Abstract
DNA cytosine methylation is a reversible epigenetic mark regulating gene expression. Aberrant methylation profiles are concomitant with developmental defects and cancer. Numerous studies in the past decade have identified enzymes and pathways responsible for active DNA demethylation both on a genome-wide as well as gene-specific scale. Recent findings have strengthened the idea that 5-methylcytosine oxidation catalyzed by members of the ten-eleven translocation (Tet1–3) oxygenases in conjunction with replication-coupled dilution of the conversion products causes the majority of genome-wide erasure of methylation marks during early development. In contrast, short and long patch DNA excision repair seems to be implicated mainly in gene-specific demethylation. Growth arrest and DNA damage-inducible protein 45 a (Gadd45a) regulates gene-specific demethylation within regulatory sequences of limited lengths raising the question of how such site specificity is achieved. A new study identified the protein inhibitor of growth 1 (Ing1) as a reader of the active chromatin mark histone H3 lysine 4 trimethylation (H3K4me3). Ing1 binds and directs Gadd45a to target sites, thus linking the histone code with DNA demethylation.
Keywords: 5-methylcytosine, DNA demethylation, reprogramming, Gadd45, Ing1, H3K4me3
Introduction
In mammalian DNA, cytosines within a CpG dinucleotide context are commonly marked by a methyl group at carbon 5 of the pyrimidine ring. By influencing, typically silencing, gene expression, the resulting modification, 5-methylcytosine (5mC), has been implicated to bear pivotal roles during embryonic development, imprinting, X-chromosome inactivation and cancer.1-3 DNA cytosine methylation permits organisms to gain an additional layer of genetic information on top of the primary DNA sequence and, hence, is classified as an epigenetic mark.
Methylation marks are commonly maintained during DNA replication by the action of the DNA methyltransferase DNMT1.4 Consequently, once set, DNA methylation has been thought to be stable even through cell divisions. Additionally, loss of methylation marks observed in dividing cells was referred to, by default, as passive DNA demethylation after several rounds of replication in the absence of DNMT1. However, research in the last decade uncovered scenarios in which methylated DNA is demethylated in a replication-independent, active manner, both at the genome-wide level as well as at specific genomic loci.
After fertilization, DNA methylation marks of the paternal pronucleus in the mouse zygote are globally erased prior to the first cell cycle.5,6 Similarly, in primordial germ cells (PGCs), the progenitor cells of gametes, methylation is lost genome-wide during their migration to the genital ridge between embryonic days E8.5–E11.5.7
A remarkable example of loci-specific active DNA demethylation in human cells was described at the estrogen receptor target gene pS2. After estrogen stimulation the pS2-promoter undergoes cycles of methylation and demethylation in less than 100 min indicating DNA methylation to be not only reversible but also highly dynamic.8,9
DNA demethylation is of particular importance for the generation of induced pluripotent stem cells (iPSCs), the artificial reprogramming of somatic cells to their pluripotent ground state. Genes for key transcription factors like Oct4 and Nanog are fully demethylated during reprogramming and this demethylation, in turn, drives their expression.10,11
Enzymes of Active DNA Demethylation
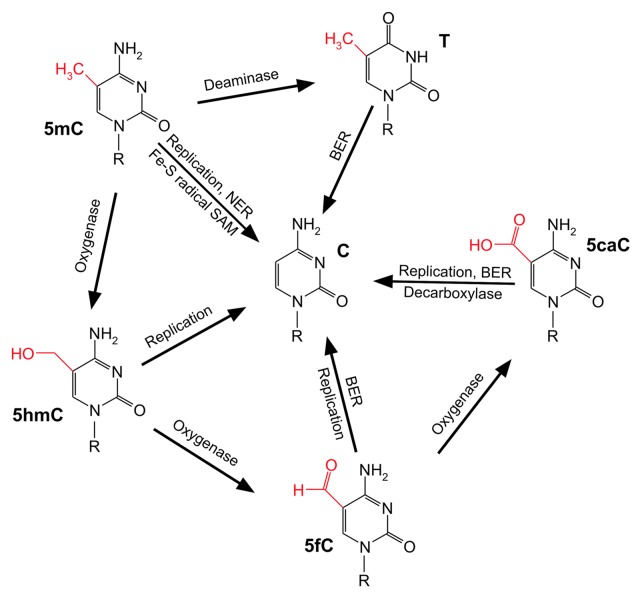
While the enzymes responsible for cytosine methylation are confined and well characterized,12 the enzymes responsible for mammalian active DNA demethylation remain rather numerous and controversial (Fig. 1).13 A single enzymatic reaction that releases the methyl group from 5mC but keeps the DNA backbone untouched (a DNA “demethylase” reaction) seems difficult to conceive. Thermodynamically it is challenging to break unpolar carbon-carbon bonds. Nevertheless, the Fe-S radical S-adenosylmethionine (SAM) domain of elongator complex protein 3 (Elp3) was shown to be involved in paternal DNA demethylation in the mouse zygote leading to the hypothesis of a direct radical SAM-mediated demethylation mechanism.14
Figure 1. The multiple faces of mammalian DNA demethylation. Schematic representation of the enzymology implicated in 5-methylcytosine (5mC) demethylation. The exocyclic group at carbon 5 of each cytosine derivative is highlighted in red. C, cytosine; T, thymine; 5hmC, 5-hydroxymethylcytosine; 5fC, 5-formylcytosine; 5caC, 5-carboxylcytosine; BER: base excision repair; NER, nucleotide excision repair. Note: Direct base excision repair of 5mC and potential deamination of 5hmC to 5-hydroxymethyluracil has not been considered due to lack of experimental confirmation. For details see main text.
On the other hand, DNA repair mechanisms that exchange complete nucleotides by canonical (unmethylated) DNA building blocks attracted attention in regard to active DNA demethylation. In fact, DNA glycosylases from the Demeter/ROS1 family initiate base excision repair (BER) of methylated cytosines in plants leading to DNA demethylation.15-17 In mammals, however, a 5mC-specific DNA glycosylase could not be confirmed.18-21 An elegant loophole was provided by the idea that 5mC is first deaminated to thymine resulting in a T/G mismatch which, in turn, is processed by thymine-specific DNA glycosylases such as TDG. PGCs deficient in activation-induced (DNA-cytosine) deaminase (AID) show a significant albeit far from complete impairment of global demethylation.22 In line with this, using an interspecies heterokaryon technology for reprogramming it was shown that AID is required to demethylate the critical genes OCT4 and NANOG in human fibroblasts.23 Moreover, reprogramming of mouse embryonic fibroblasts (MEFs) to iPSCs by the four Yamanaka factors Oct4, Sox2, c-Myc and Klf411 was demonstrated to depend on a catalytically active AID.24 Interestingly, in absence of the cofactor SAM the de novo DNA methyltransferases Dnmt3a and Dnmt3b are able to deaminate 5mC, thereby, potentially enabling dynamical methylation/demethylation cylces in short periods of time on specific genes.9
The paternal pronucleus in the mouse zygote exhibits high amounts of DNA strand breaks within the critical time frame of global demethylation indicative for an involvement of DNA repair in this process.25 Strikingly, DNA strand breaks in direct vicinity of 5mCs were found in an enhancer region of the tyrosine aminotransferase gene that is demethylated after hormone stimulation.26 The result supports short patch base excision repair as being part of the demethylation machinery.
Long patch excision repair has also been attributed to DNA demethylation. Notably, in Xenopus laevis oocytes demethylation of an oct4-reporter was shown to critically depend on the xeroderma pigmentosum complementation group G protein (XPG), the 3′-endonuclease of nucleotide excision repair (NER).27 Demethylation of the same reporter was accompanied by BrdU incorporation, indicating long patch repair synthesis instead of a one nucleotide exchange.27 Demethylation of the human rDNA promoter requires the endonuclease activity of XPG28 and the RARβ2 promoter is occupied by NER factors and exhibits DNA strand breaks, triggering DNA demethylation upon retinoic acid stimulation.29,30
Research in the field was revolutionized by two breakthrough papers in 2009 describing the (re)discovery of 5-hydroxymethylcytosine (5hmC) in human cells.31,32 5hmC is the oxidation product of 5mC catalyzed by the ten-eleven translocation (Tet1–3) family of enzymes.32,33 Tet enzymes can oxidize 5hmC further to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC).34,35 All three oxidation products have been detected in genomic DNA35-37 and their role as potential DNA demethylation intermediates has been extensively analyzed since then. Indeed, 5fC and 5caC are substrates for TDG34,38 and the embryonic lethal phenotype of Tdg null mice was explained in part by aberrant methylation during early development.39
Recently, the distribution of 5fC and 5caC in the genome of mouse ES cells (mESCs) has been achieved by high-throughput sequencing of (1) immunoprecipitated DNA using modification-specific antibodies and (2) streptavidin-captured DNA after chemical reduction and specific biotin-labeling of 5fC.40,41 Both studies demonstrated a significant accumulation of 5fC (and 5caC) residues at distal regulatory regions (enhancers) of genes across the genome in TDG-deficient mESCs, as compared with wild-type cells. Hence, 5fC and 5caC serve as intermediates of active DNA demethylation triggered by the Tet-family and TDG/BER at least at distinct genomic loci.
5hmC was postulated to be deaminated by members of the AID/APOBEC family of deaminases with the resulting 5-hydroxymethyluracil (5hmU) being a substrate for 5hmU-glycosylases such as SMUG1.42 However, neither AID nor APOBEC enzymes are able to deaminate 5hmC in vitro43,44 and 5hmU could not be detected in genomic DNA.43
The importance of Tet-mediated DNA demethylation was demonstrated in a recent study describing a modified protocol for the generation of iPSCs. One of the Yamanaka factors, Oct4, could be replaced by Tet1, highlighting the significance of the conversion of 5mC to 5hmC during reprogramming.45
Genomic stability is challenged by strand breaks, base-free sites or nucleotide gaps that necessarily occur during base or nucleotide excision repair. If coping with such toxic intermediates during demethylation at single loci might be a solvable task for cells, the same problem, likely, is of a different nature during global DNA demethylation in a limited time frame. As a matter of fact, in the paternal pronucleus of the mouse zygote, 5hmC signals appear concomitant with the loss of 5mC signals.46-48 A major fraction of 5mCs was, obviously, oxidized, and this was dependent on Tet3. Later, it was also shown that 5fC and 5caC signals occur during late pronuclear stages.36 Strikingly, the signals for 5hmC, 5fC and 5caC diminished subsequently after each round of replication by a factor of two.36,49 The majority of genome-wide DNA demethylation in the paternal pronucleus is therefore a mixture of active enzymatic conversion of 5mCs and passive, replication-coupled dilution of the oxidation products, avoiding any threat for genomic instability.50 Moreover, a similar strategy for global DNA demethylation holds true for mouse PGC reprogramming. In contrast to what happens in the zygote, Tet1 and Tet2 are responsible for the conversion of a majority of 5mCs to 5hmCs in PGCs.51
The three-step oxidation of a methylated pyrimidine, as it is the case for Tet-mediated conversion of 5mC to 5caC, has a precedent. In the thymidine salvage pathway, thymine is converted to uracil, in other words, 5-methyluracil is demethylated. Beside a consecutive three-step oxidation to iso-orotate (5-carboxyluracil), the pathway includes a final decarboxylation to gain uracil.52 In search for a parallel pathway for 5mC demethylation, a decarboxylation activity toward 5caC was indeed demonstrated in a cell-free system using nuclear extracts of mESCs.53 The result opens up a new avenue for non-toxic DNA demethylation.
In conclusion, the mechanisms of active DNA demethylation in mammals not only tend to be highly multifaceted but also offer the promise of surprising us in future research.
Gadd45 Proteins are Regulators of DNA Demethylation
Knowledge about the core enzymes acting on 5mC and its derivatives is just one side of the coin. Non-enzymatic mediators of DNA demethylation shed light on the regulation of the process upstream. Growth arrest and DNA damage-inducible protein 45 a (Gadd45a) were shown to promote DNA demethylation by DNA repair.27,54,55 Gadd45a is an 18 kDa acidic protein without obvious enzymatic activity. Gadd45a is located in the nucleus and associated with ribonucleoprotein speckles.56 Together with Gadd45b and Gadd45g, it is part of a family of histone fold stress-response proteins that modulate diverse cellular processes, one of them being DNA repair.57,58 Epigenetically, Gadd45 proteins act as scaffold proteins for downstream components of the repair machinery, thus directly regulating DNA demethylation. Gadd45 seems to promote gene-specific demethylation only.59-61 Typically, specific external stimuli lead to the upregulation of Gadd45 proteins, which, in turn, drive the demethylation and transcriptional activation of certain target genes. For instance, after electroconvulsive treatment of adult mice neurons Gadd45b is strongly upregulated leading to demethylation and expression of key genes for adult neurogenesis.62
Together with components of the NER machinery (see above) Gadd45a has been demonstrated to be required to demethylate reporter plasmids in Xenopus oocytes as well as the human rDNA and RARβ2 promoter.27,28,30 Gadd45 proteins also promote BER-mediated DNA demethylation, as has been first demonstrated for foreign methylated DNA and endogenous target genes in zebrafish embryos.63 A T/G mismatch intermediate accompanied the demethylation, suggesting deamination of 5mC prior to BER.63 GADD45a was also found to physically interact with AID and TDG in human cells.64 The protein, thus, might regulate pivotal processes during development. However, in contrast to Tdg−/− mutants, mice lacking Gadd45a (but not simultaneously Gadd45b and Gadd45g) are viable and do not show significant alterations in global DNA methylation.59 A potential involvement of Gadd45 in Tet-mediated DNA demethylation remains to be investigated (see below).
Ing1 Directs DNA Demethylation to H3K4me3
Gene-specific DNA demethylation primarily affects methylated CpGs within regulatory regions of limited lengths, whereas 5mCs within the gene body or in intergenic regions are often left untouched. This site specificity suggests a targeting mechanism that guides general demethylation factors to certain CpGs.
Mammalian DNA is wrapped around histone proteins organized in nucleosomes that consist of two H2A-H2B dimers and a H3-H4 tetramer each. Histone proteins are posttranslationally covalently modified by, e.g., methylation, acetylation or phosphorylation of distinct amino acids at the N-terminal tails, constituting the so-called histone code.65 The histone code is an important epigenetic feature regulating gene expression and DNA repair, among other processes. Trimethylation of H3 lysine 4 (H3K4me3) is typically found at promoter regions of genes that are transcriptionally active.66 Thus, the occurrence of H3K4me3 strikingly resembles hypomethylation of promoter regions after gene-specific DNA demethylation and, hence, gene activation. Are both epigenetic marks linked to each other to determine the transcriptional status of the corresponding gene? And, if yes, what is the cause and what is the effect?
The answers to those questions were provided recently by the identification of an H3K4me3 reader that directs Gadd45a and the demethylation machinery to their target 5mCs.67 The protein inhibitor of growth 1 (human ING1b, mouse Ing1) is a known interactor of Gadd45a and exhibits similar properties as both are stress-response proteins influencing the cell cycle and promoting DNA repair.68 Schäfer et al.67 at first confirmed the physical interaction of GADD45a and ING1b in the human cell line HEK293T. They identified that ING1b cooperates with and is required for GADD45a to demethylate and reactivate methylation-silenced reporter plasmids. ING1b consists of an N-terminal PCNA interacting protein domain, a partial bromodomain and a C-terminal plant homeodomain (PHD finger), enabling binding to H3K4me3.69 Importantly, the PHD finger domain was essential for GADD45a-mediated demethylation suggesting a link between the histone mark H3K4me3 and 5mC demethylation. Indeed, in RKO cells, the MAGEB2 promoter, an endogenous target of GADD45a demethylation, was occupied with H3K4me3 marks; GADD45a and ING1b were bound to the same region simultaneously. MEFs lacking Ing1 or Gadd45a had a reduced, double knockouts an abolished potential to induce expression of the Mageb1–3 genes and demethylation of the Mageb2 promoter upon UV-stimulation. Strikingly, knockdown of Wdr5, an essential subunit of the H3K4 methyltransferase complex MLL,70 prevented Mageb1–3 expression and Mageb2 demethylation induced by either UV-irradiation or GADD45a and ING1b overexpression in wild-type MEFs. Moreover, loss of H3K4me3 marks impaired GADD45a and ING1b binding to the Mageb2 locus. Global gene expression profiling of HEK293T cells overexpressing GADD45a, ING1b or both proteins as well as MEFs lacking Gadd45a, Ing1 or both genes revealed roughly a hundred additional endogenous target genes in the human and mouse genome, respectively, potentially regulated by the same mechanism, as exemplified for the Mage genes above. Of note, this does not apply for all genes affected in their gene expression by Gadd45a and Ing1, since a comparable number of genes was, against expectations, also upregulated in double knockout MEFs. However, it should not be neglected that DNA demethylation is just one of many cellular processes influenced by Gadd45a and Ing1.
The study uncovered an unknown player in mammalian gene-specific DNA demethylation upstream of the enzymatic reactions on 5mC and links the histone code with DNA demethylation by two factors: Ing1, an H3K4me3 reader, and Gadd45a, a DNA demethylation regulator. The data favors a model by which H3K4me3 serves as a determinant for DNA demethylation via Ing1 and Gadd45a (Fig. 2). Interestingly, in a pull-down assay of mESC nuclear extracts using modified oligonucleotides as bait, Ing1 was found to bind 5fC residues in vitro.71 The result hints at an involvement of Ing1 in Tet-mediated DNA demethylation. In line with this, and as already stated, Gadd45a was shown to directly interact with TDG.64 Thus, Ing1 might target Gadd45a/TDG to oxidized derivatives of 5mC. This assumption has to be clarified in upcoming studies. Additionally, future work has now to decipher the physiological relevance of the targeting process in development and disease, as well as to identify additional cofactors required for regulating demethylation. Finally, the chromatin context should be considered when analyzing the modus operandi of known or yet-to-be-identified demethylation factors.
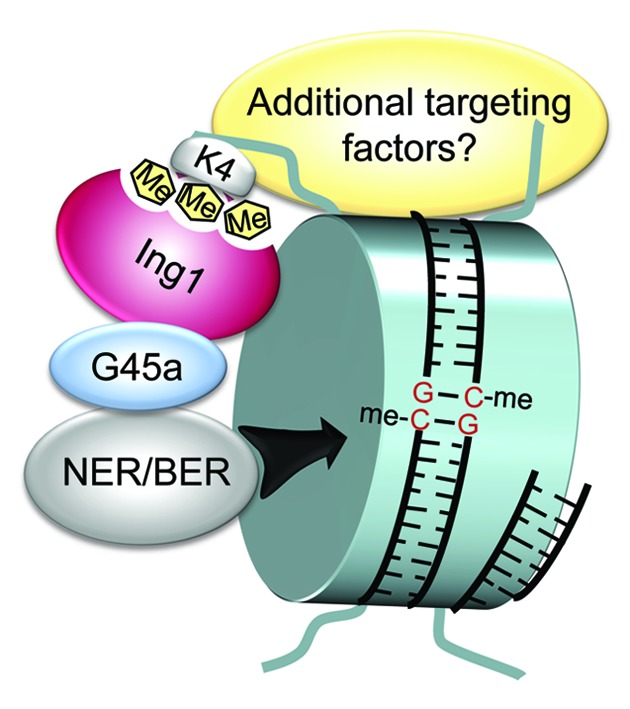
Figure 2. Ing1 directs Gadd45a and the demethylation machinery to H3K4me3. Proposed model for site-specific demethylation by Gadd45a. At a given gene promoter histone H3 trimethylated at lysine 4 (H3K4me3) is specifically recognized by Ing1. Gadd45a and the repair machinery are recruited through Ing1 binding. Subsequently, 5mCs are excised by DNA repair leading to DNA demethylation. Additional targeting factors may be required for the process. G45a, Gadd45a; NER, nucleotide excision repair; BER, base excision repair. Figure is from ref. 67 with permission of the authors.
Summary
The complex world of mammalian DNA demethylation emerges from studies identifying numerous enzymes acting on 5mC and its derivatives. Unequivocally, DNA repair mechanisms are involved in gene-specific DNA demethylation whereas cells presumably avoid hazardous intermediates of DNA repair in genome-wide erasure of methylation marks during development. Gadd45 proteins are regulators of gene-specific and repair-mediated demethylation in different contexts. Site-specific demethylation by Gadd45a is ensured by a targeting mechanism involving the histone mark H3K4me3 as determinant and the histone code reader Ing1 as transducer. The mechanism seems to be valid for the epigenetic regulation of a substantial amount of genes in humans and mice.
Acknowledgment
I thank Andrea Schäfer, IMB Mainz, for providing Figure 2 and apologize to those colleagues whose work was not cited due to space limitations. This work was supported by an ERC senior investigator grant (“DNA Demethylase”) to Christof Niehrs, IMB Mainz.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24977
References
- 1.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich M. The controversial denouement of vertebrate DNA methylation research. Biochemistry (Mosc) 2005;70:568–75. doi: 10.1007/s10541-005-0150-z. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–9. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 5.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 6.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 7.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 8.Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 9.Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–8. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci U S A. 2006;103:11796–801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci U S A. 2006;103:6853–8. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J Am Chem Soc. 2006;128:12510–9. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortázar D, Kunz C, Saito Y, Steinacher R, Schär P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, et al. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–65. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci U S A. 2000;97:5135–9. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–5. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–7. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhutani N, Decker MN, Brady JJ, Bussat RT, Burns DM, Corbel SY, et al. A critical role for AID in the initiation of reprogramming to induced pluripotent stem cells. FASEB J. 2013;27:1107–13. doi: 10.1096/fj.12-222125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–88. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci U S A. 2006;103:11112–7. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–5. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schäfer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–53. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Le May N, Fradin D, Iltis I, Bougnères P, Egly JM. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol Cell. 2012;47:622–32. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 30.Le May N, Mota-Fernandes D, Vélez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–6. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaffeneder T, Hackner B, Truss M, Münzel M, Müller M, Deiml CA, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–12. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 38.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–8. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–23. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 40.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, et al. Genome-wide Analysis Reveals TET- and TDG-Dependent 5-Methylcytosine Oxidation Dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013;153:678–91. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–8. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rangam G, Schmitz KM, Cobb AJ, Petersen-Mahrt SK. AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS One. 2012;7:e43279. doi: 10.1371/journal.pone.0043279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, et al. Replacement of Oct4 by Tet1 during iPSC Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation in Reprogramming. Cell Stem Cell. 2013;12:453–69. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–10. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 47.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–7. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 49.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Münzel M, Lischke U, Stathis D, Pfaffeneder T, Gnerlich FA, Deiml CA, et al. Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chemistry. 2011;17:13782–8. doi: 10.1002/chem.201102782. [DOI] [PubMed] [Google Scholar]
- 51.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–52. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr., Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim Biophys Acta. 2005;1723:256–64. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Schiesser S, Hackner B, Pfaffeneder T, Müller M, Hagemeier C, Truss M, et al. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew Chem Int Ed Engl. 2012;51:6516–20. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
- 54.Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Niehrs C, Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–7. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Sytnikova YA, Kubarenko AV, Schäfer A, Weber AN, Niehrs C. Gadd45a is an RNA binding protein and is localized in nuclear speckles. PLoS One. 2011;6:e14500. doi: 10.1371/journal.pone.0014500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollander MC, Fornace AJ., Jr. Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene. 2002;21:6228–33. doi: 10.1038/sj.onc.1205774. [DOI] [PubMed] [Google Scholar]
- 58.Smith ML, Kontny HU, Zhan Q, Sreenath A, O’Connor PM, Fornace AJ., Jr. Antisense GADD45 expression results in decreased DNA repair and sensitizes cells to u.v.-irradiation or cisplatin. Oncogene. 1996;13:2255–63. [PubMed] [Google Scholar]
- 59.Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, Sapienza C, et al. Conserved DNA methylation in Gadd45a(-/-) mice. Epigenetics. 2009;4:98–9. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schäfer A, Schomacher L, Barreto G, Döderlein G, Niehrs C. Gemcitabine functions epigenetically by inhibiting repair mediated DNA demethylation. PLoS One. 2010;5:e14060. doi: 10.1371/journal.pone.0014060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–7. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–12. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 66.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schäfer A, Karaulanov E, Stapf U, Döderlein G, Niehrs C. Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes Dev. 2013;27:261–73. doi: 10.1101/gad.186916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheung KJ, Jr., Mitchell D, Lin P, Li G. The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res. 2001;61:4974–7. [PubMed] [Google Scholar]
- 69.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 70.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 71.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]