Abstract
The polytypic Nicaraguan Midas cichlids ( Amphilophus cf. citrinellus) have been established as a model system for studying the mechanisms of speciation and patterns of diversification in allopatry and sympatry. The species assemblage in Crater Lake Apoyo has been accepted as a textbook example for sympatric speciation. Here, we present a first comprehensive data set of population genetic (mtDNA & AFLPs) proxies of species level differentiation for a representative set of individuals of all six endemic Amphilophus species occurring in Crater Lake Apoyo. AFLP genetic differentiation was partitioned into a neutral and non-neutral component based on outlier-loci detection approaches, and patterns of species divergence were explored with Bayesian clustering methods. Substantial levels of admixture between species were detected, indicating different levels of reproductive isolation between the six species. Analysis of neutral genetic variation revealed several A . zaliosus as being introgressed by an unknown contributor, hereby rendering the sympatrically evolving L. Apoyo flock polyphyletic. This is contrasted by the mtDNA analysis delivering a clear monophyly signal with Crater Lake Apoyo private haplotypes characterising all six described species, but also demonstrating different demographic histories as inferred from pairwise mismatch distributions.
Introduction
Speciation as the cessation of gene flow between closely related populations has traditionally been classified into the three geographic modes allopatric, parapatric or sympatric, and the debate about their respective importance, prevalence and likelihood has produced hundreds of publications [1-8]. Only recently however, it has been stipulated to stop the ‘unproductive’ debates about geographic modes of speciation as well as to replace the dogmatic arguments for characterizing speciation via sexual versus natural selection by a more differentiated exploration of the relative importance of the underlying processes, and how they possibly interact [9-11]. Promising model systems for such studies are clades nested within a strong phylogenetic proximity that can be analyzed genetically, ecologically and behaviorally in as many aspects of reproductive isolation as possible.
Without confounding effects of allopatric population differentiation, crater lake cichlid species flocks, such as the Midas cichlids endemic to L. Apoyo (Nicaragua) constitute such a model to study patterns and processes of speciation due to its accessibility and feasibility to sample all species [8,12,13]. Encompassing only six described species ( Amphilophus astorquii , A. chancho , A . flaveolus , A . globosus , A . supercilius and A . zaliosus [14-16]) a comprehensive picture of the genomic diversity and divergence between them has not been presented to date. Yet, the putative monophyly of the flock has been documented based on mitochondrial DNA and nuclear markers [17-21]. However, data left room for a secondary introgression scenario from outside L. Apoyo [12,19]. Rapid speciation in sympatry leading to the formation of A . zaliosus has been hypothesized [21,22], but the likelihood of hybridization and introgression had not been investigated. Also, the contribution of the remaining L. Apoyo Midas cichlid species in this process had not been investigated before. For A . zaliosus , ecological disruptive selection has been proposed as the main factor leading to the speciation event out of the original L. Apoyo Amphilophus stock [21].
A common generalization from various studies and simulations is that divergence with gene flow in sympatry is easy and rapid if the naturally selected ‘magic’ traits diverge and pleiotropically lead to reproductive isolation [23-25]. Traits such as body shape or coloration under disruptive natural selection could lead to sexual isolation, if they pleiotropically affect pre-zygotic isolating mechanisms (e.g. female mate choice), and do thus promote rapid speciation. Given the very recent origin of the L. Apoyo radiation (ca. 21.000 B.P. [26]) it is not unlikely that divergently selected ‘magic’ traits integrating ecological performance and mate choice, i.e. body shape and coloration, triggered reproductive isolation.
We present results from a population genetic study of L. Apoyo’s Midas cichlids including, for the first time, samples of all six formally described species, as well as from several potential hybrid individuals. Combined findings from population genetics and phylogenetics are used to (1) infer the probability of a single colonization event postulated for the formation of a complex sympatric species flock in statu nascendi. Because this is the first study claiming to include all currently recognized L. Apoyo Amphilophus species level diversity, we (2) test whether there is population genetic data supporting the presence of distinct genetic clusters within L. Apoyo and whether they coincide with the described species. We (3) use genetic differentiation as measured with F-statistics, hierarchical AMOVAs and Bayesian clustering as proxies for the strength of divergence between the species and the amount of gene flow. We (4) assess signatures of introgression and hybrid speciation among sympatric L. Apoyo Amphilophus in the face of apparent species cohesiveness. Finally (5), the demographic history of the six species is estimated based on pairwise mismatch distributions of mtDNA haplotypes and the results are compared to those obtained from earlier studies. The work presented with its – to date – unique comprehensive taxon sampling is designed to deliver descriptive baseline data to augment the hitherto published data on parts of the L. Apoyo Midas cichlid radiation (e.g. [17-22,27]).
Materials and Methods
The study was carried out under research permits from the Ministerio del Ambiente y los Recursos Naturales (MARENA), Nicaragua (Permit numbers: DT-MAS-MICA_C003-03-01-07 and No. 012-03007/DGAP). No endangered or protected species were involved and all procedures conformed to the animal behaviour society guidelines for the use of animals in research as well as to the legal requirements of Nicaragua and Germany. All included individuals were caught during three field seasons from January to April in 2007, December to March in 2007/2008 and April 2009 in Nicaragua. One individual of Amphilophus lyonsi (aquarium stock) was included as distantly related outgroup taxon, its adequacy supported by e.g. Concheiro et al. [28] or Říčan et al. [29], and two Amphilophus sp. from L. Nicaragua (Isletas) and the Rio San Juan (San Carlos) integrated as close allopatric representatives. With an estimated age of less than 23,000 years [26,30] L. Apoyo is one of the younger volcanic crater lakes in Nicaragua, situated within an almost circular caldera about 4 km west of L. Nicaragua. Its surface occupies 20.92 km2, its diameter measuring more than 4 km and its maximum depth 178 m [31]. Fishes were caught SCUBA diving with harpoon following preliminary field identification and anesthetized and killed using an overdose of clove oil. Each specimen was photographed to document coloration and preserved with pinned fins in 4-10% formalin. Individual whole body and tissue vouchers are stored permanently at Bavarian State Collection Munich (ZSM, Table S1). Species were identified according to the most recent species descriptions [15,16], and the key used is available from Figure S1. Specimens included 18 individuals of Amphilophus astorquii , 21 of A . chancho , 20 of A . flaveolus , 20 of A . globosus , 19 of A . supercilius and 22 of A . zaliosus . Seven individuals were included that could not unambiguously be assigned to any of the six species a priori - possibly due to hybrid origin.
Phylogeny reconstruction
Genomic DNA was extracted using the Quiagen® DNeasy® 96 Tissue Kit for animal tissues according to the protocol provided by the manufacturer. Part of the mitochondrial control region was amplified using previously published primers and protocols as well as one newly designed primer: L15995 [32], H00651 [33] and H00834 (5'- ATATACACATGTCACGTAAG -3'). PCR conditions were 15 min at 95°C; 39 cycles of 95°C for 30 s, 58°C for 90 s and 72°C for 90 s, followed by 72°C for 10 min. Sequencing of the ~790 bp long fragment was done at the sequencing service of the Department of Biology of the Ludwig Maximilian University (Munich), using the Big Dye v.3.1 kit and primers L15995, H00834 and H00498 (5'- GAACCCCTTGCCCGCTAGAAAGAAT -3'). The alignment was created in ClustalX (1.81) with default settings [34]. Various sites as listed in Geiger et al. [19] were eliminated from the final alignment because they contain many single nucleotide repeats that cannot be aligned reliably. Haplotype-frequencies were calculated using Collapse 1.2 [35] with default settings, i.e. treating gaps as 5th state. A median-joining haplotype network containing all shortest phylogenetic trees (all maximum parsimony or MP trees) was constructed using NETWORK 4.5.10 following Bandelt et al. [36,37] with default settings (epsilon=0).
To study the demographic history and relative timing of the population expansion, a coalescence-based pairwise-mismatch distribution was calculated in Arlequin 3.5. The θ-estimates θ0 and θ1 are the product of 2µN0 and 2µN1 with mutation rate μ and N the effective population size at times 0 and 1. Tau (τ) is a relative measure of time since population expansion [38,39]. All mtDNA control region sequences available from Genbank on L. Apoyo Amphilophus individuals identified to species (Mar, 2012) were included in the pairwise mismatch analysis; their accession numbers are listed in Table S2. Plausibility of the results was checked according to Schenekar and Weiss [40], using a range of substitution rates (0.01 to 0.1 substitutions per lineage per site per MY), taking into account the variability of reported values (e.g. [41-43]).
The AFLP genotyping with 20 selective main amplifications is based on the Vos et al. [44] protocol, modified according to Herder et al. [45]. For the tree-reconstruction based on AFLP data the software package TREECON 1.3b was used [46] and the Link et al. [47] distance-measure chosen to compute a matrix based on the binary AFLP matrix.
To assess robustness of the AFLP-based phylogenetic hypothesis, and to explore alternative branching patterns, leaf-stability (LS) and lineage-movement (LM) indices for each single taxon and selected clades were calculated in Phyutility v.2.2 [48]. The LS index measures the consistency of each taxon’s position across a chosen number of bootstrap replicates. A value of 1 indicates that the individual’s position in the topology is stable and equal in all examined trees. The LM index calculates attachment frequencies of selected branches from alternative tree topologies thus identifying where a lineage is falling alternatively to its position in the tree based on the complete (non-bootstrapped) matrix [48].
To test for homoplasy-excess possibly introduced by hybrid taxa, a tree-based method as suggested by Seehausen [49] was applied. The inclusion of a hybrid taxon introduces homoplasy with clades containing the hybrid’s parental lineage due to their mosaic composition of the genome. Removal of a hybrid should decrease the amount of homoplasy and thus increase bootstrap support for clades containing hybrid parents or their descendants. Conversely, removal of non-hybrid taxa should not affect bootstrap support of other nodes.
For visualization of conflicting phylogenetic signal the Link et al. distance matrix was used to create a phylogenetic network based on the Neighbor-Net algorithm [50] as implemented in SplitsTree [51].
Outlier locus detection
In order to discern between patterns of neutral processes and selection, a Bayesian method was applied to identify potential candidate loci under selection, implemented in BAYESCAN 1.0 using default settings [52]. Jeffreys’ scale of evidence for Bayes factors (BFs) was applied to identify candidate loci using the strictest criteria ‘decisive’, corresponding to BFs greater 100 and selection posterior probabilities above 99%. Performance of BAYESCAN was evaluated with an alternative software program to identify loci under directional selection: Dfdist (www.rubic.rdg.ac.uk/~mab/stuff/), most commonly used for AFLP markers [53].
Inference of genetic structure
First, the Bayesian algorithm of STRUCTURE 2.2 [54,55] was used to identify the number of differentiated clusters without a priori group designation based on the complete AFLP matrix. The admixture-model with correlated allele-frequencies [56] was chosen, and α (admixture parameter) and λ (allelic frequencies parameter) were set to be inferred from the data. Each run consisted of a burn-in period of 50k followed by 250k iterations for posterior-probability estimates. All runs for each single K (number of populations or clusters) were replicated at least 20 times, where K ranged from 1 to 11. Runs that did not converge during the burn-in phase were identified by log. probability vs. iteration plots and removed from further analysis. Complementary, the approach of Evanno et al. [57] was applied to detect the uppermost hierarchical level of genetic structure by calculating ΔK from the STRUCTURE output ‘LnP(D)’.
After inferring the most likely number of genetic clusters the GENSBACK (gb) option in STRUCTURE [54] was used to test for immigrant ancestors. This model makes use of prior population information for each individual assisting the clustering process and calculates the probabilities that each individual was either from its predefined population (here species) or had a parent or grandparent from any other species (i.e. gb=2). Sensitivity of the data to v (probability of individual misclassification or mixed ancestry), was evaluated by varying the parameter MIGRPRIOR (0.1, 0.05, and 0.01), as suggested by Pritchard et al. [54]. We thus tested for immigrant ancestors from the last two generations and used for the seven potential hybrids as predefined species those with which they clustered in the NJ tree. Additionally, we performed STRUCTURE runs and principal components analysis (PCA) as graphical representation of the effect of the outlier loci on population differentiation for the AFLP matrix containing outlier loci only, and on the presumably neutral AFLP matrix without outlier loci. The structure of genetic diversity was investigated using hierarchical AMOVAs as implemented in Arlequin 3.11 for both, the mtDNA and AFLP data independently. Molecular variance was estimated among and within (1) the six species and (2) the three sampling locations. Loci identified to be under directional selection were removed from the AFLP matrix and AMOVAs as well as F-statistics recalculated as a measure of presumably neutral differentiation. Differentiation between species was estimated using F-statistics on uncorrected p-distances [58] as implemented in Arlequin 3.5, and their significance tested by permutating haplotypes among populations as well as generating bootstrap confidence intervals. For the AFLP data set, additionally pairwise Ф ST based on standard Jaccard coefficient were generated after distance transformation (d=1-s) in FAMD [59].
Data Accessibility
DNA sequences: GenBank accessions GU355718- GU355726; GU355729- GU355733; GU355735; GU355737; GU355739- GU355742; GU355744- GU355748; GU355764; GU355770; GU355850; GU355851; GU362707- G362709; JF784052-JF784149.
DNA aliquots: stored permanently at ZSM DNA-Bank, available upon request from dnabank@zsm.mwn.de
Results
Phylogenetic relationships
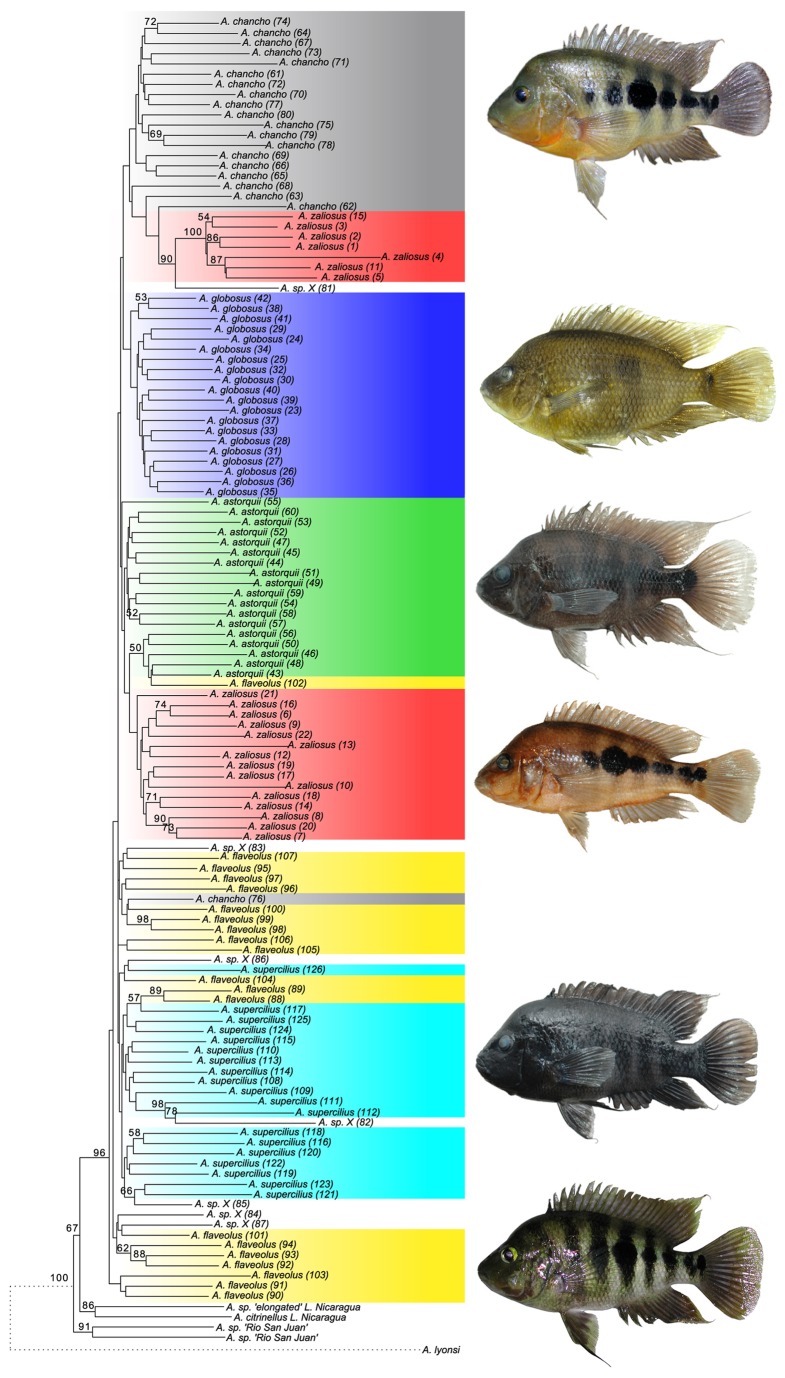
In terms of a phylogenetic species concept the recovered NJ-phylogeny delivered different but overall low levels of monophyly support for the six L. Apoyo Midas cichlid species. The topology of the NJ-tree based on the Link et al. [47] distance measure on 2297 AFLP loci contains both taxonomically homogeneous and heterogeneous clades (Figure 1).
Figure 1. Neighbor-joining tree based on Link et al. distance measure using 2297 AFLP loci with bootstrap values ≥ 50 given above nodes.
The single species forming a monophylum is A . globosus , whereas genotypes of all other species do not group monophyletic: Amphilophus chancho forms a cluster together with seven A . zaliosus individuals; A . astorquii falls into two main clusters, one including an A . flaveolus and being sister to the second A . zaliosus assemblage; A . flaveolus is situated basal to the Apoyo in-group and falls into several clusters with affinities to A . supercilius ; the latter species also falls into several clusters, although closely associated ones. Several individuals were not grouped according to our species identification and will be discussed later. Mean leaf-stability indices (Table S1) were similarly low for most species and individuals ( A . astorquii , m=0.55; A . chancho , m=0.56; A . flaveolus , m=0.55; A . supercilius , m=0.58; A . zaliosus , m=0.57, without the seven conspicuous A . zaliosus individuals which had elevated LS indices m=0.81), except for A . globosus (m=0.65). The included potential hybrid individuals did not show conspicuously in- or decreased LS indices.
As a consequence of the recovered topology with rather heterogeneous clades, the tree based tests for homoplasy-excess and lineage-movement where only performed for selected clades and not for species specific groups only. We conducted 36 removal experiments and excluded one at a time either each clade with bootstrap support (BS) >25%, all individuals of a single species or each of the potential hybrid individuals. Conspicuous results were the increase of BS for the monophyly of A . globosus from m=30.2% to 50% upon removal of A . chancho , and the monophyletic grouping of the otherwise non-monophyletically A . zaliosus upon removal of either A . chancho (BS=58%), A . astorquii (BS=39%) or A . supercilius (BS=29%). This hints to a possible hybridization component in the speciation event of the latter three species.
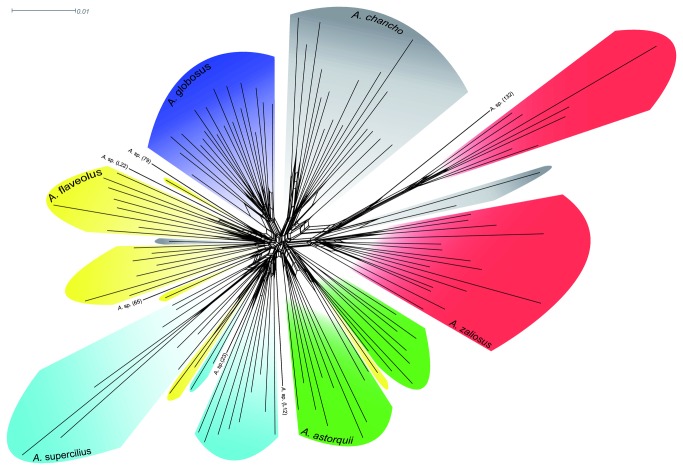
The lineage-movement exploration showed that the seven conspicuous A . zaliosus individuals appeared in 38.8% of 2000 bootstrap replicates outside the whole Midas cichlid species complex as basal sister to A . lyonsi . This was only found in 4% for A . zaliosus excluding the seven conspicuous individuals and in 14% for A . chancho but not for any remaining species. The in-group neighbor-net derived from genetic distances calculated with the Link et al. algorithm [47] shows the same major groups as the NJ-tree but also indicates that there is conflicting signal at the base of the L. Apoyo radiation as indicated by numerous reticulations (Figure 2).
Figure 2. AFLP neighbor-network based on Link’s genetic distances.
Mismatch distribution & mtDNA haplotypes
Among all 126 mtDNA control region sequences included, 38 different haplotypes were identified that are unique to L. Apoyo (NCBI Genbank, blastn search with default settings, Mar 2013). All six sampled species contain haplotypes that are closely (≤ 5 mutations) related to a central haplotype (“A”). Almost half of the 126 individuals (42.06%) from the six species carry the most common haplotype “A”. The relationships within the Apoyo Midas cichlids based on mtDNA are complex and all species share a variable amount of mitochondrial haplotypes (Figure 3). The coalescence based pairwise-mismatch distributions showed marked differences between the demographic histories of the six species (Figure 4). While A . astorquii , A . flaveolus , and A . zaliosus show rather smooth, steady sloped distributions as assumed for stable or expanding populations, there is clear multimodal, wave-like pattern in A . chancho and A . supercilius as would be predicted for populations that have experienced a sudden growth or decline [38] in the past. Rapid population expansion as deduced from large differences between θ0 and θ1 were observed in A . globosus and A . zaliosus only, and all observed mismatch distributions did not differ significantly from the distributions expected under population expansion, except in A . zaliosus (Table 1). Using the mismatch calculator provided by Schenekar and Weiss [40] we tested a range of substitution rates assuming a conservative generation time of two years to explain the observed Τ-value derived from Arlequin 3.5. We found pronounced differences in the mutation rates µ (substitutions per lineage per site per MY) in the six species, assuming that all species have evolved in L. Apoyo, i.e. within the last ca. 21.000 years (Table 1). According to this, the mutation rate in the mtDNA control region is higher in A . chancho , A . flaveolus and A . supercilius (µ=0.082 to 0.091) than in A . astorquii and A . globosus (µ=0.028 to 0.045), the latter being much closer to the reported values for different fish groups (e.g. [41] µ=0.036 [42]; µ=0.02 [43]; µ=0.022 to 0.045). However, our values do well agree with the range reported for different other Midas cichlid populations (e.g. [17] µ=0.019 to 0.0986).
Figure 3. Median-joining parsimony-network based on the mitochondrial control-region, containing nine equal parsimonious trees.
Circle size corresponds to sample size, branch length correlates with number of mutational steps.
Figure 4. Coalescence-based mtDNA control region mismatch analysis of the six Lake Apoyo Amphilophus species showing different demographic histories.
Expansions ca. two mutations ago occurred in A . chancho and A . supercilius and ongoing population expansions are detected in A. astorquii , A . globosus and A . zaliosus .
Table 1. Estimated parameters for the pairwise mismatch analysis of the mtDNA sequence variability in the control region.
| A . astorquii | A . chancho | A . flaveolus | A . globosus | A . supercilius | A . zaliosus | |
|---|---|---|---|---|---|---|
| Number of individuals | 76 | 40 | 27 | 20 | 18 | 149 |
| Mean no. of differences | 1,107 | 1,610 | 2,120 | 0,658 | 1,288 | 0,922 |
| Τ | 1,4 | 2,6 | 2,5 | 0,7 | 2,5 | 0 |
| θ0 | 0,03164 | 0 | 0,00703 | 0 | 0,5625 | 0 |
| θ1 | 2,947 | 3,062 | 6,210 | 99999 | 1,175 | 427,35 |
| SSD | 0,00024 | 0,03936 | 0,00328 | 0,00113 | 0,07397 | 0,27345 |
| Raggedness index | 0,03278 | 0,13579 | 0,02309 | 0,09172 | 0,35059 | 0,12026 |
| µ (thousand years) | 0.045 (21.021) | 0.091 (19.305) | 0.082 (20.599) | 0.028 (16.891) | 0.082 (20.599) | - |
sig. in bold.
Relative time since population expansion tau (τ), mutation parameter theta at time 0, before, and 1, after expansion, Harpending’s Raggedness index, sum of squared differences from mismatch analyses (SSD), and minimum µ (substitutions per lineage per site per MY) for time since expansion in thousands of years (in brackets).
Genetic structure with and without outlier loci
Between-species genetic differentiation based on the short mtDNA control region sequence data varied greatly and delivered significant FST’ s in nine out of 15 species-pair comparisons (Table 2). The smallest but still significant FST was calculated between A . zaliosus and A . astorquii (0.021), the highest between A . chancho and A . globosus (0.573). Investigation of within- and between-species variation using hierarchical AMOVAs with the mtDNA haplotype data showed that most (72%) variation is attributable to within-species differentiation, whereas 28% can be explained by between-species differences. Dividing the mtDNA haplotypes into groups according to sample location did not explain any amount of genetic variation (Table 3).
Table 2. Pairwise FST-estimates based on the mtDNA control region, minimum and maximum values in bold (below diagonal), and p-values assessed by 10,000 permutations.
| A . zaliosus | A . globosus | A . astorquii | A . chancho | A . flaveolus | A . supercilius | |
|---|---|---|---|---|---|---|
| A . zaliosus | - | <0.001 | <0.023 | <0.001 | 0.672 | 0.185 |
| A . globosus | 0.032 | - | 0.058 | <0.001 | <0.027 | <0.009 |
| A . astorquii | 0.021 | 0.028 | - | <0.001 | 0.530 | 0.141 |
| A . chancho | 0.514 | 0.573 | 0.530 | - | <0.001 | <0,001 |
| A . flaveolus | -0.009 | 0.037 | -0.003 | 0.502 | - | 0.459 |
| A . supercilius | 0.015 | 0.061 | 0.029 | 0.422 | -0.007 | - |
Table 3. AMOVA hierarchical genetic analysis of pairwise differences based on the mtDNA control region data.
| Source of variation | df | Percentage of variation | |
| among species | 5 | 28.23* | |
| within species | 113 | 71.77*
|
|
| among locations | 2 | -0.59ns | |
significant (10,100 permutations)
ns: not significant.
Using BAYESCAN 1.0 to identify potential candidate loci under directional selection from the AFLP matrix, 49 loci (ca. 2%) were detected to be influenced by directional selection applying Jeffreys’ scale of evidence for Bayes factors with the strictest criteria ‘decisive’ (selection posterior probabilities above 99%). Within the Dfdist analysis we used two significance levels (99% or 95%) and used as baseline FST either the average or the trimmed mean FST, both provided by the Ddatacal program (part of the Dfdist package) as suggested by e.g. Pérez-Figueroa et al. [53] or Caballero et al. [60]. The software detected between one (p<0.01, average FST) and 19 (p<0.05, trimmed FST) loci that showed higher FST values than under neutral expectations. Loci detected by Dfdist were always among the 49 hits obtained with BAYESCAN. Further analyses of the AFLP data were conducted on both, the complete matrix and on the reduced matrix (without the 49 BAYESCAN loci), which we refer to as the neutral matrix.
All pairwise FST’ s obtained from the complete and the reduced, neutral AFLP matrix were significantly greater than zero and ranged between 0.052 and 0.223 based on pairwise differences and 0.096-0.353 based on the Jaccard coefficient for the complete matrix and between 0.036-0.141 (pairwise differences) and 0.067-0.219 (Jaccard coefficient, Table 4) for the neutral matrix. Both methods delivered highly correlated results (r=0.99, p<0.001) with the Ф ST’s based on the Jaccard coefficient being on average 42 ± 2.6% higher. Exclusion of the 49 loci detected to be under selection had an equal effect on both measures (r=0.98, p<0.001): FST’ s decreased by 30.8 ± 6.8% and Ф ST’s decreased by 30.3 ± 6.7%. Pairwise FST’ s were differently strong influenced by the exclusion of the 49 loci, and a weak correlation between FST and decrease was observed (r=0.63, p<0.05). The effect ranged from a decrease by 42.8% ( A . chancho / A . zaliosus ) to 20.9% ( A . astorquii / A . flaveolus ).
Table 4. Pairwise FST-estimates based on the binary AFLP matrix.
| A . zaliosus | A . globosus | A . astorquii | A . chancho | A . flaveolus | A . supercilius | |
|---|---|---|---|---|---|---|
| A . zaliosus | - | 0.141/0.219 | 0.098/0.171 | 0.099/0.170 | 0.092/0.163 | 0.119/0.207 |
| A . globosus | 0.223/0.353 | - | 0.105/0.177 | 0.099/0.166 | 0.098/0.160 | 0.112/0.184 |
| A . astorquii | 0.134/0.233 | 0.142/0.238 | - | 0.078/0.141 | 0.049/0.092 | 0.064/0.118 |
| A . chancho | 0.173/0.293 | 0.148/0.252 | 0.101/0.182 | - | 0.071/0.129 | 0.085/0.152 |
| A . flaveolus | 0.154/0.266 | 0.128/0.212 | 0.062/0.114 | 0.105/0.188 | - | 0.036/0.067 |
| A . supercilius | 0.188/0.317 | 0.159/0.263 | 0.084/0.153 | 0.132/0.230 | 0.052/0.096 | - |
First value corresponds to FST based on pairwise differences according to Weir & Cockerham [55], second value gives the Ф ST based on standard Jaccard coefficient after distance transformation (d=1-s). All pairwise FST comparisons were significant based on 10,100 permutations following sequential Bonferroni correction. Lower left: comparisons based on all 2297 loci; upper right: neutral estimates based on the reduced AFLP matrix excluding the 49 loci identified by BAYESCAN. Minimum and maximum values in bold.
Strongest differentiation based on all loci was detected between A . zaliosus and A . globosus (FST 0.223, Ф ST 0.353) and smallest, but still significant differentiation between A . flaveolus and A . supercilius (FST 0.052, Ф ST 0.096). The discovered differentiation was not expected from the number of private alleles for each species: A . zaliosus (88), A . supercilius (53), A . chancho (48), A . astorquii (32), A . flaveolus (24) and A . globosus (21).
Results from AMOVAs with the AFLP data were similar to those obtained from mtDNA: genetic variation was partitioned as 14% among and 86% within species. Grouping individuals according to sample location delivered a small but significant amount of variation explained (1.6%, Table 5). Excluding the 49 outlier loci (2% of all loci) had a clear effect on AMOVA outcomes: among species variation decreased by one third while within species variation increased by 5%.
Table 5. AMOVA hierarchical genetic analysis of pairwise differences based on the complete AFLP matrix with 2297 loci and on the reduced matrix excluding the 49 loci identified as being under selection.
| Source of variation |
df | Percentage of variation |
||
|---|---|---|---|---|
| all loci | neutral loci only | |||
| among species | 5 | 13.77* | 9.15* | |
| within species | 113 | 86.23* | 90.85* | |
| among locations | 2 | 1.58* | 1.31* | |
significant (10,100 permutations).
Findings from the Bayesian cluster analysis with STRUCTURE v2.2 without a priori species designation suggest different levels of population integrity of the six species and point to the presence of hierarchical genetic structure. The Bayesian cluster method detected six clusters (K=6) to be most likely, which was also confirmed by a second peak of Evanno’s ΔK at K=6 (Figure S2), roughly corresponding to the six species. Of the 22 A . zaliosus , 15 were consistently grouped in one cluster with all A . astorquii (Figure 5, top). Repeating the STRUCTURE runs including only the latter two species shows that they do not form a genetically homogeneous group - the clustering method clearly differentiates between the two species - and shows that there are two clusters within A . zaliosus , one of which with affinities to A. astorquii (Figure S3), the other composed of the seven aforementioned conspicuous individuals.
Figure 5. Results of STRUCTURE clustering analyses.
Top: on complete AFLP matrix for K6 with group information, GENSBACK 2 and MIGPRIOR 0.05; Middle: same as top but on the 49 AFLP outlier loci only; Bottom: on the 49 outlier loci for K6 without group information. Species of sample origin given above, sample IDs of individuals given below.
Testing for misclassified or immigrant individuals in STRUCTURE with GENSBACK=2 and varying v (0.1, 0.05, 0.01) gave highly similar results independently of v. Mean proportions of correctly classified individuals (above 95% probability) derived from seven independent STRUCTURE runs with varying v were for: A . globosus 95%, A . astorquii 80.2%, A . chancho 80%, A . flaveolus 64.3% and A . supercilius 94%. As in the first analysis, 68% of the A . zaliosus where always misclassified as A . astorquii . Several individuals were constantly detected as being misclassified or having ancestry in one or more other species (Figure 5; Tables 6 & 7), and only one of the seven included potential hybrids demonstrated a clear signature of admixed origin while the remaining were mainly assigned to the species with which they clustered in the NJ tree reconstruction (Table 6).
Table 6. Summary table for the seven potential hybrid individuals with IDs used throughout this study.
| Sample ID | 81 | 82 | 83 | 84 | 85 | 86 | 87 |
|---|---|---|---|---|---|---|---|
| mtDNA | A . zaliosus | A | B | A | unique | unique | C |
| NJ cluster | A.chancho/zaliosus | A . supercilius | A . flaveolus | A . flaveolus | A . supercilius | A.flaveolus/supercilius | A . flaveolus |
| STRUCTURE | 97-99% A . astorquii grandparent | 100% A . zaliosus grandparent | 24-68% A . flaveolus & 30-66% A . supercilius grandparent | 97-100% A . flaveolus | 97-100% A . supercilius | 99-100% A . supercilius | 100% A . flaveolus |
| LM | A.chancho/zaliosus (stable) | A . supercilius (stable) | 45% A . flaveolus 7% A . supercilius 2% A . chancho | 40% A . flaveolus 19% A . supercilius | 54% A . supercilius | 28% A . flaveolus 13% A . supercilius | 33% A . flaveolus 6% A . astorquii |
mtDNA haplotype inferred from the parsimony network; position in the NJ tree based on Link’s distance measure; STRUCTURE results show the consensus from seven runs with varying v (0.01, 0.05 and 0.1) and GENSBACK=2; alternative clustering in 2000 bootstrap replicates identified by the lineage-movement procedure (LM).
Table 7. ‘No immigrant ancestry’ gives the probability that the ancestry of each individual is exclusively in its predefined species, following columns show the probabilities that each individual has the given amount of ancestry in the given source species.
| ID | species | no immigrant ancestry | immigrant | immigrant grandparent | NJ cluster |
|---|---|---|---|---|---|
| 42 | A . globosus | 35-83% | - | 16-61% A . chancho | A . globosus |
| 45 | A . astorquii | 69-96% | - | 28% A . supercilius | A . astorquii |
| 47 | A . astorquii | 27-66% | - | 30-66% A . supercilius | A . astorquii |
| 51 | A . astorquii | 40-99% | - | 43-60% A . chancho | A . astorquii |
| 58 | A . astorquii | 46-87% | - | 37-54% A . chancho | A . astorquii |
| 62 | A . chancho | - | - | 99-100% A . zaliosus | A . chancho |
| 63 | A . chancho | - | - | 99-100% A . zaliosus | A . chancho |
| 68 | A . chancho | 0-2% | - | 98-99% A . globosus | A . chancho |
| 76 | A . chancho | - | 86-95% A . flaveolus | 12-15% A . globosus | A . flaveolus |
| 88 | A . flaveolus | - | 100% A . supercilius | - | A . supercilius |
| 89 | A . flaveolus | - | 98-100% A . supercilius | - | A . supercilius |
| 90 | A . flaveolus | 16-70% | - | 30-84% A . astorquii | A . flaveolus |
| 102 | A . flaveolus | - | 99-100% A . astorquii | - | A . astorquii |
| 104 | A . flaveolus | 30-98% | - | 17-33% A . supercilius | A . supercilius |
| 105 | A . flaveolus | 0-14% | - | 63-87% A . chancho , 20% A . supercilius | not resolved |
| 106 | A . flaveolus | 5-71% | 10-75% A . supercilius | 12-33% A . supercilius , 13% A . astorquii | not resolved |
| 112 | A . supercilius | 0-1% | - | 99-100% A . zaliosus | A . supercilius |
Results show the consensus from seven runs with varying v (0.01, 0.05 and 0.1) and GENSBACK=2. Position in the NJ tree based on Link’s distance measure, individual’s IDs are those used throughout.
The STRUCTURE runs on the reduced AFLP matrix and on the 49 outlier loci matrix without a priori species assignation delivered extremely different results. No differentiated, coherent clusters were detected in the neutral AFLP matrix, irrespective of K. Only the seven A . zaliosus individuals which also clustered together in the NJ tree showed assignment probabilities around 50% to a non-Apoyo cluster (plot not shown). Quite to the contrary, including only the 49 outlier loci in the STRUCTURE runs was highly informative: 1) K=6 was the most likely number of genetically differentiated groups, identical to the outcome with the complete AFLP matrix; 2) all A . zaliosus individuals grouped together, while they were split based on the complete matrix; 3) a much higher percentage of individuals showed elevated admixture proportions as compared to the complete AFLP matrix and 4) only 8.9% (compared to 80.2% based on the complete matrix) of the A . astorquii individuals showed assignment probabilities above 95% to a unique cluster. Averaging over ten runs and all individuals, A . astorquii showed the following mean (+/- SD) admixture proportions: 5.97% (0.07 SD) with A . chancho , 31.5% (0.26 SD) with A . flaveolus , 6.75% (0.06 SD) with A . globosus , 14.83% (0.24 SD) with A . supercilius and 10.2% (0.05 SD) with A . zaliosus .
As with the STRUCTURE analyses on the neutral AFLP matrix and on the 49 outlier loci only, comparison of the two PCA delivered interesting insights: while there are coherent species specific clusters including all A . zaliosus individuals in a single cluster derived from the outlier loci, those clusters collapse and largely overlap except for the seven conspicuous A . zaliosus specimens that group distantly from all remaining Apoyo fishes when analysing the neutral matrix (Figure S4).
Discussion
Our results suggest five out of the six endemic L. Apoyo Midas cichlid species to be polyphyletic with substantial levels of gene flow between them. Within A . zaliosus several individuals showed putative allochthonous phylogenetic affinities and levels of neutral differentiation not related to divergent selection within L. Apoyo indicating a possible past introgression event from outside. The demographic histories estimated from the mtDNA sequences together with the differentiation estimates based on the genome scan suggest following scenarios: primary, older splits from an ancestral form for A . zaliosus and A . globosus and secondary, more recent divergence events including a hybridization component for A . astorquii , A . chancho , A . flaveolus and A . supercilius as deduced from the homoplasy-excess tests and cluster analyses. Altogether, our findings support a sympatric in situ speciation scenario for all L. Apoyo Midas cichlid species.
Outlier loci detection
Speciation is thought to begin with the rise of genomically-localised barriers to gene exchange associated with loci for local adaptation, intrinsic incompatibility or assortative mating [61]. In the early stages, gene flow will be reduced at only few loci which likely contribute to the initiation of reproductive isolation. We found that only a small proportion (ca. 2%) of the fragments show highly significant signatures of divergent selection, similarly to what had been reported for allopatric populations or ecotypes (reviewed in e.g. [62]) and also in line with a recent study on Nicaraguan Midas Cichlids including three L. Apoyo species (1.7% for L. Apoyo [20]). A comparative transcriptome study between A . astorquii and A . zaliosus identified six ESTs with signals of strong diversifying selection based on the ratio of non-synonymous to synonymous substitutions with BLAST assigned functions in biosynthetic - metabolic processes, brain development and cognition, response to hormone stimuli and in the nervous system [63]. Whether the discovered sequence differences are a product of adaptive molecular evolution or directly play a role in speciation remains yet to be tested. Resulting from the anonymity of AFLP markers used in this study, it is not possible to infer whether the detected outlier loci reside on already identified genomic regions or are linked and occur on one or few regions under selection. While the proportion of AFLP markers showing signs of selection is comparable to the aforementioned study [20], our results deviate from their finding which suggested that signatures of divergent selection in L. Apoyo are mainly driven by the divergence of A . astorquii . A similar pattern of consistently deviant allele frequencies of the detected outlier loci for A . astorquii is not present in our data. The reason for this might lie in a different sample composition with differing sample sizes when screening for outlier loci, but also in a software refinement from BAYESCAN version 1.0 to 2.01 allowing now to adjust a prior odd value for the neutral model. However, re-analysing the data with the same species sub-set as in Kautt et al. [20] and using the newer BAYESCAN 2.01 did also not result in the deviant outlier allele frequencies for A . astorquii . Kautt et al. [20] explained the pattern of deviant allele frequencies by a potential chromosomal inversion in high frequency in A . astorquii , leading to differentiation of loci within this region due to a reduction in recombination. Also, an adaptive allele on the inversion could evoke signatures of selection on multiple loci by spreading, or several adaptive alleles could reside on the same inversion, being protected from recombination and facilitating speciation [64]. As the authors state, a role of chromosomal inversions in Midas cichlid evolution is conceivable (e.g. [27,65]), but testing these genomic architectural mechanisms is only possible with markers of known position.
Signals of ancient introgression?
The seven conspicuous A . zaliosus solely showed a strong differentiation signal in the STRUCTURE runs and PCA based on the neutral AFLP matrix. We argue that those individuals carry alleles from an unknown allopatric source, reflecting a past introgression event. They showed levels of neutral differentiation apparently not related to divergent selection within L. Apoyo. Further support for this hypothesis comes from several findings: 1) the group was characterised by four, fixed private alleles while no other species carried any fixed private alleles in our sample; 2) they clustered with the remaining A . zaliosus when considering the 49 outlier loci only, but clustered distantly from the whole Apoyo radiation based on the complete and neutral AFLP matrix (PCA & STRUCTURE); 3) the lineage-movement procedure revealed a tendency for this group to appear outside the whole Apoyo clade based on bootstrapped data. An alternative explanation to introgression from outside Apoyo is the retention of ancestral alleles that have been lost in all remaining individuals, but it is hard to explain why those alleles should have persisted just in seven individuals of a single species. It is equally unlikely that they had evolved in a subset of A . zaliosus individuals only, and perished again in the majority of sampled individuals, also rendering ‘hybrid speciation’ sensu Mallet [66] as origin of A . zaliosus implausible. We further exclude the possibility that the findings are due to technical artefacts, because the seven individuals were analysed on two different deep-well plates, contain one replicate sample, and the four fixed as well as 22 private alleles stem from different primer combinations. Interestingly, all seven individuals were collected along the western shore of L. Apoyo, their distribution thus significantly deviating from the expected distribution as judged from the remaining sampled A . zaliosus (χ2=12.32, df=2, p<0.001). The observed distribution might be due to philopatric behaviour with respect to breeding site, but also spatially localized genetic drift might have played a role.
The impact of the hypothesized introgression event on the speciation propensity of the L. Apoyo radiation is not fully explorable with our data, because they cannot be related to an annotated genome. The fact that the introduced alleles have apparently increased current neutral variation only does not preclude a role for them as substrate for selection directly after the putative introgression event. Hybridization and introgression can generate and increase standing genetic variation for new adaptive trait combinations suitable for exploiting resources not utilized previously [67-70]. The same mechanisms concurrently prevent the loss of genetic variation due to strong selection prevailing in sympatric speciation scenarios and may even induce speciation [66]. The product is not necessarily the formation of a hybrid species, but rather an increase in genetic variance with the introgressing form disappearing in a hybrid swarm. Theory indicates that adaptation can be more rapid if evolution acts on standing genetic variation instead of relying on the rather rare occurrence of beneficial mutations [71,72]. Our results suggest the existence of remains of an introgression event in the neutral genetic variance of A . zaliosus , thereby having increased standing genetic variation. This is also predicted by the syngameon hypothesis [49], as there was no detectable genetic structure based on the neutral AFLP matrix, indicative of a ‘common’ neutral gene pool (disregarding the allochthonous signal of the seven A . zaliosus ). Independent of that, the finding demonstrates the possibility that introgression of allochthonous alleles into seemingly fully isolated crater lake species flocks can take place. However, we find no evidence that contradicts the sympatric nature as scenario where new species emerge within a freely breeding population without geographic isolation for the L. Apoyo species flock. Theoretically possible under a variety of more or less stringent conditions (e.g. [3-6,73-76]), it is yet still considered uncommon in nature and only very few unequivocal examples are generally accepted [24,77].
Inference of demographic history
The differences between the pairwise mismatch distributions support a scenario of stepwise species emergence – as opposed to a sudden simultaneous evolution of all six species, similar to the radiation in stages model [78], although on a much smaller scale than in the original context as model for the species rich East African cichlids [79]. However, the peculiar congruence between the strong AFLP based genetic differentiation in A . globosus and A . zaliosus and their rather steady sloped mismatch distribution which is typically for stable populations argue for a primary divergence of those two species. The clear, wavelike pattern of the pairwise mismatches in A . chancho and A . supercilius is characteristic for a sudden or exponential single growth or decline, corresponding to that event about two mutations ago. Whether this pattern was also influenced by selective sweeps in those species cannot be answered with our data. This was similarly documented for a non A . zaliosus sample from L. Apoyo of unclear species composition [18,21], where also ongoing demographic expansion was detected for A . zaliosus . In A . chancho and A . supercilius , both, the pattern of pairwise mtDNA mismatches and the AFLP based FST estimates argue for secondary divergence events after the formation of stronger isolated A . globosus & A . zaliosus populations. Notably, our and all other published demographic inferences of Midas cichlids based on the mtDNA control region (e.g. [17,18,21,80]) clearly support scenarios for in situ differentiation in different Nicaragua crater lakes, and never lead to pre lake-origin dates.
Current Gene Flow
Which L. Apoyo species have been recently interconnected via gene flow? Although in the phylogenetic reconstruction A . astorquii fell into (only) two neighbouring clusters, it is, according to STRUCTURE, PCA and FST results, the species that hybridized most often. Since it is the most abundant of the six species ([81]; pers. obs.) this may also be an effect of stochasticity. It would be interesting to test whether there is a link between the species’ genomic mosaic signatures and their abundance, which could hint to a selective advantage of hybridization, but more data on biology and demography are needed to tackle this question. Admixture proportions of A . astorquii from A . flaveolus were the highest estimated, which seems to be biologically sound since they are often found in close proximity (personal observation), hereby suggesting microhabitat utilization as a factor for reproductive isolation in sympatry. But, since also other species pairs are ecologically similar and breed in direct vicinity, but are nevertheless strongly sexually isolated from each other, divergence in microhabitat use as prominent factor loses attractiveness as explanation. Interestingly, of the seven individuals classified a priori as potential hybrids, none grouped clearly with A . astorquii . We speculate that, although A . astorquii and A . flaveolus have apparently hybridized more frequently, resulting crosses are phenotypically less conspicuous than hybrid offspring of other crosses as e.g. between A . flaveolus and A . supercilius .
Interestingly, Kautt et al. [20] investigated the genomic signatures of divergent selection in L. Apoyo Amphilophus (excluding A . globosus , A . flaveolus and A . supercilius ), and found A . astorquii most strongly differentiated. Their pairwise FST estimates (based on approximately half the number of AFLP loci studied here) are higher but similar in scale to our estimates for A . astorquii / A . zaliosus (0.164 vs. 0.134), higher for A . astorquii / A . chancho (0.145 vs. 0.101) but substantially lower for the A . chancho / A . zaliosus (0.066 vs. 0.173) comparison. As discussed above, reasons for this might be due to different sample composition and size. Kautt et al. [20] speculate that the conspicuously long phylogenetic branches and elevated gene diversity they observed in A . astorquii hints to an increased evolutionary rate in this species. While our results also show the peculiar position of A . astorquii in the L. Apoyo radiation, we would explain their findings with the observed increased admixture proportions (Figure 5) as discussed below.
Hybridization and Speciation
Traditionally, animal-hybrids are presumed to have reduced fitness, but recently the number of examples where hybridization apparently facilitated speciation and adaptive radiations in animals has increased [66,67,82]. Gene flow through hybridization may constitute a vector for advantageous alleles between species through backcrossing, or hybrids may show higher fitness relative to their genitors when transgressive segregation combines alleles creating novel hybrid traits [70,83-86]. It has also been hypothesized that hybridization is adaptive, leading to selection for weak discrimination between con- and heterospecifics under certain environmental conditions [87] while another extreme consequence of hybridization is despeciation [88-90]. Theoretically, reproductive isolation and functional divergence (i.e. speciation) are possible despite substantial levels of gene flow [4,91-94], a likely scenario also for L. Apoyo Midas cichlids.
It has repeatedly been put forward that hybridization is particularly advantageous where new ecological niches are created by changing or newly invaded environments (e.g. [49,67,86]). The colonization of L. Apoyo likely constituted such a situation, offering apart from reduced competition at least two novel dimensions, i.e. depth and higher water transparency, since the most likely source for the seeding population was L. Nicaragua (e.g. [17-19,21]) with mean depth ca. 13m and 0.25-0.35m Secchi disk transparency, contrasting L. Apoyo with mean depth ca. 142m and 3.5-9.5m Secchi disk transparency [95]. We know from lab experiments and field observations that ‘gold’ coloration of individual Midas cichlids affects mate choice (e.g. [96-99]), demonstrating that visual cues play a role during pair formation in Midas cichlids. An increase in transparency might thus have allowed for changes in mate choice patterns on visual cues. Water turbidity affects sexual selection by impairing the possibility for visually based mate choice [88,100,101] and also the reverse ‘speciation through sensory drive’ has been documented in Lake Victoria cichlids [102] underpinning the potential of this mechanism. Thus the conventional argument that ecological selection mediated by competition for resources among ecotypes is the primary initial driver of sympatric speciation [12,78] might also be challenged by the L. Apoyo Amphilophus case, where disruptive sexual selection on coloration may be a primary driver of initial reproductive isolation as recently suggested for two other prime examples for sympatric speciation [77].
Conclusions
This is the first study based on a taxonomic comprehensive data set of population genetic proxies of species level differentiation for a representative set of individuals of all six endemic Amphilophus species in L. Apoyo. In summary, we find no strong indication that any of the six species has originated from ‘homoploid hybrid speciation’ sensu Mallet [66], i.e. that hybridization played a primary role in the origin of one of the six species. If so we would expect to find a similar constant signature of introgression in any of the six species as has been described for the seven conspicuous A . zaliosus individuals above. However, we demonstrate the existence of various levels of gene flow between species pairs, underlining their incipient status and a potentially non-detrimental or even beneficial role of hybridization. The observed pronounced overall genetic differentiation of A . globosus and A . zaliosus supports a scenario of primary divergence in those species as opposed to a secondary divergence including a hybridization component at least for A . astorquii , and likely also for A . chancho , A . flaveolus and A . supercilius as deduced from the homoplasy-excess tests and cluster analyses. Our comprehensive dataset supports the hypothesis of primary divergence between a limnetic and benthic habitat as suggested before for Midas cichlids from L. Apoyo [18,20] and other fish groups. In order to test the validity of the proposed secondary, more recent divergence events for A . chancho and A . supercilius several approaches seem promising. A detailed comparison of the ecology of the six species could for example show whether there might be a relationship between the sudden population growth of the latter species and their ecology. In comparison to other crater lake cichlid assemblages it could be tested whether this is a general pattern after the repeatedly demonstrated primary split along the benthic – limnetic axis. In the near future, making use of reference cichlid genomes and mapped markers to examine in detail the genome-wide divergence patterns in Midas cichlids will replace the anonymity of AFLP markers. This will allow for quantification of genomic regions under selection, but also answer questions about the underlying functional genetic basis that triggers speciation (e.g. [103-105]). For this, Nicaraguan Midas cichlids are an excellent model system, offering parallel natural experiments in independently colonized crater lakes in a well documented geological context. Given the very recent origin of the different crater lake assemblages in Nicaragua, it is likely possible to observe evolution in action and to identify not what has shaped this diversity, but what is creating diversity.
Supporting Information
Key to the six Amphilophus species endemic in Lake Apoyo, Nicaragua.
(PDF)
Estimation criteria for the number of genetic clusters in the AFLP data set. Above: Mean LnP(D) with SD from 20 replicates for each K, calculated without ‘locprior’ model (STRUCTURE v2.2). Below: Evanno’s model choice criterion ‘ΔK’ for the uppermost level of genetic structure.
(PDF)
Results of STRUCTURE clustering analysis with A . zaliosus and A . astorquii only for K3 using STRUCTURE v2.2 without group information. Species of sample origin given above.
(PDF)
Plot of 1st and 2nd principal component scores based on the complete AFLP matrix (A), the 49 outlier loci only (B) and the neutral AFLP matrix (C). ID numbers are given for the potential hybrid individuals. Variance explained by 1st and 2nd PC for A) 7 & 4%, B) 18 & 10% and for C) 7 & 4%, respectively.
(PDF)
Sample list with individual’s ID used throughout, voucher collection number, isolate number, taxon information, sample location, leaf stability index (LS) and GenBank accession numbers.
(DOC)
Genbank accession numbers of mtDNA control region sequences included in the pairwise mismatch analysis.
(DOC)
Acknowledgments
We are grateful to Lorenzo López, Elmer Nicaragua, Reyna Membreño, and Martha Pastrano for participation in field collections. We thank Topi Lehtonen for his assistance on many dives and for helping to sink ‘Flor de Caña’ between them. We thank Isabella Stöger, Nicolas Straube, Jobst Pfänder and Fabian Herder for helpful comments on earlier versions of the manuscript. We are indebted to Dirk Neumann for the successful endeavour to ship the voucher specimens from Nicaragua to Bavaria.
Funding Statement
This work was supported in part by FUNDECI/GAIA (www.gaianicaragua.org)and by grants provided to MFG by DAAD (www.daad.de), LMU (www.lmu.de) and the Bavarian Elite Aid Act (BayEFG, www.elitenetzwerk.bayern.de). Additional support for molecular analyses was provided by the DNA Bank Network (www.dnabank-network.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mayr E (1942) Systematics and the origin of species from the viewpoint of a zoologist. New York: Columbia University Press. [Google Scholar]
- 2. Maynard Smith J (1966) Sympatric speciation. Am Nat, 100: 637–650. doi:10.1086/282457. [Google Scholar]
- 3. Turner GF, Burrows MT (1995) A model of sympatric speciation by sexual selection. Proc R Soc Lond B Biol Sci, 260: 287–292. doi:10.1098/rspb.1995.0093. [Google Scholar]
- 4. Dieckmann U, Doebeli M (1999) On the origin of species by sympatric speciation. Nature, 400: 354–357. doi:10.1038/22521. PubMed: 10432112. [DOI] [PubMed] [Google Scholar]
- 5. Kondrashov AS, Kondrashov FA (1999) Interactions among quantitative traits in the course of sympatric speciation. Nature, 400: 351–354. doi:10.1038/22514. PubMed: 10432111. [DOI] [PubMed] [Google Scholar]
- 6. Almeida CR, de Abreu FV (2003) Dynamical instabilities lead to sympatric speciation. Evol Ecol Res, 5: 739–757. [Google Scholar]
- 7. Fitzpatrick BM, Fordyce JA, Gavrilets S (2008) What, if anything, is sympatric speciation? J Evol Biol, 21: 1452–1459. doi:10.1111/j.1420-9101.2008.01611.x. PubMed: 18823452. [DOI] [PubMed] [Google Scholar]
- 8. Mallet J, Meyer A, Nosil P, Feder JL (2009) Space, sympatry and speciation. J Evol Biol, 22: 2332–2341. doi:10.1111/j.1420-9101.2009.01816.x. PubMed: 19732264. [DOI] [PubMed] [Google Scholar]
- 9. Rundle HD, Nosil P (2005) Ecological speciation. Ecol Lett, 8: 336–352. doi:10.1111/j.1461-0248.2004.00715.x. [Google Scholar]
- 10. Butlin RK, Ritchie MG (2009) Genetics of speciation. Heredity (Edinb), 102: 1–3. doi:10.1038/hdy.2008.97. PubMed: 19088743. [DOI] [PubMed] [Google Scholar]
- 11. The Marie Curie SPECIATION Network (2012) What We Need Know About Speciation? Trends Ecol Evol, 27(1)27–39. [DOI] [PubMed] [Google Scholar]
- 12. Schliewen UK, Kocher TD, McKaye KR, Seehausen O, Tautz D (2006) Evolutionary biology: Evidence for sympatric speciation?. Nature, 444: E12–E13. doi:10.1038/nature05419. PubMed: 17151605. [DOI] [PubMed] [Google Scholar]
- 13. Losos JB, Mahler DL (2010) Adaptive radiation: the interaction of ecological opportunity, adaptation, and speciation. In Bell MA, Eanes WF, Futuyma DJ, Levinton JS, Evolution After Darwin: the First 150 Years. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 14. Barlow GW, Munsey JW (1976) The Red Devil–Midas-Arrow Cichlid Species Complex in Nicaragua. In Thorsen TB. Investigations of the ichthyofauna of Nicaraguan lakes. Lincoln: University of Nebraska. [Google Scholar]
- 15. Stauffer JR, McCrary JK, Black KE (2008) Three new species of cichlid fishes (Teleostei: Cichlidae) from Lake Apoyo, Nicaragua. Proc Biol Soc Wash, 121: 117–129. doi:10.2988/06-37.1. [Google Scholar]
- 16. Geiger MF, McCrary JK, Stauffer JR (2010a) Description of two new species of the Midas cichlid complex (Teleostei: Cichlidae) from Lake Apoyo, Nicaragua. Proc Biol Soc Wash, 123: 159–173. doi:10.2988/09-20.1. [Google Scholar]
- 17. Barluenga M, Meyer A (2004) The Midas cichlid species complex: Incipient sympatric speciation in Nicaraguan cichlid fishes? Mol Ecol, 13: 2061–2076. doi:10.1111/j.1365-294X.2004.02211.x. PubMed: 15189226. [DOI] [PubMed] [Google Scholar]
- 18. Barluenga M, Meyer A (2010) Phylogeography, colonization and population history of the Midas cichlid species complex (Amphilophus spp.) in the Nicaraguan crater lakes. BMC Evol Biol 10: 326. doi:10.1186/1471-2148-10-326. PubMed: 20977752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geiger MF, McCrary JK, Schliewen UK (2010b) Not a simple case - A first comprehensive phylogenetic hypothesis for the Midas cichlid complex in Nicaragua (Teleostei: Cichlidae: Amphilophus). Mol Phylogenet Evol, 56: 1011–1124. doi:10.1016/j.ympev.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 20. Kautt AF, Elmer KR, Meyer A (2012) Genomic signatures of divergent selection and speciation patterns in a ‘natural experiment’, the young parallel radiations of Nicaraguan crater lake cichlid fishes. Mol Ecol, 21: 4770–4786. doi:10.1111/j.1365-294X.2012.05738.x. PubMed: 22934802. [DOI] [PubMed] [Google Scholar]
- 21. Barluenga M, Stölting KN, Salzburger W, Muschick M, Meyer A (2006) Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature, 439: 719–723. doi:10.1038/nature04325. PubMed: 16467837. [DOI] [PubMed] [Google Scholar]
- 22. Wilson AB, Noack-Kunnmann K, Meyer A (2000) Incipient speciation in sympatric Nicaraguan crater lake cichlid fishes: sexual selection versus ecological diversification. Proc Biol Sci, 267: 2133–2141. doi:10.1098/rspb.2000.1260. PubMed: 11413624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gavrilets S (2004) Fitness Landscapes and the Origin of Species. Monogr Popul Biol 41 Princeton, NJ.: Princeton University Press. [Google Scholar]
- 24. Bolnick DI, Fitzpatrick BM (2007) Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst, 38: 459–487. doi:10.1146/annurev.ecolsys.38.091206.095804. [Google Scholar]
- 25. Feulner PG, Plath M, Engelmann J, Kirschbaum F, Tiedemann R (2009) Magic trait electric organ discharge (EOD): Dual function of electric signals promotes speciation in African weakly electric fish. Commun Integr Biol, 2: 329–331. doi:10.4161/cib.2.4.8386. PubMed: 19721881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sussman D (1982) Apoyo caldera, Nicaragua: A major quaternary silicic eruptive center. J Vulcanology Geother Res, 24: 249–242. [Google Scholar]
- 27. Recknagel H, Elmer KR, Meyer A (2013) A hybrid genetic linkage map of two ecologically and morphologically divergent Midas cichlid fishes (Amphilophus spp.) obtained by massively parallel DNA Sequencing (ddRADSeq). G3 , 3: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pérez GA, Rícan O, Ortí G, Bermingham E, Doadrio I et al. (2007) Phylogeny and biogeography of 91 species of heroine cichlids (Teleostei: Cichlidae) based on sequences of the cytochrome b gene. Mol Phylogenet Evol, 43: 91–110. doi:10.1016/j.ympev.2006.08.012. PubMed: 17045493. [DOI] [PubMed] [Google Scholar]
- 29. Rícan O, Zardoya R, Doadrio I (2008) Phylogenetic relationships of Middle American cichlids (Cichlidae, Heroini) based on combined evidence from nuclear genes, mtDNA, and morphology. Mol Phylogenet Evol, 49: 941–957. doi:10.1016/j.ympev.2008.07.022. PubMed: 18725305. [DOI] [PubMed] [Google Scholar]
- 30. BANIC (1977) Informe Financiero 1976. Banco Nicaragüense de Industria y Comercio, Managua, Nicaragua. 46 pp., as reported in Waid et al .. Encuentro 1999: 51, 80. [Google Scholar]
- 31. CIRA–UNAN Managua (2008). Plan: Informe sobre el Lago de Apoyo de Manejo Reserva Natural Laguna de Apoyo. Catarina, Nicaragua: CLUSA, Vol. III. [Google Scholar]
- 32. Meyer A, Morrissey JM, Schartl M (1994) Recurrent origin of a sexually selected trait in Xiphophorus fishes inferred from a molecular phylogeny. Nature, 368: 39–542. doi:10.1038/368039a0. [DOI] [PubMed] [Google Scholar]
- 33. Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S et al. (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A, 86: 6196–6200. doi:10.1073/pnas.86.16.6196. PubMed: 2762322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res, 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Posada D (2004) Collapse ver. 1.2. A tool for collapsing sequences to haplotypes. [Online]. Available: http://darwin.uvigo.es.
- 36. Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141: 743–753. PubMed: 8647407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol, 16: 37–48. doi:10.1093/oxfordjournals.molbev.a026036. PubMed: 10331250. [DOI] [PubMed] [Google Scholar]
- 38. Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol, 9(3): 552–569. PubMed: 1316531. [DOI] [PubMed] [Google Scholar]
- 39. Gaggiotti O, Excoffier (2000) A simple method of removing the effect of a bottleneck and unequal population sizes on pairwise genetic distancesL. Proc R Soc Lond B Biol Sci, 267: 81–87. doi:10.1098/rspb.2000.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schenekar T, Weiss S (2011) High rate of calculation errors in mismatch distribution analysis results in numerous false inferences of biological importance. Heredity (Edinb) (advance online publication). doi:10.1038/hdy.2011.48. PubMed: 21731052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donaldson KA, Wilson RR Jr. (1999) Amphi-panamic geminates of snook (Percoidei: Centropomidae) provide a calibration of the divergence rate in the mitochondrial DNA control region of fishes. Mol Phylogenet Evol, 13: 208–213. doi:10.1006/mpev.1999.0625. PubMed: 10508553. [DOI] [PubMed] [Google Scholar]
- 42. Nagl S, Tichy H, Mayer WE, Takahata N, Klein J (1998) Persistence of neutral polymorphisms in Lake Victoria cichlid fish. Proc Natl Acad Sci U S A, 95: 14238–14243. doi:10.1073/pnas.95.24.14238. PubMed: 9826684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sato A, Takezaki N, Tichy H, Figueroa F, Mayer WE et al. (2003) Origin and speciation of haplochromine fishes in East African crater lakes investigated by the analysis of their mtDNA, Mhc genes, and SINEs. Mol Biol Evol, 20: 1448–1462. doi:10.1093/molbev/msg151. PubMed: 12777512. [DOI] [PubMed] [Google Scholar]
- 44. Vos P, Hogers R, Bleeker M, Reijans M, van de lee T et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res, 23: 4407–4414. doi:10.1093/nar/23.21.4407. PubMed: 7501463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Herder F, Pfaender J, Schliewen UK (2008) Adaptive sympatric speciation of polychromatic ‘roundfin’ sailfin silverside fish in Lake Matano (Sulawesi). Evolution, 62: 2178–2195. doi:10.1111/j.1558-5646.2008.00447.x. PubMed: 18616575. [DOI] [PubMed] [Google Scholar]
- 46. Van de Peer Y, De Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci, 10: 569–570. PubMed: 7828077. [DOI] [PubMed] [Google Scholar]
- 47. Link W, Dixkens C, Singh M, Schwall M, Melchinger AE (1995) Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theor Appl Genet, 90: 27–32. [DOI] [PubMed] [Google Scholar]
- 48. Smith SA, Dunn CW (2008) Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics, 24: 715–716. doi:10.1093/bioinformatics/btm619. PubMed: 18227120. [DOI] [PubMed] [Google Scholar]
- 49. Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol, 19: 198–207. doi:10.1016/j.tree.2004.01.003. PubMed: 16701254. [DOI] [PubMed] [Google Scholar]
- 50. Bryant D, Moulton V (2004) Neighbor-Net: An Agglomerative Method for the Construction of Phylogenetic Networks. Mol Biol Evol, 21: 255–265. PubMed: 14660700. [DOI] [PubMed] [Google Scholar]
- 51. Huson DH (1998) SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics, 14: 68–73. doi:10.1093/bioinformatics/14.1.68. PubMed: 9520503. [DOI] [PubMed] [Google Scholar]
- 52. Foll M, Gaggiotti OE (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics, 179: 927–939. doi:10.1534/genetics.107.084541. PubMed: 18505879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pérez-Figueroa A, García-Pereira MJ, Saura M, Rolán-Alvarez E, Caballero A (2010) Comparing three different methods to detect selective loci using dominant markers. J Evol Biol, 23: 2267–2276. doi:10.1111/j.1420-9101.2010.02093.x. PubMed: 20796133. [DOI] [PubMed] [Google Scholar]
- 54. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155: 945–959. PubMed: 10835412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes, 7: 574–578. doi:10.1111/j.1471-8286.2007.01758.x. PubMed: 18784791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164: 1567–1587. PubMed: 12930761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol, 14: 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x. PubMed: 15969739. [DOI] [PubMed] [Google Scholar]
- 58. Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358–1370. doi:10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 59. Schlüter PM, Harris SA (2006) Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes, 6: 569–572. doi:10.1111/j.1471-8286.2006.01225.x. [Google Scholar]
- 60. Caballero A, Quesada H, Rolán-Alvarez E (2008) Impact of amplified fragment length polymorphism size homoplasy on the estimation of population genetic diversity and the detection of selective loci. Genetics, 179: 539–554. doi:10.1534/genetics.107.083246. PubMed: 18493070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Butlin RK (2010) Population genomics and speciation. Genetica, 138: 409–418. doi:10.1007/s10709-008-9321-3. PubMed: 18777099. [DOI] [PubMed] [Google Scholar]
- 62. Oleksiak MF (2010) Genomic approaches with natural fish populations. J Fish Biol, 76: 1067–1093. doi:10.1111/j.1095-8649.2010.02563.x. PubMed: 20409163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Elmer KR, Fan S, Gunter HM, Jones JC, Boekhoff S et al. (2010a) Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Mol Ecol, 19 Suppl 1: 197–211. doi:10.1111/j.1365-294X.2009.04488.x. PubMed: 20331780. [DOI] [PubMed] [Google Scholar]
- 64. Kirkpatrick, Barton (2006) Chromosome inversions, local adaptation and speciation. Genetics, 173: 419–434. doi:10.1534/genetics.105.047985. PubMed: 16204214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hoffmann AA, Rieseberg LH (2008) Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst, 39: 21–42. doi:10.1146/annurev.ecolsys.39.110707.173532. PubMed: 20419035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mallet J (2007) Hybrid speciation. Nature, 446: 279–283. doi:10.1038/nature05706. PubMed: 17361174. [DOI] [PubMed] [Google Scholar]
- 67. Dowling TE, Secor CL (1997) The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Evol Syst, 28: 593–620. doi:10.1146/annurev.ecolsys.28.1.593. [Google Scholar]
- 68. Burke JM, Arnold ML (2001) Genetics and the Fitness of Hybrids. Annu Rev Genet, 35: 31–52. doi:10.1146/annurev.genet.35.102401.085719. PubMed: 11700276. [DOI] [PubMed] [Google Scholar]
- 69. Baack EJ, Rieseberg LH (2007) A genomic view of introgression and hybrid speciation. Curr Opin Genet Dev, 17: 513–518. doi:10.1016/j.gde.2007.09.001. PubMed: 17933508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stelkens RB, Schmid C, Selz O, Seehausen O (2009) Phenotypic novelty in experimental hybrids is predicted by the genetic distance between species of cichlid fish. BMC Evol Biol, 9: 283. doi:10.1186/1471-2148-9-283. PubMed: 19961584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Barrett RD, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evol, 23: 38–44. doi:10.1016/j.tree.2007.09.008. PubMed: 18006185. [DOI] [PubMed] [Google Scholar]
- 72. Wolf JB, Lindell J, Backström N (2010) Speciation genetics: Current status and evolving approaches. Philos Trans R Soc Lond B Biol Sci, 365: 1717–1733. doi:10.1098/rstb.2010.0023. PubMed: 20439277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Coyne JA, Orr HA (2004) Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 74. Kawata M, Yoshimura J (2000) Speciation by sexual selection in hybridizing populations without viability selection. Evol Ecol Res, 2: 897–909. [Google Scholar]
- 75. Ripa J (2009) When is sympatric speciation truly adaptive? An analysis of the joint evolution of resource utilization and assortative mating. Evol Ecol, 23: 31–52. doi:10.1007/s10682-008-9267-z. [Google Scholar]
- 76. van Doorn GS, Edelaar P, Weissing FJ (2009) On the origin of species by natural and sexual selection. Science, 326: 1704–1707. doi:10.1126/science.1181661. PubMed: 19965377. [DOI] [PubMed] [Google Scholar]
- 77. Martin CH (2012) Weak disruptive selection and incomplete phenotypic divergence in two classic examples of sympatric speciation: cameroon crater lake cichlids. Am Nat, 180(4): E90–E109. doi:10.1086/667586. PubMed: 22976018. [DOI] [PubMed] [Google Scholar]
- 78. Streelman JT, Danley PD (2003) The stages of vertebrate evolutionary radiation. Trends Ecol Evol, 18: 126–131. doi:10.1016/S0169-5347(02)00036-8. [Google Scholar]
- 79. Kocher TD (2004) Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet 5: 288–298. doi:10.1038/nrg1316. PubMed: 15131652. [DOI] [PubMed] [Google Scholar]
- 80. Elmer KR, Lehtonen TK, Fan S, Meyer A (2013) Crater lake colonization by neotropical cichlid fishes. Evolution, 67: 281–288. doi:10.1111/j.1558-5646.2012.01755.x. PubMed: 23289578. [DOI] [PubMed] [Google Scholar]
- 81. McCrary JK, López LJ (2008) El monitoreo de las mojarras (Amphilophus spp.) en Nicaragua con aportes sobre su ecología y estado de conservación en la Laguna de Apoyo. Rev Nicaragüense Biodiversidad, 1: 43–50. [Google Scholar]
- 82. Salazar C, Baxter SW, Pardo-Diaz C, Wu G, Surridge A et al. (2010) Genetic Evidence for Hybrid Trait Speciation in Heliconius Butterflies. PLOS Genet, 6, 1000930 PubMed: 20442862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity (Edinb), 83 ( Pt 4): 363–372. doi:10.1038/sj.hdy.6886170. PubMed: 10583537. [DOI] [PubMed] [Google Scholar]
- 84. Ellstrand NC, Schierenbeck KA (2000) Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci U S A, 97: 7043–7050. doi:10.1073/pnas.97.13.7043. PubMed: 10860969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gross BL, Kane NC, Lexer C, Ludwig F, Rosenthal DM et al. (2004) Reconstructing the origin of Helianthus deserticola: Survival and selection on the desert floor. Am Nat, 164: 145–156. doi:10.1086/422223. PubMed: 15278840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nolte A, Freyhof J, Stemshorn K, Tautz D (2005) An invasive lineage of sculpins, Cottus sp. (Pisces, Teleostei) in the Rhine with new habitat adaptations has originated by hybridization between old phylogeographic groups. Proc R Soc Lond B Biol Sci, 272: 2379–2387. doi:10.1098/rspb.2005.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pfennig KS (2007) Facultative mate choice drives adaptive hybridization. Science, 318: 965–967. doi:10.1126/science.1146035. PubMed: 17991861. [DOI] [PubMed] [Google Scholar]
- 88. Seehausen O, van Alphen JM, Witte F (1997) Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science, 277: 1808–1811. doi:10.1126/science.277.5333.1808. [Google Scholar]
- 89. Seehausen O (2006) Conservation: Losing Biodiversity by Reverse Speciation. Curr Biol, 16: 337 16682344. [DOI] [PubMed] [Google Scholar]
- 90. Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D et al. (2006) Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol, 15: 343–355. PubMed: 16448405. [DOI] [PubMed] [Google Scholar]
- 91. Smith TB, Wayne RK, Girman DJ, Bruford MW (1997) A role for ecotones in generating rainforest biodiversity. Science, 276: 1855–1857. doi:10.1126/science.276.5320.1855. [Google Scholar]
- 92. Crandall KA, Bininda-Emonds OR, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol, 15: 290–295. doi:10.1016/S0169-5347(00)01876-0. PubMed: 10856956. [DOI] [PubMed] [Google Scholar]
- 93. Barreto FS, McCartney MA (2008) Extraordinary AFLP fingerprint similarity despite strong assortative mating between reef fish color morphospecies. Evolution, 62: 226–233. PubMed: 18053072. [DOI] [PubMed] [Google Scholar]
- 94. Nielsen EE, Hemmer-Hansen J, Poulsen NA, Loeschcke V, Moen T et al. (2009) Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua). BMC Evol Biol, 9: 276–287. doi:10.1186/1471-2148-9-276. PubMed: 19948077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Barlow GW (1976) The Midas cichlid in Nicaragua. In Thorsen TB. Investigations of the ichthyofauna of Nicaraguan lakes. Lincoln: University of Nebraska. [Google Scholar]
- 96. Barlow GW, Rogers W (1978) Female Midas cichlids’ choice of mate in relation to parents’ and to own color. Biol Behav, 3: 137–145. [Google Scholar]
- 97. Barlow GW, Rogers W, Cappeto RV (1977) Incompatability and assortative mating in the Midas cichlid. Behav Ecol Sociobiol, 2: 49–59. doi:10.1007/BF00299288. [Google Scholar]
- 98. McKaye KR, Barlow GW (1976) Competition between color morphs of the midas cichlid, Cichlasoma citrinellum in Lake Jiloa, Nicaragua. In Thorson TB. Investigations of the Ichthyofauna of Nicargauan Lakes. Lincoln: University of Nebraska. . [Google Scholar]
- 99. Elmer KR, Lehtonen TK, Meyer A (2009) Color assortative mating contributes to sympatric divergence of neotropical cichlid fish. Evolution, 63: 2750–2757. doi:10.1111/j.1558-5646.2009.00736.x. PubMed: 19490078. [DOI] [PubMed] [Google Scholar]
- 100. Engström-Öst J, Candolin U (2007) Human-induced water turbidity alters selection on sexual displays in sticklebacks. Behav Ecol, 18: 393–398. doi:10.1093/beheco/arl097. [Google Scholar]
- 101. Järvenpää M, Lindström K (2004) Water turbidity by algal blooms causes mating system breakdown in a shallow-water fish, the sand goby Pomatoschistus minutus. Proc R Soc Lond B Biol Sci, 271: 2361–2365. doi:10.1098/rspb.2004.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HD et al. (2008) Speciation through sensory drive in cichlid fish. Nature, 455: 620–626. doi:10.1038/nature07285. PubMed: 18833272. [DOI] [PubMed] [Google Scholar]
- 103. Elmer KR, Lehtonen TK, Kautt AF, Harrod C, Meyer A (2010b) Rapid sympatric ecological differentiation of crater lake cichlid fishes within historic times. BMC Biol, 8: 60. doi:10.1186/1741-7007-8-60. PubMed: 20459869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Elmer KR, Meyer A (2011) Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol Evol, 26: 298–306. doi:10.1016/j.tree.2011.02.008. PubMed: 21459472. [DOI] [PubMed] [Google Scholar]
- 105. Henning F, Jones JC, Franchini P, Meyer A (2013) Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genomics, 14: 171. doi:10.1186/1471-2164-14-171. PubMed: 23497064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Key to the six Amphilophus species endemic in Lake Apoyo, Nicaragua.
(PDF)
Estimation criteria for the number of genetic clusters in the AFLP data set. Above: Mean LnP(D) with SD from 20 replicates for each K, calculated without ‘locprior’ model (STRUCTURE v2.2). Below: Evanno’s model choice criterion ‘ΔK’ for the uppermost level of genetic structure.
(PDF)
Results of STRUCTURE clustering analysis with A . zaliosus and A . astorquii only for K3 using STRUCTURE v2.2 without group information. Species of sample origin given above.
(PDF)
Plot of 1st and 2nd principal component scores based on the complete AFLP matrix (A), the 49 outlier loci only (B) and the neutral AFLP matrix (C). ID numbers are given for the potential hybrid individuals. Variance explained by 1st and 2nd PC for A) 7 & 4%, B) 18 & 10% and for C) 7 & 4%, respectively.
(PDF)
Sample list with individual’s ID used throughout, voucher collection number, isolate number, taxon information, sample location, leaf stability index (LS) and GenBank accession numbers.
(DOC)
Genbank accession numbers of mtDNA control region sequences included in the pairwise mismatch analysis.
(DOC)
Data Availability Statement
DNA sequences: GenBank accessions GU355718- GU355726; GU355729- GU355733; GU355735; GU355737; GU355739- GU355742; GU355744- GU355748; GU355764; GU355770; GU355850; GU355851; GU362707- G362709; JF784052-JF784149.
DNA aliquots: stored permanently at ZSM DNA-Bank, available upon request from dnabank@zsm.mwn.de