Abstract
CpG repression in RNA viruses has been known for decades, but a reasonable explanation has not yet been proposed to explain this phenomenon. In this study, we calculated the CpG odds ratio of all RNA viruses that have available genome sequences and analyzed the correlation with their genome polarity, base composition, synonymous codon usage, phylogenetic relationship, and host. The results indicated that the viral base composition, synonymous codon usage and host selection were the dominant factors that determined the CpG bias in RNA viruses. CpG usage variation between the different viral groups was caused by different combinations of these pressures, which also differed from each other in strength. The consistent under-representation of CpG usage in −ssRNA viruses is determined predominantly by base composition, which may be a consequence of the U/A preferred mutation bias of −ssRNA viruses, whereas the CpG usage of +ssRNA viruses is affected greatly by their hosts. As a result, most +ssRNA viruses mimic their hosts' CpG usage. Unbiased CpG usage in dsRNA viruses is most likely a result of their dsRNA genome, which allows the viruses to escape from the host-driven CpG elimination pressure. CpG was under-represented in all reverse-transcribing viruses (RT viruses), suggesting that DNA methylation is an important factor affecting the CpG usage of retroviruses. However, vertebrate-infecting RT viruses may also suffer host' CpG elimination pressure that also acts on +ssRNA viruses, which results in further under-representation of CpG in the vertebrate-infecting RT viruses.
Introduction
The relative abundance of neighbor nucleotides (dinucleotides) has been known as a genome signature of species since the 1960s and has been studied extensively in genomic DNA samples from many organisms, including vertebrates, invertebrates, plants, and prokaryotes [1]–[4]. These studies have demonstrated that TpA is under-represented in almost all organisms tested, whereas CpG is differentially represented in the genomes of eukaryotic organisms [2]. TpA depletion is believed to be caused by its presence in two out of three canonical stop codons and in transcriptional regulatory motifs (e.g., the TATA box sequence). Therefore, TpA avoidance reduces the risk of nonsense mutations and minimizes improper transcription [5]. CpG under-representation has been directly linked to cytosine DNA methylation, an epigenetic modification that plays important roles in diverse biological processes, such as gene and transposon silencing, genetic imprinting and X chromosome inactivation [6]. Methylated cytosines are prone to mutate into thymines through spontaneous deamination, resulting in the dinucleotide TpG and the subsequent presence of a CpA on the opposite strand after DNA replication [7]. This result is consistent with the concomitant CpA and TpG over-representation in CpG-suppressed organisms. Thus, the over-representation of CpA and TpG is considered to be a consequence of the under-representation of CpG.
Interestingly, CpG has also been observed to be predominantly under-represented in RNA viruses (in both retroviruses and riboviruses) [8], [9]. CpG deficiency in retroviruses may be due to host cytosine methylation [10], [11]. However, the precise mechanism that contributes to CpG under-representation in riboviruses is still largely unknown. Because riboviruses do not form DNA intermediates during genome replication, the methylation-deamination model is unlikely to apply. To date, two non-exclusive explanations have been suggested to explain the prevalence of CpG under-representation in riboviruses: the nucleotide-stacking energy model and the host innate immunity evasion model. The nucleotide-stacking energy model is based on the fact that in DNA duplexes, CpG has a much higher stacking energy than other dinucleotides, which may reduce the rate of transcription and replication of viruses [12]. However, this hypothesis has been challenged by the subsequent finding that the free energy of RNA duplexes of CpGs lies in the middle of all 16 possible dinucleotides [13].
The second hypothesis is based on the observation that the CpG odds ratio values of mammal-infecting riboviruses are lower than the riboviruses infecting other taxa. Influenza A virus, which originated from an avian reservoir, has undergone significant CpG reduction since its introduction into humans [14]. This hypothesis is further reinforced by the fact that the CpG motif in an AU-rich oligonucleotide can significantly stimulate the immune response of plasmacytoid dendritic cells [15], [16]. Furthermore, the replicative fitness of poliovirus decreases sharply with increased frequencies of the dinucleotides CpG and UpA in the capsid region [17]. Nevertheless, this hypothesis cannot explain the normal distribution of CpG in riboviruses infecting other taxa, such as invertebrates [18] and the under-representation of CpG in some plant viruses [8], [9]. In this study, we examined CpG usage in RNA viruses and their hosts to further address CpG under-representation in RNA viruses.
Results
Data characterization
We downloaded all available full genomic sequences of RNA viruses [including both riboviruses and reverse-transcribing viruses (RT viruses)] from the RefSeq database. After removing inaccurate sequences, a total of 1,955 sequences was obtained, which represented the genome of 1120 RNA viruses. These viruses belong to 61 different viral families or unassigned genera, which include 95.3% of the recognized viral families or unassigned genera from the Ninth Report of the International Committee on Taxonomy of Viruses (ICTV) (Dataset S1). The downloaded genome sequences covering virtually all types of RNA viruses identified thus far, as well as the host categories that they infect.
Overall CpG variation in RNA viruses
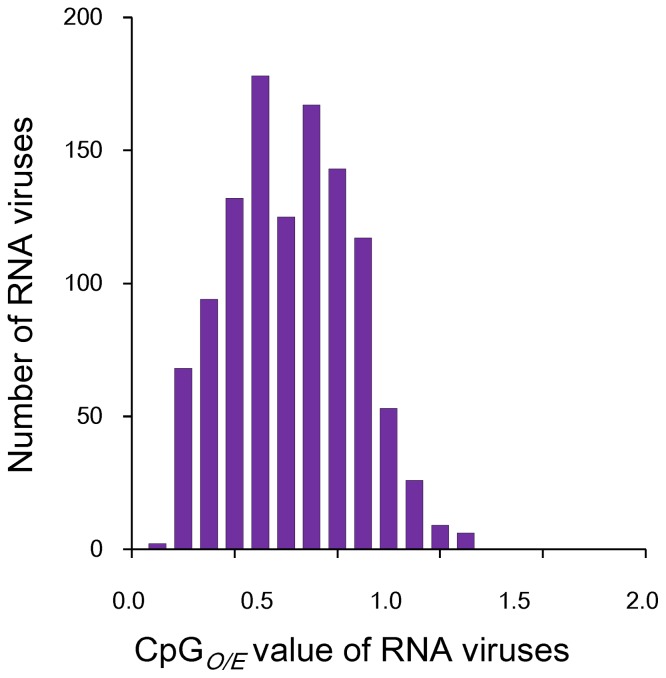
To obtain an overall view of CpG usage in RNA viruses, we examined the CpG odds ratio, the observed CpG incidence normalized to the expected CpG frequency (CpGO/E), of each viral genome (Dataset S2). When there is no selection (i.e., when all 16 dinucleotide pairs are randomly used), the CpG frequency should be similar to its expected frequency, and the CpGO/E value should approach 1. A CpGO/E value of a virus ≤0.78 or ≥1.23 indicates that it is significantly under-represented or over-represented in that virus [19]. Accordingly, a CpGO/E value between 0.79–1.12 can be recognized as being in the normal frequency range. The mean CpGO/E value of these RNA viruses was 0.67±0.240 (Table 1), with 0.15 and 1.35 as the minimum and maximum CpGO/E values, respectively. CpG was found to be significantly under-represented in 744 RNA viruses and over-represented in 12 RNA viruses, whereas it was normally distributed in the rest 364 viruses. These results suggest that CpG is under-represented in most of RNA viruses examined.
Table 1. CpG usage of RNA viruses.
| CpGO/E | CpGO/E _CDS | CpGO/E _CDS_12 | CpGO/E _CDS_23 | CpGO/E _CDS_31 | |
| Mean odd ratio | 0.67±0.240 | 0.65±0.248 | 0.50±0.214 | 0.58±0.266 | 0.90±0.395 |
| CpGO/E ≤0.78 | 744 | 749 | 1019 | 879 | 489 |
| CpGO/E 0.79–1.22 | 364 | 358 | 101 | 221 | 394 |
| CpGO/E ≥1.23 | 12 | 13 | 0 | 20 | 237 |
To obtain further insight into the variation of CpG bias in RNA viruses, we produced a CpGO/E distribution profile (Figure 1). The CpGO/E distribution profile deviated negatively from the normal range, which also suggests that CpG is under-represented in most of the examined RNA viruses. Instead of a unimodal pattern, the CpG distribution profile appears to be composed of more than one normal distribution. To test whether this special distribution pattern is unique to CpG, we further analyzed the distribution patterns of the other 15 dinucleotides (Figure S1). The distribution patterns of the other 15 dinucleotides displayed an apparently unimodal distribution pattern (Figure S1). In addition, the distribution range of CpG (ranging from 0.15 to 1.35) was broader than that of other dinucleotides. These results suggest huge variations of CpG bias in RNA viruses.
Figure 1. CpG usage pattern of RNA viruses.
The y-axis depicts the number of viruses with the specific CpGO/E values given on the x-axis.
Effect of base composition
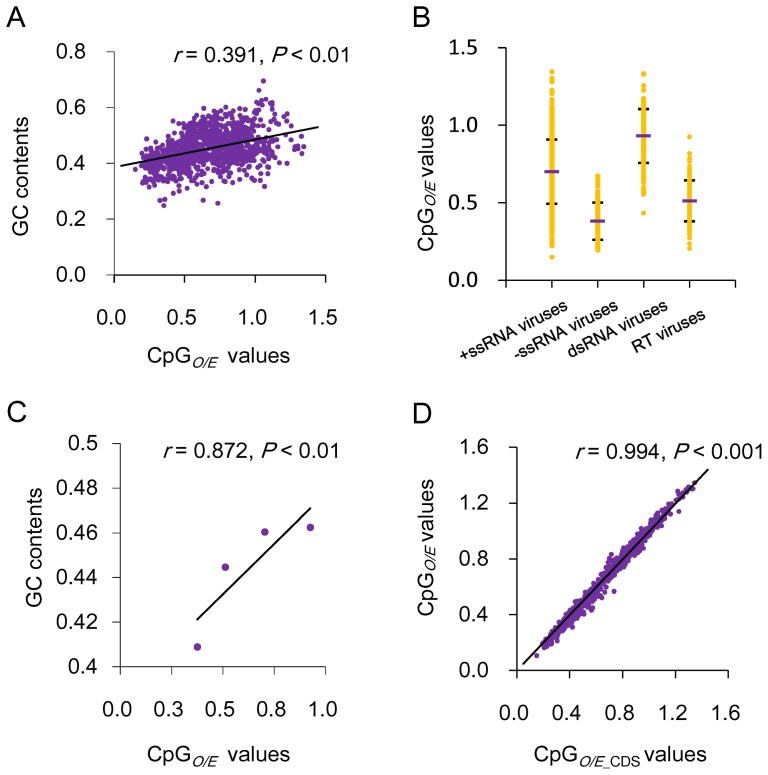
To understand the possible factors that affect CpG bias in RNA viruses, we first examined the correlation between viral CpGO/E values and base composition, an important indicator of viral mutational bias [20]. As shown in Figure 2A, the viral genomic G and C content (GC content) positively correlated with their CpGO/E values (Spearman's rho correlation coefficient r = 0.391, P<0.01), suggesting that the base composition has a role in shaping the CpG usage of RNA viruses. Previous research has demonstrated that different viral groups may differ from each other in mutation bias [21]. Thus, we suspect that different viral groups may also differ in their CpG usages. Therefore, the 1120 RNA viruses were divided into four groups, namely, double-stranded RNA (dsRNA) viruses, positive single-stranded (+ssRNA) viruses, negative single-stranded (−ssRNA) viruses and RT viruses, which contained 130, 724, 152, and 114 viruses, respectively. As shown in Figure 2B, the −ssRNA viruses have the lowest mean CpGO/E value (0.38±0.12), followed by RT and +ssRNA viruses (0.51±0.132 and 0.70±0.207), whereas dsRNA viruses have the highest mean CpGO/E value (0.93±0.174). This is consistent with the mean GC contents of the four viral groups (Table S1). In fact, a strong positive correlation was observed between the mean CpGO/E and GC content for the four viral groups (Figure 2C). The four viral groups also differed from each other in the distribution range of CpG odds ratio. The −ssRNA viruses have the most convergent distribution range, followed by RT and dsRNA viruses, whereas the +ssRNA viruses have the most divergent CpG distribution range. A variance analysis demonstrated a significant difference in CpG usage between the four groups (Table 2). These results suggest that different viral groups differ in their CpG usage.
Figure 2. The influence of GC content on viral CpG usage.
(A) Correlation between CpG odds ratios and GC contents of RNA viruses. (B) CpG usage variation between the four groups of RNA viruses. CpGO/E distribution range of each viral group is shown by yellow dots, and the mean CpGO/E value of each viral group is indicated by the purple bar. The standard deviations of the mean CpGO/E values are also indicated. (C) Correlation between the mean CpGO/E and mean GC content values. (D) Correlation between the CpGO/E and CpGO/E _CDS values.
Table 2. Pair-wised variance analysis CpG usage between viral groups.
| −ssRNA virus | +ssRNA virus | dsRNA virus | RT virus | |
| −ssRNA virus | - | 26.049** | 30.027** | 8.408** |
| +ssRNA virus | - | 12.913** | 13.256** | |
| dsRNA virus | - | 21.046** | ||
| RT virus | - |
Note: F statistic of one-way ANOVA analysis is 239.252 (P<0.001). The pair-wised comparisons were performed based on the CpGO/E values of each viral group, and the resulting T values of the independent T-test are shown. ** indicates P<0.0001. For the detailed analysis procedure, please refer to the Materials and Methods section.
To further investigate CpG variation between viral families, we calculated the mean CpGO/E value of each viral family or unassigned genus. The 1120 viruses belong to 61 viral families or unassigned genera, which can be further classed into 9 dsRNA, 37 +ssRNA, 12 −ssRNA, and 3 RT viral families or unassigned genera. CpG was significantly under-represented in all −ssRNA and RT viral families, independent of the phylogenetic relationships of these viral families (Table S2). On the other hand, the viral family or unassigned genus of dsRNA and +ssRNA groups displayed huge variation in CpG usage. For example, the mean CpGO/E value fluctuated from 0.44 (Astroviridae) to 1.16 (Alphatetraviridae) in the +ssRNA viral families, and fluctuated from 0.63 (Birnaviridae) to 1.18 (Cystoviridae) in the dsRNA viral families. Furthermore, +ssRNA viral families in the same order (Nidovirales, Picornavirales, or Tymovirales) also differed from each other greatly in the mean CpGO/E value (Table S2). These results suggest that there is no obvious relationship between viral CpG content and their phylogeny.
CpG bias in coding and noncoding regions
The fact that the distribution profile of GpCO/E which has the same C and G composition as CpG and is regularly used as an indicator of nucleotide composition bias [22], was unimodal (Figures 1 and S1) suggests that there are other factors affecting CpG usage in RNA viruses besides mutational bias. To explore this possibility, we investigated the influence of specific secondary structures and/or base composition in the viral non-coding regions on viral CpG usage. For example, rhinoviruses contains a 3′ U/A rich untranslated region (UTR) that completely lacks CpG [23]. Thus, we calculated the CpGO/E odds ratios in the coding regions (referred to as CpGO/E _CDS) of these RNA viruses (Dataset S3). The genomic CpGO/E value is very close to the CpGO/E _CDS value of the same RNA virus (Datasets S2 and S3). A correlation analysis indicated that the CpGO/E values were highly correlated with the CpGO/E _CDS values (Spearman's rho correlation coefficient r = 0.994; Figure 2D). These results suggest that biased CpG usage in the non-coding region only has a small influence on overall CpG usage. In other words, the observed under-representation of CpG in RNA viruses is not caused by the biased CpG usage in the non-coding regions but determined mainly by the coding regions.
Effect of synonymous codon usage
Because CpG usage in RNA viruses is determined mainly by the coding regions, we further analyzed the constraints of synonymous codon usage on CpG usage (Figure 2D). The distribution of CpG in a coding region can be found in three locations, two locations within a codon (CGN or NCG) and one across codon boundaries (the third codon position of the first codon and the first codon position of the following codon). One would expect that if CpG suppression were driven solely by the selection of non CpG-containing synonymous codons, the CpG dinucleotide should be suppressed only within the codons, but not at the location across codon boundaries. However, if CpG were under-represented in all three locations, we could not conclude that CpG bias was under the selection of viral mutational pressure because host-driven CpG elimination pressure may also be involved. Nevertheless, comparing CpG usage at the three locations between a virus and its host may help us to distinguish between host selective pressure and viral mutation bias.
The CpG odds ratios at the three locations of these RNA viruses were calculated separately and designated CpGO/E _CDS_12, CpGO/E _CDS_23, and CpGO/E _CDS_31, respectively (Dataset S3 and Table 1). The mean CpGO/E _CDS_12, CpGO/E _CDS_23, and CpGO/E _CDS_31 values of these RNA viruses were 0.50±0.214, 0.58±0.266, and 0.90±0.395, respectively. Furthermore, 237 viruses were determined to have CpG significantly over-represented at the location across codon boundaries, whereas none or very few viruses (20 viruses) were determined to have CpG significantly over-represented within codons. These results suggest that CpG is consistently under-represented in most RNA viruses at the locations within codons, but varied greatly at the location across codon boundaries.
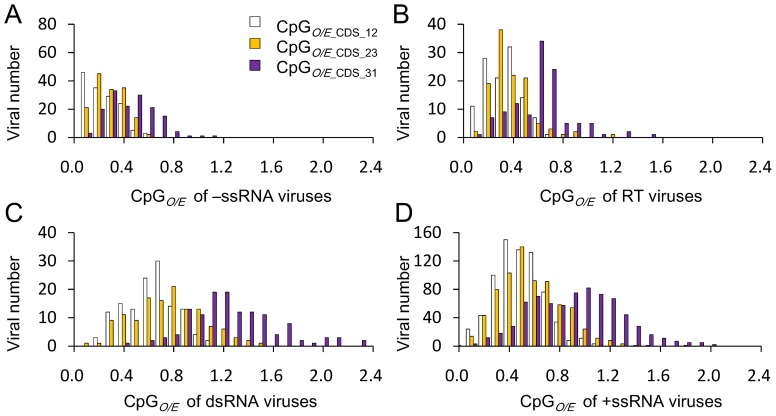
To gain further insight into the effect of synonymous codon on CpG bias, we further produced CpG distribution profiles covering each location of the four viral groups (Figure 3). In all viral groups, the viral CpGO/E values at the two locations within codons are apparently lower than the CpGO/E value at the location across codon boundaries of the same viral groups, suggesting that selection of CpG non-containing synonymous codon usage may be an important evolutionary force driving the CpG usage in RNA viruses. At the location across codon boundaries, CpG was under-represented in most −ssRNA and RT viruses and over-represented in most dsRNA viruses, whereas CpG varied greatly in +ssRNA viruses. Furthermore, the CpG distribution profile of +ssRNA viruses at the position across codon boundaries was bimodal, with one peak representing +ssRNA viruses with under-represented CpG odds ratios and the other peak representing +ssRNA viruses with over-represented CpG odds ratios, suggesting great variation of CpG usage in +ssRNA viruses at the location across codon boundaries.
Figure 3. CpG usage pattern of RNA viruses within coding region.
(A–D) Distribution of CpG at the three locations in the coding regions of −ssRNA, RT, dsRNA, and +ssRNA viruses, respectively.
We further compared CpG usage variation among codons between viral families or unassigned genera (Table S2). Within codons, CpG was under-represented in most viral families or unassigned genera. Conversely, different viral families displayed huge variations in CpG usage at the location across codon boundaries: CpG was significantly under-represented in all −ssRNA and RT viral families and normal or over-represented in all dsRNA viral families, whereas CpG was significantly under-represented in a few +ssRNA viral families and normal to over-represented in other families. These results also suggest that the selection of CpG non-containing synonymous codons may be an important factor affecting viral CpG usage. CpG was consistently under-represented in most −ssRNA and RT viruses but differed greatly among +ssRNA viruses at the location across codon boundaries, which suggests that other factor(s) beside mutation bias and synonymous codon usage may be involved in shaping the CpG usage of RNA viruses.
Influence of host
Previous studies have shown that the CpGO/E values of human-infecting riboviruses are lower than that of riboviruses infecting other taxa, suggesting the possible influence of the host on viral CpG usage [14], [18]. To further explore this possibility, the entire referenced mRNA sequence data of 70 species, including 17 species of vertebrates, 14 invertebrates, 9 plants, 12 fungi, and 18 bacteria were downloaded (Dataset S4), and the coding regions were extracted to calculate the CpG odds ratio. In our analysis, the entire mRNA sequence data were used instead of the full genome sequence because host mRNAs are physically present and translated in the cytoplasm where the replication of most RNA viruses takes place. In such a case, the host mRNA should suffer similar selection pressure as the viral RNAs. The coding region was used instead of the entire mRNA sequence to avoid errors induced by the incomplete 5′ and/or 3′ UTRs of some mRNAs and to reduce the possible CpG bias caused by specific RNA structures in host mRNA compared with viral CpG usage. CpGO/E _CDS, CpGO/E _CDS_12, CpGO/E _CDS_23, and CpGO/E _CDS values for each host was calculated. As shown in Dataset S4, the CpG odds ratios of hosts in the same group are very similar, whereas these ratios differed greatly for hosts between groups. These results suggest that hosts in the same group may have similar CpG usage and therefore may exert similar selective pressures on the viruses that infect them. Based on mean CpGO/E _CDS values, it is clear that CpG was under-represented in vertebrates and plants, whereas was normally distributed in fungi, invertebrates, and bacteria (Table 3). In detailed analysis of CpG usage at the three locations separately, it is obvious that different host groups differ greatly in CpG usage, especially at the location across codon boundaries (Table 3). At the location across codon boundaries, CpG was significantly under-represented in vertebrates, normally distributed in plants, whereas it was significantly over-represented in fungi, invertebrates, and bacteria. As mentioned above, CpG usage at this location is independent of synonymous codon usage. Thus, if the hosts have selective pressure on viral CpG usage besides synonymous codon selection, the viruses infecting them should have similar CpG usage at the same location. For instance, if vertebrates have CpG selection pressure on viruses infecting them besides synonymous codon choice, CpG would also be under-represented at the location across codon boundaries of the viruses infecting them. Likewise, CpG should not be under-represented at the location between neighbor codons in fungus-, invertebrate-, and bacterium-infecting RNA viruses.
Table 3. CpG usage of RNA viruses and their respective hosts.
| Typea | Viral Number | Mean CpGO/E | Mean CpGO/E _CDS_12 | Mean CpGO/E _CDS_23 | Mean CpGO/E _CDS_31 | |
| Hosts | B | 18 | 1.00±0.167 | 0.62±0.288 | 0.93±0.197 | 1.43±0.409 |
| F | 12 | 0.88±0.126 | 0.62±0.072 | 0.79±0.149 | 1.24±0.209 | |
| I | 14 | 1.02±0.090 | 0.60±0.074 | 1.07±0.163 | 1.41±0.179 | |
| P | 9 | 0.69±0.206 | 0.47±0.031 | 0.64±0.223 | 0.95±0.372 | |
| V | 17 | 0.47±0.042 | 0.46±0.176 | 0.37±0.048 | 0.58±0.079 | |
| +ssRNA viruses | B | 9 | 1.10±0.056 | 1.00±0.104 | 0.94±0.118 | 1.47±0.143 |
| F | 21 | 0.74±0.186 | 0.61±0.272 | 0.63±0.230 | 0.94±0.409 | |
| I | 40 | 0.87±0.154 | 0.72±0.162 | 0.76±0.286 | 1.10±0.397 | |
| P | 432 | 0.75±1.83 | 0.54±0.165 | 0.68±0.232 | 1.03±0.314 | |
| V | 223 | 0.57±0.183 | 0.44±0.176 | 0.46±0.191 | 0.78±0.339 | |
| −ssRNA viruses | I | 1 | 0.57 | 0.47 | 0.55 | 0.71 |
| P | 24 | 0.35±0.108 | 0.18±0.086 | 0.33±0.124 | 0.49±0.159 | |
| V | 126 | 0.38±0.120 | 0.28±0.140 | 0.33±0.122 | 0.49±0.186 | |
| dsRNA viruses | B | 3 | 0.97±0.106 | 0.58±0.149 | 0.90±0.265 | 1.51±0.260 |
| F | 50 | 0.98±0.174 | 0.70±0.177 | 0.80±0.224 | 1.46±0.420 | |
| I | 14 | 0.91±0.193 | 0.64±0.257 | 0.90±0.281 | 1.21±0.210 | |
| P | 26 | 0.88±0.158 | 0.67±0.208 | 0.63±0.237 | 1.31±0.303 | |
| V | 37 | 0.88±0.168 | 0.65±0.217 | 0.88±0.332 | 1.13±0.250 | |
| RT viruses | P | 50 | 0.50±0.126 | 0.32±0.136 | 0.38±0.114 | 0.73±0.236 |
| V | 64 | 0.52±0.137 | 0.42±0.124 | 0.46±0.191 | 0.59±0.218 |
B, indicates bacteria or bacterium-infecting RNA viruses; F, indicates fungi or fungus-infecting RNA viruses; I, indicates invertebrates or invertebrate-infecting RNA viruses; P, indicates plants or plant-infecting RNA viruses; V, indicates vertebrates or vertebrate-infecting RNA viruses.
The host ranges of the 1120 RNA viruses were determined (Dataset S1). Clearly, some RNA viruses can be categorized into two host sets (e.g., many viruses in the family Flaviviridae and Bunyaviridae can replicate in both invertebrates and vertebrates). These viruses were categorized into the viral group infecting the host class with the lower CpGO/E value because the host class with the lower CpGO/E value should exert stronger selection pressure on the viruses that infect them. For example, the arthropod-borne flaviviruses were classified as vertebrate-infecting viruses because the CpG odds ratio of vertebrates is much lower than that of invertebrates [2], [24].
The CpG usage of RNA viruses was compared with that of their respective hosts. CpG was consistently under-represented at the location across codon boundaries in all −ssRNA viruses and RT viruses and over-represented at the same location in all dsRNA viruses, and these results were independent of the host type infected (Table 3). The genomes of vertebrates and plants are highly methylated, and, as a result, the under-representation of CpG in RT viruses may be due to host DNA methylation, because these organisms produce DNA intermediates during the replication of their genome [25], [26]. However, −ssRNA and dsRNA viruses do not produce DNA intermediates during replication and should not be affected by host DNA methylation. Therefore, the consistent under-representation of CpG in −ssRNA viruses and over-representation of CpG in dsRNA viruses at the location between neighboring codons suggest that the CpG usage of −ssRNA and dsRNA viruses at the location may be not deeply affected by their hosts.
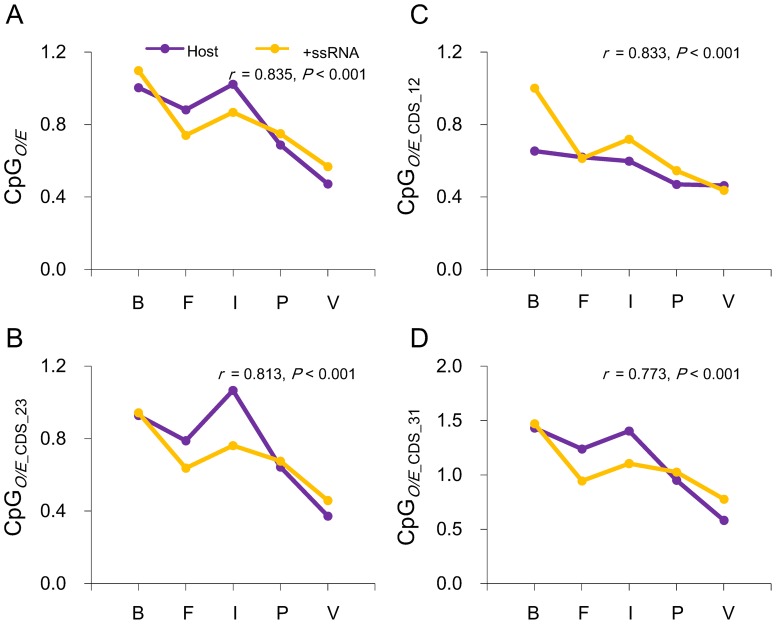
Interestingly, CpG was exclusively under-represented in vertebrate +ssRNA viruses, over-represented in bacterial +ssRNA viruses and normally distributed in +ssRNA viruses that infect other host types at the location across codon boundaries, which is consistent with the CpG usage in different host groups at the same location (Table 3). These results indicate that the host may have a role in shaping the CpG usage of +ssRNA viruses. Moreover, CpG usage in +ssRNA viruses within codons was also very similar to that of their respective host. For example, vertebrate +ssRNA viruses have the lowest CpG frequency within codon locations, whereas bacterium +ssRNA viruses have the highest CpG frequency. In fact, the mean CpGO/E value of +ssRNA viruses at each location was highly correlated with that of its host (Figure 4). Furthermore, within those phylogenetically related +ssRNA viral families (i.e., viral families of the same order), the mean CpGO/E value of vertebrate-infecting viral families is also significantly lower than that of other viral families that infect other hosts (Table S2). For example, the mean CpGO/E values of the two vertebrae-infecting families, Arteriviridae and Coronaviridae, within the order Nidovirales are significantly lower than the values of the invertebrate-infecting families within the same order (Roniviridae). The CpGO/E values of the vertebrate-infecting family of the order Picornavirales (Picornaviridae) is also significantly lower than other viral families that infect other hosts. Taken together, these results clearly suggest that the host drives the CpG usage in +ssRNA viruses.
Figure 4. +ssRNA viruses mimic the CpG usage of their respective host.
Correlation between +ssRNA viral and host's mean CpGO/E (A), mean CpGO/E _CDS_12 (B), mean CpGO/E _CDS_23 (C), and mean CpGO/E _CDS_31 (D). The abbreviations at the bottom of each chart (B, F, I, P, and V) represent bacteria or bacterial-infecting +ssRNA viruses, fungi or fungus-infecting +ssRNA, invertebrates or invertebrate-infecting +ssRNA viruses, plants or plant-infecting +ssRNA viruses, and vertebrates or vertebrate-infecting +ssRNA viruses, respectively.
Further support for this hypothesis comes from an analysis of viruses from Flaviviridae that infect both vertebrate and invertebrate. Previous studies have demonstrated that the overall CpG odds ratio of vertebrate-infecting flaviruses are lower than that of flaviruses that only infect invertebrates, suggesting that the two classes of flaviruses may suffer different CpG selection pressures from their respective host [18], [27]. To further investigate this possibility, we compared the CpG usage of the two groups of flaviruses at the three codon positions separately. The mean CpG odds ratio values of the vertebrate-infecting flaviviruses at the three codon positions were significantly lower than that of flaviviruses that only infect invertebrates (Table 4; pair-wised t-test, P<0.001). As mentioned above, the CpG usage at the location across codon boundaries is independent of synonymous codon usage or amino acid usage. Thus, differences in CpG bias should arise from host selection instead of mutation bias because viruses of the same family have evolved from the same ancestor and have similar genomic structure and replication mechanisms.
Table 4. CpG usages of flaviviruses that infect different types of hosts.
| Invertebrate-infecting flaviviruses | Vertebrate-infecting flaviviruses | |
| Mean CpGO/E | 0.85±0.074 | 0.50±0.113 |
| Mean CpGO/E _CDS | 0.87±0.076 | 0.49±0.117 |
| Mean CpGO/E _CDS_12 | 0.72±0.044 | 0.33±0.126 |
| Mean CpGO/E _CDS_23 | 0.74±0.100 | 0.47±0.116 |
| Mean CpGO/E _CDS_31 | 1.16±0.147 | 0.68±0.178 |
Discussion
In this study, we re-examined the CpG frequencies of RNA viruses in an attempt to uncover the mechanism of CpG under-representation in RNA viruses. Our analysis included all available genome sequences of RNA viruses in the RefSeq database, which represents a broader sequence pool than previous studies [8], [9], [14]. Hence, the observations obtained from our dataset are more detailed and more completely reflect the real nature of RNA viruses than previous studies. Our results indicated that CpG under-representation is prevalent in RNA viruses when considering the mean CpGO/E value (0.67±0.240), which is consistent with earlier observations [8], [9], [14]. Nevertheless, more than 33.6% of the riboviruses (376 of 1,120) analyzed in the present study have normal or over-represented CpG frequencies (CpGO/E>0.79). This proportion was much higher than in previous studies [8], [9]. This difference is likely caused by the limited number of genomic sequences that were available to previous studies and the subsequently biased data composition; most of the sequences included in previous analyses were human-infecting RNA viruses [8], [9]. Together with the multimodality of the CpGO/E distribution profiles (Figures 1 and S1), our results clearly demonstrate the existence of huge level of CpG usage diversity in RNA viruses.
Our data revealed that base composition, synonymous codon usage and host selection are the three important factors that affect CpG usage in RNA viruses. Other factors, such as RNA secondary structure in noncoding regions, may also play a minor role in affecting CpG usage in RNA viruses. However, our results indicate that the base composition is the core factor that determines riboviral CpG usage and establishes the “keynote” of riboviral CpG bias. With the exception of bacterium-infecting RNA viruses, all RNA viruses tend to use CpG non-containing synonymous codons, suggesting that the selection of CpG non-containing synonymous codon also plays an important role in shaping CpG usage in RNA viruses. However, our data do not prove that this selection is caused by host driving CpG elimination or amino acid composition bias because base composition also significantly affects viral synonymous codon usage [28], [29], and consistent CpG usage at the locations within codons was only observed between +ssRNA viruses and their respective hosts (Table 3). Based on the analysis of CpG usage at the location across codon boundaries between viral and host groups, host-driving CpG elimination pressure plays an important role in shaping the CpG usage of +ssRNA and RT viruses but not −ssRNA or dsRNA viruses. Taken together, our results indicate that the tremendous variation in CpG usage between −ssRNA, +ssRNA, dsRNA and RT viruses is caused by different combinations of the above mentioned selective pressures, which also differ from each other in strength.
CpG was consistently under-represented in all −ssRNA viruses at all locations within the coding regions. This under-representation of CpG is independent of the infected host or their phylogenetic relationship (Table 3). Together with the fact that −ssRNA viruses are not producing DNA intermediates during the replication of their genome, it is reasonable to believe that the CpG usage of −ssRNA viruses may not be affected greatly by the host they infect. The base composition of all −ssRNA viruses included in our analysis is biased toward UA content (Dataset S1), suggesting that this group of viruses may have a UA mutation bias. A variance analysis demonstrated that the base composition of −ssRNA viruses was significantly different from that of other viral groups (Table 5). These results indicate that −ssRNA viruses have a UA preferred mutation bias, which may result in consistent under-representation of CpG in −ssRNA viruses.
Table 5. Pair-wised variance analysis the GC contents between viral groups.
| −ssRNA virus | +ssRNA virus | dsRNA virus | RT virus | |
| −ssRNA viruses | - | 11.950** | 7.161** | 4.517** |
| +ssRNA viruses | - | 0.285 | 2.257* | |
| dsRNA viruses | - | 1.925* | ||
| RT virus | - |
Note: F statistic of one-way ANOVA analysis is 33.343 (P<0.001). The pair-wised comparisons were performed based on the GC contents of each viral group and the resulting T values of the independent T-test are shown. * indicates P<0.05, ** indicates P<0.0001.
The most interesting finding of our analysis was that the CpG usage of +ssRNA viruses varied greatly: significantly under-represented in all locations in the +ssRNA viruses infecting vertebrates; under-represented within the codon locations in plants, fungi, and invertebrate +ssRNA viruses; and normal at all locations in bacterium-infecting viruses (Figure 3 and Table 3). Interestingly, this variation was consistent with the infected hosts, which suggest that the host exerts a significant influence on CpG usage in +ssRNA viruses. Vertebrates appear to have the strongest influence on the +ssRNA viruses that infect them, which results in the vertebrate-infecting +ssRNA viruses mimicking vertebrate CpG usage in all coding region locations. Plant-, fungus-, and invertebrate-infecting +ssRNA viruses have CpG under-represented within codons but not at locations between neighboring codons, which suggests that these hosts affect CpG usage of +ssRNA viruses, most likely through synonymous codon selection. This result is consistent with our previous finding that synonymous codon usage by citrus tristeza virus, a woody plant-infecting +ssRNA virus within the Closteroviridae family, highly adapts to its citrus host [30]. Bacteria do not present a negative selection toward CpG representation in their genomes [2], [24], [31]. Thus, bacteria may impart no CpG selection pressure on the +ssRNA viruses that infect them. Our results support this conclusion.
In contrast to −ssRNA and +ssRNA viruses, CpG was normal or over-represented in dsRNA viruses at the location across codon boundaries (Tables 3 and S1). Moreover, this characteristic of CpG usage in dsRNA viruses is also independent of the host they infect and their phylogenetic relationship. We believe that this normal frequency usage of CpG in dsRNA viruses is caused by their specific life cycles, in which they produce rare ssRNAs during their genome replication. In eukaryotes, there are two interrelated host antiviral systems, innate immunity and RNA silencing. However, none of the receptors that specifically recognize dsRNA in the two antiviral systems (e. g., Toll-like receptor 3 [TLR3] and RIG-I-like RNA helicases in innate immunity and dsRNA binding protein [DRB] in RNA silencing) display any CpG preference [32]–[35]. Consequently, CpG elimination pressure from the host perhaps cannot act on dsRNA viruses.
RT viruses represent a special group of RNA viruses in our analysis, because they produce DNA intermediates during their genome replication. DNA methylation has been shown to play an important role in the antiviral response against RT viruses and endogenous retrotransposons [25], [26], [36]. These results are consistent with the consistent under-representation of CpG in RT viruses, especially at the location across codon boundaries. Interestingly, the CpG odds ratios of the vertebrate-infecting RT viruses were also significantly lower than that of plant-infecting RT viruses, suggesting that in addition to cytosine methylation, there is other CpG selection pressure from vertebrates acting on the RT viruses to infect them. In other words, vertebrate-infecting RT viruses may suffer two types of CpG selection pressures from their host, one most likely through DNA methylation, and the other may be the same as the CpG elimination pressure that acts on +ssRNA viruses at the RNA level.
Based on our results, host driving of CpG elimination at the RNA level appears unique to vertebrates. Studies have shown that ssRNAs with specific sequence motifs (e.g., motifs of AU- or GU-rich and CpG flanked by AU) can significantly stimulate the antiviral immune responses by promoting the secretion of type I interferon [15], [16]. Furthermore, exogenous or abnormal ssRNA is believed to be detected by toll-like receptors (TLRs), such as toll-like receptor 7/8 (TLR7/8) in mammal cells [37]–[40]. However, there is no direct clue to support the idea that host driving of CpG elimination at the RNA level is dived directly by TLRs, such as TLR7/8. In addition, TLRs and TLR-like proteins have been found in almost all eukaryotic organisms, including vertebrates, insects and plants [32], [33], [41]–[44]. Thus, further research is needed to uncover the mechanism of CpG elimination in RNA viruses.
Methods
Data acquisition and treatment
All viral sequence data were downloaded from the RefSeq database available at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov) in GenBank format (accessed on Mar. 05, 2013). The genomic sequences of RNA viruses were identified using a BioPython script, which also filtered inaccurate sequences (containing ambiguous nucleotides, ORFs not in a multiple of three, or non-valid start and/or stop codons). The final dataset contained 1,955 sequences, which represented the full genome of 1120 RNA viruses.
All reference mRNA sequences of viral hosts were either directly downloaded from the NCBI RefSeq database using the taxonomic name of the host species as the query, the molecular type limited to mRNA, and the gene location limited to the nucleus or fetched from complete genome sequences downloaded from GenBank using a BioPython script. Each data set was then applied to a BioPython pipeline to extract the open reading frames (ORFs) and remove any erroneous sequences using the same conditions. Sequences containing ambiguous nucleotides or fewer than 300 nucleotides were excluded from our analysis. The final complete mRNA sequence set for all hosts can be provided upon request.
Dinucleotide odds ratio analysis
The dinucleotide odds ratio was defined as the observed frequency of a dinucleotide pair in a given sequence divided by the frequencies of the two mononucleotides that form the dinucleotide pair in the same sequence. The dinucleotide odds ratio for RNA viruses was calculated based on the following equation:
where fX and fY denote the frequencies of the mononucleotides X and Y in a given sequence, and fXY denotes the frequency of dinucleotide XY in the same sequence. Using statistical theory, the dinucleotide relative abundance may be conservatively described as significantly low if ρXY ≤0.78 and significantly high if ρXY ≥1.23 [5].
In the case of double-stranded DNA (dsDNA), the frequency of each dinucleotide must be calculated in a symmetric manner considering the complementary sequence. Thus, a symmetric version of the dinucleotide odds ratio, ρ*XY, can be calculated based on the following equation:
where X and Y denote the two dinucleotides, and Z and W indicate the two complementary nucleotides of Y and X, respectively.
All calculations were accomplished using BioPython scripts that are available upon request.
Viral hosts range mapping
The viral host range was determined based on the information provided with the sequence file, from the Ninth Report of the International Committee on Taxonomy of Viruses, ViralZone database (http://viralzone.expasy.org/), or Wikipedia (http://en.wikipedia.org/). However, for some viruses, the host ranges were verified by a literature search.
Statistical analysis
Correlation analyses were performed using SPSS (Statistical Package for the Social Sciences) 16.0 software (SPSS Inc., Chicago, Illinois, USA). For variance analyses of CpGO/E values and GC content between the different viral groups, we first performed one-way analysis of variance (one-way ANOVA) F-tests using Fisher's Least Significant Difference (LSD) method. There were significant differences between different viral groups when the resulting P value was <0.05. Additionally, the pair-wised independent t-tests were performed to analyze whether the viral groups were significantly different from one another. All variance analyses were performed using SPSS 16.0 software.
Supporting Information
Dinucleotide usage patterns of RNA viruses. The y-axis depicts the number of viruses with the specific CpGO/E values given on the x-axis. (A–O) Distribution patterns of ApA, ApT, ApG, ApC, TpA, TpT, TpG, TpC, CpA, CpT, CpC, GpA, GpT, GpG and GpC, respectively. Note that the distribution pattern of TpA negatively deviates from the normal frequency range (0.79–1.22), whereas the distribution patterns of TpG and CpA positively deviate from the normal frequency range, suggesting TpA was under-represented in most RNA viruses and TpG and CpA were over-represented in most RNA viruses.
(PDF)
Mean GC content of different viral groups.
(DOCX)
CpG usage variation between different viral families.
(DOCX)
RNA virus information.
(XLSX)
Genomic dinucleotide odd ratio values and base compositions of RNA viruses.
(XLSX)
CpG odd ratio values of coding regions of RNA viruses.
(XLSX)
CpG odd ratio values of hosts.
(XLSX)
Acknowledgments
We are grateful to Professor Xueping Zhou (Zhejiang University) and Xin-Shun Ding (The Samuel Roberts Noble Foundation) for their helpful suggestions on the manuscript.
Funding Statement
This study was supported by the Natural Science Foundation of China (Project No. 31101417 and 31101415) and the Natural Science Foundation of Zhejiang Province (Project No: Y3110175 and Y3110277). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. De Amicis F, Marchetti S (2000) Intercodon dinucleotides affect codon choice in plant genes. Nucl Acids Res 28: 3339–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karlin S, Burge C (1995) Dinucleotide relative abundance extremes: a genomic signature. Trends Genet 11: 283–290. [DOI] [PubMed] [Google Scholar]
- 3. Simmen MW (2008) Genome-scale relationships between cytosine methylation and dinucleotide abundances in animals. Genomics 92: 33–40. [DOI] [PubMed] [Google Scholar]
- 4. Elango N, Hunt BG, Goodisman MA, Yi SV (2009) DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A 106: 11206–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karlin S, Mrázek J (1997) Compositional differences within and between eukaryotic genomes. Proc Natl Acad Sci U S A 94: 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucl Acids Res 8: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rima BK, McFerran NV (1997) Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J Gen Virol 78: 2859–2870. [DOI] [PubMed] [Google Scholar]
- 9. Karlin S, Doerfler W, Cardon LR (1994) Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol 68: 2889–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shpaer EG, Mullins JI (1990) Selection against CpG dinucleotides in lentiviral genes: a possible role of methylation in regulation of viral expression. Nucl Acid Res 18: 5793–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Kuyl A, Berkhout B (2012) The biased nucleotide composition of the HIV genome: a constant factor in a highly variable virus. Retrovirology 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breslauer KJ, Frank R, Blocker H, Marky LA (1986) Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A 83: 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Serra MJ, Turner DH (1995) Predicting thermodynamic properties of RNA. Methods Enzymol 259: 242–261. [DOI] [PubMed] [Google Scholar]
- 14. Greenbaum BD, Levine AJ, Bhanot G, Rabadan R (2008) Patterns of evolution and host gene mimicry in Influenza and other RNA viruses. PLoS Pathog 4: e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenbaum BD, Rabadan R, Levine AJ (2009) Patterns of oligonucleotide sequences in viral and host cell RNA identify mediators of the host innate immune system. PLoS ONE 4: e5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jimenez-Baranda S, Greenbaum B, Manches O, Handler J, Rabadán R, et al. (2011) Oligonucleotide motifs that disappear during the evolution of influenza virus in humans increase alpha interferon secretion by plasmacytoid dendritic cells. J Virol 85: 3893–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burns CC, Campagnoli R, Shaw J, Vincent A, Jorba J, et al. (2009) Genetic inactivation of Poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J Virol 83: 9957–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lobo FP, Mota BEF, Pena SDJ, Azevedo V, Macedo AM, et al. (2009) Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS ONE 4: e6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollander M, Wolfe DA (1999) Nonparametric statistical methods 2nd edition. New York: Wiley.
- 20. Eyre-Walker A (1999) Evidence of Selection on Silent Site Base Composition in Mammals: Potential Implications for the Evolution of Isochores and Junk DNA. Genetics 152: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Auewarakul P (2005) Composition bias and genome polarity of RNA viruses. Virus Res 109: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elango N, Yi SV (2008) DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol 25: 1602–1608. [DOI] [PubMed] [Google Scholar]
- 23. Megremis S, Demetriou P, Makrinioti H, Manoussaki AE, Papadopoulos NG (2012) The genomic signature of human rhinoviruses A, B and C. PLoS One. 7: e44557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karlin S (1998) Global dinucleotide signatures and analysis of genomic heterogeneity. Curr Opin Microbiol 1: 598–610. [DOI] [PubMed] [Google Scholar]
- 25. Ellis J, Hotta A, Rastegar M (2007) Retrovirus silencing by an epigenetic TRIM. Cell 131: 13–14. [DOI] [PubMed] [Google Scholar]
- 26. Leung DC, Lorincz MC (2012) Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci 37: 127–133. [DOI] [PubMed] [Google Scholar]
- 27. Schubert AM, Putonti C (2010) Evolution of the sequence composition of flaviviruses. Infect Genet Evol 10: 129–136. [DOI] [PubMed] [Google Scholar]
- 28. Adams MJ, Antoniw JF (2004) Codon usage bias amongst plant viruses. Arch Virol 149: 113–135. [DOI] [PubMed] [Google Scholar]
- 29. Jenkins GM, Holmes EC (2003) The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res 92: 1–7. [DOI] [PubMed] [Google Scholar]
- 30. Cheng XF, Wu XY, Wang HZ, Sun YQ, Qian YS, et al. (2012) High codon adaptation in citrus tristeza virus to its citrus host. Virol J 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y, Rocha EPC, Leung FCC, Danchin A (2004) Cytosine methylation is not the major factor inducing CpG dinucleotide deficiency in bacterial genomes. J Mol Evol 58: 692–700. [DOI] [PubMed] [Google Scholar]
- 32. Thompson AJV, Locarnini SA (2007) Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol 85: 435–445. [DOI] [PubMed] [Google Scholar]
- 33. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413: 732–738. [DOI] [PubMed] [Google Scholar]
- 34. Malathi K, Dong B, Gale M, Silverman RH (2007) Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448: 816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hohn T, Vazquez F (2011) RNA silencing pathways of plants: Silencing and its suppression by plant DNA viruses. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms 1809: 588–600. [DOI] [PubMed] [Google Scholar]
- 37. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531. [DOI] [PubMed] [Google Scholar]
- 38. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, et al. (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529. [DOI] [PubMed] [Google Scholar]
- 39. Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, et al. (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 101: 5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, et al. (2008) Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol 180: 3729–3738. [DOI] [PubMed] [Google Scholar]
- 41. Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, et al. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115. [DOI] [PubMed] [Google Scholar]
- 42. Xi Z, Ramirez JL, Dimopoulos G (2008) The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog 4: e1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zambon RA, Nandakumar M, Vakharia VN, Wu LP (2005) The Toll pathway is important for an antiviral response in Drosophila . Proc Natl Acad Sci U S A 102: 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawai T, Akira S (2008) Toll-like receptor and RIG-1-like receptor signaling. Ann N Y Acad Sci 1143: 1–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dinucleotide usage patterns of RNA viruses. The y-axis depicts the number of viruses with the specific CpGO/E values given on the x-axis. (A–O) Distribution patterns of ApA, ApT, ApG, ApC, TpA, TpT, TpG, TpC, CpA, CpT, CpC, GpA, GpT, GpG and GpC, respectively. Note that the distribution pattern of TpA negatively deviates from the normal frequency range (0.79–1.22), whereas the distribution patterns of TpG and CpA positively deviate from the normal frequency range, suggesting TpA was under-represented in most RNA viruses and TpG and CpA were over-represented in most RNA viruses.
(PDF)
Mean GC content of different viral groups.
(DOCX)
CpG usage variation between different viral families.
(DOCX)
RNA virus information.
(XLSX)
Genomic dinucleotide odd ratio values and base compositions of RNA viruses.
(XLSX)
CpG odd ratio values of coding regions of RNA viruses.
(XLSX)
CpG odd ratio values of hosts.
(XLSX)