Abstract
Major histocompatibility complex (MHC) genes are highly polymorphic and informative in disease association, transplantation, and population genetics studies with particular importance in the understanding of human population diversity and evolution. The aim of this study was to describe the HLA diversity in Mexican admixed individuals. We studied the polymorphism of MHC class I (HLA-A, -B, -C), and class II (HLA-DRB1, -DQB1) genes using high-resolution sequence based typing (SBT) method and we structured the blocks and conserved extended haplotypes (CEHs) in 234 non-related admixed Mexican individuals (468 haplotypes) by a maximum likelihood method. We found that HLA blocks and CEHs are primarily from Amerindian and Caucasian origin, with smaller participation of African and recent Asian ancestry, demonstrating a great diversity of HLA blocks and CEHs in Mexicans from the central area of Mexico. We also analyzed the degree of admixture in this group using short tandem repeats (STRs) and HLA-B that correlated with the frequency of most probable ancestral HLA-C/−B and -DRB1/−DQB1 blocks and CEHs. Our results contribute to the analysis of the diversity and ancestral contribution of HLA class I and HLA class II alleles and haplotypes of Mexican admixed individuals from Mexico City. This work will help as a reference to improve future studies in Mexicans regarding allotransplantation, immune responses and disease associations.
Introduction
The human major histocompatibility complex (MHC) is located within chromosomal region 6p21.3 and spans at least 3.4 Mb of DNA containing as many as 420 genes, including the HLA system, other immune related genes and pseudogenes [1]. The extensive polymorphism of the HLA genes within populations could have resulted from selective pressures including functional adaptation particularly to bacteria, viruses and parasites [2]–[5]. Also, the hypothesis of heterozygote advantage proposed that individuals with heterozygosity at HLA loci would be more efficient to respond against pathogens in pathogen-enriched environments [6]. Nevertheless, studies of genetics of infectious diseases are difficult to replicate due to the complex nature of the environmental factors and the degree of genetic diversity among human populations. In this regard, MHC genes are important because they are involved in immune responses, and are essential markers to study genetic diversity, disease susceptibility and allotransplantation [7].
Different studies using DNA polymorphic markers such as short tandem repeats (STRs), low and intermediate resolution HLA typing, ABO, MN and Rr-Hr blood groups, serum haptoglobin, albumin, and Factor Bf types have described the complexity of the genetic admixture of Mexican populations. These studies have revealed a non-homogeneus combination of Amerindian, Caucasian, and African genes in Mexican admixed individuals [8]–[10]. In this context, an important role of ethnicity in the susceptibility to different inflammatory and infectious diseases has been attributable to the incorporation of MHC alleles by admixture with Caucasian, Asian and African populations [11].
An important aspect of the MHC genetics is the inheritance of non-random associated alleles known as linkage disequilibrium (LD) [12]. Extensive studies on the existence of small blocks and other relatively fixed genetic fragments within the human MHC have been conducted [7], [13]. Specific DNA blocks with specific alleles of two or more MHC loci are often haplospecific for particular conserved extended haplotypes (CEHs). The frequency of CEHs and specific block combinations varies between major ethnic groups and/or in different geographic locations; these variations in the frequency of CEHs and blocks can be used as measurements of genetic diversity of the MHC [13]; however, little is known about the MHC blocks distribution and conserved haplotypes combination in Latin-American admixed human groups. Thus, the aim of the present study is to describe the distribution of HLA class I and class II blocks and the HLA CEHs using high resolution typing in a group of Mexican admixed individuals from Mexico City.
Results
HLA-A, -B, -C, -DRB1, and -DQB1 Allelic Frequencies in Mexican Admixed Individuals
The distribution of HLA-A, -B, -C, -DRB1 and -DQB1 alleles are listed in Table 1 . We detected 34 HLA-A, 64 HLA-B, 28 HLA-C, 39 HLA-DRB1, and 15 HLA-DQB1 alleles. The most frequent alleles were: 1) HLA-A : A*02∶01, A*24∶02, A*02∶06. A*68∶01, and A*31∶01, 2) HLA-B : B*39∶05, B*39∶06, B*51∶01, B*35∶01, and B*40∶02; 3) HLA-C : C*07∶02, C*04∶01, C*01∶02, C*03∶04, C*06∶02, and C*07∶01; 4) HLA-DRB1 : DRB1*08∶02, DRB1*04∶07, DRB1*14∶06, DRB1*07∶01, DRB1*04∶04, and DRB1*16∶02 and 5) HLA-DQB1 : DQB1*03∶01, DQB1*03∶02, DQB1*04∶02, DQB1*05∶01, and DQB1*02∶02 with frequencies higher than 5%. A significant deviation from Hardy-Weinberg equilibrium (HWE) was detected at the HLA-DRB1 locus (p<0.05).
Table 1. Allelic frequencies of HLA-A, -B, -C, -DRB1, and -DQB1 in 234 Mexicans.
| HLA-A | F | HLA-B | F | HLA-C | F | HLA-DRB1 | F | HLA-DQB1 | F |
| A*02∶01 | 0.2286 | B*39∶05 | 0.0791 | C*07∶02 | 0.2073 | DRB1*08∶02 | 0.1944 | DQB1*03∶01 | 0.2479 |
| A*24∶02 | 0.1688 | B*39∶06 | 0.0684 | C*04∶01 | 0.1859 | DRB1*04∶07 | 0.1175 | DQB1*03∶02 | 0.2457 |
| A*02∶06 | 0.0962 | B*51∶01 | 0.0598 | C*01∶02 | 0.0897 | DRB1*14∶06 | 0.1004 | DQB1*04∶02 | 0.2051 |
| A*68∶01 | 0.0791 | B*35∶01 | 0.0577 | C*03∶04 | 0.0662 | DRB1*07∶01 | 0.0705 | DQB1*05∶01 | 0.0684 |
| A*31∶01 | 0.0791 | B*40∶02 | 0.0534 | C*06∶02 | 0.0598 | DRB1*04∶04 | 0.0662 | DQB1*02∶02 | 0.0598 |
| A*01∶01 | 0.0363 | B*48∶01 | 0.0427 | C*07∶01 | 0.0534 | DRB1*16∶02 | 0.0641 | DQB1*06∶02 | 0.0363 |
| A*68∶03 | 0.0342 | B*07∶02 | 0.0406 | C*08∶01 | 0.0470 | DRB1*15∶01 | 0.0363 | DQB1*02∶01 | 0.0321 |
| A*03∶01 | 0.0321 | B*35∶17 | 0.0385 | C*08∶02 | 0.0406 | DRB1*03∶01 | 0.0321 | DQB1*03∶03 | 0.0214 |
| A*68∶02 | 0.0299 | B*35∶12 | 0.0385 | C*03∶05 | 0.0342 | DRB1*13∶01 | 0.0256 | DQB1*06∶04 | 0.0214 |
| A*29∶02 | 0.0256 | B*14∶02 | 0.0321 | C*03∶03 | 0.0299 | DRB1*01∶02 | 0.0235 | DQB1*05∶03 | 0.0171 |
| A*11∶01 | 0.0214 | B*15∶15 | 0.0321 | C*16∶01 | 0.0256 | DRB1*14∶02 | 0.0235 | DQB1*06∶03 | 0.0150 |
| A*26∶01 | 0.0192 | B*44∶03 | 0.0278 | C*12∶03 | 0.0256 | DRB1*04∶03 | 0.0214 | DQB1*06∶01 | 0.0107 |
| A*23∶01 | 0.0171 | B*52∶01 | 0.0214 | C*15∶09 | 0.0235 | DRB1*13∶02 | 0.0214 | DQB1*03∶19 | 0.0107 |
| A*02∶05 | 0.0171 | B*15∶01 | 0.0214 | C*05∶01 | 0.0214 | DRB1*04∶02 | 0.0214 | DQB1*05∶02 | 0.0064 |
| A*30∶02 | 0.0150 | B*39∶02 | 0.0214 | C*15∶02 | 0.0192 | DRB1*04∶11 | 0.0192 | DQB1*05:new† | 0.0021 |
| A*30∶01 | 0.0128 | B*35∶43 | 0.0192 | C*02∶02 | 0.0128 | DRB1*01∶01 | 0.0192 | ||
| A*33∶01 | 0.0128 | B*49∶01 | 0.0192 | C*14∶02 | 0.0085 | DRB1*11∶04 | 0.0171 | ||
| A*66∶01 | 0.0128 | B*18∶01 | 0.0171 | C*08∶03 | 0.0085 | DRB1*14∶01 | 0.0171 | ||
| A*32∶01 | 0.0085 | B*15∶30 | 0.0171 | C*16∶04 | 0.0043 | DRB1*11∶01 | 0.0128 | ||
| A*24∶25 | 0.0064 | B*57∶01 | 0.0150 | C*02∶10 | 0.0043 | DRB1*10∶01 | 0.0128 | ||
| A*25∶01 | 0.0064 | B*35∶14 | 0.0150 | C*03∶02 | 0.0043 | DRB1*15∶02 | 0.0107 | ||
| A*68∶05 | 0.0064 | B*13∶02 | 0.0128 | C*12∶02 | 0.0043 | DRB1*11∶02 | 0.0085 | ||
| A*03∶02 | 0.0064 | B*53∶01 | 0.0128 | C*04∶07 | 0.0043 | DRB1*12∶02 | 0.0064 | ||
| A*01∶02 | 0.0043 | B*38∶01 | 0.0128 | C*16∶02 | 0.0021 | DRB1*13∶03 | 0.0064 | ||
| A*33∶03 | 0.0043 | B*58∶02 | 0.0128 | C*07∶04 | 0.0021 | DRB1*04∶01 | 0.0064 | ||
| A*02∶211 | 0.0021 | B*39∶01 | 0.0107 | C*15∶05 | 0.0021 | DRB1*01∶03 | 0.0064 | ||
| A*34∶01 | 0.0021 | B*40∶05 | 0.0107 | C*17∶01 | 0.0021 | DRB1*16∶01 | 0.0043 | ||
| A*01∶03 | 0.0021 | B*35∶03 | 0.0107 | C*03:new§ | 0.0021 | DRB1*04∶10 | 0.0043 | ||
| A*34∶02 | 0.0021 | B*41∶01 | 0.0107 | DRB1*08∶04 | 0.0043 | ||||
| A*30∶04 | 0.0021 | B*44∶02 | 0.0107 | DRB1*12∶01 | 0.0043 | ||||
| A*74∶01 | 0.0021 | B*35∶24 | 0.0085 | DRB1*13∶05 | 0.0021 | ||||
| A*26∶17 | 0.0021 | B*37∶01 | 0.0085 | DRB1*04∶05 | 0.0021 | ||||
| A*02∶02 | 0.0021 | B*14∶01 | 0.0085 | DRB1*15∶03 | 0.0021 | ||||
| A*02∶24 | 0.0021 | B*50∶01 | 0.0085 | DRB1*08∶03 | 0.0021 | ||||
| B*40∶27 | 0.0085 | DRB1*04∶08 | 0.0021 | ||||||
| B*55∶01 | 0.0064 | DRB1*03∶02 | 0.0021 | ||||||
| B*58∶01 | 0.0064 | DRB1*08∶01 | 0.0021 | ||||||
| B*45∶01 | 0.0064 | DRB1*09∶01 | 0.0021 | ||||||
| B*27∶05 | 0.0064 | DRB1*13∶04 | 0.0021 | ||||||
| B*08∶01 | 0.0064 | ||||||||
| B*39∶08 | 0.0064 | ||||||||
| B*35∶08 | 0.0064 | ||||||||
| B*35∶16 | 0.0064 | ||||||||
| B*15∶17 | 0.0064 | ||||||||
| B*51∶02 | 0.0043 | ||||||||
| B*15∶02 | 0.0043 | ||||||||
| B*15∶03 | 0.0043 | ||||||||
| B*39∶10 | 0.0043 | ||||||||
| B*15∶39 | 0.0043 | ||||||||
| B*15∶31 | 0.0043 | ||||||||
| B*35∶02 | 0.0043 | ||||||||
| B*40∶04 | 0.0021 | ||||||||
| B*15∶16 | 0.0021 | ||||||||
| B*40∶20 | 0.0021 | ||||||||
| B*40∶08 | 0.0021 | ||||||||
| B*15∶18 | 0.0021 | ||||||||
| B*15∶10 | 0.0021 | ||||||||
| B*57∶03 | 0.0021 | ||||||||
| B*27∶03 | 0.0021 | ||||||||
| B*07∶14 | 0.0021 | ||||||||
| B*35∶20 | 0.0021 | ||||||||
| B*56∶01 | 0.0021 | ||||||||
| B*35∶40N | 0.0021 | ||||||||
| B*35:new* | 0.0021 |
Similar to B*35∶01 with a mutation at codon 207 ggc>tgc (Gly>Cys).
Similar to C*03∶04 with a mutation at codon 189 gtg>atg (Val>Met).
Similar to DQB1*05∶02 with a silent mutation at codon 133 cgg>cga.
Distribution of HLA-C/−B and -DRB1/−DQB1 Blocks in Mexican Admixed Individuals
HLA-C/−B blocks found in this group of Mexican admixed individuals are grouped in Table 2 . Twenty-six Amerindian (Native American) most probable ancestry (MPA) HLA-C/−B blocks (41.3%) were found. The most frequent (frequency ≥3.0%) Amerindian HLA-C/−B blocks were: C*07∶02/B*39∶05 (Haplotype Frequency (HF) = 0.0726), C*07∶02/B*39∶06 (HF = 0.0619), C*04∶01/B*35∶17 (HF = 0.0363), C*04∶01/B*35∶12 (HF = 0.0341) and C*08∶01/B*48∶01 (HF = 0.0320).
Table 2. Frequencies of HLA-C-B blocks in 234 admixed Mexican individuals (468 haplotypes).
| -C-B block | n | H.F. | Δ′ | t | -C-B block | n | H.F. | Δ′ | t | |||||||||
| Amerindian | C * 07∶02 | B * 39∶05 | 34 | 0.0726 | 0.8975 | 6.36 | Asian | C * 04∶01 | B * 35∶16 | 3 | 0.0064 | 1.0000 | 1.80 | |||||
| C * 07∶02 | B * 39∶06 | 29 | 0.0619 | 0.8025 | 5.40 | C * 14∶02 | B * 51∶01 | 3 | 0.0064 | 0.7339 | 1.71 | |||||||
| C * 04∶01 | B * 35∶17 | 17 | 0.0363 | 1.0000 | 4.43 | C * 08∶01 | B * 15∶02 | 2 | 0.0042 | 1.0000 | 1.42 | |||||||
| C * 04∶01 | B * 35∶12 | 16 | 0.0341 | 0.8632 | 4.14 | C * 07∶04 | B * 15∶18 | 1 | 0.0021 | 1.0000 | 1.00 | |||||||
| C * 08∶01 | B * 48∶01 | 15 | 0.0320 | 0.7376 | 4.02 | C * 03∶03 | B * 35∶01 | 1 | 0.0021 | 0.0141 | 0.19 | |||||||
| C * 03∶04 | B * 40∶02 | 11 | 0.0235 | 0.4196 | 3.20 | C * 01∶02 | B * 35∶01 | 1 | 0.0021 | −0.5908 | −1.21 | |||||||
| C * 03∶05 | B * 40∶02 | 10 | 0.0213 | 0.6045 | 3.19 | C * 12∶03 | B * 35∶03 | 1 | 0.0021 | 0.1788 | 0.90 | |||||||
| C * 01∶02 | B * 15∶30 | 8 | 0.0170 | 1.0000 | 2.92 | C * 07∶02 | B * 40∶02 | 1 | 0.0021 | −0.8007 | −2.17 | |||||||
| C * 01∶02 | B * 15∶01 | 7 | 0.0149 | 0.6701 | 2.61 | C * 04∶01 | B * 40∶05 | 1 | 0.0021 | 0.0154 | 0.06 | |||||||
| C * 03∶04 | B * 39∶02 | 5 | 0.0106 | 0.4642 | 2.13 | C * 01∶02 | B * 55∶01 | 1 | 0.0021 | 0.2670 | 0.79 | |||||||
| B * 35∶01 | C * 03∶05 | 5 | 0.0106 | 0.2700 | 1.98 | C * 03∶02 | B * 58∶01 | 1 | 0.0021 | 0.4967 | 1.00 | |||||||
| C * 04∶01 | B * 35∶03 | 4 | 0.0085 | 0.7538 | 1.93 | Total | 16 | 0.0338 | ||||||||||
| C * 04∶01 | B * 35∶24 | 4 | 0.0085 | 1.0000 | 2.10 | |||||||||||||
| C * 07∶02 | B * 39∶01 | 4 | 0.0085 | 0.7471 | 1.93 | Unknown | C * 01∶02 | B * 15∶15 | 13 | 0.0277 | 0.8534 | 3.71 | ||||||
| C * 07∶02 | B * 39∶02 | 4 | 0.0085 | 0.2414 | 1.12 | C * 01∶02 | B * 35∶43 | 9 | 0.0192 | 1.0000 | 3.10 | |||||||
| C * 03∶04 | B * 40∶05 | 3 | 0.0064 | 0.5714 | 1.66 | C * 15∶09 | B * 51∶01 | 9 | 0.0192 | 0.8065 | 3.06 | |||||||
| C * 07∶01 | B * 15∶17 | 3 | 0.0064 | 1.0000 | 1.75 | C * 04∶01 | B * 35∶14 | 6 | 0.0128 | 1.0000 | 2.58 | |||||||
| C * 07∶02 | B * 39∶08 | 3 | 0.0064 | 1.0000 | 1.83 | C * 03∶03 | B * 52∶01 | 6 | 0.0128 | 0.5876 | 2.47 | |||||||
| C * 08∶03 | B * 48∶01 | 3 | 0.0064 | 0.7387 | 1.72 | C * 04∶01 | B * 35∶08 | 3 | 0.0064 | 1.0000 | 1.80 | |||||||
| C * 02∶02 | B * 27∶05 | 2 | 0.0042 | 0.6623 | 1.41 | C * 03∶04 | B * 40∶27 | 3 | 0.0064 | 0.7321 | 1.71 | |||||||
| C * 08∶01 | B * 51∶02 | 2 | 0.0042 | 1.0000 | 1.42 | C * 06∶02 | B * 07∶02 | 2 | 0.0042 | 0.0540 | 0.68 | |||||||
| C * 07∶02 | B * 35∶01 | 1 | 0.0021 | −0.8228 | −2.37 | C * 08∶01 | B * 14∶01 | 2 | 0.0042 | −1.0000 | −1.28 | |||||||
| C * 03∶04 | B * 35∶01 | 1 | 0.0021 | −0.4456 | −0.73 | C * 04∶07 | B * 15∶31 | 2 | 0.0042 | 1.0000 | 1.42 | |||||||
| C * 03∶05 | B * 39∶06 | 1 | 0.0021 | −0.0938 | −0.10 | C * 03∶03 | B * 15∶39 | 2 | 0.0042 | 1.0000 | 1.42 | |||||||
| C * 03∶04 | B * 40∶08 | 1 | 0.0021 | 1.0000 | 1.00 | C * 06∶02 | B * 35∶02 | 2 | 0.0042 | 1.0000 | 1.42 | |||||||
| C * 03∶04 | B * 51∶01 | 1 | 0.0021 | −0.4654 | −0.79 | C * 07∶02 | B * 51∶01 | 2 | 0.0042 | −0.6583 | −1.85 | |||||||
| Total | 194 | 0.4133 | C * 15∶05 | B * 07∶02 | 1 | 0.0021 | 1.0000 | 1.00 | ||||||||||
| C * 07∶01 | B * 07∶14 | 1 | 0.0021 | 1.0000 | 1.00 | |||||||||||||
| Caucasian | C * 07∶02 | B * 07∶02 | 15 | 0.0320 | 0.7893 | 3.91 | C * 03∶03 | B * 13∶02 | 1 | 0.0021 | 0.1407 | 0.85 | ||||||
| C * 16∶01 | B * 44∶03 | 8 | 0.0170 | 0.6571 | 2.89 | C * 05∶01 | B * 14∶02 | 1 | 0.0021 | 0.0699 | 0.70 | |||||||
| C * 12∶03 | B * 38∶01 | 6 | 0.0128 | 1.0000 | 2.50 | C * 08∶03 | B * 14∶02 | 1 | 0.0021 | 0.2249 | 0.90 | |||||||
| C * 05∶01 | B * 18∶01 | 5 | 0.0106 | 0.6167 | 2.26 | C * 02∶10 | B * 14∶02 | 1 | 0.0021 | 0.4833 | 0.97 | |||||||
| C * 05∶01 | B * 44∶02 | 4 | 0.0085 | 0.7956 | 2.03 | C * 03∶03 | B * 15∶01 | 1 | 0.0021 | 0.0720 | 0.72 | |||||||
| C * 06∶02 | B * 50∶01 | 4 | 0.0085 | 1.0000 | 2.04 | C * 08∶02 | B * 15∶03 | 1 | 0.0021 | 0.4787 | 0.96 | |||||||
| C * 07∶01 | B * 08∶01 | 3 | 0.0064 | 1.0000 | 1.75 | C * 03∶02 | B * 15∶10 | 1 | 0.0021 | 1.0000 | 1.00 | |||||||
| C * 06∶02 | B * 37∶01 | 3 | 0.0064 | 0.7339 | 1.71 | C * 04∶01 | B * 15∶15 | 1 | 0.0021 | −0.6444 | −1.31 | |||||||
| C * 04∶01 | B * 44∶03 | 3 | 0.0064 | 0.0533 | 0.34 | C * 07∶02 | B * 15∶15 | 1 | 0.0021 | −0.6811 | −1.43 | |||||||
| C * 03∶04 | B * 15∶01 | 2 | 0.0042 | 0.1427 | 1.00 | C * 08∶02 | B * 18∶01 | 1 | 0.0021 | 0.0876 | 0.70 | |||||||
| C * 12∶03 | B * 18∶01 | 2 | 0.0042 | 0.2301 | 1.32 | C * 01∶02 | B * 27∶05 | 1 | 0.0021 | 0.2670 | 0.79 | |||||||
| C * 04∶01 | B * 39∶06 | 2 | 0.0042 | −0.6667 | −1.94 | C * 16∶01 | B * 35∶01 | 1 | 0.0021 | 0.0267 | 0.31 | |||||||
| C * 06∶02 | B * 57∶01 | 2 | 0.0042 | 0.2398 | 1.19 | C * 15∶09 | B * 35∶01 | 1 | 0.0021 | 0.0347 | 0.37 | |||||||
| C * 02∶02 | B * 14∶02 | 1 | 0.0021 | 0.1388 | 0.84 | C * 07∶01 | B * 35∶01 | 1 | 0.0021 | −0.3126 | −0.43 | |||||||
| C * 17∶01 | B * 41∶01 | 1 | 0.0021 | 1.0000 | 1.00 | C * 03∶03 | B * 35∶12 | 1 | 0.0021 | 0.0340 | 0.47 | |||||||
| C * 07∶01 | B * 51∶01 | 1 | 0.0021 | −0.3371 | −0.48 | C * 07∶02 | B * 35∶12 | 1 | 0.0021 | −0.7342 | −1.71 | |||||||
| C * 04∶01 | B * 56∶01 | 1 | 0.0021 | 1.0000 | 1.04 | C * 04∶01 | B * 35∶20 | 1 | 0.0021 | 1.0000 | 1.04 | |||||||
| C * 07∶02 | B * 57∶01 | 1 | 0.0021 | −0.3166 | −0.41 | C * 15∶09 | B * 35∶40N | 1 | 0.0021 | 1.0000 | 1.00 | |||||||
| Total | 64 | 0.1359 | C * 03∶04 | B * 35:New * | 1 | 0.0021 | 1.0000 | 1.00 | ||||||||||
| C * 03∶03 | B * 37∶01 | 1 | 0.0021 | 0.2267 | 0.91 | |||||||||||||
| Caucasian shared with other populations | C * 04∶01 | B * 35∶01 | 15 | 0.0320 | 0.4530 | 3.23 | C * 03∶04 | B * 39∶01 | 1 | 0.0021 | 0.1427 | 0.71 | ||||||
| C * 08∶02 | B * 14∶02 | 11 | 0.0235 | 0.7219 | 3.40 | C * 03:New § | B * 39∶02 | 1 | 0.0021 | 1.0000 | 1.00 | |||||||
| C * 15∶02 | B * 51∶01 | 9 | 0.0192 | 1.0000 | 3.09 | C * 04∶01 | B * 39∶05 | 1 | 0.0021 | −0.8559 | −2.83 | |||||||
| C * 07∶01 | B * 49∶01 | 6 | 0.0128 | 0.6477 | 2.45 | C * 01∶02 | B * 39∶05 | 1 | 0.0021 | −0.7014 | −1.75 | |||||||
| C * 06∶02 | B * 13∶02 | 5 | 0.0106 | 0.8226 | 2.26 | C * 02∶02 | B * 39∶05 | 1 | 0.0021 | 0.0945 | 0.55 | |||||||
| C * 08∶02 | B * 14∶01 | 4 | 0.0085 | 1.0000 | 2.04 | C * 03∶04 | B * 39∶06 | 1 | 0.0021 | −0.5323 | −0.99 | |||||||
| C * 07∶01 | B * 41∶01 | 4 | 0.0085 | 0.7886 | 2.01 | C * 07∶01 | B * 39∶06 | 1 | 0.0021 | −0.4200 | −0.67 | |||||||
| C * 12∶02 | B * 52∶01 | 2 | 0.0042 | 1.0000 | 1.42 | C * 04∶01 | B * 40∶02 | 1 | 0.0021 | −0.7778 | −2.06 | |||||||
| C * 02∶02 | B * 40∶02 | 1 | 0.0021 | 0.1212 | 0.72 | C * 07∶02 | B * 40∶04 | 1 | 0.0021 | 1.0000 | 1.05 | |||||||
| C * 03∶03 | B * 55∶01 | 1 | 0.0021 | 0.3126 | 0.94 | C * 07∶02 | B * 40∶05 | 1 | 0.0021 | −0.0433 | −0.04 | |||||||
| Total | 58 | 0.1235 | C * 03∶04 | B * 40∶20 | 1 | 0.0021 | 1.0000 | 1.00 | ||||||||||
| C * 16∶04 | B * 44∶02 | 1 | 0.0021 | 0.4946 | 0.99 | |||||||||||||
| African | C * 04∶01 | B * 53∶01 | 6 | 0.0128 | 1.0000 | 2.58 | C * 08∶01 | B * 44∶03 | 1 | 0.0021 | 0.0310 | 0.40 | ||||||
| C * 06∶02 | B * 58∶02 | 6 | 0.0128 | 1.0000 | 2.51 | C * 16∶02 | B * 44∶03 | 1 | 0.0021 | 1.0000 | 1.00 | |||||||
| C * 07∶01 | B * 57∶01 | 3 | 0.0064 | 0.3960 | 1.62 | C * 16∶01 | B * 48∶01 | 1 | 0.0021 | 0.0420 | 0.50 | |||||||
| C * 12∶03 | B * 39∶10 | 2 | 0.0042 | 1.0000 | 1.42 | C * 08∶02 | B * 48∶01 | 1 | 0.0021 | 0.0100 | 0.18 | |||||||
| C * 06∶02 | B * 45∶01 | 2 | 0.0042 | 0.6453 | 1.38 | C * 01∶02 | B * 49∶01 | 1 | 0.0021 | 0.0226 | 0.19 | |||||||
| C * 02∶10 | B * 15∶03 | 1 | 0.0021 | 0.4978 | 1.00 | C * 06∶02 | B * 49∶01 | 1 | 0.0021 | 0.0540 | 0.47 | |||||||
| C * 14∶02 | B * 15∶16 | 1 | 0.0021 | 1.0000 | 1.00 | C * 07∶02 | B * 49∶01 | 1 | 0.0021 | −0.4685 | −0.72 | |||||||
| C * 02∶02 | B * 27∶03 | 1 | 0.0021 | 1.0000 | 1.00 | C * 08∶02 | B * 51∶01 | 1 | 0.0021 | −0.1278 | −0.14 | |||||||
| C * 16∶01 | B * 45∶01 | 1 | 0.0021 | 0.3156 | 0.95 | C * 08∶01 | B * 51∶01 | 1 | 0.0021 | −0.2468 | −0.32 | |||||||
| C * 16∶01 | B * 51∶01 | 1 | 0.0021 | 0.0245 | 0.28 | C * 16∶04 | B * 52∶01 | 1 | 0.0021 | 0.4890 | 0.98 | |||||||
| C * 07∶01 | B * 57∶03 | 1 | 0.0021 | 1.0000 | 1.00 | C * 06∶02 | B * 52∶01 | 1 | 0.0021 | 0.0422 | 0.41 | |||||||
| C * 07∶01 | B * 58∶01 | 1 | 0.0021 | 0.2954 | 0.89 | C * 04∶01 | B * 55∶01 | 1 | 0.0021 | 0.1795 | 0.49 | |||||||
| C * 0801 | B * 4027 | 1 | 0.0021 | 0.2127 | 0.85 | |||||||||||||
| Total | 26 | 0.0551 | C * 12∶03 | B * 57∶01 | 1 | 0.0021 | 0.1201 | 0.85 | ||||||||||
| C * 04∶01 | B * 58∶01 | 1 | 0.0021 | 0.1795 | 0.49 | |||||||||||||
| Total | 110 | 0.2326 | ||||||||||||||||
Blocks of each ancestry (Amerindian, Caucasian, Caucasian shared with other populations, African, and Asian) were defined as those found in original populations with H.F. >1,0%, and not found in other native human groups in frequencies higher than 1,0%. We consider t value must be ≥2.0 to denote statistically significant association and thus validate Δ′ (shaded values).
Similar to B*35∶01 with a mutation at codon 207 ggc>tgc (Gly>Cys).
Similar to C*03∶04 with a mutation at codon 189 gtg>atg (Val>Met).
Eighteen HLA-C/−B blocks (13.2%) were of Caucasian MPA, the most frequent being: C*07∶02/B*07∶02 (HF = 0.0320), C*16∶01/B*44∶03 (HF = 0.0170), C*12∶03/B*38∶01 (HF = 0.0128), and C*05∶01/B*18∶01 (HF = 0.0106). The most common predominantly Caucasian -C/−B blocks shared with other ethnic groups were C*04∶01/B*35∶01 (HF = 0.0320), C*08∶02/B*14∶02 (HF = 0.0235), C*15∶02/B*51∶01 (HF = 0.0192), and C*06∶02/B*13∶02 (HF = 0.0106).
We also found 12 blocks (5.5%) from African MPA, being C*07∶01/B*49∶01 (HF = 0.0128), C*04∶01/B*53∶01 (HF = 0.0128) and C*06∶02/B*58∶02 (HF = 0.0128) were the most representative. Also 11 blocks (3.3%) of Asian MPA were found in our sample, all of them were uncommon, with frequencies below 1.0%. HLA-C/B blocks that were not previously reported numbered 57 (19.4%) –including two haplotypes harboring HLA-B and HLA-C new alleles-, even though the vast majority of them did not reach frequencies above 1.0%. C*01∶02/B*15∶15 (HF = 0.0277), C*01∶02/B*35∶43 (HF = 0.0192), C*15∶09/B*51∶01 (HF = 0.0192), C*03∶03/B*52∶01 (HF = 0.0129) and C*04∶01/B*35∶14 (HF = 0.0128) were the main non-previously described HLA-C/−B associations.
The frequencies of HLA-DRB1/−DQB1 blocks are sumarized in Table 3 . Eight HLA-DRB1/−DQB1 blocks (n = 240 of 468, 51.2%) were from Amerindian MPA. The most frequent blocks were: DRB1*08∶02/DQB1*04∶02 (HF = 0.1902), DRB1*04∶07/DQB1*03∶02 (HF = 0.1153), DRB1*14∶06/DQB1*03∶01 (HF = 0.0983), DRB1*16∶02/DQB1*03∶01 (HF = 0.0641), and DRB1*14∶02/DQB1*03∶01 (HF = 0.0235). In addition, 10 Caucasian MPA and 19 predominantly Caucasian HLA-DRB1/−DQB1 blocks were frequent in this sample. For example: DRB1*03∶01/DQB1*02∶01 (HF = 0.0320); DRB1*15∶01/DQB1*06∶02 (HF = 0.0320) and DRB1*04∶02/DQB1*03∶02 (HF = 0.0214) were the most frequent Caucasian MPA haplotypes in our group, while DRB1*04∶04/DQB1*03∶02 (HF = 0.0620); DRB1*07∶01/DQB1*02∶02 (HF = 0.0598); DRB1*01∶02/DQB1*05∶01 (HF = 0.0235) and DRB1*04∶03/DQB1*03∶02 (HF = 0.0214) were the most common blocks that are usually found in European populations. All these haplotypes exhibited significant LD with Δ′ values higher than 0.85. We found 8 African and seven Asian MPA blocks with frequencies lower than 1.0%. Six -DRB1/−DQB1 haplotypes not found in autochthonous populations [14], including one haplotype bearing DRB1*16∶01 in association with a new DQB1*05 allele, are reported in our group. All the above mentioned HLA-C/−B ( Table 2 ) and HLA-DRB1/DQB1 ( Table 3 ) blocks were in LD represented by significant Δ′ values and were demonstrated to be statistically relevant as they have t values ≥2.0 and p values <0.0005.
Table 3. Frequencies of HLA-DRB1-DQB1 blocks in 234 Mexican admixed individuals (468 haplotypes).
| DRB1-DQB1 block | n | H.F. | Δ′ | t | DRB1-DQB1 block | n | H.F. | Δ′ | t | ||||
| Amerindian | DRB1*08∶02 | DQB1*04∶02 | 89 | 0.1902 | 0.9723 | 12.33 | African | DRB1*10∶01 | DQB1*05∶01 | 5 | 0.0107 | 0.8211 | 2.24 |
| DRB1*04∶07 | DQB1*03∶02 | 54 | 0.1153 | 0.9518 | 8.51 | DRB1*13∶01 | DQB1*03∶03 | 4 | 0.0086 | 0.3942 | 1.96 | ||
| DRB1*14∶06 | DQB1*03∶01 | 46 | 0.0983 | 0.9717 | 7.88 | DRB1*08∶04 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.55 | ||
| DRB1*16∶02 | DQB1*03∶01 | 30 | 0.0641 | 1.0000 | 6.25 | DRB1*03∶02 | DQB1*04∶02 | 1 | 0.0022 | 1.0000 | 1.05 | ||
| DRB1*14∶02 | DQB1*03∶01 | 11 | 0.0235 | 1.0000 | 3.68 | DRB1*12∶01 | DQB1*05∶01 | 1 | 0.0022 | 0.4632 | 0.93 | ||
| DRB1*04∶11 | DQB1*03∶02 | 8 | 0.0171 | 0.8526 | 2.93 | DRB1*13∶01 | DQB1*05∶01 | 1 | 0.0022 | 0.0159 | 0.18 | ||
| DRB1*04∶10 | DQB1*04∶02 | 1 | 0.0022 | 0.3706 | 0.71 | DRB1*13∶04 | DQB1*03∶01 | 1 | 0.0022 | 1.0000 | 1.10 | ||
| DRB1*04∶11 | DQB1*04∶02 | 1 | 0.0022 | −0.4595 | −0.70 | DRB1*15∶03 | DQB1*06∶02 | 1 | 0.0022 | 1.0000 | 1.00 | ||
| Total | 240 | 0.5129 | Total | 16 | 0.0346 | ||||||||
| Caucasian | DRB1*03∶01 | DQB1*02∶01 | 15 | 0.0320 | 1.0000 | 4.02 | Asian | DRB1*11∶02 | DQB1*03∶01 | 3 | 0.0064 | 0.6674 | 1.60 |
| DRB1*15∶01 | DQB1*06∶02 | 15 | 0.0320 | 0.8779 | 4.01 | DRB1*04∶04 | DQB1*04∶02 | 2 | 0.0043 | −0.6862 | −2.03 | ||
| DRB1*04∶02 | DQB1*03∶02 | 10 | 0.0214 | 1.0000 | 3.50 | DRB1*09∶01 | DQB1*03∶03 | 1 | 0.0022 | 1.0000 | 1.00 | ||
| DRB1*11∶04 | DQB1*03∶01 | 8 | 0.0171 | 1.0000 | 3.13 | DRB1*12∶01 | DQB1*03∶01 | 1 | 0.0022 | 0.3348 | 0.63 | ||
| DRB1*13∶01 | DQB1*06∶03 | 6 | 0.0128 | 1.0000 | 2.49 | DRB1*13∶01 | DQB1*06∶02 | 1 | 0.0022 | 0.0487 | 0.59 | ||
| DRB1*07∶01 | DQB1*03∶03 | 5 | 0.0107 | 0.4620 | 2.10 | DRB1*13∶02 | DQB1*05∶01 | 1 | 0.0022 | 0.0338 | 0.33 | ||
| DRB1*04∶01 | DQB1*03∶02 | 3 | 0.0064 | 1.0000 | 1.90 | DRB1*15∶01 | DQB1*05∶01 | 1 | 0.0022 | −0.1415 | −0.16 | ||
| DRB1*11∶01 | DQB1*03∶01 | 2 | 0.0043 | 0.1130 | 0.40 | ||||||||
| DRB1*04∶07 | DQB1*03∶01 | 1 | 0.0022 | −0.9268 | −3.80 | Total | 10 | 0.0217 | |||||
| DRB1*08∶03 | DQB1*03∶01 | 1 | 0.0022 | 1.0000 | −0.60 | ||||||||
| Total | 66 | 0.1411 | |||||||||||
| Caucasian shared with other populations | DRB1*04∶04 | DQB1*03∶02 | 29 | 0.0620 | 0.9144 | 5.92 | DRB1*04∶07 | DQB1*06∶04 | 1 | 0.0022 | −0.1509 | −0.17 | |
| DRB1*07∶01 | DQB1*02∶02 | 28 | 0.0598 | 1.0000 | 5.66 | DRB1*10∶01 | DQB1*05∶02 | 1 | 0.0022 | 0.3247 | 0.98 | ||
| DRB1*01∶02 | DQB1*05∶01 | 11 | 0.0235 | 1.0000 | 3.41 | DRB1*11∶02 | DQB1*03∶19 | 1 | 0.0022 | 0.2419 | 0.98 | ||
| DRB1*04∶03 | DQB1*03∶02 | 10 | 0.0214 | 1.0000 | 3.50 | DRB1*14∶06 | DQB1*04∶02 | 1 | 0.0022 | −0.8965 | −3.37 | ||
| DRB1*01∶01 | DQB1*05∶01 | 9 | 0.0192 | 1.0000 | 3.07 | DRB1*16∶01 | DQB1*05:New† | 1 | 0.0022 | 1.0000 | 1.00 | ||
| DRB1*13∶02 | DQB1*06∶04 | 9 | 0.0192 | 0.8978 | 3.07 | Total | 7 | 0.0153 | |||||
| DRB1*14∶01 | DQB1*05∶03 | 8 | 0.0171 | 1.0000 | 2.89 | ||||||||
| DRB1*15∶02 | DQB1*06∶01 | 5 | 0.0107 | 1.0000 | 2.27 | ||||||||
| DRB1*11∶01 | DQB1*03∶19 | 4 | 0.0086 | 0.7974 | 2.02 | ||||||||
| DRB1*01∶03 | DQB1*05∶01 | 3 | 0.0064 | 1.0000 | 1.75 | ||||||||
| DRB1*12∶02 | DQB1*03∶01 | 3 | 0.0064 | 1.0000 | 1.90 | ||||||||
| DRB1*13∶03 | DQB1*03∶01 | 3 | 0.0064 | 1.0000 | 1.90 | ||||||||
| DRB1*04∶05 | DQB1*03∶02 | 1 | 0.0022 | 1.0000 | 1.09 | ||||||||
| DRB1*04∶08 | DQB1*03∶01 | 1 | 0.0022 | 1.0000 | 1.10 | ||||||||
| DRB1*04∶10 | DQB1*03∶02 | 1 | 0.0022 | 0.3366 | 0.63 | ||||||||
| DRB1*08∶01 | DQB1*04∶02 | 1 | 0.0022 | 1.0000 | 1.05 | ||||||||
| DRB1*13∶05 | DQB1*03∶01 | 1 | 0.0022 | 1.0000 | 1.10 | ||||||||
| DRB1*15∶01 | DQB1*05∶02 | 1 | 0.0022 | 0.3081 | 0.93 | ||||||||
| DRB1*16∶01 | DQB1*05∶02 | 1 | 0.0022 | 0.4968 | 1.00 | ||||||||
| Total | 129 | 0.2761 | |||||||||||
Blocks of each ancestry (Amerindian, Caucasian, Caucasian shared with other populations, African and Asian) were defined as those found in original populations with H.F. >1,0%, and not found in other native human groups in frequencies higher than 1,0%. We consider t value must be ≥2.0 to denote statistically significant association and thus validate Δ′ (shaded values).† Similar to DQB1*05∶02 with a silent mutation at codon 133 cgg>cga.
Conserved Extended HLA Haplotypes
We listed known CEHs in Table 4 . A total of 23 Amerindian, 10 Caucasian, 8 Caucasian-shared with other populations, 1 African, 1 Asian, and 37 not previously reported (unknown) HLA-C/−B/−DRB1/−DQB1 haplotypes were found in our admixed Mexican sample. Amerindian CEHs with frequencies higher than 1.0% were C*07∶02/B*39∶05/DRB1*04∶07/DQB1*03∶02 (HF = 0.0406); C*07∶02/B*39∶06/DRB1*14∶06/DQB1*03∶01 (HF = 0.0342); C*04∶01/B*35∶17/DRB1*08∶02/DQB1*04∶02 (HF = 0.0299); C*01∶02/B*15∶15/DRB1*08∶02/DQB1*04∶02 (HF = 0.0171); C*08∶01/B*48∶01/DRB1*08∶02/DQB1*04∶02 (HF = 0.0171), and C*04∶01/B*35∶12/DRB1*08∶02/DQB1*04∶02 (HF = 0.015). Caucasian MPA blocks with frequencies above 1.0% include: C*07∶02/B*07∶02/DRB1*15∶01/DQB1*06∶02 (HF = 0.0150), C*16∶01/B*44∶03/DRB1*07∶01/DQB1*02∶02 (HF = 0.0128); and C*08∶02/B*14∶02/DRB1*01∶02/DQB1*05∶01 (HF = 0.0107). Neither African, Asian, Caucasian-shared with other populations, nor not previously reported haplotypes were found in frequencies above 1.0%, except for haplotype C*07∶02/B*39∶05/DRB1*08∶02/DQB1*04∶02 (HF = 0.0107), although it appears not to be in LD. t value for this haplotype does not reach statistical significance. For the rest of the CEHs detected, t values were ≥2.0.
Table 4. HLA Conserved Extended Haplotypes in 234 Mexican admixed individuals (468 haplotypes).
| C-B-DRB1-DQB1 haplotype | n | H.F. | Δ′ | t | C-B-DRB1-DQB1 haplotype | n | H.F. | Δ′ | t | ||||||||
| Amerindian | C*07∶02 | B*39∶05 | DRB1*04∶07 | DQB1*03∶02 | 19 | 0.0406 | 0.5025 | 4.15 | African | C*06∶02 | B*58∶02 | DRB1*13∶01 | DQB1*03∶03 | 4 | 0.0086 | 1.0000 | 2.02 |
| C*07∶02 | B*39∶06 | DRB1*14∶06 | DQB1*03∶01 | 16 | 0.0342 | 0.5482 | 3.89 | Total | 4 | 0.0086 | |||||||
| C*04∶01 | B*35∶17 | DRB1*08∶02 | DQB1*04∶02 | 14 | 0.0299 | 0.7256 | 3.64 | Asian | C*08∶01 | B*15∶02 | DRB1*12∶02 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.42 | |
| C*01∶02 | B*15∶15 | DRB1*08∶02 | DQB1*04∶02 | 8 | 0.0171 | 0.5251 | 2.45 | Total | 2 | 0.0043 | |||||||
| C*08∶01 | B*48∶01 | DRB1*08∶02 | DQB1*04∶02 | 8 | 0.0171 | 1.0000 | 2.23 | Unknown | C*07∶02 | B*39∶05 | DRB1*08∶02 | DQB1*04∶02 | 5 | 0.0107 | −0.2267 | −0.14 | |
| C*04∶01 | B*35∶12 | DRB1*08∶02 | DQB1*04∶02 | 7 | 0.0150 | 0.3054 | 1.78 | C*04∶01 | B*53∶01 | DRB1*13∶02 | DQB1*06∶04 | 4 | 0.0086 | 0.6601 | 2.01 | ||
| C*01∶02 | B*15∶30 | DRB1*08∶02 | DQB1*04∶02 | 4 | 0.0086 | 0.3826 | 1.49 | C*04∶01 | B*35∶14 | DRB1*16∶02 | DQB1*03∶01 | 4 | 0.0086 | 0.6438 | 1.95 | ||
| C*03∶05 | B*40∶02 | DRB1*04∶07 | DQB1*03∶02 | 4 | 0.0086 | 0.3234 | 1.61 | C*01∶02 | B*35∶43 | DRB1*04∶03 | DQB1*03∶02 | 4 | 0.0086 | 0.4323 | 1.98 | ||
| C*15∶02 | B*51∶01 | DRB1*08∶02 | DQB1*04∶02 | 4 | 0.0086 | 0.3140 | 1.36 | C*03∶04 | B*40∶02 | DRB1*16∶02 | DQB1*03∶01 | 4 | 0.0086 | 0.3200 | 1.78 | ||
| C*03∶05 | B*35∶01 | DRB1*04∶07 | DQB1*03∶02 | 3 | 0.0064 | 0.5489 | 1.59 | C*07∶02 | B*39∶06 | DRB1*04∶07 | DQB1*03∶02 | 4 | 0.0086 | 0.0394 | −0.66 | ||
| C*04∶01 | B*35∶17 | DRB1*16∶02 | DQB1*03∶01 | 3 | 0.0064 | 0.1096 | 1.14 | C*01∶02 | B*15∶01 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.2944 | 1.13 | ||
| C*07∶02 | B*39∶05 | DRB1*16∶02 | DQB1*03∶01 | 3 | 0.0064 | 0.0295 | 0.49 | C*04∶01 | B*35∶01 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.0121 | 0.09 | ||
| C*03∶05 | B*40∶02 | DRB1*04∶04 | DQB1*03∶02 | 3 | 0.0064 | 0.2538 | 1.48 | C*07∶02 | B*39∶05 | DRB1*14∶02 | DQB1*03∶01 | 3 | 0.0064 | 0.2158 | 1.37 | ||
| C*03∶03 | B*52∶01 | DRB1*14∶06 | DQB1*03∶01 | 3 | 0.0064 | 0.4455 | 1.55 | C*08∶01 | B*48∶01 | DRB1*04∶04 | DQB1*03∶02 | 3 | 0.0064 | 0.1472 | 1.28 | ||
| C*01∶02 | B*15∶01 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.2368 | 1.17 | C*15∶09 | B*51∶01 | DRB1*04∶07 | DQB1*03∶02 | 3 | 0.0064 | 0.2482 | 1.27 | ||
| C*01∶02 | B*15∶30 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | 0.1682 | 0.93 | C*07∶02 | B*07∶02 | DRB1*11∶04 | DQB1*03∶01 | 2 | 0.0043 | 0.2235 | 1.28 | ||
| C*04∶01 | B*35∶24 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.4658 | 1.32 | C*01∶02 | B*15∶01 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 0.2385 | 1.18 | ||
| C*01∶02 | B*35∶43 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.0396 | 0.21 | C*04∶07 | B*15∶31 | DRB1*14∶01 | DQB1*05∶03 | 2 | 0.0043 | 1.0000 | 1.42 | ||
| C*07∶02 | B*39∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.3826 | 1.05 | C*05∶01 | B*18∶01 | DRB1*11∶01 | DQB1*03∶19 | 2 | 0.0043 | 0.4946 | 1.41 | ||
| C*03∶04 | B*39∶02 | DRB1*04∶11 | DQB1*03∶02 | 2 | 0.0043 | 0.3896 | 1.39 | C*04∶01 | B*35∶01 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.0740 | 0.77 | ||
| C*07∶02 | B*39∶02 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.3826 | 1.05 | C*06∶02 | B*35∶02 | DRB1*11∶04 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.42 | ||
| C*03∶04 | B*40∶02 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | −0.0439 | −0.06 | C*04∶01 | B*35∶12 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 0.0672 | 0.75 | ||
| C*08∶03 | B*48∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.5884 | 1.25 | C*04∶01 | B*35∶12 | DRB1*04∶07 | DQB1*03∶02 | 2 | 0.0043 | 0.0133 | 0.14 | ||
| Total | 117 | 0.2504 | C*04∶01 | B*35∶12 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | 0.0296 | 0.31 | |||||||
| C*01∶02 | B*35∶43 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 0.1708 | 1.09 | ||||||||||
| Caucasian | C*07∶02 | B*07∶02 | DRB1*15∶01 | DQB1*06∶02 | 7 | 0.0150 | 0.4478 | 2.62 | C*06∶02 | B*37∶01 | DRB1*01∶03 | DQB1*05∶01 | 2 | 0.0043 | 0.6645 | 1.42 | |
| C*16∶01 | B*44∶03 | DRB1*07∶01 | DQB1*02∶02 | 6 | 0.0128 | 0.7341 | 2.44 | C*03∶04 | B*39∶02 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 0.3604 | 1.28 | ||
| C*08∶02 | B*14∶02 | DRB1*01∶02 | DQB1*05∶01 | 5 | 0.0107 | 0.4414 | 2.22 | C*07∶02 | B*39∶02 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.4658 | 1.32 | ||
| C*06∶02 | B*13∶02 | DRB1*07∶01 | DQB1*02∶02 | 4 | 0.0086 | 0.7873 | 1.99 | C*07∶02 | B*39∶05 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | −0.4015 | −0.85 | ||
| C*05∶01 | B*18∶01 | DRB1*03∶01 | DQB1*02∶01 | 3 | 0.0064 | 0.5868 | 1.71 | C*07∶02 | B*39∶06 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 0.0129 | 0.24 | ||
| C*07∶01 | B*57∶01 | DRB1*07∶01 | DQB1*03∶03 | 3 | 0.0064 | 1.0000 | 1.74 | C*07∶02 | B*39∶08 | DRB1*04∶07 | DQB1*03∶02 | 2 | 0.0043 | 0.6241 | 1.33 | ||
| C*07∶01 | B*08∶01 | DRB1*03∶01 | DQB1*02∶01 | 2 | 0.0043 | 0.6556 | 1.40 | C*12∶03 | B*39∶10 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 1.0000 | 1.42 | ||
| C*12∶03 | B*38∶01 | DRB1*04∶02 | DQB1*03∶02 | 2 | 0.0043 | 0.3188 | 1.37 | C*03∶04 | B*40∶02 | DRB1*04∶03 | DQB1*03∶02 | 2 | 0.0043 | 0.1807 | 1.30 | ||
| C*12∶03 | B*38∶01 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 0.2909 | 1.24 | C*03∶04 | B*40∶02 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | 0.0926 | 0.69 | ||
| C*05∶01 | B*44∶02 | DRB1*04∶02 | DQB1*03∶02 | 2 | 0.0043 | 0.4891 | 1.40 | C*03∶05 | B*40∶02 | DRB1*14∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.1807 | 1.30 | ||
| Total | 36 | 0.0771 | C*07∶01 | B*41∶01 | DRB1*08∶04 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.42 | |||||||
| C*06∶02 | B*45∶01 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 1.0000 | 1.42 | ||||||||||
| Caucasian shared with other populations | C*08∶02 | B*14∶01 | DRB1*07∶01 | QDB1*02∶02 | 3 | 0.0064 | 0.7341 | 1.71 | C*07∶02 | B*51∶01 | DRB1*15∶01 | DQB1*06∶02 | 2 | 0.0043 | 1.0000 | 1.42 | |
| C*07∶01 | B*49∶01 | DRB1*13∶02 | DQB1*06∶04 | 3 | 0.0064 | 0.4902 | 1.72 | C*15∶09 | B*51∶01 | DRB1*04∶11 | DQB1*03∶02 | 2 | 0.0043 | 0.2353 | 1.35 | ||
| C*04∶01 | B*35∶01 | DRB1*04∶04 | DQB1*03∶02 | 3 | 0.0064 | 0.1472 | 1.28 | C*15∶09 | B*51∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.0396 | 0.21 | ||
| C*08∶02 | B*14∶02 | DRB1*03∶01 | DQB1*02∶01 | 2 | 0.0043 | 0.1547 | 1.22 | C*03∶03 | B*52∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.1768 | 0.69 | ||
| C*04∶01 | B*35∶01 | DRB1*11∶04 | DQB1*03∶01 | 2 | 0.0043 | 0.2252 | 1.29 | Total | 92 | 0.1975 | |||||||
| C*04∶01 | B*35∶01 | DRB1*14∶01 | DQB1*05∶03 | 2 | 0.0043 | 0.2252 | 1.29 | ||||||||||
| C*07∶02 | B*07∶02 | DRB1*15∶02 | DQB1*06∶01 | 2 | 0.0043 | 0.3788 | 1.35 | ||||||||||
| C*06∶02 | B*50∶01 | DRB1*03∶01 | DQB1*02∶01 | 2 | 0.0043 | 0.4834 | 1.38 | ||||||||||
| Total | 19 | 0.0407 | |||||||||||||||
Extension of Conserved Extended HLA Haplotypes to HLA-A
In Table 5 we show the preferential association of the common HLA-C/−B/−DRB1/−DQB1 CEH with HLA-A alleles in Mexican admixed population. It is remarkable that CEHs were common in this sample, with five haplotypes found with HF ≥1.0%: A*24∶02/C*07∶02/B*39∶06/DRB1*14∶06/DQB1*03∶01 (HF = 0.0256), A*02∶01/C*04∶01/B*35∶17/DRB1*08∶02/DQB1*04∶02 (HF = 0.0150), A*68∶03/C*07∶02/B*39∶05/DRB1*04∶07/DQB1*03∶02 (HF = 0.0107), A*02∶06/C*07∶02/B*39∶05/DRB1*04∶07/DQB1*03∶02 (HF = 0.0107), and A*02∶01/C*07∶02/B*39∶05/DRB1*04∶07/DQB1*03∶02 (HF = 0.0128), the first four of them being identified within samples of Native American people from all over the Americas, and the last one not found yet in other populations. Importantly, six Caucasian and one African CEHs were found. A set of 38 haplotypes was classified as not previously reported (unknown), some of them resulted from recombination between Caucasian and Amerindian blocks. Interestingly, one CEH which is frequent in Askenazi Jewish population was also observed in our sample (A*26∶01/C*12∶03/B*38∶01/DRB1*04∶02/DQB1*03∶02).
Table 5. Extension of HLA Conserved Extended Haplotypes to HLA-A in 234 Mexican Admixed individuals.
| A-C-B-DRB1-DQB1 haplotype | n | H.F. | Δ′ | t | |||||
| Amerindian | A*24∶02 | C*07∶02 | B*39∶06 | DRB1*14∶06 | DQB1*03∶01 | 12 | 0.0256 | 0.6992 | 2.76 |
| A*02∶01 | C*04∶01 | B*35∶17 | DRB1*08∶02 | DQB1*04∶02 | 7 | 0.0150 | 0.3518 | 1.46 | |
| A*68∶03 | C*07∶02 | B*39∶05 | DRB1*04∶07 | DQB1*03∶02 | 5 | 0.0107 | 0.2834 | 2.02 | |
| A*02∶06 | C*07∶02 | B*39∶05 | DRB1*04∶07 | DQB1*03∶02 | 5 | 0.0107 | 0.1848 | 1.46 | |
| A*02∶01 | C*04∶01 | B*35∶12 | DRB1*08∶02 | DQB1*04∶02 | 4 | 0.0086 | 0.4444 | 1.21 | |
| A*02∶01 | C*01∶02 | B*15∶15 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.1898 | 0.68 | |
| A*68∶01 | C*01∶02 | B*15∶15 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.3214 | 1.38 | |
| A*02∶01 | C*08∶01 | B*48∶01 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.1898 | 0.68 | |
| A*02∶06 | C*08∶01 | B*48∶01 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.3085 | 1.31 | |
| A*24∶02 | C*04∶01 | B*35∶12 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.1407 | 0.58 | |
| A*68∶01 | C*04∶01 | B*35∶17 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.0693 | 0.64 | |
| A*02∶06 | C*03∶04 | B*40∶02 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 1.0000 | 1.28 | |
| A*02∶06 | C*04∶01 | B*35∶17 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.5170 | 0.47 | |
| A*02∶01 | C*03∶04 | B*39∶02 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*07∶02 | B*39∶05 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.5679 | 0.93 | |
| A*31∶01 | C*04∶01 | B*35∶17 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.0693 | 0.64 | |
| Total | 59 | 0.1263 | |||||||
| Caucasian | A*02∶01 | C*07∶02 | B*07∶02 | DRB1*15∶01 | DQB1*06∶02 | 4 | 0.0086 | 0.4444 | 1.21 |
| A*30∶02 | C*05∶01 | B*18∶01 | DRB1*03∶01 | DQB1*02∶01 | 3 | 0.0064 | 1.0000 | 1.72 | |
| A*29∶02 | C*16∶01 | B*44∶03 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 0.3158 | 1.32 | |
| A*02∶01 | C*16∶01 | B*44∶03 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 0.1357 | 0.45 | |
| A*01∶01 | C*07∶01 | B*57∶01 | DRB1*07∶01 | DQB1*03∶03 | 2 | 0.0043 | 0.6541 | 1.35 | |
| A*33∶01 | C*08∶02 | B*14∶02 | DRB1*01∶02 | DQB1*05∶01 | 2 | 0.0043 | 0.3922 | 1.38 | |
| Total | 15 | 0.0322 | |||||||
| African | A*66∶01 | C*06∶02 | B*58∶02 | DRB1*13∶01 | DQB1*03∶03 | 4 | 0.0086 | 1.0000 | 1.99 |
| Total | 4 | 0.0086 | |||||||
| Unknown | A*02∶01 | C*07∶02 | B*39∶05 | DRB1*04∶07 | DQB1*03∶02 | 6 | 0.0128 | 0.1130 | 0.68 |
| A*24∶02 | C*04∶01 | B*35∶14 | DRB1*16∶02 | DQB1*03∶01 | 4 | 0.0086 | 1.0000 | 1.67 | |
| A*30∶01 | C*06∶02 | B*13∶02 | DRB1*07∶01 | DQB1*02∶02 | 4 | 0.0086 | 1.0000 | 1.99 | |
| A*68∶02 | C*04∶01 | B*53∶01 | DRB1*13∶02 | DQB1*06∶04 | 4 | 0.0086 | 1.0000 | 1.96 | |
| A*02∶01 | C*01∶02 | B*15∶30 | DRB1*08∶02 | DQB1*04∶02 | 3 | 0.0064 | 0.6759 | 1.21 | |
| A*23∶01 | C*07∶01 | B*41∶01 | DRB1*08∶04 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.40 | |
| A*66∶01 | C*12∶03 | B*39∶10 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 1.0000 | 1.40 | |
| A*26∶01 | C*06∶02 | B*37∶01 | DRB1*01∶03 | DQB1*05∶01 | 2 | 0.0043 | 1.0000 | 1.39 | |
| Table 5 . | Cont. | ||||||||
| A*26∶01 | C*06∶02 | B*45∶01 | DRB1*07∶01 | DQB1*02∶02 | 2 | 0.0043 | 1.0000 | 1.39 | |
| A*03∶01 | C*06∶02 | B*35∶02 | DRB1*11∶04 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.38 | |
| A*68∶01 | C*07∶02 | B*39∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 1.0000 | 1.31 | |
| A*68∶01 | C*07∶02 | B*39∶02 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.31 | |
| A*31∶01 | C*03∶03 | B*52∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 1.0000 | 1.31 | |
| A*02∶06 | C*01∶02 | B*15∶30 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.28 | |
| A*02∶06 | C*04∶01 | B*35∶24 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.28 | |
| A*31∶01 | C*03∶05 | B*35∶01 | DRB1*04∶07 | DQB1*03∶02 | 2 | 0.0043 | 0.6381 | 1.25 | |
| A*31∶01 | C*03∶05 | B*40∶02 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 0.6381 | 1.25 | |
| A*02∶06 | C*15∶09 | B*51∶01 | DRB1*04∶07 | DQB1*03∶02 | 2 | 0.0043 | 0.6312 | 1.22 | |
| A*31∶01 | C*03∶04 | B*40∶02 | DRB1*04∶07 | DQB1*03∶02 | 2 | 0.0043 | 0.4571 | 1.20 | |
| A*31∶01 | C*03∶05 | B*40∶02 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.4571 | 1.20 | |
| A*24∶02 | C*04∶01 | B*35∶01 | DRB1*14∶01 | DQB1*05∶03 | 2 | 0.0043 | 1.0000 | 1.18 | |
| A*24∶02 | C*04∶01 | B*35∶12 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 1.0000 | 1.18 | |
| A*24∶02 | C*01∶02 | B*35∶43 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 1.0000 | 1.18 | |
| A*24∶02 | C*03∶04 | B*40∶02 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.18 | |
| A*02∶06 | C*03∶04 | B*40∶02 | DRB1*16∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.4468 | 1.15 | |
| A*02∶01 | C*07∶02 | B*07∶02 | DRB1*11∶04 | DQB1*03∶01 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*01∶02 | B*15∶01 | DRB1*04∶04 | DQB1*03∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*01∶02 | B*35∶43 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*03∶04 | B*39∶02 | DRB1*04∶11 | DQB1*03∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*05∶01 | B*44∶02 | DRB1*04∶02 | DQB1*03∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*08∶03 | B*48∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*02∶01 | C*15∶09 | B*51∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 1.0000 | 1.10 | |
| A*24∶02 | C*01∶02 | B*15∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.5990 | 1.06 | |
| A*24∶02 | C*07∶02 | B*39∶06 | DRB1*04∶07 | DQB1*03∶02 | 2 | 0.0043 | 0.3985 | 0.94 | |
| A*02∶01 | C*03∶03 | B*52∶01 | DRB1*14∶06 | DQB1*03∶01 | 2 | 0.0043 | 0.5679 | 0.93 | |
| A*02∶01 | C*04∶01 | B*35∶01 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.5679 | 0.93 | |
| A*02∶01 | C*07∶02 | B*39∶05 | DRB1*14∶02 | DQB1*03∶01 | 2 | 0.0043 | 0.5679 | 0.93 | |
| A*24∶02 | C*07∶02 | B*39∶05 | DRB1*08∶02 | DQB1*04∶02 | 2 | 0.0043 | 0.2782 | 0.82 | |
| Total | 87 | 0.1869 | |||||||
Blocks of each ancestry (Amerindian, Caucasian, Caucasian shared with other populations, African and Asian) were defined as those found in original populations with H.F. >1,0%, and not found in other native human groups in frequencies higher than 1,0%. We consider t value must be ≥2.0 to denote statistically significant association and thus validate Δ′ (shaded values).
HLA Genetic Diversity in Mexicans
The extensive polymorphism of the HLA loci in this group of Mexicans was confirmed using polymorphism information content (PIC) values >0.5. HLA-B and -DRB1 loci were the most polymorphic with PIC values of 0.9544 and 0.9123, respectively. HLA-C and –A loci were relatively less polymorphic with PIC values of 0.8845 and 0.8776, respectively, and the less polymorphic locus was HLA-DQB1 (PIC = 0.8020). The degree of polymorphism of HLA loci was also corroborated by the power of discrimination (PD) values. A lower observed heterozygosity (OH) than expected heterozygosity (EH) was found for HLA-DRB1 locus, Table 6 .
Table 6. Measures of genetic diversity at the allele level for HLA system in a Mexican admixed population.
| HLA Allele | O.H. | E.H. | p value | PIC | PD |
| HLA-A | 0.8718 | 0.8919 | 0.5303 | 0.8776 | 0.7382 |
| HLA-B | 0.9487 | 0.9668 | 0.2942 | 0.9544 | 0.8531 |
| HLA-C | 0.9009 | 0.8947 | 0.2001 | 0.8845 | 0.7972 |
| HLA-DRB1 | 0.9013 | 0.9193 | 0.0061 | 0.9123 | 0.7981 |
| HLA-DQB1 | 0.8205 | 0.8256 | 0.2682 | 0.8020 | 0.6376 |
O.H.: Observed heterozygosity. E.H.: Expected Heterozygosity. p values <0.05 are considered statistically significative and thus reflect differences between O.H. and E.H. PIC: Polymorphism Information Contents. PD: Power of Discrimination.
Mexican Admixed Individuals have a Significant Proportion of Amerindian and Caucasian Genetic Components
The admixture estimations using HLA-B revealed an Amerindian contribution of 59.97%; Caucasian contribution of 25.71%; African contribution of 14.13%; and Asian contribution of 0.18%. These results were similar to the estimations obtained using STRs: Amerindian contribution: 60.5%; Caucasian: 25.9% and African: 13.6%.
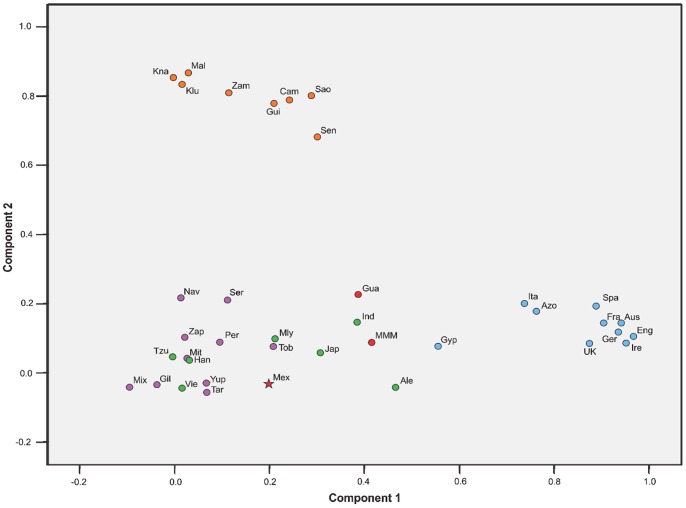
In addition, the results using the ABF revealed a frequency of Amerindian HLA-C/−B blocks of 41.3%, followed by Caucasian 25.8%, African 5.5% and Asian 3.3% blocks. The ABF of MHC class II blocks were as follows: Amerindian 51.2%, Caucasian 41.7%, African 3.4% and Asian 2.1%. Further evidence of the distribution of immunogenetic diversity can be observed in the principal component analysis (PCA) plot ( Figure 1 ), in which our Mexican admixed sample (Mex) clusters together with Native American and Asian populations (which can not be clearly differentiated from each other when HLA-B frequencies are taken as the variable of the factor analysis), and not with the African or European clusters.
Figure 1. Principal component analysis (PCA) plot reveals a close genetic relationship of Mexican admixed individuals from Mexico City to Native American groups.
Orange dots refer to African populations. Blue dots represent European samples. Green dots correspond to Asian human groups. Native American populations are represented by purple dots. Red figures are admixed populations from Mexico, with a star locating our Mexico City admixed sample. Proper references of each population group included in the analysis are given in the Materials and Methods section. Ire: Ireland, Eng: England, Ger: Germany, Aus: Austria; Spa: Spain, Ita: Italy, UK: United Kingdom, Fra: France, Gyp: Gypsy, Azo: Azores, Sao: São Tomé Island, Cam: Cameroon, Mal: Mali, Zam: Zambia, KLu: Luo from Kenia, KNa: Nandi from Kenia, Sen: Senegal, Gui: Guinea Bissau, Ale: Aleut from Bering Island, Jap: Japan, Tzu: Taiwan, Han: south China, Ind: north India, Mly: Malasya, Vie: Vietnam, Tar: Tarahumara, Gil: Native Americans from Gila River, Yup: Yu’pik from Alaska, Mit: Mixtec from Oaxaca, Zap: Zapotec from Oaxaca, Mix: Mixe from Oaxaca, Ser: Seri from Sonora, Nav: Navajo from New Mexico, Uro: Uro from Titikaka Lake, Tob: Toba from Rosario, MMM: “Mexican Mestizo” sample, Gua: Guadalajara City, Mex: this study.
Discussion
Here, we analyzed MHC class I (HLA-C/B) and class II (HLA-DRB1/DQB1) blocks diversity, ancestry, and the frequency of CEHs from HLA-C/B/DRB1/DQB1 and their extension to HLA-A in a total number of 468 haplotypes of individuals from Mexico City. We found that 41.0% of the HLA-C/−B blocks in our group were from Amerindian origin. In addition, some of these HLA-C/−B blocks also have been described in Asian populations (e.g: C*08∶01/B*48∶01) including Ivatan from Philippines [15] and several ethnic groups from Taiwan [14]. These findings may indicate that those haplotypes could be frequent in an ancestral group from which both Amerindians and South-East Asians originated from. Amerindian HLA-C/−B blocks observed in the present study, have been also reported, with high frequencies, among Amerindian groups including Zapotecs, Mixe, and Mixtec from Oaxaca State in the southeast of Mexico [16]; Tarahumara from Chihuahua State in the north of Mexico [17]; Native Americans from US [18]; and Yucpa from Venezuela [19]. Genetic admixture estimations were similar to those previously reported data from Mexico City [9]. We detected 13.2% of haplotypes of Caucasian MPA and 11.1% were predominantly Caucasian but shared with other populations including the haplotypes C*07∶02/B*07∶02, C*16∶01/B*44∶03, C*12∶03/B*38∶01 and C*05∶01/B*18∶01 [13], [18], [20], [21], [22].
In the PCA, our Mexican admixed sample (Mex) clearly separated from the European and African clusters and located within a loose cluster including populations from Asia and Native human groups from America. Notably, the “Mestizo” sample from Mexico (MMM) and the sample from Guadalajara (Gua) showed to be more proximate to the European cluster; Guadalajara population samples have shown a high degree of European genetic component in other works [23], [24]. Differences in admixed populations show the importance of not taking “Mestizo” as a global grouping category for individuals or populations with shared ancestry derived from demographic history of the colonial period. Also, lack of available data with high resolution HLA typing is evident in Native American groups.
Regarding MHC class II blocks, 51.2% of them were from Amerindian MPA, the most common being DRB1*08∶02/DQB1*04∶02 and DRB1*04∶07/DQB1*03∶02, whereas 40% of the -DRB1/−DQB1 blocks were from Caucasian MPA. These haplotypes are common in Mexican Amerindians, as well as in Xavante from Central Brazil, Toba from Argentina, Athabaskan from Canada, and Mayans from Guatemala [25]–[28]. Interestingly, HLA class II blocks show a restricted diversity as it was pointed out by the fact that eleven CEH (HF >0.5%; t ≥2.0) are associated with only three HLA class II blocks: DRB1*04∶07/DQB1*03∶02, DRB1*14∶06/DQB1*03∶01, and mainly DRB1*08∶02/DQB1*04∶02. This trend is also shown in CEH extended to the HLA-A locus. Less than 5% of class II blocks from African or Asian probable ancestry were detected.
Genetic diversity parameters confirm the high degree of polymorphism of the HLA genes in the studied sample. HLA-B and HLA-DRB1 were the most polymorphic loci according to PIC and PD values, followed by HLA-C locus. However, lower OH than EH was found for HLA-DRB1 locus. This may indicate that selective forces are acting on the HLA-DRB1 locus in Mexicans, as well as in the Mexican Amerindian populations, resulting in low class II diversity. Also, low class II diversity may have been produced by the limited -DRB1 allelic diversity that the first human settlers carried with them into the Americas [29]–[31] and their incorporation into the admixed Mexican genetic pool. Deviation from neutral expectations tends to ocurr by an excess of heterozygotes; however, homozygous excess has also been observed [31]. Migration patterns into Mexico City in the last 60 years also have to be taken into account to adecuately address an explanation for the low number of heterozygous individuals, as they represent an important source of incorporation of alleles and haplotypes –mainly from indigenous populations-, hence modifying the allelic diversity.
In our study the admixture estimations using STRs confirm the greater contribuition of Amerindian and Caucasian and a small contribution of African and Asian genes. The results obtained using the ABF of HLA-C/−B blocks also demonstrated a greater contribution of Amerindian (41.3%), followed by Caucasian (24.6%), African (6.7%), and Asian (3.0%) genes in the admixed Mexicans. Also, the estimations using the ABF of MHC class II blocks revealed that 51.2% of them were from Amerindian and 40.4% from Caucasian MPA. These findings suggest that ABF method is applicable to analyze the genetic diversity and ancestral structure of admixed populations. In this perspective, the genetic admixture of Mexicans could have resulted from the Spaniards, which arrived to Mexico early in the 16th century. Caucasian component consisted in conquerors and colonizers from Andalucía, Leon, Extremadura, and the Castillas, as well as Portugal and Genoa. Spaniards settled extensively all over the Viceroyalty of the New Spain and a massive migration of colonizers begun on the 17th century and prevailed through the next two centuries. Presence of Caucasian-MPA or Caucasian-shared blocks or haplotypes may be explained by these demographic traits. The preponderance of haplotypes commonly found in Caucasian populations may be due to the fact that more Caucasian human groups than African or Asian ones have been studied, or may simply reflect a lower genetic diversity among Caucasians. Another hypothesis is that population replacement, together with the collapse of Native American groups that took place due to infectious diseases [32] and the conquest wars, may explain the high prevalence of Caucasian genetic blocks within Mexican admixed individuals [33]. African contribution, although subtle, is present in admixed Mexicans due to slaves introduced to Mexico from Africa during the first three centuries of Spanish colonial domination. All African specific associations present in this study are found in Sub Saharan Africa [14], [18], [34,], the place where slaves were extracted from by colonial slave traders [18], [35]. For example, C*07∶01/B*49∶01, C*04∶01/B*53∶01, C*06∶02/B*58∶02, DRB1*13∶01/DQB1*03∶03, and DRB1*08∶04/DQB1*03∶01 blocks have been found in Africa, for instance in Bandiagara from Mali, Bantu from Congo, Bioko from Equatorial Guinea, Luo and Nandi from Kenya, Lusaka from Zambia, Ugandans and Kampala from Uganda, and Yaounde from Cameroon, [18], [36]–[39] and have been reported also in African American population from the US [40].
On the other hand, the presence of Asian genes in Mexican population possibly resulted from relative recent immigration of Chinese traders and slaves by transpacific travels from the oriental shores of Asia to the western coasts of Mexico, mainly disembarking in the port of Acapulco. Thus, the Não de China (the Manila Galleon) route, together with a foreign investment policy starting in the 19th century, helped the Chinese community to become the largest non-Spaniard community in Mexico by mid-1920s [41]. The Asian contribution to the genetic pool conformation of Mexico is modest, mainly due to lack of admixture between Asian immigrants and Mexicans; however, classical Asian associations were found in our sample such as C*04∶01/B*35∶16 [42] and C*08∶01/B*15∶02 [18], [27], [43]. The admixture estimations using different indicators support a tryhybrid model of Amerindian, Caucasian and African ancestry in Mexicans. But we were able to detect also a small Asian component in Mexicans.
It is well known that MHC diversity influences the susceptibility or resistance to a wide variety of autoimmune disorders and infectious diseases caused by viruses, yeasts, bacteria and parasites. It has been suggested that pathogen-mediated selection might explain the maintenance of MHC diversity at population level [44], [45]. However, the role of MHC diversity associated to the admixture between different ethnic groups in the resistance or susceptibility to autoimmune or infectious disease remains unclear. Furthermore, recent studies have suggested that genes that confer susceptibility to autoimmune diseases might be maintained in specific ethnic groups because they primarily confer protection against infectious agents, the major factor driving selection and influencing human adaptation to local environments [2]–[6], [46]–[48]. Functional studies are necessary to define whether the genetic diversity of HLA is influenced in pathogen-enriched environments. The analyses of HLA diversity in the context of pathogen richness have shown a positive correlation between HLA class I allele diversity and pathogen richness and a negative correlation of HLA class II diversity, particularly HLA-DQB1 loci, and pathogen richness, suggesting that HLA class I and class II genes have disctint evolutionary strategies to confer immunity against infectious agents [5]. In this context, the higher diversity of HLA class I genes may result from the high mutation rate of intracelular pathogens, particularly viruses. In contrast, the lower diversity of MHC class II genes might result from the fixation of some alleles that provide efficient immune protection against highly prevalent extracelular pathogens in specific populations (e.g. parasites). In Mexicans, we found a high frequency of some MHC class II alleles that predispose to rheumatoid arthritis (RA) (DRB1*04∶04, DRB1*14∶02, and DRB1*01∶02), to systemic lupus erythematosus (SLE) (DRB1*03∶01) [11] and to systemic sclerosis (SSc) (DRB1*11∶04) (Rodriguez-Reyna TS et al., Unpublished data). It is possible that class II MHC alleles associated with autoimmunity, together with alleles found in Native American populations may have increased their frequencies due to past selective processes or infectious and parasitic diseases developed in different environments and thus explain in part the susceptiblity to develop autoimmune diseases in Mexico or the clinical characteristics of these diseases in Mexican population.
In summary, Mexican admixed individuals from the central area of Mexico have an important component of Amerindian and Caucasian MHC class I (HLA-C/−B) and class II (HLA-DRB1/−DQB1) blocks and HLA CEHs. A relatively low frequency of African and Asian HLA blocks and CEHs were detected. In line with these results, admixture estimations using STRs and HLA-B revealed a greater proportion of Amerindian, followed by Caucasian and African ancestry in this population. The high frequency of certain relatively fixed haplotypes might result from many possible mechanisms, including recent population bottlenecks, recombination suppression, preferential transmission, migration and admixture, and/or genetic drift or natural selection. Our findings suggest that the study of HLA class I and class II blocks and CEHs diversity might be useful to characterize the ancestral contributions in admixed populations, as well as to perform studies of disease susceptibility and transplantation.
Materials and Methods
Subjects
A total of 234 unrelated Mexican admixed individuals were studied, including a group of 80 Mexican admixed participants belonging to 40 families. A total number of 468 haplotypes were analyzed in this study. Every participant came from Mexico City and had a Mexican ancestry whose parents and grandparents were born in Mexico. Age mean of studied individuals was 38.2±15.3 years. There were 120 females (51%) and 114 males (49%).
Ethics Statement
The Institutional Review Board of the National Institute of Respiratory Diseases (INER) reviewed and approved the protocols for genetic studies. All subjects provided written informed consent for these studies, and they authorized the storage of their DNA samples at INER repositories for this and future studies. In this study we did not collected samples from minors/children, only young adults older than 17 years were included.
HLA Typing
Genomic DNA was obtained from peripheral blood mononuclear cells (PBMC), using the QIAamp DNA mini kit (Qiagen, Valencia, CA, USA). High-resolution HLA class I and class II typing was performed by a sequence-based method (SBT) as previously described [49]. Briefly, we amplified exon 2 and 3 from HLA-A, -B and -C and exon 2 for HLA-DRB1 and -DQB1. Polymerase chain reaction (PCR) contained 1.5 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl (pH = 8.3), 200 mM concentrations of each dATP, dTTP, dGTP, and dCTP; 10 pM concentration of each primer, 30 ng of DNA and 0.5 U of Taq DNA polymerase in a final volume of 25 µl. Amplification was done on a PE9700 thermal cycler (Applied Biosystems, Foster City, CA, USA) using the following cycling conditions: 95°C for 30 s, 65°C for 30 s, 72°C for 1 min, preceded by 5 min at 95°C, and followed by a final elongation at 72°C for 5 min. Amplified products were sequenced independently in both directions using BigDye Terminator ™ chemistry in an ABI PRISM® 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Data were analyzed using match tools allele assignment software (Applied Biosystems, Foster City, CA,USA) using the IMGT/HLA sequence database alignment tool (http://www.ebi.ac.uk/imgt/hla/align.html) [50]. Ambiguities were solved using group-specific sequencing primers (GSSPs) that have been reported and validated previously [49].
HLA Blocks and Conserved Extended Haplotypes Assignment
HLA allelic and haplotypic frequencies were obtained by gene counting; one hundred and sixty of the 468 haplotypes were obtained by direct observation because they were obtained by HLA typing in the parents and siblings of 40 families, while the rest were acquired from HLA genotyping of 154 non-related individuals. Haplotypes were estimated by maximum likelihood methods using the computer program Arlequin ver. 3.0 [51]. This software was also used to calculate HWE, OH, and EH at a locus-by-locus level with 1×106 steps in the Markov chain and 1×105 dememorization steps. p-values ≤0.05 indicated statistical difference between OH and EH and thus a deviation from HWE. Listed HLA-C/B, HLA-DRB1/DQB1 and CEHs and their extension to the HLA-A locus of Mexican origin were estimated by the maximum likelihood method based on the Δ′ between alleles of two loci and between the two blocks and/or the extension to the HLA-A region, as previously described [18]. Haplotypes or DNA blocks of African, Asian and Caucasian MPA were assigned based on previous reported frequencies [7], [13], [18]. Estimation of delta (Δ) and relative delta (Δ′) values to measure LD, nonrandom association of alleles at two or more loci, and their statistical significance, were calculated using previously described methods [18]. Absolute Δ′ values of 1 indicates complete LD; 0 corresponds to no LD. As many of this associations may return |Δ′| values of 1.000 -even though that value may be result of a random association between two infrequent alleles- we used the statistic parameter t, to validate all Δ′ data adjusted by sample size and number of times that each allele appeared in the sample [52]. Only t values ≥2.0 were considered significant.
HLA Genetic Diversity Calculations
Genetic diversity of each HLA loci was assessed by two previously described forensic parameters: PIC and PD [53]–[55] that were computed using the PowerStat ver.1.2 spreadsheet (Promega Corporation, Fitchburg, WI, USA) as described elsewhere [56]. PIC measures the strength of a genetic marker for linkage studies by indicating the degree of polymorphism of a locus. PIC >0.5 is considered as highly polymorphic [54]. PD is defined as the probability of finding two random individuals with different genotypes for that locus in the studied population, and values higher than 0.8 indicate high polymorphism in the studied population context [57]. The OH and EH of all HLA loci was also calculated [53].
Admixture Estimations in Mexicans using HLA Genes
Admixture estimates were obtained by maximum-likelihood method using the Leadmix software [58], with k = 4 parental populations (Africa, America, Asia, and Europe) and HLA-B as the genetic estimator. Caucasian component was estimated with a pooled sample (N = 315) consisting of data from southern Portugal [34] and an European population sample from USA [18]; African Nandi from Kenia [59] served as the African parental component (N = 239); a pooled Native American sample (N = 146) was used, which consisted of data from Mixtec of Oaxaca, SE Mexico [16] and Tarahumara from Chihuahua, north of Mexico [17]; finally, Han from southern China data (N = 281) were used to estimate the Asian contribution [60]. Principal Components Analysis (PCA) for 38 populations with HLA-B data available was performed using the IBM SPSS Statistics 19 software (IBM Corporation, Armonk, NY, USA) to analyse the distribution of HLA-B alleles in human groups of the proposed ancestries, Figure 1 . PCA included population data of Ireland [20], NW of England [61], Germany [62], Austria [63]; Spain, Italy, United Kingdom [64], France [65], Gypsy from Andalucía (Spain; data collected by López-Nevot et al.) [14], Azores Terceira Island [66], Forro from São Tomé Island [67], Beti from Cameroon [68], Bandiagara from Mali, Lusaka from Zambia, Luo and Nandi from Kenia [69], Mandeka from Senegal [70], Guinea Bissau [71], Aleut from Bering Island (Russia) [72], center of Japan [73], a cord blood bank of Tzu Chi Foundation (Taiwan) [74], Han from southern China [60], north India [75], Kensiu from Malasya [76], Kinh from Vietnam [77], Tarahumara from northern Mexico [17], Native Americans from Gila River (USA) [78], Yu’pik from Alaska (USA) [79], Mixtec, Zapotec, and Mixe from Oaxaca (Mexico) [16], Seri from Sonora (Mexico) [80], Navajo from New Mexico (USA) [81], Uro from Titikaka Lake (Peru) [82], and Toba from Rosario (Argentina; data collected by Cintia Marcos et al.) [14]. Also, two admixed populations from Mexico were included: a “Mexican Mestizo” sample [83] and a sample from Guadalajara City, western Mexico [23]. As an approach to estimate the diversity and contribution of previously described [7], [13] Caucasian, Asian, and African HLA blocks in our population, we also calculated the aggregate block frequencies (ABF) [7], [13] adding the frequencies of those HLA clas I and II blocks with frequencies greater than 1% in our study population.
Admixture Estimations in Mexicans using STRs
We genotyped fifteen autosomal STR markers (CSF1PO, FGA, THO1, TPOX, VWA, D3S11358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, D19S433, and D2S1338) along with amelogenin using the Applied Biosystems AmpFl STR Identifiler Kit (Applied Biosystems, Foster City, CA, USA). PCR amplification was carried out on a Gene Amp 7500 thermocycler (Applied Biosystems, Foster City, CA, USA) using 1 ng of DNA according to the manufacturer’s protocol. The PCR conditions were: 95°C during 11 min followed by 28 cycles of 94°C for 1 min, 59°C for 1 min, 72°C for 1 min followed by a hold at 60°C for 60 min. PCR products were diluted 1∶15 in Hi-Di™ formamide and GS500-LIZ internal size standard (Applied Biosystems, Foster City, CA, USA) and analyzed on the ABIPrism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Allele calls were made using Genotype 3.7 software by comparison with kit allelic ladders (Applied Biosystems, Foster City, CA, USA). We performed an admixture estimation using the STR’s data by a model-based clustering method with the Structure software v. 2.3.4 [84], assuming k = 3 populations and 1×104 dememorisation steps, using Spaniards [85], Fang Africans [86], and a Native-American pool of Huastecos [87] and Tepehuas [88] from the central region of Mexico, as parental populations.
Funding Statement
No current external funding sources for this study.
References
- 1. Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, et al. (2004) Gene map of the extended human MHC. Nat Rev Genet 12: 889–899. [DOI] [PubMed] [Google Scholar]
- 2. de Vries RR, Meera Khan P, Bernini LF, van Loghem E, van Rood JJ (1979) Genetic control of survival in epidemics. J Immunogenet 6: 271–287. [DOI] [PubMed] [Google Scholar]
- 3. Gilbert SC, Plebanski M, Gupta S, Morris J, Cox M, et al. (1998) Association of malaria parasite population structure, HLA, and immunological antagonism. Science 279: 1173–1177. [DOI] [PubMed] [Google Scholar]
- 4. Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, et al. (2005) Pathogen-driven selection and worldwide HLA class I diversity. Current Biology 15: 1022–1027. [DOI] [PubMed] [Google Scholar]
- 5. Sanchez-Mazas A, Lemaître JF, Currat M (2012) Distinct evolutionary strategies of human leucocyte antigen loci in pathogen-rich environments. Philos Trans R Soc Lond B Biol Sci 367: 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Penn DJ, Damjanovich K, Potts WK (2002) MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci USA 99: 11260–11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yunis EJ, Zuniga J, Larsen CE, Fernandez-Vina M, Granados J, et al.. (2005) Single Nucleotide Polymorphism blocks and haplotypes: Human MHC block diversity. In: Meyers RA, editor. Encyclopedia of Molecular Cell Biology and Molecular Medicine. 2nd ed. Weinheim: Wiley-VCH Verlag GmbH & Co. 191–215.
- 8. Lisker R, Ramírez E, Briceño RP, Granados J, Babinsky V (1990) Gene frequencies and admixture estimates in four Mexican urban centers. Hum Biol 62: 791–801. [PubMed] [Google Scholar]
- 9. Barquera R, Zúñiga J, Hernández-Díaz R, Acuña-Alonzo V, Montoya-Gama K, et al. (2008) HLA class I and class II haplotypes in admixed families from several regions of Mexico. Mol Immunol 45: 1171–1178. [DOI] [PubMed] [Google Scholar]
- 10. Juárez-Cedillo T, Zúñiga J, Acuña-Alonzo V, Pérez-Hernández N, Rodríguez-Pérez JM, et al. (2008) Genetic admixture and diversity estimations in the Mexican Mestizo population from Mexico City using 15 STR polymorphic markers. Forensic Sci Int Genet 2: e37–e39. [DOI] [PubMed] [Google Scholar]
- 11. Granados J, Vargas-Alarcón G, Andrade F, Melín-Aldana H, Alcocer-Varela J, et al. (1996) The role of HLA-DR alleles and complotypes through the ethnic barrier in systemic lupus erythematosus in Mexicans. Lupus 5: 184–189. [DOI] [PubMed] [Google Scholar]
- 12.Ceppellini R, Curtoni ES, Mattiuz PL, Miggiano V, Scudeller G, et al.. (1967) Genetics of leukocyte antigens: A family study of segregation and linkage. In: Curtoni ES, Mattiuz PL, Tosi RM, editors. Histocompatibility. Copenhagen: Munksgaard. 189–202.
- 13. Yunis EJ, Larsen CE, Fernandez-Viña M, Awdeh ZL, Romero T, et al. (2003) Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue Antigens 62: 1–20. [DOI] [PubMed] [Google Scholar]
- 14. González-Galarza FF, Christmas S, Middleton D, Jones AR (2011) Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acid Res 39: D913–D919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu CC, Trejaut J, Lee HL, Chang SL, Lin M (2007) Ivatan from Bantanes, Philippines. Anthropology/human genetic diversity population reports. In: Hansen JA (ed) Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference, Volume I. Seattle: IHWG Press. 611–615.
- 16. Hollenbach JA, Thomson G, Cao K, Fernández-Viña M, Erlich HA, et al. (2001) HLA diversity, differentiation, and haplotype evolution in Mesoamerican Natives. Hum Immunol 62: 378–390. [DOI] [PubMed] [Google Scholar]
- 17. García-Ortíz JE, Sandoval-Ramírez L, Rangel-Villalobos H, Maldonado-Torres H, Cox S, et al. (2006) High-resolution molecular characterization of the HLA class I and class II in the Tarahumara Amerindian population. Tissue Antigens 68: 135–146. [DOI] [PubMed] [Google Scholar]
- 18. Cao K, Hollenbach J, Shi X, Shi W, Chopek M, et al. (2001) Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol 62: 1009–1030. [DOI] [PubMed] [Google Scholar]
- 19. Layrisse Z, Guedez Y, Domínguez E, Paz N, Montagnani S, et al. (2001) Extended HLA haplotypes in a Carib Amerindian population: the Yucpa of the Perija Range. Hum Immunol 62: 992–1000. [DOI] [PubMed] [Google Scholar]
- 20. Dunne C, Crowley J, Hagan R, Rooney G, Lawlor E (2008) HLA-A, B, Cw, DRB1, DQB1 and DPB1 alleles and haplotypes in the genetically homogenous Irish population. Int J Immunogenet 35: 295–302. [DOI] [PubMed] [Google Scholar]
- 21. Middleton D, Williams F, Hamill MA, Meenagh A (2000) Frequency of HLA-B alleles in a Caucasoid population determined by a two-stage PCR-SSOP typing strategy. Hum Immunol 61: 1285–1297. [DOI] [PubMed] [Google Scholar]
- 22. Schmidt AH, Baier D, Solloch UV, Stahr A, Cereb N, et al. (2009) Estimation of high-resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Hum Immunol 70: 895–902. [DOI] [PubMed] [Google Scholar]
- 23. Leal CA, Mendoza-Carrera F, Rivas F, Rodriguez-Reynoso S, Portilla-de Buen E (2005) HLA-A and HLA-B allele frequencies in a mestizo population from Guadalajara, Mexico, determined by sequence-based typing. Tissue Antigens 66: 666–673. [DOI] [PubMed] [Google Scholar]
- 24. Martínez-Cortés G, Salazar-Flores J, Haro-Guerrero J, Rubi-Castellanos R, Velarde-Félix JS, et al. (2013) Maternal admixture and population structure in Mexican-Mestizos based on mtDNA haplogroups. Am J Phys Anthropol 151: 526–537. [DOI] [PubMed] [Google Scholar]
- 25. Cerna M, Falco M, Friedman H, Raimondi E, Maccango A, et al. (1993) Differences in HLA class II alleles of isolated South American Indian populations from Brazil and Argentina. Hum Immunol 37: 213–220. [DOI] [PubMed] [Google Scholar]
- 26. Fernández-Viña MA, Lázaro AM, Marcos CY, Nulf C, Raimondi E, et al. (1997) Dissimilar evolution of B-locus versus A-locus and class II of the HLA region in South American Indian tribes. Tissue Antigens 50: 233–250. [DOI] [PubMed] [Google Scholar]
- 27. Monsalve MV, Edin G, Devine DV (1998) Analysis of HLA class I and class II in Na-Dene and Amerindian populations from British Columbia, Canada. Hum Immunol 59: 48–55. [DOI] [PubMed] [Google Scholar]
- 28. Gómez-Casado E, Martínez-Laso J, Moscoso J, Zamora J, Martin-Villa M, et al. (2003) Origin of Mayans according to HLA genes and the uniqueness of Amerindians. Tissue Antigens 61: 425–436. [DOI] [PubMed] [Google Scholar]
- 29. Erlich HA, Mack SJ, Bergström T, Gyllensten UB (1997) HLA class II alleles in Amerindian populations: implications for the evolution of HLA polymorphism and the colonization of the Americas. Hereditas 127: 19–24. [DOI] [PubMed] [Google Scholar]
- 30. Tsuneto LT, Probst CM, Hutz MH, Salzano FM, Rodríguez-Delfín LA, et al. (2003) HLA class II diversity in seven Amerindian populations. Clues about the origins of the Aché. Tissue Antigens 62: 512–526. [DOI] [PubMed] [Google Scholar]
- 31. Sánchez-Mazas A, Fernández-Viña M, Middleton D, Hollenbach JA, Buhler S, et al. (2011) Immunogenetics as a tool in anthropological studies. Immunology 133: 143–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marr JS, Kiracofe JB (2000) Was the Huey Cocoliztli a haemorrhagic fever? Med Hist 44: 341–362. [PMC free article] [PubMed] [Google Scholar]
- 33. Arrieta-Bolaños E, Madrigal JA, Shaw BE (2012) Human Leukocyte Antigen profiles of Latin American populations: differential admixture and its potential impact on hematopoietic stem cell transplantation. Bone Marrow Res 2012: 136087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spínola H, Brehm A, Williams F, Jesus J, Middleton D (2002) Distribution of HLA alleles in Portugal and Cabo Verde. Relationships with the slave trade route. Ann Hum Genet 66: 285–296. [DOI] [PubMed] [Google Scholar]
- 35.Barquera R, Acuña-Alonzo V (2012) The African colonial migration into Mexico: History and biological consequences. In: Crawford MH & BC Campbell (ed.). Causes and Consequences of Human Migration. An evolutionary perspective. Cambridge University Press. 201–223.
- 36. de Pablo R, García-Pacheco JM, Vilches C, Moreno ME, Sanz L, et al. (1997) HLA class I and class II allele distribution in the Bubi population from the island of Bioko (Equatorial Guinea). Tissue Antigens 50: 593–601. [DOI] [PubMed] [Google Scholar]
- 37. Ellis JM, Mack SJ, Leke RF, Quakyi I, Johnson AH, et al. (2000) Diversity is demonstrated in class I HLA-A and HLA-B alleles in Cameroon, Africa: description of HLA-A*03012, *2612, *3006 and HLA-B*1403, *4016, *4703. Tissue Antigens 56: 291–302. [DOI] [PubMed] [Google Scholar]
- 38. Renquin J, Sánchez-Mazas A, Halle L, Rivalland S, Jaeger G, et al. (2001) HLA class II polymorphism in Aka Pygmies and Bantu Congolese and a reassessment of HLA-DRB1 African diversity. Tissue Antigens 58: 211–222. [DOI] [PubMed] [Google Scholar]
- 39. Kijak GH, Walsh AM, Koehler RN, Moqueet N, Eller LA, et al. (2009) HLA class I allele and haplotype diversity in Ugandans supports the presence of major east African genetic cluster. Tissue Antigens 73: 262–269. [DOI] [PubMed] [Google Scholar]
- 40. Maiers M, Gragert L, Klitz W (2007) High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 68: 779–788. [DOI] [PubMed] [Google Scholar]
- 41.Hu-Dehart E (1995) The Chinese of Peru, Cuba, and Mexico. In: Cohen R (ed.). The Cambridge survey of world migration. Cambridge. Cambridge University Press. 220–391.
- 42. Shankarkumar U (2004) HLA-A, -B, and -Cw allele frequencies in a Parsi population from Western India. Hum Immunol 65: 992–993. [Google Scholar]
- 43. Shi L, Shi L, Yao YF, Matsushita M, Yu L, et al. (2010) Genetic link among Hani, Bulang, and other Southeast Asian populations: evidence from HLA -A, -B, -C, -DRB1 genes and haplotypes distribution. Int J Immunogenet 37: 467–475. [DOI] [PubMed] [Google Scholar]
- 44. Doherty P, Zinkernagel R (1975) A biological role for the major histocompatibility antigens. Lancet 1: 1406–1409. [DOI] [PubMed] [Google Scholar]
- 45. Jeffery KJ, Bangham CR (2000) Do infectious diseases drive MHC diversity? Microbes Infect 2: 1335–1341. [DOI] [PubMed] [Google Scholar]
- 46. Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admettla A, Pattini L, et al. (2011) Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet 7: e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finch CE, Crimmins EM (2004) Inflammatory exposure and historical changes in human life-Spans. Science 305: 1736–1739. [DOI] [PubMed] [Google Scholar]
- 48. Gluckman PD, Hanson MA (2004) Living with the past: evolution, development, and patterns of disease. Science 305: 1733–1736. [DOI] [PubMed] [Google Scholar]
- 49. Lebedeva TV, Mastromarino SA, Lee E, Ohashi M, Alosco SM, et al. (2011) Resolution of HLA class I sequence-based typing ambiguities by group-specific sequencing primers. Tissue Antigens 77: 247–250. [DOI] [PubMed] [Google Scholar]
- 50. Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG (2001) IMGT/HLA Database–a sequence database for the human major histocompatibility complex. Nucleic Acid Res 29: 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Excoffier L, Laval G, Schneider S (2007) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 52. Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav. Genet. 2: 3–19. [DOI] [PubMed] [Google Scholar]
- 53. Yasuda N (1988) HLA polymorphism information content (PIC). Jinrui Idengaku Zasshi 33: 385–387. [DOI] [PubMed] [Google Scholar]
- 54. Shen CM, Zhu BF, Deng YJ, Ye SH, Yan JW, et al. (2010) Allele polymorphism and haplotype diversity of HLA-A, -B and -DRB1 loci in sequence based typing for Chinese Uyghur ethnic group. PLoS ONE 5: e13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yan C, Wang R, Li J, Deng Y, Wu D, et al. (2003) HLA-A gene polymorphism defined by high-resolution sequence-based typing in 161 Northern Chinese Han people. Genomics Proteomics Bioinformatics 1: 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tereba A (1999) Tools for analysis of population statistics. Profiles DNA 2: 3–5. [Google Scholar]
- 57. Zhu BF, Yang G, Shen CM, Qin HX, Liu SZ, et al. (2010) Distributions of HLA-A and –B alleles and haplotypes in the Yi ethnic minority of Yunnan, China: relationship to other populations. J Zhejiang Univ Sci B 11: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J (2003) Maximum Likelihood estimation of admixture proportions from genetic data. Genetics 164: 747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cao K, Moormann AM, Lyke KE, Masaberg C, Sumba OP, et al. (2004) Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63: 293–325. [DOI] [PubMed] [Google Scholar]
- 60.Tratchenberg E, Vinson M, Hayes E, Hsu YM, Houtchens K, et al.. (2007) Southern Han Chinese from People’s Republic of China. Anthropology/human genetic diversity population reports. In: Hansen JA (ed.). Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference. Seattle: IHWG Press. 616–617.
- 61. Alfirevic A, Gonzalez-Galarza F, Bell C, Martinsson K, Platt V, et al. (2012) In silico analysis of HLA associations with drug-induced liver injury: use of a HLA-genotyped DNA archive from healthy volunteers. Genome Med 6: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmidt AH, Baier D, Solloch UV, Stahr A, Cereb N, et al. (2009) Estimation of high-resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Hum Immunol 11: 895–902. [DOI] [PubMed] [Google Scholar]
- 63. Rosenmayr A, Pointner-Prager M, Winkler M, Mitterschiffthaler A, Pelzmann B, et al. (2011) The Austrian Bone Marrow Donor Registry: Providing Patients in Austria with Unrelated Donors for Transplant - a Worldwide Cooperation. Transfus Med Hemother 5: 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pingel J, Solloch UV, Hofmann JA, Lange V, Ehninger G, et al. (2013) High-resolution HLA haplotype frequencies of stem cell donors in Germany with foreign parentage: how can they be used to improve unrelated donor searches? Hum Immunol 74: 330–340. [DOI] [PubMed] [Google Scholar]
- 65. Loiseau P, Busson M, Balere ML, Dormoy A, Bignon JD, et al. (2007) HLA Association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant 13: 965–974. [DOI] [PubMed] [Google Scholar]
- 66. Spínola H, Brehm A, Bettencourt B, Middleton D, Bruges-Armas J (2005) HLA class I and II polymorphisms in Azores show different settlements in Oriental and Central islands. Tissue Antigens 66: 217–30. [DOI] [PubMed] [Google Scholar]
- 67. Saldanha N, Spínola C, Santos MR, Simões JP, Bruges-Armas J, et al. (2009) HLA polymorphisms in Forros and Angolares from São Tomé Island (West Africa): Evidence for the Population Origin. Journal of Genetic Genealogy 5: 76–85. [Google Scholar]
- 68. Torimiro JN, Carr JK, Wolfe ND, Karacki P, Martin MP, et al. (2006) HLA class I diversity among rural rainforest inhabitants in Cameroon: identification of A*2612-B*4407 haplotype. Tissue Antigens 67: 30–37. [DOI] [PubMed] [Google Scholar]
- 69. Cao K, Moormann AM, Lyke KE, Masaberg C, Sumba OP, et al. (2004) Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63: 293–325. [DOI] [PubMed] [Google Scholar]
- 70. Sanchez-Mazas A, Steiner QG, Grundschober C, Tiercy JM (2000) The molecular determination of HLA-Cw alleles in the Mandenka (West Africa) reveals a close genetic relationship between Africans and Europeans. Tissue Antigens 56: 303–312. [DOI] [PubMed] [Google Scholar]
- 71. Spínola H, Bruges-Armas J, Middleton D, Brehm A (2005) HLA polymorphisms in Cabo Verde and Guiné-Bissau inferred from sequence-based typing. Hum Immunol 66: 1082–1092. [DOI] [PubMed] [Google Scholar]
- 72. Moscoso J, Crawford MH, Vicario JL, Zlojutro M, Serrano-Vela JI, et al. (2008) HLA genes of Aleutian Islanders living between Alaska (USA) and Kamchatka (Russia) suggest a possible southern Siberia origin. Mol Immunol 45: 1018–1026. [DOI] [PubMed] [Google Scholar]
- 73. Saito S, Ota S, Yamada E, Inoko H, Ota M (2000) Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens 56: 522–529. [DOI] [PubMed] [Google Scholar]
- 74. Wen SH, Lai MJ, Yang KL (2008) Human leukocyte antigen-A, -B, and -DRB1 haplotypes of cord blood units in the Tzu Chi Taiwan Cord Blood Bank. Hum Immunol 69: 430–436. [DOI] [PubMed] [Google Scholar]
- 75. Rani R, Marcos C, Lazaro AM, Zhang Y, Stastny P (2007) Molecular diversity of HLA-A, -B and -C alleles in a North Indian population as determined by PCR-SSOP. Int J Immunogenet 34: 201–208. [DOI] [PubMed] [Google Scholar]
- 76. Jinam TA, Saitou N, Edo J, Mahmood A, Phipps ME (2010) Molecular analysis of HLA Class I and Class II genes in four indigenous Malaysian populations. Tissue Antigens 75: 151–158. [DOI] [PubMed] [Google Scholar]
- 77. Hoa BK, Hang NT, Kashiwase K, Ohashi J, Lien LT, et al. (2008) HLA-A, -B, -C, -DRB1 and -DQB1 alleles and haplotypes in the Kinh population in Vietnam. Tissue Antigens 71: 127–134. [DOI] [PubMed] [Google Scholar]
- 78. Williams R, Chen YF, Endres R, Middleton D, Trucco M, et al. (2009) Molecular variation at the HLA-A, B, C, DRB1, DQA1, and DQB1 loci in full heritage American Indians in Arizona: private haplotypes and their evolution. Tissue Antigens 74: 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Leffell MS, Fallin MD, Erlich HA, Fernandez-Viña M, Hildebrand WH, et al. (2002) HLA antigens, alleles and haplotypes among the Yup’ik Alaska natives: report of the ASHI Minority Workshops, Part II. Hum Immunol 63: 614–625. [DOI] [PubMed] [Google Scholar]
- 80.Infante E, Alaez C, Flores H, Gorodezky C (2007) Seri from Sonora, Mexico. In Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference, ed by Hansen JA, Seattle: IHWG Press, pp633–634.
- 81.Mack SJ, Crawford MH, Saha N, Jani AJ, Geyer LN, et al.. (2007) North Indians from New Delhi, India. Vol. I, In Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference, ed by Hansen JA,. Seattle: IHWG Press, pp605–607.
- 82. Arnaiz-Villena A, Gonzalez-Alcos V, Serrano-Vela JI, Reguera R, Barbolla L, et al. (2009) HLA genes in Uros from Titikaka Lake, Peru: origin and relationship with other Amerindians and worldwide populations. Int J Immunogenet 36: 159–167. [DOI] [PubMed] [Google Scholar]
- 83. Middleton D, Williams F, Meenagh A, Daar AS, Gorodezky C, et al. (2000) Analysis of the distribution of HLA-A alleles in populations from five continents. Hum Immunol 61: 1048–1052. [DOI] [PubMed] [Google Scholar]
- 84. Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sanz P, Prieto V, Flores I, Torres Y, López-Soto M, et al. (2001) Population data of 13 STRS in Southern Spain (Andalusia). Forensic Sci Int 119: 113–115. [DOI] [PubMed] [Google Scholar]
- 86. Calzada P, Suárez I, García S, Barrot C, Sánchez C, et al. (2005) The Fang population of Equatorial Guinea characterized by 15 STR-PCR polymorphisms. Int J Legal Med 119: 107–110. [DOI] [PubMed] [Google Scholar]
- 87. Barrot C, Sánchez C, Ortega M, González-Martín A, Brand-Casadevall C, et al. (2005) Characterisation of three Amerindian populations from Hidalgo State (Mexico) by 15 STR-PCR polymorphisms. Int J Legal Med 119: 111–115. [DOI] [PubMed] [Google Scholar]
- 88. González-Martín A, Gorostiza A, Rangel-Villalobos H, Acunha V, Barrot C, et al. (2008) Analyzing the genetic structure of the Tepehua in relation to other neighbouring Mesoamerican populations. A study based on allele frequencies of STR markers. Am J Hum Biol 20: 605–613. [DOI] [PubMed] [Google Scholar]