 1999). Meiotic maturation/ ovulation and oocyte production is regulated by the MSP signaling molecule secreted from sperm (Miller et al. 2003; Miller et al. 2001). In the presence of sperm (younger adult hermaphrodite or mated animal), an oocyte undergoes maturation and ovulation every ~23 minutes and oogenesis is continuous (Govindan et al. 2006; Govindan et al. 2009; Lee et al. 2007; McCarter et al. 1999). In the absence of sperm (adult hermaphrodites that have exhausted their self-sperm, sex determination mutant females), oocytes arrest in diakinesis and ongoing oogenesis is inhibited. Thus, oocyte production and utilization only occurs in the presence of sperm. Fertilization triggers completion of meiosis and the initiation of zygotic development. Marecello et al (2012) discuss fertilization in Chapter 11 and Robertson and Lin (2012), in Chapter 12, describe events in the oocyte-to-embryo transition that are essential for launching zygotic development and for the early events that distinguish the somatic and germline lineages, thus continuing the cycle of the germline into the next generation.
5. Physiology and the Environment
How C. elegans maintain reproductive fitness in their natural environment is a newer area of study. In contrast to most of experimental analysis of germline development in the laboratory, where there are unlimited nutrients, C. elegans in the wild live a boom or bust lifestyle, where one moment there is abundant bacteria in their environment, followed by little or no food. This lifestyle puts significant pressure on the organism to delay progeny production until the environment is optimal (Felix and Braendle 2010). In fact, three different diapauses have been identified that reduce metabolism and increase stress resistance in response to starvation conditions (Figure 5). These diapauses keep the worm alive and allow the it to resume normal growth, development, and/or reproduction when environmental conditions become favorable. When newly-hatched L1 larvae are faced with hardship, L1 diapause blocks initiation of post-embryonic development, including blocking somatic gonad (Z1 & Z4) and germ cell (Z2 & Z3) divisions, and larvae becomes metabolically quiescent. This is the major form of C. elegans that is resistant to freezing temperatures, a feature that is exploited in the laboratory to store frozen wild-type and mutant strains. If hardship occurs during L2, an alternate developmental stage called dauer occurs. Dauer stage worms can survive with little to no food for months, where somatic gonad and germ cell development are arrested at the L2 stage. When hardship occurs in the L4/adult stage, adult reproductive diapause can maintain a small group of proliferative zone cells adjacent to the DTC (Angelo and Van Gilst 2009; Seidel and Kimble 2011). The remaining germ cells degrade and no more productive oogenesis or embryogenesis occurs until conditions improve (Figure 5). Upon return of food, the proliferative zone cells resume cell division and differentiation to repopulate the gonad, supporting the view that the proliferative zone contains a true stem cell population. Starved L4/adults have another option to improve the possibility of their offspring surviving. In a process called bagging, the mother stops laying embryos. The existing embryos within the hermaphrodite hatch inside their mother, using her body as a food source. This process is terminal for the parental hermaphrodite, but gives the progeny the ability to survive through the starvation period. An understanding of what determines whether the outcome is bagging or adult reproductive diapause remains to be uncovered. However, the more mechanisms a worm can utilize to survive hardship, the more likely it will be able to propagate from one generation to the next. Hubbard et al. (2012) discuss how physiology and the availability of nutrients control germline development in Chapter 5.
6. Gene expression
Transcription profiling identified significant differences in mRNA content between the germline and the soma, as well as oogenesis versus spermatogenesis: ~1650 genes have oogenesis enriched expression, ~1350 genes have spermatogenesis enriched expression and ~1250 with germline intrinsic expression (Reinke et al. 2004). Epigenetic control mechanisms, which are discussed by Van Wynsberghe and Maine (2012) in Chapter 13, play an import role in a regulation of gene expression in the germline. For example, the X-chromosome is depleted of spermatogenesis expressed genes and is silenced in the germline of XO males, while the few oogenesis genes that reside on the X-chromosome are silenced except for a small burst of expression in late pachytene and diplotene of the hermaphrodite (Kelly et al. 2002). Translational regulation is used extensively in temporal/ spatial control of germline gene expression in C. elegans, as is true in other organisms. For example, Merritt et al. (2008) created fusion genes, which each contain the same germline specific promoter, a histone H2B fused to GFP for visualization, and a unique 3′ UTR from 30 known germline expressed genes. Of the 30 constructs containing different 3′UTRs, 24 had the same expression pattern as the endogenous gene from which the 3′UTR was obtainined. Since the 3′ UTRs are is the only difference among the 24 transcripts, the results suggest that correct protein expression pattern is largely controlled via translational regulation through their 3′UTR. In Chapter 8, Nousch and Eckmann (2012) present the current state of knowledge for translational regulation and its implications for germ cell development.
Self-fertile hermaphroditism in C. elegans is a recently evolved character as most sister species have a male / female mode of reproduction. Haag and Liu (2012), in Chapter 14, discuss how translational control and the rapid evolution of binding sites in mRNA targets, for RNA binding protein regulators, likely contributed to evolutionary differences in germline sex determination in the Caenorhabditis clade.
7. Conclusions
The germline, from one generation to the next, is essential for the continuum of metazoan life. This volume presents a series of reviews that provide our current understanding of the various aspects of germ cell development that are necessary for this continuum in C. elegans. The information, insights and questions posed that are yet to be answered provide a foundation upon which future avenues of research will further our understanding of the cycle of the germline.
Acknowledgments
We thank Dave Hansen for comments. Nanette Pazdernik is supported by NIH T32 HD 49305. Research in the Schedl laboratory is supported by R01 GM085150.
References
- Ahringer J. Reverse genetics. In: Ambros V, WormBook, editor. The C. elegans Research Community. 2006. doi:doi/10.1895/wormbook.1.47.1. [Google Scholar]
- Angelo G, Van Gilst MR. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science. 2009;326(5955):954–958. doi: 10.1126/science.1178343. doi:1178343 [pii] 10.1126/science.1178343. [DOI] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51(4):589–599. doi: 10.1016/0092-8674(87)90128-0. doi:0092-8674(87)90128-0 [pii] [DOI] [PubMed] [Google Scholar]
- Bailly A, Gartner A. this volume of Advances in Experimental Medicine and Biology. 2012. Germ cell apoptosis and DNA damage responses. Chapter 9. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Moerman DG. C. elegans deletion mutant screening. Methods Mol Biol. 2006;351:51–58. doi: 10.1385/1-59745-151-7:51. doi:1-59745-151-7:51 [pii] 10.1385/1-59745-151-7:51. [DOI] [PubMed] [Google Scholar]
- Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124(4):925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- Brenner S. The Genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Stretton AOW, Kaplan S. Genetic Code: The /‘Nonsense/’ Triplets for Chain Termination and their Suppression. Nature. 1965;206(4988):994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chu DS, Shakes DC. this volume of Advances in Experimental Medicine and Biology. 2012. Meiotic development in C. elegans. Chapter 7. [Google Scholar]
- Felix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol. 2010;20(22):R965–969. doi: 10.1016/j.cub.2010.09.050. doi:S0960-9822(10)01168-1 [pii] 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40(11):1375–1383. doi: 10.1038/ng.248. doi:ng.248 [pii] 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert MA, Starck J, Beguet B. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol Cell. 1984;50(1):77–85. doi: 10.1111/j.1768-322x.1984.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Cheng H, Harris JE, Greenstein D. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr Biol. 2006;16(13):1257–1268. doi: 10.1016/j.cub.2006.05.020. doi:S0960-9822(06)01562-4 [pii] 10.1016/j.cub.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development. 2009;136(13):2211–2221. doi: 10.1242/dev.034595. doi:136/13/2211 [pii] 10.1242/dev.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10(12):4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Kao H-L, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, Wang S, Starr DA, Hyman AA, Schedl T, Desai A, Piano F, Gunsalus KC, Oegema K. A High-Resolution C. elegans Essential Gene Network Based on Phenotypic Profiling of a Complex Tissue. Cell. 2011;145(3):470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag ES, Liu Q. this volume of Advances in Experimental Medicine and Biology. 2012. Using Caenorhabditis to explore the evolution of the germ line. Chapter 14. [DOI] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural Features of the Adult Hermaphrodite Gonad of Caenorhabditis elegans: Relations between the Germ Line and Soma. Developmental Biology. 1999;212(1):101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Hansen D, Schedl T. Advances in Experimental Medicine and Biology. 2012. Stem cell proliferation versus meiotic fate decision in C. elegans. Chapter 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH, Stern BD. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100(3):365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Developmental Biology. 1976;49(1):200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hobert O. The impact of whole genome sequencing on model system genetics: get ready for the ride. Genetics. 2010;184(2):317–319. doi: 10.1534/genetics.109.112938. doi:184/2/317 [pii] 10.1534/genetics.109.112938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Barnes TM. More is Not Better: Brood Size and Population Growth in a Self-Fertilizing Nematode. Proceedings of the Royal Society of London Series B: Biological Sciences. 1991;246(1315):19–24. doi: 10.1098/rspb.1991.0119. doi:10.1098/rspb.1991.0119. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Korta DZ, óDalf D. this volume of Advances in Experimental Medicine and Biology. 2012. Physiological control of germline development. Chapter 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski J, Kornfeld K. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics. 1999;153(2):743–752. doi: 10.1093/genetics/153.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol. 2007;308(1):206–221. doi: 10.1016/j.ydbio.2007.05.019. doi:S0012-1606(07)01043-3 [pii] 10.1016/j.ydbio.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125(13):2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee MH, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129(2):479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian DJ, Hubbard EJ. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev Biol. 2005;279(2):322–335. doi: 10.1016/j.ydbio.2004.12.021. doi:S0012-1606(04)00883-8 [pii] 10.1016/j.ydbio.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Kim S, Spike CA, Greenstein D. this volume of Advances in Experimental Medicine and Biology. 2012. Control of Oocyte Growth and Meiotic Maturation in C. elegans. Chapter 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Developmental Biology. 1979;70(2):396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81(2):208–219. doi: 10.1016/0012-1606(81)90284-0. doi:0012-1606(81)90284-0 [pii] [DOI] [PubMed] [Google Scholar]
- Lee MH, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007;177(4):2039–2062. doi: 10.1534/genetics.107.081356. doi:177/4/2039 [pii] 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui DY, Colaiácovo MP. this volume of Advances in Experimental Medicine and Biology. 2012. Meiotic development in C. elegans. Chapter 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox AS, Habermann B, Desai A, Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132(12):2837–2848. doi: 10.1242/dev.01828. doi:132/12/2837 [pii] 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- Marcello MR. this volume of Advances in Experimental Medicine and Biology. Fertilization. Chapter 11. [Google Scholar]
- Maupas Modes et formes de reproduction des nematodes. Translation by Marie-Anne Felix. Archives de Zoologie Experimentale et Generale. 1900;8:463–624. [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181(2):121–143. doi: 10.1006/dbio.1996.8429. doi:10.1006/dbio.1996.8429 S0012-1606(96)98429-8 [pii] [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205(1):111–128. doi: 10.1006/dbio.1998.9109. doi:S0012-1606(98)99109-6 [pii] 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18(19):1476–1482. doi: 10.1016/j.cub.2008.08.013. doi:S0960-9822(08)01053-1 [pii] 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Seydoux G. Transgenic solutions for the germline. WormBook. 2010:1–21. doi: 10.1895/wormbook.1.148.1. doi:10.1895/wormbook.1.148.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291(5511):2144–2147. doi: 10.1126/science.1057586. doi:10.1126/science.1057586 291/5511/2144 [pii] [DOI] [PubMed] [Google Scholar]
- Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17(2):187–200. doi: 10.1101/gad.1028303. doi:10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. doi: 10.1016/j.cell.2008.01.038. doi:S0092-8674(08)00139-6 [pii] 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S, Govindan JA, McGovern M, Hubbard EJ, Greenstein D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development. 2009;136(13):2223–2234. doi: 10.1242/dev.034603. doi:136/13/2223 [pii] 10.1242/dev.034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M, Eckmann CR. this volume of Advances in Experimental Medicine and Biology. 2012. Translational control in the C. elegans germ line. Chapter 8. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131(2):311–323. doi: 10.1242/dev.00914. doi:10.1242/dev.00914 dev.00914 [pii] [DOI] [PubMed] [Google Scholar]
- Robertson S, Lin R. this volume of Advances in Experimental Medicine and Biology. 2012. The oocyte-to-embryo transition. Chapter 12. [DOI] [PubMed] [Google Scholar]
- Sarin S, Prabhu S, O’Meara MM, Pe’er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods. 2008;5(10):865–867. doi: 10.1038/nmeth.1249. doi:nmeth.1249 [pii] 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa JA, Pitt JN, Priess JR. Analysis of RNA associated with P granules in germ cells of C. Development. 2001;128(8):1287–1298. doi: 10.1242/dev.128.8.1287. [DOI] [PubMed] [Google Scholar]
- Seidel HS, Kimble J. The oogenic germline starvation response in C. elegans. PLoS One. 2011;6(12):e28074. doi: 10.1371/journal.pone.0028074. doi:10.1371/journal.pone.0028074 PONE-D-11-15470 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Maine EM. this volume of Advances in Experimental Medicine and Biology. 2012. Epigenetic Control of Germline Development. Chapter 13. [DOI] [PubMed] [Google Scholar]
- Wang JT, Seydoux S. this volume of Advances in Experimental Medicine and Biology. 2012. Germ cell specification. Chapter 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Argon Y, Nelson GA. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol. 1981;91(1):26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematodeCaenorhabditis elegans. Developmental Biology. 1979;73(2):304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28(2):160–164. doi: 10.1038/88878. doi:10.1038/88878 88878 [pii] [DOI] [PubMed] [Google Scholar]
- Wolke U, Jezuit EA, Priess JR. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 2007;134(12):2227–2236. doi: 10.1242/dev.004952. doi:dev.004952 [pii] 10.1242/dev.004952. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. doi: 10.1126/science.1207773. doi:science.1207773 [pii] 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Herman RK. Investigating C. elegans development through mosaic analysis. Development. 2003;130(20):4761–4768. doi: 10.1242/dev.00701. doi:10.1242/dev.00701 130/20/4761 [pii] [DOI] [PubMed] [Google Scholar]
- Zanetti S, Puoti A. this volume of Advances in Experimental Medicine and Biology. 2012. Sex determination in the C. elegans germline. Chapter 3. [DOI] [PubMed] [Google Scholar]