Abstract
Drosophila heart development is an invaluable system to study the orchestrated action of numerous factors that govern cardiogenesis. Cardiac progenitors arise within specific dorsal mesodermal regions that are under the influence of temporally coordinated actions of multiple signaling pathways. The Drosophila Iroquois complex (Iro-C) consists of the three homeobox transcription factors araucan (ara), caupolican (caup) and mirror (mirr). The Iro-C has been shown to be involved in tissue patterning leading to the differentiation of specific structures, such as the lateral notum and dorsal head structures and in establishing the dorsal-ventral border of the eye. A function for Iro-C in cardiogenesis has not been investigated yet. Our data demonstrate that loss of the whole Iro complex, as well as loss of either ara/caup or mirr only, affect heart development in Drosophila . Furthermore, the data indicate that the GATA factor Pannier requires the presence of Iro-C to function in cardiogenesis. Furthermore, a detailed expression pattern analysis of the members of the Iro-C revealed the presence of a possibly novel subpopulation of Even-skipped expressing pericardial cells and seven pairs of heart-associated cells that have not been described before. Taken together, this work introduces Iro-C as a new set of transcription factors that are required for normal development of the heart. As the members of the Iro-C may function, at least partly, as competence factors in the dorsal mesoderm, our results are fundamental for future studies aiming to decipher the regulatory interactions between factors that determine different cell fates in the dorsal mesoderm.
Introduction
One of the fundamental questions in developmental biology is how multiple recurring signal inputs are interpreted and integrated to generate different cell fates. The dorsal mesoderm of Drosophila embryos is an ideal system to study the molecular mechanisms that determine the different developmental fates of dorsal mesodermal cells. The dorsal mesoderm initially becomes subdivided into the cardiac, visceral and somatic mesodermal domains, which harbor progenitors for the heart (myocardial and pericardial cells), the circular midgut muscles and for some somatic, dorsolateral body-wall muscles [1]. The determination of these three mesodermal primordia, as well as the specification and differentiation of the different mesodermal cell types require complex interactions between a number of factors including signaling pathways initiated by the Wnt family member Wingless (Wg), the TGF-β family member Decapentaplegic (Dpp), the Notch signaling pathway and the Ras/mitogen-activated protein kinase (MAPK) pathway, which can be activated by the epidermal (EGFR) or fibroblast growth factor receptor (FGFR) [2,3,4,5,6,7,8]. Previous studies have established a hierarchy of these pathways where the ectodermal Wg and Dpp signals prepattern the mesoderm. Dpp is expressed along the dorsal edge of the ectoderm and its signal is required for the induction of the visceral mesoderm [4,9]. Dpp maintains the mesodermal expression of tinman (tin), a homeobox transcription factor crucial for rendering cells competent to respond to the signaling pathways mentioned above [4,10]. Wg is expressed in stripes perpendicular to dpp and is restricted to the anterior domain of each trunk segment. The combined activity of Dpp and Wg distinguishes the cardiac and somatic mesoderm from visceral mesoderm [11,12]. Tin is indispensable but not sufficient for the formation of the different dorsal mesodermal primordia [10,13,14]. In addition to tin, cardiac mesoderm formation depends on the GATA factor pannier (pnr), the T-box factors Dorsocross1-3 (referred to as Doc) and the LIM-homeodomain transcription factor tailup (tup) [15,16,17,18,19]. Once the cardiac progenitors have been specified they differentiate into myocardial and pericardial cells that build the fly heart.
In Drosophila , the Iroquois complex (Iro-C) of homeobox transcription factors consists of three highly conserved TALE (three amino acid loop extension) homeodomain proteins Araucan (Ara), Caupolican (Caup) and Mirror (Mirr). In addition to the conserved homeodomain, the amino acid sequence of Iro-C members harbors an EGF-like motif [20,21,22], which represents a putative protein-protein interaction domain that is similar to the interaction domain of Notch proteins [20,21]. Two members, Ara and Caup, also contain two putative MAPK phosphorylation sites, suggesting that their transcriptional activity can be regulated by the MAPK pathway [20]. Indeed, such a regulation has been described for Caup [23] and for vertebrate Irx2 [24]. Alignments of the three proteins show that Ara and Caup are more similar to each other than they are to Mirr, however, all three proteins share a high sequence identity in the homeodomain and in the EGF-like motif [20,21]. Furthermore, Ara and Caup have highly similar expression patterns and were shown to function redundantly in different tissues [20,22,23]. Previous studies have identified two distinct functions for Iro-C. First, Iro-C is involved in establishing domains in which the cells acquire competence to properly interpret and respond to the signaling pathways they are exposed to, hence the cells acquire a particular differentiation potential. This has been studied with respect to notum patterning, specification of dorsal head structures and the dorsal-ventral subdivision of the eye [25,26,27,28,29]. Second, Iro-C is required for the specification and/or differentiation of specific structures or cell types such as wing veins, sensory organs of the notum and some lateral transverse muscles [20,23]. In vertebrates, the Iro complex comprises six members (irx1-6) that are expressed both in distinct and overlapping domains including the developing mammalian heart [30,31]. Owing to their expression in specific structures of the heart and possible functional redundancy, the overall heart phenotypes of single irx knockout mice are not dramatic, yet a missing irx gene does impact proper heart function [31]. For example, irx3 is required for the normal electrophysiological properties of the heart [32] and adult irx4-deficient mice develop cardiomyopathy [33].
We were interested to determine whether Iro-C functions in Drosophila heart development since this process also requires the establishment of a domain that is competent to respond to cardiac signals. To this end we investigated the expression pattern of Ara/Caup and Mirr during embryogenesis and analyzed the heart phenotypes in different Iro-C mutant embryos. At early embryonic stages, the expression pattern suggests an involvement of Iro-C in patterning the dorsal mesoderm along the anterior-posterior (AP) axis. Analyses of cardiac markers in different Iro-C mutants at early stages confirmed this hypothesis. Interestingly, the expression of all crucial cardiac transcription factors was affected except for the GATA factor pnr. Analyses of heart markers at late embryonic stages revealed distinct phenotypes showing a disorder in the stereotypic pattern of the heart cells. By stage 15 we detected the expression of Ara/Caup in the most anterior Even-skipped (Eve) pericardial cells. Interestingly, we also observed Ara/Caup in cells distributed evenly along the heart tube that appear to be distinct from any of the heart cells known so far. These newly identified Ara/Caup positive heart-associated cells also expressed the third Iro-C member, Mirr. Mirr protein was additionally expressed in a cell adjacent to the Ara/Caup/Mirr-positive cell along the heart tube. Taken together, our data describe the cardiac mesoderm as an additional domain where members of the Iro-C may function as competence factors, which are also involved in the differentiation of heart cell types.
Materials and Methods
Drosophila stocks and crosses
The following mutant fly stocks were used: Df(3L)iro-2 (stock# 4507), Df(3L)iro DFM3 (stock# 36531), tup isl-1 (stock# 3556), pnr VX6 (stock# 6334) (all from The Bloomington Stock Center), mirr e48 [21], iro DFM1 [20], tin 346 [13] and Df(3L)DocA [34]. Df(3L)iro-2, Df(3L)iro DFM3, mirr e48 and iro DFM1 flies were rebalanced with TM3, ftz-lacZ to identify homozygous mutant embryos. CantonS (stock#1) served as a wild-type stock. Of note, the deficiency Df(3L)iro-2 lacks several other genes in addition to ara, caup and mirr. Although none of these genes have been associated with heart development, a few of them are involved in modulating different signaling pathways, e.g. the Wg and Notch pathways. The following Gal4 and UAS-lines were used: twi-Gal4;24B-Gal4 [17], UAS-caup, UAS-ara [20], UAS-mirr [21].
Immunohistochemistry and in situ hybridization
Antibody staining (single and double labeling) was performed essentially as described [35]. Primary antibodies that did not require amplification of the signal were detected with a DyLight594-conjugated AffiniPure donkey anti-rabbit IgG (H+L) (1:200) (Dianova, Hamburg, Germany). If amplification of the signal was necessary, biotinylated secondary antibodies were used (1:200) (Dianova) in combination with the Tyramide Signal Amplification System (Perkin Elmer, Rodgau, Germany) and dichlorotriazinylamino fluorescin (DTAF) (1:200) (Dianova). Embryos were mounted in Vectashield (Vector Laboratories). Embryos from immunostainings and in situ hybridizations were analyzed using the Keyence BZ-8000K epifluorescence microscope, with the image-analyzing software BZ-Analyzer (Keyence, Neu-Isenburg, Germany) and on the Leica TCS SP5II confocal microscope. Statistical computing was performed using GraphPad Prism (Prism-graphpad.com).
Primary antibodies were used at the following dilutions: rat anti-Caup (1:200), this antibody recognizes Ara as well [36,37], rabbit anti-Mirr (1:500) [38], rabbit anti-Dmef2 (1: 2000) [39], rabbit anti-Tin (1:500) [40], rabbit anti-Tin (1:1000) [41], rabbit anti-β3Tubulin (1:5000) [42], rabbit anti-Odd (1:100) [43], rabbit anti-Doc 2 (1:2000) [34], rabbit anti-Eve (1:1000) [44], rabbit anti-Zfh-1 (1:5000) [45]. The following primary antibodies were from the Developmental Studies Hybridoma Bank (DSHB): mouse anti-En (1:1), mouse anti-Wg (1:1), mouse anti-chicken Isl1 (Tup) (1:20) [19], mouse anti-Eve (1:20), mouse anti-FasIII (1:100), mouse anti-Prc (EC11) (1:20).
Fluorescent in situ hybridization for caup, dpp and pnr was performed according to standard procedures. The dpp antisense RNA was generated from the 2.9 kb dpp E55 fragment [46], the pnr antisense RNA from a 2.6 kb fragment [47]. The plasmid containing caup cDNA (RE64213) was obtained from the Drosophila Genomics Resource Center (DGRC). For generating an in situ RNA probe, a 982 bp fragment of the caup cDNA (EcoRI / HindIII) was subcloned into pBKS. Double fluorescent in situ hybridization and immunostaining was adapted from Knirr et al. (1999) [48]. The digoxigenin-labeled RNA in situ probes were generated using the DIG RNA Labeling mix from Roche (Mannheim, Germany).
Results
Embryonic expression pattern of the members of the Iro-C
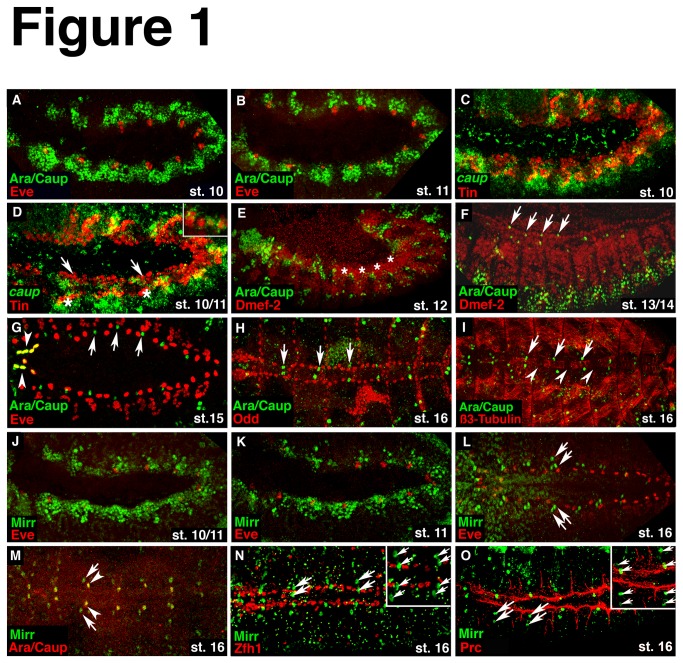
We first evaluated the expression pattern of Ara/Caup and Mirr with respect to their possible involvement in heart development. Of note, since the only available and widely used antibody against Caup also detects Ara, the immunostainings are labeled as Ara/Caup [23,36,37]. At stage 10 Ara/Caup-expressing cells are detected along the dorsal edge of the ectoderm (data not shown and [27,37]) and in an undulating pattern in the dorsal mesoderm (Figure 1A). During stage 11 the initially continuous expression of Ara/Caup in the dorsal mesoderm becomes restricted to segmental patches with the Ara/Caup-positive clusters abutting the Eve expressing cells that are progenitors for pericardial cells and dorsal somatic muscles [44,49] (Figure 1B). An in situ hybridization for caup mRNA transcripts coupled with an immunostaining for Tin confirms the presence of caup mRNA in the dorsal mesoderm that encompasses cardiac and visceral mesoderm (Figure 1C). Ara mRNA transcripts are detected in a similar pattern albeit at a lower level compared to the mesodermal expression of caup mRNA transcripts (data not shown). When Tin expression becomes restricted to dorsal and ventral clusters that represent cardiac and visceral mesodermal cells, respectively, we detect a stronger expression of caup mRNA around the ventral clusters of Tin-positive cells (Figure 1D). During stage 12, Ara/Caup-positive cells disappear in the dorsal region where heart progenitors have been specified and begin to differentiate (Figure 1E). During stage 13 we detect single Ara/Caup-expressing cells alongside the row of Dmef2-positive myocardial cells (Figure 1F). A double immunostaining for Ara/Caup and the pericardial cell marker Eve shows that the Ara/Caup-expressing cells are located between the Eve pericardial cells by stage 15. The Eve pericardial cells at the anterior tip of the heart co-express Ara/Caup (Figure 1G). To determine whether the Ara/Caup-positive cells that appear to be embedded between the Eve pericardial cells belong to one of the known pericardial cell types, we performed double immunostainings for Ara/Caup and Odd (Figure 1H), Ara/Caup and Tin (Figure S1A) and a double staining for caup mRNA and Zfh-1 protein (data not shown). We did not detect a co-expression of Ara/Caup with any of these factors. A double immunostaining for Ara/Caup and the structural protein β-Tubulin, a marker that specifically labels the four Tin-positive myocardial cells in each hemisegment [50], shows that the Ara/Caup-expressing cells are located laterally to the β-Tubulin-expressing (and therefore also Tin-positive) cells (Figure 1I). The expression pattern of Mirr is similar to that of Ara/Caup in that Mirr protein is detected in the dorsal mesoderm by stage 10/11 with a more pronounced expression around the Eve cell clusters (Figure 1J) and in the dorsal ectoderm (data not shown and [21]). Similar to Ara/Caup, the initial continuous expression along the dorsal side becomes restricted to segmental patches during stage 11 (Figure 1K) and vanishes from the dorsal mesoderm during mid-embryogenesis (data not shown). By stage 15/16 Mirr is expressed segmentally in pairs of cells along the heart tube, however, unlike Ara/Caup, Mirr is not expressed in any of the Eve pericardial cells (Figure 1L). Moreover, one of the two Mirr positive cells co-expresses Ara/Caup (Figure 1M). From this observation we conclude that the Ara/Caup expressing cells embedded between the Eve pericardial cells shown in Figure 1G are also positive for Mirr. We did not observe a co-localization of Mirr and Zfh-1 (Figure 1N) or of Mirr and Tin (Figure S1B) in pericardial cells. To obtain more information on the localization of the Mirr-expressing cell pairs, we performed a double-immunostaining for Mirr and the pericardial cell marker, Pericardin (Prc) (Figure 1O). Prc is an extracellular matrix protein that is detected around pericardial cells and that strongly labels the basal membrane of the epithelial myocardial cells [51]. Additionally, Prc is expressed along the seven alary muscles projecting from the dorsal vessel. The Mirr-expressing cell pairs lie approximately in the middle between the Prc-positive extensions with one cell being almost adjacent to the Prc-expressing basal membrane of the myocardial cells. In summary, the early expression pattern of members of the Iro-C in the dorsal mesoderm suggests a role for these factors in establishing territories with different cell fates. The co-expression of Ara/Caup and Eve in the anterior Eve pericardial cells suggests a function for Ara/Caup in the diversification of pericardial cells. Lastly, the Ara/Caup/Mirr-positive cells, as well as the Mirr-only positive cells detected along the forming heart tube may represent novel heart-associated cells.
Figure 1. Expression pattern of Ara/Caup and Mirr during embryogenesis in wild-type embryos.
(A) Double immunostaining for Ara/Caup and Eve shows the mesodermal expression of Ara/Caup as a continuous band along the dorsal side of the embryo. (B) During stage 11 Ara/Caup proteins are present in segmental clusters abutting the Eve cell clusters. (C) In situ hybridization for caup transcripts and immunostaining for Tin further demonstrates the presence of caup in the dorsal mesoderm. (D) Starting at stage 11 Tin-positive cells segregate into cardiac cells (arrows) and cells of the visceral mesoderm (asterisks). There is still a considerable overlap of Tin and caup mRNA expression. The inset in (D) show a higher magnification to better visualize caup mRNA transcripts around Tin protein that is located in the nucleus. (E) By stage 12 Ara/Caup expression has vanished from the dorsal mesoderm (asterisks) where heart cells begin to differentiate. (F) Starting at mid-embryogenesis, single Ara/Caup-positive cells arise along the Dmef2 expressing myocardial cell row (arrows). (G) Double labeling for Ara/Caup and Eve at stage 15 shows their co-expression in the most anterior Eve-positive pericardial cells (arrowheads), as well as Ara/Caup-only expressing cells located between the Eve pericardial cells (arrows). (H) The Ara/Caup-expressing cells (arrows) are not positive for the pericardial cell marker Odd. (I) β3-Tubulin that labels the four Tin-positive myocardial cells in each hemisegment by stage16 and allows a more accurate localization of the Ara/Caup expressing cells (arrows) along the heart tube. The arrowheads point to the β3-Tubulin-negative myocardial cells in each segment. These cells express Seven-up and Doc. (J) Mirr expression in the dorsal mesoderm is similar to the expression of Ara/Caup at early embryonic stages. (K) During stage 11, Mirr protein accumulates around the Eve-positive cell clusters. (L) At stage 16, pairs of Mirr-only expressing cells are detected between the Eve pericardial cells (arrows). (M) Double labeling for Ara/Caup and Mirr reveals the co-expression of all three factors in one cell (arrowheads) of the segmentally arranged Mirr expressing (arrows) cell pairs. (N) None of the Zfh-1-expressing pericardial cells co-expresses Mirr. (O) Double immunostaining for Mirr (arrows) and the extracellular matrix molecule Pericardin (Prc) that labels pericardial cells and is expressed along the extensions of the alary muscles, shows that the Mirr-positive cells are located between these extensions. Additionally, one of the two Mirr-positive cells lies adjacent to the Prc-expressing basal membrane of the myocardial cells. All embryos are oriented with the anterior to the left. Embryos in A-F, J, K are shown from the lateral side. A dorsal view of the embryos in G, H, I, L-O is shown.
Loss of Iro-C affects expression of crucial cardiac transcription factors except for pnr
The early expression pattern of the Iro-C factors suggests that they are involved in patterning the dorsal mesoderm thereby ensuring the normal development of its derivatives. In the present study we wanted to determine the impact of the loss of Iro-C on heart development. We initially analyzed embryos that contain a deletion on the third chromosome (Df(3L)iro-2) that eliminates all three members of the Iro-C (ara, caup and mirr) [20,52,53]. The phenotypes of an additional deficiency line that lacks ara, caup and mirr (iro DFM3) [36] are shown in Figure S2. Since Ara and Caup have been indicated to act redundantly in different tissues, we chose a second mutant that lacks ara and caup but still expresses mirr (iro DFM1) [20,53,54]. To validate the contribution of ara and caup to the observed heart phenotypes we analyzed embryos with the genetic background Df(3L)iro-2/iro DFM1. These embryos are homozygous mutant for ara and caup and heterozygous for mirr. Since mirr is more divergent from ara and caup and could either not play a role in cardiogenesis at all or elicit a different heart phenotype we investigated embryos mutant for mirr (mirr e48) separately. The mirr e48 allele contains a 1kb deletion in the mirr promoter region and therefore mirr e48 mutants are devoid of the gene product [21,55].
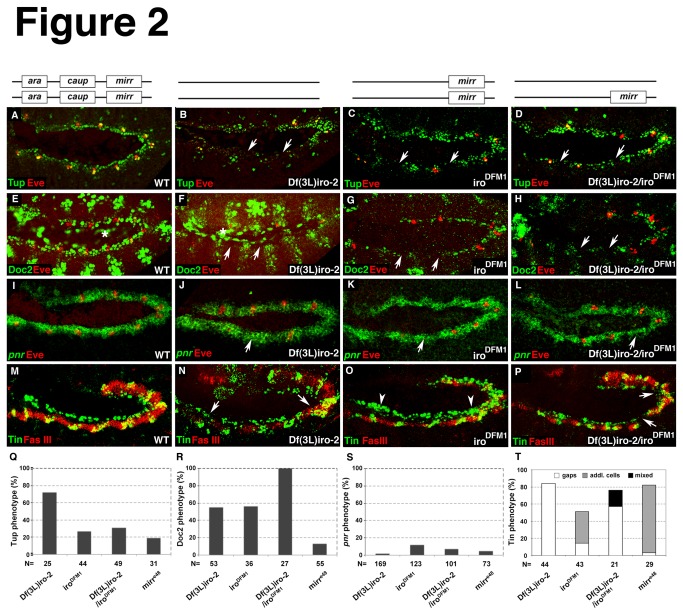
The transcription factors tin, pnr, tup, and Doc are well-characterized components of the transcriptional network that is crucial for the formation of the cardiac mesoderm and are required for the specification and differentiation of heart progenitors. To determine whether their expression depends on Iro-C, we analyzed the expression of Tup, Doc and Tin protein, as well as of pnr mRNA in the different Iro mutant genetic backgrounds at early stages. The bar graphs in Figure 2Q-T depict the percentage of embryos that exhibited a phenotype. The graphs include the quantification of heart phenotypes observed in mirr mutants shown in Figure 3. For both Tup and Doc, we observed a strong downregulation in the dorsal margin of the mesoderm, which corresponds to the cardiac region (Figure 2A-H and Figure 3A-D). A comparison of the penetrance of the phenotypes shows that unlike Tup, Doc is more strongly affected in mutants that lack ara and caup (iro DFM1) (Figure 2Q, R). Since Doc 2 was downregulated only in 13% of the embryos mutant for mirr, it seems that mirr is less important for maintaining Doc (Figure 2R and Figure 3C, D). The expression of pnr transcripts was unaffected in the majority (85-95%) of embryos of all investigated mutants (Figure 2I-L, S and Figure 3E, F). Our current understanding of pnr function in the dorsal mesoderm is that pnr is required for maintaining Doc and Tup expression [18,19]. Since we observed a downregulation of Doc 2 in almost 60% of the iro DFM1 embryos, this finding indicates that ara/caup are required in addition to Pnr to regulate Doc 2 expression. Analyses of Tin expression revealed that the majority of embryos mutant either for ara/caup or mirr exhibit an increased number of Tin-expressing cells (Figure 2M, O, T and Figure 3G, H). Df(3L)iro-2 embryos lacking all three Iro-C members were characterized by small gaps in the Tin-expressing domains at stages 11/12 (Figure 2M, N, T). It may be that in this particular deficiency additional genes are deleted, which influences the phenotype. In fact, 76% of the iro DFM3 mutant embryos that carry a smaller deletion and lack expression of all three Iro-C factors were also characterized by additional Tin-positive cells at stages 11/12 (Figure S2D, E). The majority of heterozygous Df(3L)iro-2/iro DFM1 embryos showed a mild reduction of Tin and approximately 20% of the embryos showed a mixed phenotype consisting of small gaps and accumulation of Tin-positive cells (Figure 2M, P, T). It should be noted that the expression levels of ara, caup and mirr can differ in different Iro-C mutant genetic backgrounds [29]. Different expression levels of individual members of the Iro-C could influence the phenotype since Iro-C proteins were shown to act as homo- and heterodimers [56]. Hence, the proper stoichiometry can be disturbed to various extents in the different mutants. Embryos that only lack mirr are almost exclusively characterized by an overproduction of Tin-expressing cells (Figure 2T and Figure 3G, H). Tin is expressed in cardiac progenitors as well as in precursor cells of the visceral mesoderm (Figure 1D asterisks). Since Ara/Caup expression encompasses the visceral mesoderm we performed a double immunostaining for Tin and Fasciclin III (FasIII) to also label visceral mesodermal cells. FasIII and Tin expression was reduced to variable extents in the majority of Df(3L)iro-2 embryos (Figure 2M, N), in iro DFM1 mutant embryos (Figure 2M, O) and in embryos with the genetic background Df(3L)iro-2/iro DFM1 (Figure 2M, P). In contrast to these mutants, the phenotype for FasIII was striking in iro DFM3 mutants. 88% of the iro DFM3 embryos (n =25) exhibited a strong reduction in FasIII expression (Figure S2E) and the remaining 12% were devoid of FasIII. Since the Iro-C factors are not only expressed in the mesoderm but also in the ectoderm ( [27,37] and data not shown), we tested whether Iro-C regulates the two ectodermal factors, Dpp and Wg that are crucial for heart development. If the expression of Dpp and/or Wg is affected in Iro-C mutants, this could also account for or contribute to the observed heart phenotypes. We did not detect changes in expression of dpp mRNA or Wg protein (Figure 4A-D). Consistent with our findings in the mesodermal layer, expression of pnr in the ectoderm is independent of Iro-C (Figure 4E, F) whereas ectodermal Tup expression requires Iro-C (Figure 4G, H). Taken together these findings indicate a role for Iro-C in the correct patterning of the dorsal mesoderm and in the differentiation of its derivatives.
Figure 2. Iro-C is required for the normal expression of early cardiac transcription factors.
Phenotypes were analyzed in different Iro-C mutants. The genetic background is depicted in the drawing above each column. The boxes illustrate the presence of the indicated factor. (A-D).
Embryos mutant for Iro-C exhibit a reduction of Tup-expressing cells (arrows). (E-H) Doc 2- expressing cells are strongly reduced along the dorsal edge of the mesoderm (arrows). Asterisks in E and F indicate Doc 2 expression in the amnioserosa. (I-L) pnr mRNA expression is unaffected in 85-95% of the different Iro-C mutants. Arrows point to missing Eve cell clusters. (M-P) Tin expression is affected in the different Iro-C mutants. Images are merged optical sections of double immunostainings for Tin and Fasciclin III (FasIII) in which Tin-positive cells in the visceral mesoderm labeled by FasIII appear yellow. (N) Df(3L)iro-2 embryos are characterized by a reduction of Tin-expressing cells (arrows) and reduced FasIII expression. (O) Embryos mutant for ara/caup only (iroDFM1) are characterized by an increase in Tin-positive cells in the cardiac region (arrowheads). (P) The predominant phenotype in Df(3L)iro-2/ iro DFM1 heterozygotes is a slight reduction of Tin and FasIII expression (arrows). (Q-T) Quantification of the phenotypes observed. The bar graphs depict the percentage of embryos that exhibited a phenotype. The bar graphs in Q-S depict the percentage of embryos that had reduced expression of the indicated markers. All images show lateral views of stage10/11 embryos; anterior is to the left.
Figure 3. Loss of mirr affects the normal expression of early cardiac transcription factors.
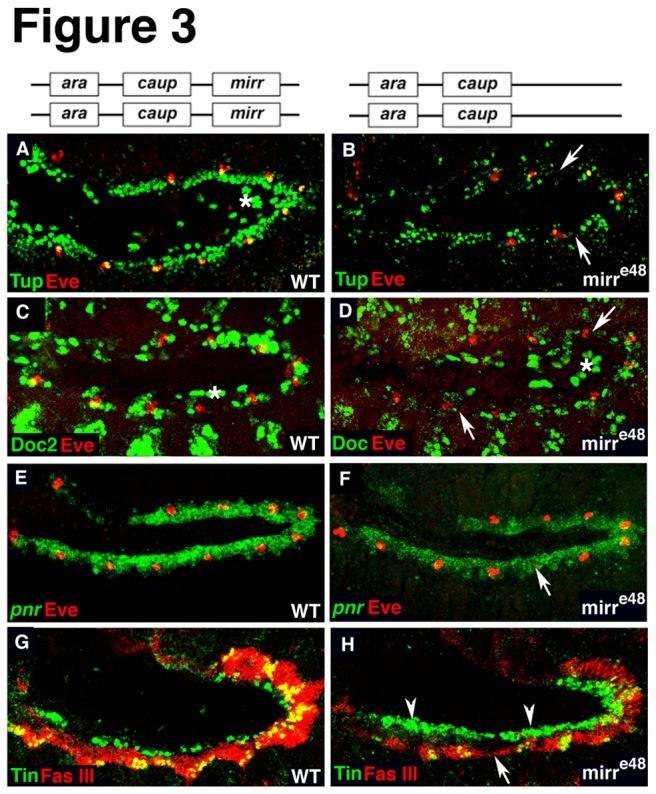
(A-D) mirr mutants are characterized by a reduction of (B) Tup- and (D) Doc-expressing cells (arrows). Asterisks in A, C and D indicate expression of Tup or Doc 2 in the amnioserosa. (E, F) pnr mRNA expression is not affected in mirr mutants. The arrow in F points to a missing Eve cell cluster. (G, H) Images are merged optical sections of double immunostainings for FasIII and Tin-expressing cells that appear yellow in the visceral mesoderm. mirr mutants exhibit an increase in Tin-expressing cells in the cardiac region (arrowheads). FasIII expression is slightly reduced in some embryos (arrow). All images show lateral views of stage10/11 embryos. The genetic background is depicted in the drawing above each column.
Figure 4. Wg and dpp expression in the ectoderm is independent of Iro-C.
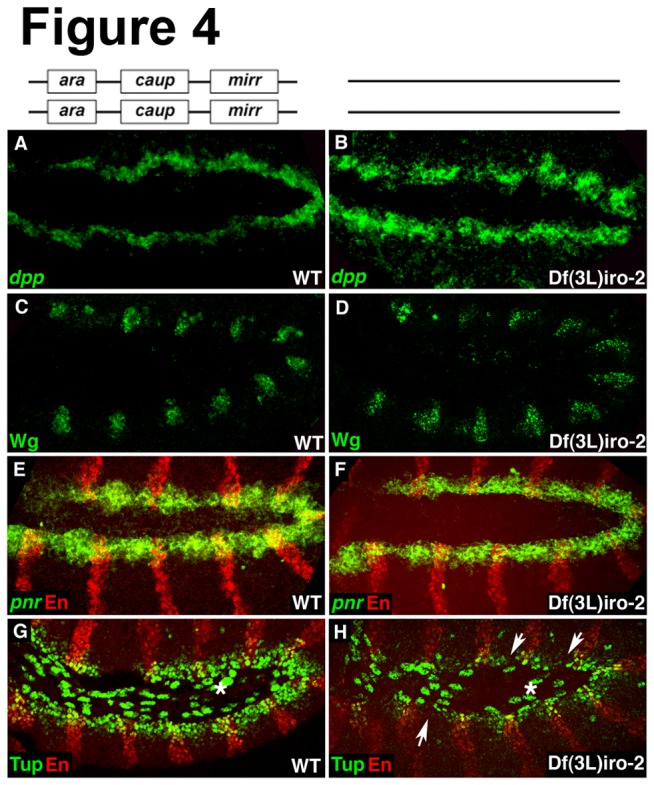
(A-D) Expression of the two crucial cardiac growth factors secreted from the ectoderm, dpp (A, B) and Wg (C, D) is unaffected in Iro-C mutants at early stages. (E, F) Ectodermal pnr mRNA expression does not depend on Iro-C. (G, H) Iro-C is required for ectodermal Tup expression (arrows). Asterisks in G and H indicate Tup expression in the amnioserosa. Images in E-H are confocal images of double stainings using Engrailed (En) as a marker for the ectodermal layer.
Embryos mutant for tin, pnr, Doc or tup show reduced expression of the Iro-C
Having shown that except for pnr, all crucial cardiac factors are affected in embryos mutant for either the whole Iro-C, for ara/caup or mirr we were interested to determine whether tup, Doc, pnr and tin are required for the expression of Ara/Caup and Mirr. Indeed, the expression of Ara/Caup depends on tup, Doc, pnr and tin with the most dramatic loss seen in tin (tin 346) and Doc (Df(3L)DocA) mutants (Figure 5A, C, E, F). Embryos mutant for tup (tup isl-1) exhibit a strong reduction of Ara/Caup-expressing cells (Figure 5A, B). The pnr (pnr VX6) mutants were also characterized by a strong loss of Ara/Caup-expressing cells, however, we frequently observed residual Ara/Caup-positive cells in a segmentally arranged “stripe-like” pattern along the trunk (Figure 5A, D). In contrast to Ara/Caup, Mirr expression appears not as strongly reduced in tup mutants (Figure 5G, H, compare with B). Initiation and/or maintenance of Mirr expression requires Doc, pnr and tin since these mutants are characterized by a dramatic reduction of Mirr-expressing cells (Figure 5I, J, K, L). The quantification of the phenotypes is illustrated in Figure 5M, which shows the percentage of tup, Doc, pnr and tin mutant embryos in which the expression of Ara/Caup and Mirr was downregulated.
Figure 5. Mesodermal expression of Ara/Caup and Mirr depends on the crucial cardiac transcription factors tup, Doc, pnr and tin.
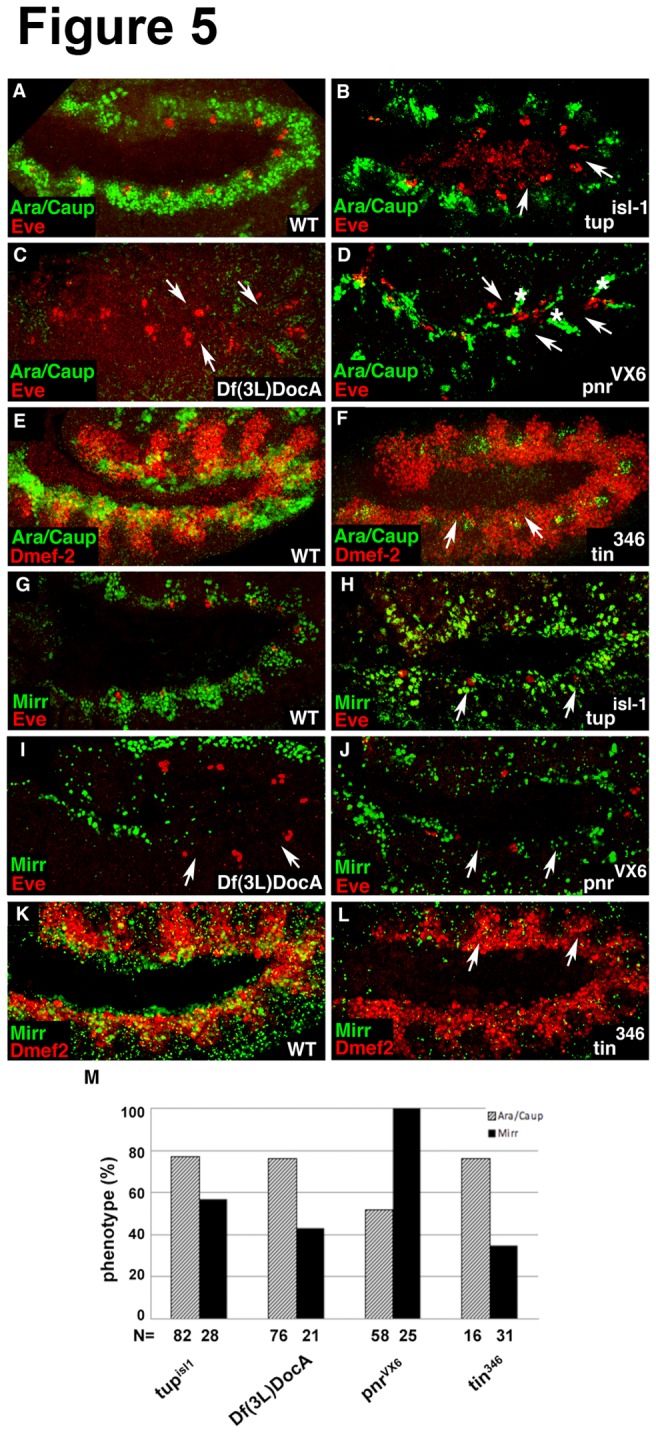
(A, B) Ara/Caup expression is strongly reduced at the dorsal edge of the mesoderm (arrows) in tup isl-1 mutants. (C) Doc mutants (Df(3L)DocA) show a dramatic loss of Ara/Caup expression in the dorsal mesoderm (arrows). (D) In pnr VX6 mutants, Ara/Caup expression is severely reduced (arrows), but some Ara/Caup-positive cells remain in a striped pattern along the dorsal side of the embryo (asterisks). (E, F) A double immunostaining for Ara/Caup and Dmef2 reveals the absence of Ara/Caup in tin mutants (arrows). (G-J) Mirr-expressing cells are mildly reduced in tup isl-1 mutants (arrows in H) whereas a strong reduction of Mirr-positive cells is observed in Doc mutants (Df(3L)DocA) (arrows in I) and in pnr VX6 mutants (arrows in J). (K, L) tin mutants show a strong reduction of Mirr-expressing cells in the dorsal mesoderm (arrows in L). (M) Quantification of the phenotypes. The bar graph shows the percentage of embryos that were characterized by a reduction either of Ara/Caup or Mirr. Lateral views of stage 10/11 embryos are shown.
Loss of Iro-C affects normal heart cell diversification and formation of the heart tube
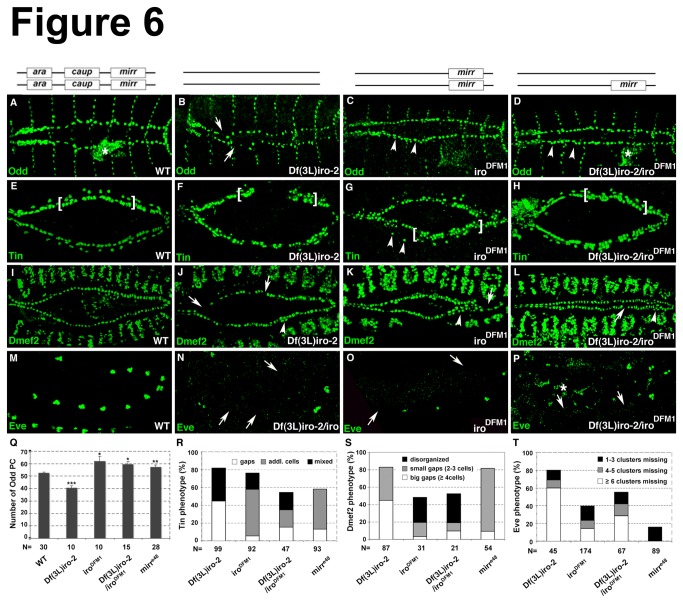
Since the analysis of embryos that are mutant for either the whole Iro-C (Df(3L)iro-2), for ara/caup only (iro DFM1) or for mirr only (mirr e48) revealed interesting phenotypes for crucial heart markers at early stages when the heart cells become specified, we wanted to determine how the early phenotypes affect heart formation at later stages. At stage 16, iro DFM1 and Df(3L)iro-2/iro DFM1 mutants appeared to have additional Odd-expressing pericardial cells (Figure 6A, C, D) and a statistical analysis confirmed this observation (Figure 6Q). Of note, the bar graphs in Figure 6Q-T include the quantification of heart phenotypes analyzed in mirr mutants shown in Figure 7. Embryos mutant for mirr were also characterized by an increase in Odd-expressing pericardial cells (Figure 7C, D). In contrast, Df(3L)iro-2 embryos exhibited a reduced number of Odd-positive pericardial cells (Figure 6A, B). Immunostainings for Tin revealed an abnormal expression pattern at stage 14/15 (Figure 6E-H, R and Figure 7E, F). In wild-type embryos Tin is expressed in four out of the six myocardial cells in each hemisegment and in four pericardial cells (not all of which are always nicely detected in immunostainings before stage 16). As observed in iro DFM1 and mirr e48 embryos at stage 11/12, these embryos were also characterized by the presence of additional Tin-positive cells at stage 14/15 (Figure 6R). A smaller percentage of iro DFM1 and mirr e48 embryos had small gaps and some embryos exhibited both phenotypes, gaps and additional cells (denoted “mixed” in the histogram). Embryos of the deficiency Df(3L)iro-2 exhibited either gaps or a combination of gaps in some hemisegments and accumulation of Tin-positive cells in other hemisegments (denoted “mixed” in the histogram). An additional phenotype observed in approximately 30% of iro DFM1 and mirr e48 embryos (not included in the histogram) indicates a detachment of Tin-positive pericardial cells (Figure 6G and Figure 8). To determine whether the additional Tin-positive cells are pericardial cells, a double immunostaining for Tin and Prc was performed. Indeed, Prc expression is detected around the additional Tin-positive cells (Figure 8A-C). As to the myocardial marker Dmef2, the phenotype encompassed the loss of generally a few Dmef2-expressing cells and a disorganization of the two myocardial cell rows (Figure 6I-L, S). A “disorganized” phenotype is characterized by small gaps and apparently additional cells as seen for example in the embryo shown in Figure 6K. Since the number of Dmef2 cells was not significantly changed in embryos with a “disorganized Dmef2 phenotype” (101 cells in iro DFM1 embryos versus 104 cells in wild-type embryos), we conclude that this is simply a misalignment of the two myocardial cell rows. The majority of embryos mutant for mirr were characterized by a mild loss of Dmef2-expressing myocardial cells and did not exhibit defects in the alignment of the two myocardial cell rows (Figure 7G, H). It should be noted that embryos that carry the smaller deletion of the whole Iro-C (iro DFM3) did not exhibit dramatic heart phenotypes at late stages. We also analyzed the phenotype for Eve at stage 10/11. At this stage Eve is expressed in 11 cell clusters that harbor precursors of Eve-positive pericardial cells and dorsal somatic muscles. In general, embryos of each investigated mutant background were characterized by a loss of Eve cell clusters (Figure 6M-P). Since the phenotypes differed in their severity, they were categorized according to the number of Eve cell clusters that were missing in the different mutants (Figure 6T). A comparison of the phenotypes shows that of all Iro-C factors mirr has the least impact on Eve-expressing cells. Only 15% of mirr mutants showed a phenotype for Eve (Figure 7A, B) and only up to three Eve cell clusters were missing. In contrast, 40% of the embryos mutant for ara/caup (iro DFM1) exhibited a phenotype with approximately 15% of the embryos missing more than 6 Eve cell clusters. Taken together these results demonstrate that the loss of Iro-C affects heart formation at least in part by interfering with the normal diversification of heart cell types.
Figure 6. Heart phenotypes embryos mutant for Iro-C.
The genetic background is depicted in the drawing above each column. The boxes illustrate the presence of the indicated factor. (A-D, Q) Loss of ara/caup results in an increase in Odd-expressing pericardial cells (arrowheads in C, D), whereas embryos carrying the larger deletion exhibited a reduced number of Odd-positive cells (arrows in B). Asterisks in A and D demarcate Odd expression in the gut. Q) The numbers of Odd-positive cells in each Iro-C mutant were statistically tested for significant differences from wild-type embryos using the Mann-Whitney test (* p < 0.01, ** p < 0.001, *** p < 0.0001). Odd-positive cells that are part of the lymph glands were not included in the counting. (E-H) Iro-C mutants exhibit an abnormal pattern of Tin-expressing cells. The brackets highlight some hemisegments in which the normal pattern of four Tin-positive myocardial cells and two Tin-expressing pericardial cells is disorganized. The arrowheads in G point to Tin-positive cells that are detached from the heart. (I-L) Phenotypes for Dmef2 varied in their severity and included loss of Dmef2-positive cells (arrows) as well as misalignment of the two myocardial cell rows (arrowheads). (M-P) Loss of Iro-C results in a reduction of Eve clusters. (R-T) Quantification of the phenotypes observed. The bar graphs depict the percentage of embryos that exhibited a particular phenotype for the indicated marker. All embryos are oriented with the anterior to the left. Dorsal views of embryos at stages 14-16 are shown in A-L. Lateral views of embryos at stages 10/11 are shown in M-P.
Figure 7. Heart phenotypes in embryos mutant for mirr.
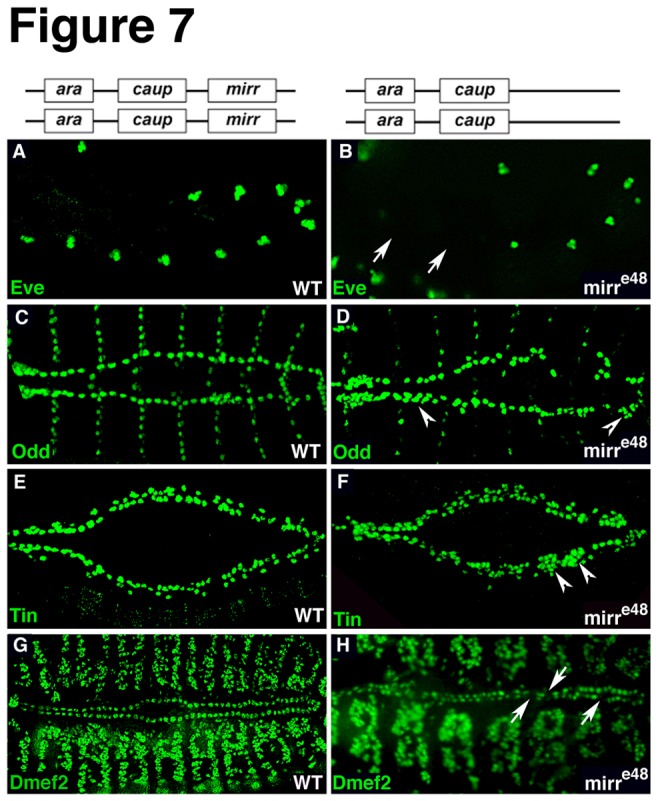
(A, B) mirr mutants have a reduced number of Eve clusters (arrows). (C, D) The number of Odd-expressing pericardial cells is increased in mirr mutants (arrowheads). (E, F) mirr mutants are characterized by an increased number of Tin-positive cells (arrowheads). (G, H) Myocardial Dmef2-expressing cells are slightly reduced in mirr mutants (arrows). Quantification of the phenotypes is included in the bar graphs shown in Figure 6 Q-T.
Figure 8. Double immunostainings for Tin and the pericardial marker Pericardin (Prc) confirm heart phenotypes affecting pericardial cells in iro DFM1 and mirr e48 embryos.
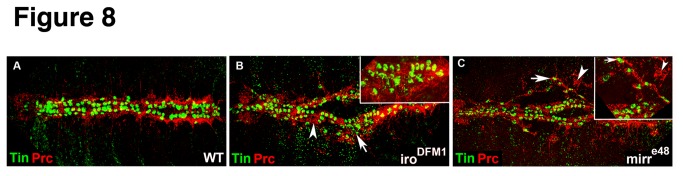
(A) Wild-type expression of Tin and Prc at stage 16. (B) Prc expression is expanded in iro DFM1 embryos (arrowhead) and includes additional Tin-positive cells (arrow). The inset is a higher magnification of the area around the arrow. (C) A mirr e48 embryo showing the detachment phenotype of Tin-expressing pericardial cells (arrows) and ectopic Prc expression (arrowheads).
Mesodermal overexpression of Iro-C elicits different effects on heart formation
Finally, we analyzed changes in the expression of heart markers after overexpressing each factor throughout the mesoderm. Generally, the percentage of embryos exhibiting a phenotype after overexpressing UAS-ara were weaker compared to the effects elicited by overexpressing UAS-caup. We cannot exclude that the difference in the severity of effects may be due to different expression levels of the cDNA constructs. Since ara/caup mutants are characterized by a loss of Eve-expressing cells we expected the opposite effect when overexpressing UAS-ara or UAS-caup. However, overexpression of either factor also resulted mainly in a reduction of Eve-positive cells in approximately 30% of the embryos (n = 150) (Figure S3A-C). Consistent with the results obtained from the analyses of ara/caup and mirr mutants, the effect of overexpressing UAS-mirr on Eve was only observed in 14% (n = 43) of the embryos and less severe compared to the effects elicited by UAS-ara or UAS-caup (Figure S3D). Consistent with the Eve phenotype analyzed in mirr mutants, this finding indicates that mirr has a lower impact on Eve expression as compared to ara/caup. Analyses of Odd-expressing pericardial cells revealed phenotypes that are complementary to the phenotypes observed in ara/caup and mirr mutants. Whereas the mutants were characterized by the presence of additional Odd-pericardial cells, overexpression of UAS-caup or UAS-mirr resulted in the loss of Odd-pericardial cells (Figure S3E, G, H). As to Tin, the phenotype is a dramatic disorganization after overexpression of either UAS-caup or UAS-mirr (Figure S3I, K, L). Interestingly, overexpression of either factor resulted in an ectopic accumulation of Tin-positive cells in the anterior part of the heart where the lymph glands are located.
In summary, our results add the Iro-C to the network of factors that play a role in dorsal mesoderm patterning. Our comprehensive analyses of changes in expression of heart marker genes demonstrate that all three members of the Iro-C are required for normal heart development, although their exact function remains to be studied in more detail. Moreover, the finding that pnr expression is unchanged yet Tin, Doc and Tup expression is not maintained at the normal level is an interesting finding suggesting that Pnr may only fully function in the presence of Iro-C.
Discussion
Tissue patterning requires the spatial and temporal coordinated action of signals providing instructive or permissive cues that result in the specification of different cell types and their subsequent differentiation into different lineages. Our analyses of Iro-C deficient embryos demonstrate that ara/caup and mirr are required in the dorsal mesoderm for normal heart development. The heart phenotypes could be caused by alterations of the fine balance of the interactions between factors of the cardiac signaling network. In early stage Drosophila embryos the mesoderm is patterned along the anterior-posterior (AP) axis with cardiac and somatic mesodermal domains alternating with visceral mesodermal domains. The tin-positive mesoderm is specified as cardiac and somatic mesoderm under the influence of combined Dpp and Wg signaling [3,11,57]. Subsequently, the cardiac and somatic mesodermal domains are further subdivided by the action of the Notch pathway and MAPK signaling activated by EGFR and FGFR [6,7,58,59]. The Eve-expressing cell clusters that give rise to pericardial and DA1 somatic muscle cells [44,49], as well as the Doc expression pattern, distinguish the cardiac and somatic mesodermal domain from the visceral mesodermal domain [18]. The early expression pattern of Ara/Caup and Mirr at stages 10/11 suggests a role for Iro-C in patterning the dorsal mesoderm along the AP axis. Consistent with their previously described functions in other developmental contexts, members of the Iro-C may integrate signaling inputs and interact with other transcription factors to specify different dorsal mesodermal derivatives. Activation of the Iro-C by the EGFR pathway is required for the specification of the notum [60,61]. Mirr was shown to interpret EGFR signaling by eliciting a specific cellular response required for patterning the follicular epithelium [62]. During Drosophila eye development, mirr expression can be regulated by Unpaired, a ligand that activates JAK/Stat signaling [63]. In fact, the JAK/Stat signaling pathway has only recently been added to the signaling pathways that function in Drosophila cardiogenesis [64]. In chromatin immunoprecipitation experiments performed by Johnson et al. [64] caup was identified as a target of Stat92E, which is the sole transcriptional effector of the JAK/Stat signaling pathway in Drosophila [64,65]. Interestingly, the increase of Odd-pericardial cells and the additional Tin-expressing cells that were the characteristic phenotypes in ara/caup (iro DFM1) and in mirr (mirr e48) mutants are highly similar to the phenotypes in stat92E mutants described by Johnson et al. [64]. Also, as described for stat92E mutants, we noticed cell adhesion defects in a number of embryos as determined by the distant location of some Tin-expressing cells from the forming heart tube. As for establishing a possible link between JAK/Stat and Iro-C in the dorsal mesoderm and specifically in cardiogenesis, it would be necessary to determine for example whether caup and mirr can rescue the heart phenotype of stat92E mutants. Also, it would be interesting to compare the expression of the other crucial heart marker genes, Tup, Doc and Pnr, in stat92E mutants at early stages to determine to what extent the phenotypes of embryos mutant for Iro-C and for JAK/Stat signaling are similar.
Members of the Iro-C were shown to be positively or negatively regulated by signaling pathways that play crucial roles in heart development. Conversely, the Iro-C factors can also regulate the activity of at least one of these pathways. Specifically, Ara/Caup, as well as Mirr were shown to regulate the expression of the glycosyltransferase fringe and as a consequence modulate Notch signaling activity in the eye [25,63,66,67]. In the dorsal mesoderm, the lateral inhibitory function of Notch signaling establishes the proper number of heart and muscle progenitors [7,68]. Given the fact that Iro-C can regulate Notch activity it may be that the loss of Iro-C leads to an imbalance of progenitor cell specification resulting in an abnormal number of heart cells. Further studies are required to decipher the molecular mechanism by which Iro-C could integrate diverse signaling inputs and thereby function in the specification and differentiation of the different dorsal mesodermal derivatives.
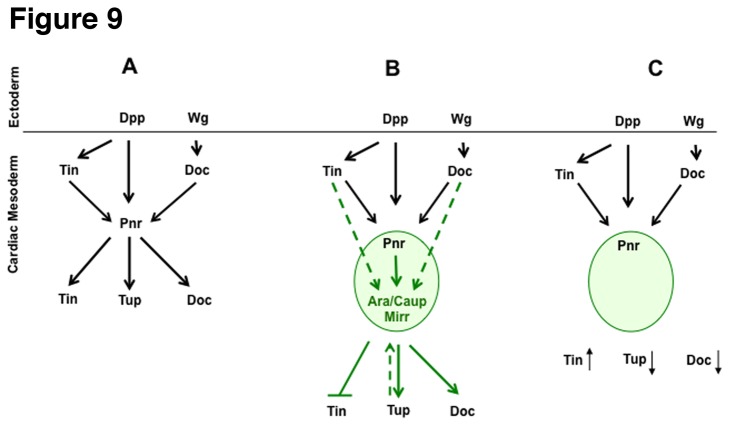
To determine whether Iro-C can be positioned into the early transcriptional network that determines a cardiac lineage, we investigated the interdependency between crucial cardiac factors and Iro-C during cardiogenesis. Analyses of the expression of Ara/Caup and Mirr in tin 346, Df(3L)DocA, pnr VX6 and tup isl-1 embryos demonstrated the dependency of Ara/Caup and Mirr on all four factors. The strongest loss of Ara/Caup and Mirr expression was observed in tin 346 and Df(3L)DocA mutants, which clearly places tin and Doc upstream of Ara/Caup and Mirr. In tup isl-1 and in pnr VX6 mutant embryos, Ara/Caup and Mirr were strongly downregulated, however regarding Ara/Caup, some expression remained in segmental patches suggesting a different level of regulation. The currently available data indicates a positive and a negative regulatory effect of pnr on Iro-C. Whereas pnr restricts Iro-C expression in the dorsal ectoderm and in the wing disc, there is also evidence that pnr can positively regulate Iro-C in the wing disc [26,37]. Whether Pnr activates or represses Iro-C appears to depend on the presence of U-shaped (Ush), a protein that modulates the transcriptional activity of Pnr [37,69]. In the wing disc it was shown that an Iro-C-lacZ (IroRE2-lacZ) construct was activated in cells that contained Pnr but were devoid of Ush [37]. Our data demonstrate that in the dorsal mesoderm, the expression of Ara/Caup and Mirr depends on pnr. Additionally our analyses show that pnr expression is independent of Iro-C, an observation that was previously mentioned by Herranz and Morata [70]. This finding is intriguing with respect to the downregulation of Tup and Doc in Iro-C mutants. Pnr is required for the maintenance of Doc and for the initiation and/or maintenance of Tup [18,19]. Since Iro-C mutants exhibit a reduction in Doc-positive cells despite the presence of pnr, members of the Iro-C appear to be required independently or in addition to pnr to maintain expression of Doc. This could be investigated by expressing ara, caup and/or mirr in the mesoderm of pnr mutants to determine whether these factors are able to restore Doc expression. Alternatively, it may be that Iro-C is required indirectly meaning that its main function is to provide a molecular context in which Pnr can be active. For example, it is known that Ush can bind to Pnr thereby inactivating Pnr function [69]. It is conceivable that the absence of Iro-C affects the spatial expression of Ush. If, in the absence of Iro-C, the expression domain of Ush shifts into the Pnr expression domain, Ush could bind to Pnr and inactivate it in the region where Pnr is required to maintain the expression of Tup and Doc. Adding to the complexity of the interpretation of the observed phenotypes is our finding that the majority of embryos that are mutant for ara/caup or for mirr were characterized by supernumerary Tin-positive cells in the cardiac region by stage 11/12. This phenotype could still be observed at later stages when the heart tube forms. The additional Tin-positive cells are pericardial cells as determined by the expression of Prc around the Tin-expressing cells. Also, we did not observe an increase of Dmef2-positive myocardial cells. Hence, our data suggests a different level of regulation of Tin by the Iro-C. Similar to the findings of Johnson et al. [64], it may be that Iro-C is normally required to restrict Tin expression at an early stage. The regulation of Tin expression can be divided into four phases [64,71]. The phenotype we observe occurs when Tin expression becomes restricted to the myo- and pericardial cells in the cardiac region. In summary, our data adds Iro-C to tin, pnr, Doc and tup whose concerted actions establish the cardiac domains in the dorsal mesoderm (Figure 9). Further studies are required to re-evaluate our current understanding of the interactions between factors of the cardiac transcriptional network.
Figure 9. The cartoons depict regulatory interactions between the cardiac transcription factors including the members of the Iro-C based on our initial analyses on their function in the early dorsal mesoderm.
For the sake of simplicity the cartoons neglect, for example, the different impacts of Ara/Caup and Mirr on the expression of the tested cardiac marker genes as well as additional regulatory interactions between the previously characterized factors. (A) In the previous model pnr was shown to be required for the maintenance of Doc, Tin and Tup in the cardiac mesoderm during stage 11. (B) Our data show that ara/caup and mirr act downstream of pnr or in combination with Pnr protein to regulate cardiac gene expression (specifically, to maintain Tup and Doc expression). Since Iro-C mutants were characterized by the presence of additional Tin-expressing cells, it appears that Iro-C is normally required to restrict the number of Tin-positive cells at stages 11/12. (C) In the absence of Iro-C Pnr is not sufficient to maintain Tup and Doc expression. The fact that we observe additional Tin-expressing cells could result from an earlier pnr-independent event in which Iro-C is involved. (A-C) Black arrows depict known interactions, green arrows depict our results, dashed arrows indicate the possibility that Tin, Doc and Tup could regulate Ara/Caup and Mirr directly without the involvement of Pnr.
According to the expression pattern of Ara/Caup and Mirr we can distinguish between an early and late role for these factors, the latter being a role in the differentiation of heart cells.
Our analyses of the expression of Ara/Caup and Mirr during embryogenesis led to the identification of hitherto unknown heart-associated cells. We detected seven pairs of Ara/Caup and Mirr expressing cells and seven pairs of Mirr only expressing cells located along the dorsal vessel. We did not detect a co-expression with any of the known pericardial cell markers. Because there are seven pairs of these cells segmentally arranged, it was tempting to speculate that these cells may function, for example, as additional attachment sites for the seven pairs of alary muscles. The alary muscles attach the heart to the dorsal epidermis and their extensions can be visualized by Prc. Due to the lack of markers little is known about the development of the alary muscles [72]. Previous work by LaBeau et al. [72] demonstrated that the alary muscles attach to the dorsal vessel in the vicinity of the Svp pericardial cells and, in addition, more laterally to one of two distinct locations on the body wall. Maybe it is the Mirr-positive cells that identify the more lateral locations. Clearly, a detailed analysis is needed to identify the function of the Ara/Caup- and Mirr- as well as Mirr-expressing cells that are positioned along the heart tube and whose existence has now been revealed. Additionally, we identified on each side at the anterior end of the dorsal vessel four pericardial cells that co-express Ara/Caup and Eve. Their location at the anterior tip of the heart is intriguing. Further analysis is required to unambiguously determine whether these cells are, for example, the wing heart progenitor cells [73] or the newly identified heart anchoring cells [74]. It is also possible that they represent a yet undefined, novel subpopulation of pericardial cells. In any case, this finding suggests that Ara/Caup plays a role in the diversification of pericardial heart cell types. Future experiments aim to determine the developmental fate of these cells.
Taken together, our initial investigation of a role for Iro-C in heart development introduces ara/caup and mirr as additional components of the transcriptional network that acts in the dorsal mesoderm and as novel factors that function in the diversification of heart cell types.
Our results show that the role of the Iro-C and its individual members, respectively, appears to be rather complex and awaits in-depth analyses. Nevertheless, this work raises important questions regarding our current understanding of interactions between the well-characterized transcription factors that will be addressed in future studies.
Supporting Information
Ara/Caup and Mirr demarcate a novel heart or heart-associated cell type. (A) Ara/Caup protein is not co-expressed with Tin. Arrows point to Ara/Caup expressing cells. (B) A double immunostaining for Mirr and Tin shows no co-expression of these factors. Arrows point to the pairs of Mirr-expressing cells that are adjacent to Tin-positive cells.
(TIF)
Heart phenotypes in embryos mutant for the Iro-C (iroDFM3). (A-C) Embryos lacking the three members of the Iro-C show a downregulation of Doc in the cardiac region (arrows). (D-F) Almost 80% of the iro DFM3 embryos are characterized by additional Tin-expressing cells at stage 12 (arrowhead). The strong downregulation of FasIII (asterisk) demonstrates the impact of Iro-C on visceral mesoderm development. (G-I) pnr mRNA expression is unaffected in the majority (86%) of iro DFM3 embryos. The arrows point to missing Eve cell clusters. (J-L) Loss of Iro-C results in a reduction of Eve cell clusters (arrow). (M-O) iro DFM3 embryos were characterized by missing Odd-positive cells in some hemisegments (arrow) and additional Odd-expressing cells in other hemisegments (brackets). The overall number of Odd-expressing pericardial cells was not significantly changed compared to wild-type embryos.
(TIF)
Heart phenotypes observed after mesodermal overexpression of individual members of the Iro-C. (A, E, I) Wild-type expression of Eve (A), Odd (E) and Tin (I). (B, F, J) Overexpression of Ara results in (B) the loss of some Eve clusters, (F) a mild disorganization of Odd-expressing pericardial cells and (J) a mild disorganization of Tin-positive cells. The ectopic accumulation of Tin-expressing cells at the anterior end of the heart (arrowhead) appears to be in the region where the lymph glands are located (compare with (E) showing normal Odd expression in the lymph glands (lg)). (C, G, K) Overexpression of Caup not only leads to a loss of Eve clusters but also to a slight expansion of these clusters (C), (G) a dramatic loss of Odd-expressing pericardial cells and (K) a severe heart tube defect as can be seen by the disorganized arrangement of Tin-positive cells. (D, H, L) Embryos overexpressing Mirr are characterized by (D) a rather mild loss of Eve clusters, (H) a dramatic loss of Odd-expressing cells including pericardial and lymph gland (lg) cells and (L) accumulations of Tin-positive cells along the region where the heart tube forms.
In all images arrows point to missing cells whereas arrowheads point to additional cells or cell accumulation. Lateral views of stage 10/11 embryos are shown in A-D. Dorsal views of stage 16 embryos are shown in E-L.
(TIF)
Acknowledgments
We are indebted to S. Campuzano for sharing the Ara/Caup antibody and fly lines. We appreciate receiving antibodies and fly lines from R. Bodmer, M. Frasch, J. Skeath, H. McNeill and B. Paterson. We thank the Developmental Studies Hybridoma Bank, the Bloomington Stock Center and the Drosophila Genomics Resource Center for providing antibodies, fly lines and cDNAs. We thank J. Schirner for her contribution and are very grateful to C. Donow for technical assistance.
Funding Statement
This work was supported by a grant to P.P. from the Deutsche Forschungsgemeinschaft (SFB497, TP8). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Frasch M, Nguyen HT (1999) Genetic control of mesoderm patterning and differentiation during Drosophila embryogenesis. Adv Dev Biochem: 1-47. [Google Scholar]
- 2. Bate M (1993) The mesoderm and its derivatives. In The development of Drosophila melanogaster. New York: Cold Spring Harbor Laboratory Press; pp. 1013-1090. [Google Scholar]
- 3. Wu X, Golden K, Bodmer R (1995) Heart development in Drosophila requires the segment polarity gene wingless. Dev Biol 169: 619-628. doi:10.1006/dbio.1995.1174. [DOI] [PubMed] [Google Scholar]
- 4. Frasch M (1995) Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature 374: 464-467. doi:10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- 5. Riechmann V, Irion U, Wilson R, Grosskortenhaus R, Leptin M (1997) Control of cell fates and segmentation in the Drosophila mesoderm. Development 124: 2915-2922. [DOI] [PubMed] [Google Scholar]
- 6. Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F et al. (2000) Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103: 63-74. doi:10.1016/S0092-8674(00)00105-7. PubMed: 11051548. [DOI] [PubMed] [Google Scholar]
- 7. Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F et al. (2002) Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol 244: 226-242. doi:10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- 8. Jagla T, Bidet Y, Da Ponte JP, Dastugue B, Jagla K (2002) Cross-repressive interactions of identity genes are essential for proper specification of cardiac and muscular fates in Drosophila. Development 129: 1037-1047. [DOI] [PubMed] [Google Scholar]
- 9. Azpiazu N, Lawrence PA, Vincent JP, Frasch M (1996) Segmentation and specification of the Drosophila mesoderm. Genes Dev 10: 3183-3194. doi:10.1101/gad.10.24.3183. PubMed: 8985186. [DOI] [PubMed] [Google Scholar]
- 10. Yin Z, Frasch M (1998) Regulation and function of tinman during dorsal mesoderm induction and heart specification in Drosophila. Dev Genet 22: 187-200. doi:10.1002/(SICI)1520-6408(1998)22:3. PubMed: 9621427. [DOI] [PubMed] [Google Scholar]
- 11. Lockwood WK, Bodmer R (2002) The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech Dev 114: 13-26. doi:10.1016/S0925-4773(02)00044-8. PubMed: 12175486. [DOI] [PubMed] [Google Scholar]
- 12. Lee HH, Frasch M (2005) Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development 132: 1429-1442. doi:10.1242/dev.01687. PubMed: 15750188. [DOI] [PubMed] [Google Scholar]
- 13. Azpiazu N, Frasch M (1993) tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev 7: 1325-1340. doi:10.1101/gad.7.7b.1325. PubMed: 8101173. [DOI] [PubMed] [Google Scholar]
- 14. Bodmer R (1993) The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118: 719-729. PubMed: 7915669. [DOI] [PubMed] [Google Scholar]
- 15. Gajewski K, Fossett N, Molkentin JD, Schulz RA (1999) The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 126: 5679-5688. PubMed: 10572044. [DOI] [PubMed] [Google Scholar]
- 16. Alvarez AD, Shi W, Wilson BA, Skeath JB (2003) pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development 130: 3015-3026. doi:10.1242/dev.00488. PubMed: 12756183. [DOI] [PubMed] [Google Scholar]
- 17. Klinedinst SL, Bodmer R (2003) Gata factor Pannier is required to establish competence for heart progenitor formation. Development 130: 3027-3038. doi:10.1242/dev.00517. PubMed: 12756184. [DOI] [PubMed] [Google Scholar]
- 18. Reim I, Frasch M (2005) The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132: 4911-4925. doi:10.1242/dev.02077. PubMed: 16221729. [DOI] [PubMed] [Google Scholar]
- 19. Mann T, Bodmer R, Pandur P (2009) The Drosophila homolog of vertebrate Islet1 is a key component in early cardiogenesis. Development 136: 317-326. doi:10.1242/dev.022533. PubMed: 19088091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferré-Marcó D, Modolell J (1996) Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85: 95-105. doi:10.1016/S0092-8674(00)81085-5. PubMed: 8620542. [DOI] [PubMed] [Google Scholar]
- 21. McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA (1997) mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev 11: 1073-1082. doi:10.1101/gad.11.8.1073. PubMed: 9136934. [DOI] [PubMed] [Google Scholar]
- 22. Cavodeassi F, Modolell J, Gómez-Skarmeta JL (2001) The Iroquois family of genes: from body building to neural patterning. Development 128: 2847-2855. PubMed: 11532909. [DOI] [PubMed] [Google Scholar]
- 23. Carrasco-Rando M, Tutor AS, Prieto-Sánchez S, González-Pérez E, Barrios N et al. (2011) Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLOS Genet 7: e1002186 PubMed: 21811416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumoto K, Nishihara S, Kamimura M, Shiraishi T, Otoguro T et al. (2004) The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nature Neurocsience 7: 605-612. doi:10.1038/nn1249. PubMed: 15133517. [DOI] [PubMed] [Google Scholar]
- 25. Cavodeassi F, Diez Del Corral R, Campuzano S, Domínguez M (1999) Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development 126: 4933-4942. PubMed: 10529412. [DOI] [PubMed] [Google Scholar]
- 26. Cavodeassi F, Modolell J, Campuzano S (2000) The Iroquois homeobox genes function as dorsal selectors in the Drosophila head. Development 127: 1921-1929. PubMed: 10751180. [DOI] [PubMed] [Google Scholar]
- 27. Calleja M, Herranz H, Estella C, Casal J, Lawrence P et al. (2000) Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development 127: 3971-3980. PubMed: 10952895. [DOI] [PubMed] [Google Scholar]
- 28. Calleja M, Renaud O, Usui K, Pistillo D, Morata G et al. (2002) How to pattern an epithelium: lessons from achaete-scute regulation on the notum of Drosophila. Gene 292: 1-12. doi:10.1016/S0378-1119(02)00628-5. PubMed: 12119094. [DOI] [PubMed] [Google Scholar]
- 29. Ikmi A, Netter S, Coen D (2008) Prepatterning the Drosophila notum: the three genes of the iroquois complex play intrinsically distinct roles. Dev Biol 317: 634-648. doi:10.1016/j.ydbio.2007.12.034. PubMed: 18394597. [DOI] [PubMed] [Google Scholar]
- 30. Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF (2000) Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol 224: 263-274. doi:10.1006/dbio.2000.9801. PubMed: 10926765. [DOI] [PubMed] [Google Scholar]
- 31. Kim KH, Rosen A, Bruneau BG, Hui CC, Backx PH (2012) Iroquois homeodomain transcription factors in heart development and function. Circ Res 110: 1513-1524. doi:10.1161/CIRCRESAHA.112.265041. PubMed: 22628575. [DOI] [PubMed] [Google Scholar]
- 32. Zhang SS, Kim KH, Rosen A, Smyth JW, Sakuma R et al. (2011) Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci U S A 108: 13576-13581. doi:10.1073/pnas.1106911108. PubMed: 21825130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D et al. (2001) Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol 21: 1730-1736. doi:10.1128/MCB.21.5.1730-1736.2001. PubMed: 11238910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reim I, Lee HH, Frasch M (2003) The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development 130: 3187-3204. doi:10.1242/dev.00548. PubMed: 12783790. [DOI] [PubMed] [Google Scholar]
- 35. Liu J, Qian L, Wessells RJ, Bidet Y, Jagla K et al. (2006) Hedgehog and RAS pathways cooperate in the anterior-posterior specification and positioning of cardiac progenitor cells. Dev Biol 290: 373-385. doi:10.1016/j.ydbio.2005.11.033. PubMed: 16387294. [DOI] [PubMed] [Google Scholar]
- 36. Diez del Corral R, Aroca P, Gómez-Skarmeta JL, Cavodeassi F, Modolell J (1999) The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes Dev 13: 1754-1761. doi:10.1101/gad.13.13.1754. PubMed: 10398687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Letizia A, Barrio R, Campuzano S (2007) Antagonistic and cooperative actions of the EGFR and Dpp pathways on the iroquois genes regulate Drosophila mesothorax specification and patterning. Development 134: 1337-1346. doi:10.1242/dev.02823. PubMed: 17329358. [DOI] [PubMed] [Google Scholar]
- 38. Yang CH, Simon MA, McNeill H (1999) mirror controls planar polarity and equator formation through repression of fringe expression and through control of cell affinities. Development 126: 5857-5866. PubMed: 10572059. [DOI] [PubMed] [Google Scholar]
- 39. Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA et al. (1995) Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267: 688-693. doi:10.1126/science.7839146. PubMed: 7839146. [DOI] [PubMed] [Google Scholar]
- 40. Venkatesh TV, Park M, Ocorr K, Nemaceck J, Golden K et al. (2000) Cardiac enhancer activity of the homeobox gene tinman depends on CREB consensus binding sites in Drosophila. Genesis 26: 55-66. doi:10.1002/(SICI)1526-968X(200001)26:1. PubMed: 10660673. [PubMed] [Google Scholar]
- 41. Yin Z, Xu XL, Frasch M (1997) Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124: 4971-4982. PubMed: 9362473. [DOI] [PubMed] [Google Scholar]
- 42. Kremser T, Gajewski K, Schulz RA, Renkawitz-Pohl R (1999) Tinman regulates the transcription of the beta3 tubulin gene (betaTub60D) in the dorsal vessel of Drosophila. Dev Biol 216: 327-339. doi:10.1006/dbio.1999.9425. PubMed: 10588882. [DOI] [PubMed] [Google Scholar]
- 43. Ward EJ, Skeath JB (2000) Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 127: 4959-4969. PubMed: 11044409. [DOI] [PubMed] [Google Scholar]
- 44. Frasch M, Hoey T, Rushlow C, Doyle H, Levine M (1987) Characterization and localization of the even-skipped protein of Drosophila. EMBO J 6: 749-759. PubMed: 2884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Broihier HT, Moore LA, Van Doren M, Newman S, Lehmann R (1998) zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development 125: 655-666. PubMed: 9435286. [DOI] [PubMed] [Google Scholar]
- 46. Padgett RW, St Johnston RD, Gelbart WM (1987) A transcript from a Drosophila pattern gene predicts a protein homologous to the transforming growth factor-beta family. Nature 325: 81-84. doi:10.1038/325081a0. PubMed: 3467201. [DOI] [PubMed] [Google Scholar]
- 47. Ramain P, Heitzler P, Haenlin M, Simpson P (1993) pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development 119: 1277-1291. PubMed: 7916679. [DOI] [PubMed] [Google Scholar]
- 48. Knirr S, Azpiazu N, Frasch M (1999) The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development 126: 4525-4535. PubMed: 10498687. [DOI] [PubMed] [Google Scholar]
- 49. Su MT, Fujioka M, Goto T, Bodmer R (1999) The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development 126: 3241-3251. PubMed: 10375513. [DOI] [PubMed] [Google Scholar]
- 50. Leiss D, Hinz U, Gasch A, Mertz R, Renkawitz-Pohl R (1988) Beta 3 tubulin expression characterizes the differentiating mesodermal germ layer during Drosophila embryogenesis. Development 104: 525-531. PubMed: 3077351. [DOI] [PubMed] [Google Scholar]
- 51. Chartier A, Zaffran S, Astier M, Sémériva M, Gratecos D (2002) Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development 129: 3241-3253. PubMed: 12070098. [DOI] [PubMed] [Google Scholar]
- 52. Leyns L, Gómez-Skarmeta JL, Dambly-Chaudière C (1996) iroquois: a prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech Dev 59: 63-72. doi:10.1016/0925-4773(96)00577-1. PubMed: 8892233. [DOI] [PubMed] [Google Scholar]
- 53. Kehl BT, Cho KO, Choi KW (1998) mirror, a Drosophila homeobox gene in the Iroquois complex, is required for sensory organ and alula formation. Development 125: 1217-1227. PubMed: 9477320. [DOI] [PubMed] [Google Scholar]
- 54. Pichaud F, Casares F (2000) homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev 96: 15-25. doi:10.1016/S0925-4773(00)00372-5. PubMed: 10940621. [DOI] [PubMed] [Google Scholar]
- 55. Zhao D, Woolner S, Bownes M (2000) The Mirror transcription factor links signalling pathways in Drosophila oogenesis. Dev Genes Evol 210: 449-457. doi:10.1007/s004270000081. PubMed: 11180850. [DOI] [PubMed] [Google Scholar]
- 56. Bilioni A, Craig G, Hill C, McNeill H (2005) Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc Natl Acad Sci U S A 102: 14671-14676. doi:10.1073/pnas.0502480102. PubMed: 16203991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Park M, Wu X, Golden K, Axelrod JD, Bodmer R (1996) The wingless signaling pathway is directly involved in Drosophila heart development. Dev Biol 177: 104-116. doi:10.1006/dbio.1996.0149. PubMed: 8660881. [DOI] [PubMed] [Google Scholar]
- 58. Carmena A, Gisselbrecht S, Harrison J, Jiménez F, Michelson AM (1998) Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev 12: 3910-3922. doi:10.1101/gad.12.24.3910. PubMed: 9869644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grigorian M, Mandal L, Hakimi M, Ortiz I, Hartenstein V (2011) The convergence of Notch and MAPK signaling specifies the blood progenitor fate in the Drosophila mesoderm. Dev Biol 353: 105-118. doi:10.1016/j.ydbio.2011.02.024. PubMed: 21382367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zecca M, Struhl G (2002) Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development 129: 1369-1376. PubMed: 11880346. [DOI] [PubMed] [Google Scholar]
- 61. de Navascués J, Modolell J (2007) tailup, a LIM-HD gene, and Iro-C cooperate in Drosophila dorsal mesothorax specification. Development 134: 1779-1788. doi:10.1242/dev.02844. PubMed: 17409113. [DOI] [PubMed] [Google Scholar]
- 62. Fuchs A, Cheung LS, Charbonnier E, Shvartsman SY, Pyrowolakis G (2012) Transcriptional interpretation of the EGF receptor signaling gradient. Proc Natl Acad Sci U S A 109: 1572-1577. doi:10.1073/pnas.1115190109. PubMed: 22307613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zeidler MP, Perrimon N, Strutt DI (1999) Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling. Genes Dev 13: 1342-1353. doi:10.1101/gad.13.10.1342. PubMed: 10346822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Johnson AN, Mokalled MH, Haden TN, Olson EN (2011) JAK/Stat signaling regulates heart precursor diversification in Drosophila. Development 138: 4627-4638. doi:10.1242/dev.071464. PubMed: 21965617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sotillos S, Espinosa-Vázquez JM, Foglia F, Hu N, Hombría JC (2010) An efficient approach to isolate STAT regulated enhancers uncovers STAT92E fundamental role in Drosophila tracheal development. Dev Biol 340: 571-582. doi:10.1016/j.ydbio.2010.02.015. PubMed: 20171201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Domínguez M, de Celis JF (1998) A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396: 276-278. doi:10.1038/24402. PubMed: 9834035. [DOI] [PubMed] [Google Scholar]
- 67. Haltiwanger RS, Stanley P (2002) Modulation of receptor signaling by glycosylation: fringe is an O-fucose-beta1,3-N-acetylglucosaminyltransferase. Biochim Biophys Acta 1573: 328-335. doi:10.1016/S0304-4165(02)00400-2. PubMed: 12417415. [DOI] [PubMed] [Google Scholar]
- 68. Park M, Yaich LE, Bodmer R (1998) Mesodermal cell fate decisions in Drosophila are under the control of the lineage genes numb, Notch, and sanpodo. Mech Dev 75: 117-126. doi:10.1016/S0925-4773(98)00098-7. PubMed: 9739121. [DOI] [PubMed] [Google Scholar]
- 69. Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y et al. (1997) Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev 11: 3096-3108. doi:10.1101/gad.11.22.3096. PubMed: 9367990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Herranz H, Morata G (2001) The functions of pannier during Drosophila embryogenesis. Development 128: 4837-4846. PubMed: 11731463. [DOI] [PubMed] [Google Scholar]
- 71. Zaffran S, Reim I, Qian L, Lo PC, Bodmer R et al. (2006) Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development 133: 4073-4083. doi:10.1242/dev.02586. PubMed: 16987868. [DOI] [PubMed] [Google Scholar]
- 72. LaBeau EM, Trujillo DL, Cripps RM (2009) Bithorax complex genes control alary muscle patterning along the cardiac tube of Drosophila. Mech Dev 126: 478-486. doi:10.1016/j.mod.2009.01.001. PubMed: 19272319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tögel M, Pass G, Paululat A (2008) The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev Biol 318: 29-37. doi:10.1016/j.ydbio.2008.02.043. PubMed: 18430414. [DOI] [PubMed] [Google Scholar]
- 74. Zmojdzian M, Jagla K (2013) Tailup plays multiple roles during cardiac outflow assembly in Drosophila. Cell Tissue Res. doi:10.1007/s00441-013-1644-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ara/Caup and Mirr demarcate a novel heart or heart-associated cell type. (A) Ara/Caup protein is not co-expressed with Tin. Arrows point to Ara/Caup expressing cells. (B) A double immunostaining for Mirr and Tin shows no co-expression of these factors. Arrows point to the pairs of Mirr-expressing cells that are adjacent to Tin-positive cells.
(TIF)
Heart phenotypes in embryos mutant for the Iro-C (iroDFM3). (A-C) Embryos lacking the three members of the Iro-C show a downregulation of Doc in the cardiac region (arrows). (D-F) Almost 80% of the iro DFM3 embryos are characterized by additional Tin-expressing cells at stage 12 (arrowhead). The strong downregulation of FasIII (asterisk) demonstrates the impact of Iro-C on visceral mesoderm development. (G-I) pnr mRNA expression is unaffected in the majority (86%) of iro DFM3 embryos. The arrows point to missing Eve cell clusters. (J-L) Loss of Iro-C results in a reduction of Eve cell clusters (arrow). (M-O) iro DFM3 embryos were characterized by missing Odd-positive cells in some hemisegments (arrow) and additional Odd-expressing cells in other hemisegments (brackets). The overall number of Odd-expressing pericardial cells was not significantly changed compared to wild-type embryos.
(TIF)
Heart phenotypes observed after mesodermal overexpression of individual members of the Iro-C. (A, E, I) Wild-type expression of Eve (A), Odd (E) and Tin (I). (B, F, J) Overexpression of Ara results in (B) the loss of some Eve clusters, (F) a mild disorganization of Odd-expressing pericardial cells and (J) a mild disorganization of Tin-positive cells. The ectopic accumulation of Tin-expressing cells at the anterior end of the heart (arrowhead) appears to be in the region where the lymph glands are located (compare with (E) showing normal Odd expression in the lymph glands (lg)). (C, G, K) Overexpression of Caup not only leads to a loss of Eve clusters but also to a slight expansion of these clusters (C), (G) a dramatic loss of Odd-expressing pericardial cells and (K) a severe heart tube defect as can be seen by the disorganized arrangement of Tin-positive cells. (D, H, L) Embryos overexpressing Mirr are characterized by (D) a rather mild loss of Eve clusters, (H) a dramatic loss of Odd-expressing cells including pericardial and lymph gland (lg) cells and (L) accumulations of Tin-positive cells along the region where the heart tube forms.
In all images arrows point to missing cells whereas arrowheads point to additional cells or cell accumulation. Lateral views of stage 10/11 embryos are shown in A-D. Dorsal views of stage 16 embryos are shown in E-L.
(TIF)