SUMMARY
PUMA is a pro-apoptotic BCL-2 family member that drives the apoptotic response to a diversity of p53-dependent and independent cellular insults. Deciphering the spectrum of PUMA interactions that confer its context-dependent pro-apoptotic properties remains a high priority goal. Here, we report the synthesis of PUMA SAHBs, structurally-stabilized PUMA BH3 helices that, in addition to broadly targeting anti-apoptotic proteins, directly bind to BAX. NMR, photocrosslinking, and biochemical analyses revealed that PUMA SAHBs engage an α1/α6 trigger site on BAX to initiate its functional activation. We further demonstrated that a cell-permeable PUMA SAHB analog induces apoptosis in neuroblastoma cells and, like expressed PUMA protein, engages BCL-2, MCL-1 and BAX. Thus, we find that PUMA BH3 is a dual anti-apoptotic inhibitor and pro-apoptotic direct activator, and its mimetics may serve as effective pharmacologic triggers of apoptosis in resistant human cancers.
INTRODUCTION
The cellular decision to live or die is adjudicated by members of the BCL-2 protein family, which executes the activation or suppression of mitochondrial apoptosis(Llambi et al., 2011). BCL-2 proteins are classified into three groups based on sequence homology and function. Anti-apoptotic members such as BCL-2 contain up to four BCL-2 Homology (BH) domains, whereas the multidomain pro-apoptotic proteins, including BAX and BAK, contain three BH domains. A heterogeneous group of proteins that contain only the BH3 motif function as afferent sensors of stress. These so called “BH3-only” proteins relay pro-apoptotic signals to the multidomain members, which ultimately render a life or death decision based upon the overall balance between the degree of stress and the anti-apoptotic reserve. PUMA (p53-Upregulated Modulator of Apoptosis) is one such BH3-only protein that was first identified as a transcriptional target of p53(Han et al., 2001; Nakano and Vousden, 2001; Yu et al., 2001). p53 deletion and mutagenesis can effectively blunt PUMA upregulation, which may contribute to the pathogenesis, maintenance, and chemoresistance of human cancer; reconstituting PUMA function in this context can effectively reactivate apoptosis, either alone or in combination with other agents(Yu et al., 2006; Yu et al., 2001). Although oncogenesis was not observed in Puma−/− mice (Erlacher et al., 2006), PUMA is frequently downregulated across a broad spectrum of human cancers(Beroukhim et al., 2010), suggesting that it may function as a tumor suppressor in combination with other predisposing factors. Indeed, deletion of even one Puma allele in Bim−/− mice increases the incidence of lymphoma(Erlacher et al., 2006). Puma−/− cells have been shown to manifest reduced sensitivity to a variety of p53-dependent and independent insults, including irradiation, DNA-damaging agents, cytokine withdrawal, hypoxia, and endoplasmic-reticulum stress(Jeffers et al., 2003; Luo et al., 2005; Reimertz et al., 2003; Villunger et al., 2003; Yu and Zhang, 2008; Yu et al., 2001). These data highlight the importance of PUMA’s role in apoptosis regulation in health and disease, and the potential of PUMA-based therapeutics to alternatively enhance chemo- and radiosensitivity in the context of cancer treatment or mitigate damage to host tissues through targeted PUMA inhibition(Mustata et al., 2011). Thus, deciphering the spectrum of PUMA interactions that confer its context-dependent pro-apoptotic properties remains a high priority goal.
The BH3-only protein interaction circuit is believed to induce apoptosis by two complementary mechanisms. The first is by BH3-only protein-mediated “inhibition of the inhibitors” of cell death(Uren et al., 2007; Willis et al., 2007). That is, the BH3 motif of BH3-only proteins engages the canonical BH3-binding groove of anti-apoptotic targets to neutralize their capacity to bind and block the multidomain pro-apoptotic effectors BAX and BAK. In addition, select members of the BH3-only class of apoptotic proteins have been shown to directly bind and activate BAK and BAX at discrete canonical(Czabotar et al., 2013; Dai et al., 2011; Leshchiner et al., 2013; Moldoveanu et al., 2013) and, in the case of BAX, non-canonical(Gavathiotis et al., 2010; Gavathiotis et al., 2008; Leshchiner et al., 2013) BH3-binding sites. Whereas structural and biochemical data support direct and functional interactions for the BH3 domains of BIM and BID with BAX and BAK(Czabotar et al., 2013; Gavathiotis et al., 2010; Gavathiotis et al., 2008; Leshchiner et al., 2013; Moldoveanu et al., 2013; Moldoveanu et al., 2006; Walensky et al., 2006), the direct binding capability of the PUMA BH3 helix is unresolved.
A series of studies that employed in vitro functional assays, and cellular and in vivo analyses, have yielded conflicting results regarding the existence and potential mechanistic role of direct PUMA interactions with BAX and/or BAK. A physical association between PUMA protein and BAX has been shown in bacterial two-hybrid assays(Cartron et al., 2004), yeast cells(Gallenne et al., 2009), in vitro and mammalian cell co-immunoprecipitation studies(Kim et al., 2009; Yee and Vousden, 2008; Zhang et al., 2009), and by FRET analysis(Zhang et al., 2009), indicating that the two proteins can interact. Puma−/−Bim−/−Bid−/− knockout (TKO) mice show developmental defects that are reminiscent of, although perhaps less severe than(Villunger et al., 2011), those observed in Bax−/−Bak−/− mice, suggesting that eliminating key direct activators may be tantamount to knocking out Bax and Bak altogether(Ren et al., 2010). However, a series of studies document that the pro-apoptotic activity of PUMA instead derives from exclusive anti-apoptotic inhibition, citing the lack of direct interaction between PUMA and BAX upon co-immunoprecipitation from cells exposed to discrete stress stimuli(Callus et al., 2008; Jabbour et al., 2009; Willis et al., 2007). In addition, a C-terminally truncated form of PUMA that manifests decreased BAX-binding activity retained pro-apoptotic potency equivalent to that of full-length PUMA, suggestive that the contribution of PUMA-BAX engagement, at least in this experimental context, was not essential(Yee and Vousden, 2008). Such cellular analyses are made all the more challenging by the proposed “hit and run” nature of the transient interactions between direct BH3 activators and BAX/BAK, potentially confounding co-immunoprecipitation-based studies. Even liposomal release assays, which probe functional activation of BAX and BAK upon administration of discrete agonists/antagonists, have had mixed results with PUMA BH3(Du et al., 2011; Kuwana et al., 2005). Thus, the purpose of this biochemical, structural, proteomic, and cellular study is to interrogate the capacity of PUMA BH3 to directly and functionally engage BAX, and thereby contribute to its pro-apoptotic activity in vitro and in cells.
RESULTS
Design, Synthesis, and Anti-Apoptotic Binding Activity of Hydrocarbon-Stapled PUMA BH3 Helices
Whereas select BH3-only proteins, like BID, are highly structured and maintain an α-helical BH3 domain in the unbound state, others are intrinsically disordered(Hinds et al., 2007) but undergo α-helical folding of the BH3 domain upon binding to compatible anti-apoptotic grooves(Day et al., 2008). In dissecting the mechanism of BH3-mediated BAX activation, we determined that a helical BID or BIM BH3 peptide, but not unfolded ones, were capable of directly engaging full-length BAX(Walensky et al., 2006). As synthetic PUMA BH3 peptides are predominantly unstructured in solution(Letai et al., 2002) (Figure S1A), we first sought to stabilize the physiologically-relevant α helical structure by insertion of an all-hydrocarbon crosslink. By replacing i, i+4 pairs of native residues with non-natural amino acids bearing olefin tethers followed by ruthenium-catalyzed olefin metathesis, we previously generated a variety of stabilized alpha-helices of BCL-2 domains (SAHBs) modeled after the BH3 motifs of BCL-2 family proteins(Pitter et al., 2008; Stewart et al., 2010; Walensky et al., 2004). In designing stapled peptides - whether for in vitro binding studies, cellular localization analyses, structure determination, cellular signal transduction studies, or in vivo activity analyses - synthetic iteration is required to achieve optimal solubility, structural stability, cell permeability, and biological activity for the desired application. In the case of our prototype BIM SAHBA(Walensky et al., 2006), which potently bound to and activated BAX, adjustments in both sequence and charge (e.g. 146-IWIAQELRXIGDXNAYYARR-166 [+2] to 145-EIWIAQELRXIGDXNAYYA-164 [-1]) were required to enhance solubility and weaken binding potency in order to capture the transient “hit and run” interaction by NMR analysis, which is performed at high micromolar concentration(Gavathiotis et al., 2010; Gavathiotis et al., 2008). In contrast, for cellular work performed in the low micromolar range, the prototype BIM SAHBA (+2 charge) was optimal (Gavathiotis et al., 2008; LaBelle et al., 2012), given its enhanced target binding potency and capacity to penetrate intact cells.
Thus, as for our development of BIM and MCL-1 SAHBs for a diversity of applications(Gavathiotis et al., 2008; LaBelle et al., 2012; Stewart et al., 2010; Walensky et al., 2006), we initiated our synthetic efforts here by generating a panel of PUMA SAHB constructs (designated PUMA SAHBs A1-A8) of differential sequence composition, length and charge. In a screening fluorescence polarization binding assay against anti-apoptotic BCL-XLΔC, we observed a binding affinity range of 2.6–13 nM for this panel of PUMA SAHBA peptides (Figure 1A). Guided by our learnings from BIM SAHBA, we selected a compound with mid-range affinity bearing the most negative charge for in vitro biochemical and structural analyses, given our goal of evaluating functional interactions with BAX, in addition to the anti-apoptotic targets. For optimal comparison, we aimed to examine a PUMA SAHB bearing a staple in the same location used for our prior studies with BIM SAHBA. However, as for BIM SAHBs(Gavathiotis et al., 2008), we also wanted to confirm that any single staple location did not independently impact biochemical activity. Therefore, we also generated a panel of PUMA SAHBs A-L with differential localization of the hydrocarbon staple (Figure 1B). This “staple scanning” approach not only serves to dissociate any individual staple location from overall observed biochemical activity but also identifies the best binders and, importantly, those key residues and interfaces to avoid for staple placement, providing valuable structure-activity relationship information(Gavathiotis et al., 2008; Stewart et al., 2010). In each case, PUMA SAHBs with hydrocarbon staple placements that avoid the core hydrophobic binding interface and highly conserved BH3 residues, exhibit nanomolar BCL-XLΔC binding affinity in the 4–21 nM range (Figures 1B, S1B). In contrast, staples that replace key hydrophobic or charged residues of the binding interface, as found in PUMA SAHBs C, E, I, and J, manifest relatively impaired binding activity. As would be predicted, positioning the staple directly at the binding interface while also replacing the universally conserved leucine residue, as in PUMA SAHBH, is most detrimental. Finally, we sought to generate a negative control construct for PUMA SAHBA1 by swapping the location of an alanine residue at position 139 with the universally conserved leucine at position 141. Importantly, PUMA SAHBA1 and its A139L/L141A double point mutant, designated PUMA SAHBA1mut, exhibit similar, high percentage α-helical content, as assessed by circular dichroism (Figure 1C). When the corresponding FITC-derivatized analogs were subjected to anti-apoptotic protein binding analyses, PUMA SAHBA1 engaged BCL-2ΔC, BCL-XLΔC, BCL-wΔC, MCL-1ΔNΔC, and BFL-1/A1ΔC with dissociation constants in the 20–35 nM range, whereas A139L/L141A mutagenesis uniformly impaired binding activity, consistent with disruption of a major BH3 binding determinant. Thus, we find that hydrocarbon stapling of the PUMA BH3 domain can potently enforce α-helical structure (Figures 1C, S1A) and the library of constructs obey predicted structure-activity relationships based upon BH3 sequence composition. Notably, PUMA SAHBA1 engages in high affinity, sequence-specific binding interactions with all anti-apoptotic targets.
Figure 1. Sequence Composition and Anti-apoptotic Binding Activity of Hydrocarbon-stapled PUMA BH3 Peptides.
(A, B) A series of FITC-PUMA SAHBA peptides of differential BH3 domain length (A) and FITC-PUMA BH3 (131–153) constructs with varying staple positions (as reflected by the “staple walk” along the pictured helical wheel) (B) were synthesized and screened for BCL-XLΔC binding activity by fluorescence polarization assay. See also Figure S1B for separated views of the PUMA SAHB A1-L1 plots.
(C) Circular dichroism of PUMA SAHBA1 and its A139L/L141A mutant demonstrated the marked α-helical structure of both stapled peptides in aqueous solution. *, exceeds the calculated ideal α-helicity of an undecapeptide standard.
(D) FITC-PUMA SAHBA1 (solid lines) exhibits high affinity binding to the diversity of anti-apoptotic BCL-2 family proteins, whereas A139L/L141A point mutagenesis (dashed lines) markedly impairs binding activity, highlighting the BH3 sequence-dependence of PUMA SAHBA1 engagement. Data are mean ± S.D. for experiments performed in triplicate. X, stapling amino acid; B, norleucine.
See also Figure S1.
PUMA SAHB Directly Binds to BAX by Initial Interaction at the α1/α6 Trigger Site
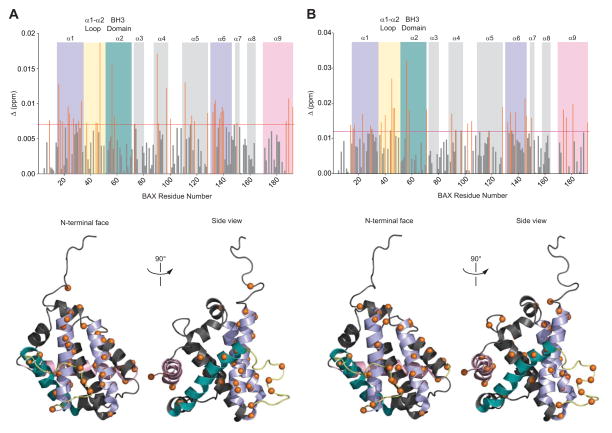
To determine if a PUMA BH3 helix could likewise directly bind to the pro-apoptotic class of BCL-2 family proteins, as previously observed for BIM SAHBs, we undertook an NMR analysis of 15N-BAX upon PUMA SAHB titration. Of note, the BAX-activating BIM BH3 interaction is transient, making the structural analysis of this dynamic process especially challenging(Gavathiotis et al., 2010; Gavathiotis et al., 2008). We thus employed PUMA SAHBA1, a relatively weaker binding PUMA SAHBA that also possessed the most negative charge (−2) to enhance solubility for high concentration NMR experiments. We observed weak but reproducible dose-responsive backbone amide chemical shift changes in a discrete subset of BAX residues localized within α1, the α1-α2 loop, and α6, corresponding to the previously identified binding site for a stapled BIM BH3 helix(Gavathiotis et al., 2008) (Figure 2A). Upon increasing the PUMA SAHBA1:15N-BAX ratio from 1:1 to 4:1, a series of chemical shift changes became more prominent in the α1-α2 loop (yellow), α2 (BH3) (teal), and α9 (pink) (Figure 2B), three regions previously implicated in BIM BH3-triggered N-terminal loop opening, BAX BH3 exposure, and C-terminal helix mobilization, respectively(Gavathiotis et al., 2010). These PUMA SAHB-induced allosteric changes are consistent with induction of a major conformational change that is transmitted from the N-terminal face through the hydrophobic core to the C-terminal face, resulting in functional activation of BAX.
Figure 2. NMR Analysis of 15N-BAX upon PUMA SAHBA1 Titration.
(A, B) At a 1:1 ratio of 15N-PUMA SAHBA1/BAX, a series of chemical shift changes localize to surface residues of α1 and α6, corresponding to the previously defined trigger site (purple) for BIM BH3-induced direct BAX activation (A). Allosteric changes involving residues of the α1-α2 loop (yellow), α2 (teal), and α9 (pink), as transmitted through the hydrophobic core (α4, α5), are also evident (A), and become more prominent upon titration up to a PUMA SAHBA1/BAX ratio of 4:1 (B). Cα atoms of BAX residues that undergo significant chemical shift change upon exposure to PUMA SAHBA1 are represented as orange bars on the plot (top) and orange spheres on the ribbon diagrams of monomeric BAX protein (calculated significance threshold >0.007 p.p.m. for 1:1 titration and >0.011 for 4:1 titration).
To probe the PUMA SAHB interaction site on BAX by a second and completely distinct experimental approach, we developed and applied photoreactive PUMA SAHBs (pSAHBs). pSAHBs contain differentially localized benzophenone moieties, which covalently trap both static and dynamic protein interactors upon UV irradiation and enable the explicit localization of intercalation sites using mass spectrometry methods, as previously reported(Braun et al., 2010). To validate the binding specificity of PUMA pSAHBs 1 and 2, we first conducted crosslinking analysis with anti-apoptotic BFL-1/A1ΔC, for which definitive structures are known. We found that the differentially-placed benzophenone moieties in each PUMA pSAHB crosslinked to discrete subregions of the canonical BH3 binding pocket (Figure S2), precisely corresponding to the established structure of a PUMA BH3/BFL-1/A1ΔC complex(Smits et al., 2008) (PDB ID# 2VOF). Having validated the fidelity of PUMA pSAHBs 1 and 2, we subjected full-length BAX to the crosslinking analysis. We found that PUMA pSAHB-1 almost exclusively crosslinked to BAX residues of the α1/α6 trigger site (Figure 3A), whereas PUMA pSAHB-2 reacted with residues at the α1/α6 trigger site and α3 residues of the canonical BH3-binding pocket (Figure 3B). BAX α9 effectively occludes the canonical BH3-binding pocket in the protein’s inactive state(Suzuki et al., 2000) and our NMR study only detected a confluence of protein surface chemical shift changes at the α1/α6 site upon ligand binding (Figure 2). Thus, the identified canonical pocket crosslinks are most likely indicative of a compatible secondary binding interaction that ensues upon allosteric release of α9 from the canonical pocket, which is induced by initial BH3 engagement of the α1/α6 trigger site. To explore this hypothesis, we repeated the PUMA pSAHB crosslinking studies on a mutant form of BAX in which the C-terminus was deleted (BAXΔC), thus exposing the canonical BH3-binding pocket from the outset. Strikingly, the balance of crosslinks completely shifted for both PUMA pSAHBs in favor of canonical pocket residues (Figures 3C, 3D). These data support a model in which exposure of the canonical BH3-binding pocket upon allosteric release of α9 reveals a secondary PUMA BH3 interaction site that may be reflective of a sequential BH3-mediated direct BAX activation mechanism.
Figure 3. BAX Interaction Site Analysis by PUMA SAHB Photoaffinity Labeling and Mass Spectrometry.
(A) PUMA pSAHB-1 was incubated with full-length BAX and the mixture subjected to UV irradiation, streptavidin pulldown, electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. A single arginine substitution (A150R) was made in PUMA pSAHB-1 to facilitate tryptic digestion into shorter and more identifiable fragments by MS. The plot (left) depicts the frequency of crosslinked sites identified across the BAX polypeptide sequence, with crosslinked residues mapped onto the solution structure of monomeric BAX (PDB ID 2K7W) (right) and colored according to the frequency of occurrence. The C-terminal α9 helix of BAX has been removed from the structure to better visualize the crosslinked residues of the canonical BH3-binding pocket.
(B–D) The corresponding analysis was performed for PUMA pSAHB-2 with full length BAX (B), PUMA pSAHB-1 with C-terminal helix-deleted BAX (BAXΔC) (C), and PUMA pSAHB-2 with BAXΔC (D). X, stapling amino acid; B, norleucine; U, 4-benzoyl-phenylalanine (Bpa).
See also Figure S2.
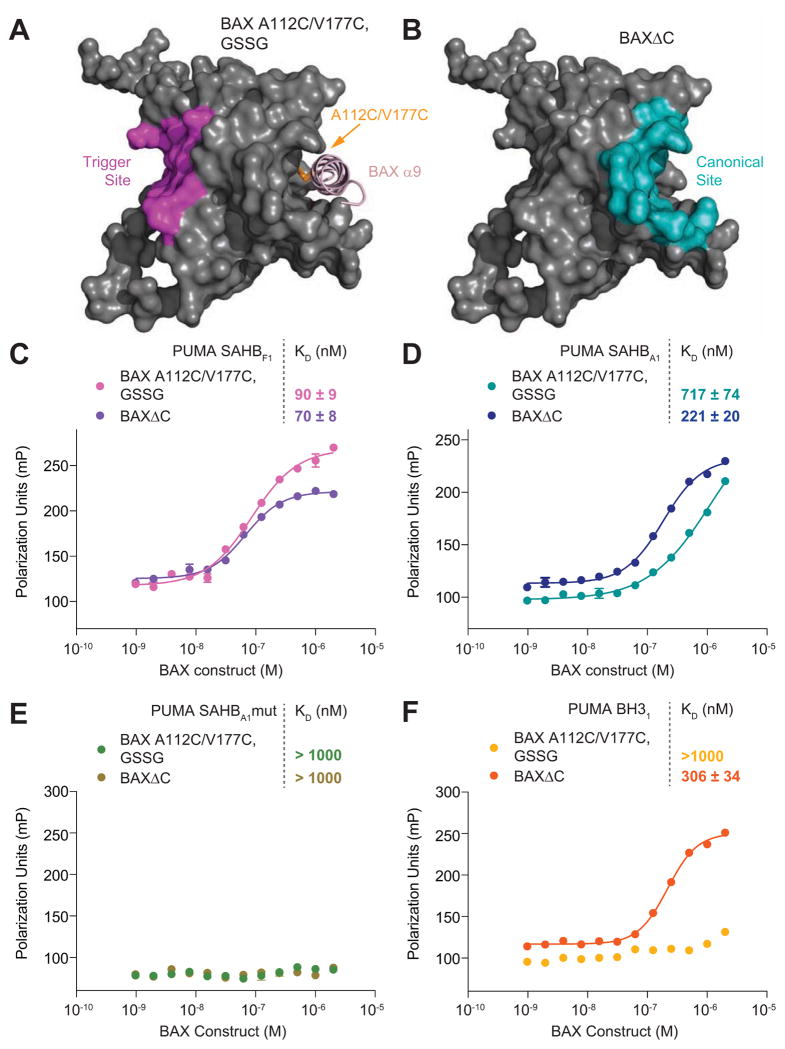
To further dissect the capacity of PUMA SAHBs to engage two discrete BH3-binding sites on BAX, we developed BAX constructs for independent evaluation of the α1/α6 and canonical BH3-binding sites. To isolate the α1/α6 trigger site at the N-terminal face of BAX, we made use of our previously described BAX A112C/V177C construct, which under oxidizing conditions locks the C-terminal α9 helix of BAX in place with an installed disulfide tether, occluding the canonical BH3-binding site (Figure 4A). We used BAXΔC to evaluate BH3-binding at the C-terminal site (Figure 4B), since we essentially observed no α1/α6 site crosslinking when the C-terminal helix is removed (Figure 3C, 3D), which is likely due to ready access to the canonical pocketas stably captured by X-ray crystallography(Czabotar et al., 2013) - and bypass of the native N-terminal triggering mechanism.
Figure 4. PUMA SAHBs Bind to Two Geographically Distinct Sites on BAX.
(A) Covalent tethering of the BAX C-terminal helix to its binding groove by an installed A112C/V177C disulfide bridge (GSSG, 0.5 mM) occludes the canonical BH3-binding surface of full-length BAX. In this context, only the α1/α6 trigger site is available for BH3 interaction.
(B) Both structural and MS analyses indicate that activator BH3 helices bind to the exposed canonical pocket of BAXΔC, a truncated BAX construct in which the C-terminal helix is removed. In this context, the canonical BH3-binding site on BAX is preferred.
(C) FITC-PUMA SAHBF1 binds to BAX A112C/V177C and BAXΔC with KDs of 90 and 70 nM, respectively, as measured by FP assay.
(D) FITC-PUMA SAHBA1 binds to BAX A112C/V177C and BAXΔC with KDs of 717and 221 nM, respectively.
(E) A139A/L141A mutagenesis of FITC-PUMA SAHBA1 abrogates BAX-binding activity, highlighting the sequence specificity of the measured FITC-PUMA SAHBA1 interactions with BAX A112C/V177C and BAXΔC.
(F) FITC-PUMA BH3 (aa 131–153) exhibited no binding activity toward BAX A112C/V177C, but engaged BAXΔC with a KD of 306 nM, suggesting that the exposed canonical pocket is more amenable to induced folding of an unstructured PUMA BH3 peptide.Data are mean ± S.D. for experiments performed in triplicate.
Each of the BAX constructs was applied in fluorescence polarization (FP) assays to probe the binding activities of FITC-PUMA SAHBs F1 and A1, which contain the F and A staples found in PUMA pSAHBs 1 and 2, respectively. Both FITC-PUMA SAHBs bound to BAX A112C/V177C and BAXΔC, indicating dual compatibility for the geographically distinct BH3-binding sites. Whereas FITC-PUMA SAHBF1 exhibited similar nanomolar affinity for each of the constructs (KDs: 90 nM, BAX A112C/V177C; 70 nM, BAXΔC), FITC-PUMA SAHBA1 showed a 3-fold preference for BAXΔC (KDs: 717 nM, BAX A112C/V177C; 221 nM, BAXΔC). Yet, in crosslinking experiments, the corresponding F- and A-stapled pSAHBs both crosslinked to α1/α6 residues in full-length BAX, with these N-terminal interactions nearly eliminated in favor of canonical site crosslinking upon removal of the C-terminus. These data support a mechanism in which activator BH3 engagement of cytosolic, full-length BAX occurs at the N-terminal site first, followed by a secondary compatible interaction at the canonical pocket once the C-terminal helix is released from its binding groove.
Negative control FP studies further highlighted the specificity of the observed FITC-PUMA SAHB binding activities, with FITC-PUMA SAHBA1 A139L/L141A showing no interaction with either BAX A112C/V177C or BAXΔC (Figure 4E). Interestingly, the unmodified FITC-PUMA BH3 peptide (aa 131–153) demonstrated no binding activity toward BAX A112C/V177C, but displayed nanomolar binding affinity for BAXΔC (Figure 4F). These data mirror the results of our very first comparative binding analyses of unmodified and hydrocarbon-stapled BID BH3 peptides with full-length and C-terminally deleted BAX(Walensky et al., 2006). The unstructured BID BH3 peptide could readily bind by induced folding into the exposed C-terminal deep pocket of BAXΔC, but a prefolded BH3 helix in the form of BID SAHB was required for interaction with full-length BAX. This unexpected result suggested that BH3-binding to full-length BAX was somehow different than that observed for BAXΔC. We considered the possibility that the prefolded BID SAHB could be more effective than the unfolded peptide at displacing the BAX C-terminus to achieve canonical site binding(Walensky et al., 2006). If such competition with an intramolecular helix was not energetically feasible, as suggested by the original NMR analysis of full-length BAX(Suzuki et al., 2000), we alternatively hypothesized that a different interaction site could exist, which ultimately led to our discovery of the more shallow BH3-compatible binding site at the N-terminal surface of BAX(Gavathiotis et al., 2008). Importantly, the inability of the unmodified FITC-PUMA BH3 peptide to engage oxidized BAX A112C/V177C, but readily bind to BAXΔC, provides further evidence that the two BAX constructs indeed present two geographically distinct BH3-binding surfaces.
PUMA SAHB Directly Activates BAX-mediated Pore Formation
To link the direct interaction between PUMA SAHB and BAX to its functional activation, we employed a reductionist liposomal system that measures BAX pore formation in a membrane environment upon the addition of discrete reagents. Whereas treatment with BAX, PUMA SAHBA1, or its A139L/L141A mutant alone had little to no effect on the liposomes, the combination of BAX and PUMA SAHBA1 yielded dose-responsive liposomal release of entrapped fluorophore (Figure 5A). Point mutagenesis of the PUMA BH3 interacting surface completely abrogated the effect (Figure 5B), underscoring the sequence-based specificity of PUMA SAHBA1 activity. Likewise, in the context of BAX/BAK-deficient mitochondria isolated from Alb-creposBaxflox/−Bak−/− mice, dose-responsive release of cytochrome c was only observed upon incubation with the combination of BAX and PUMA SAHBA1. Again, the specificity of PUMA SAHBA1 activity was confirmed by impairment of BAX-mediated cytochrome c release upon SAHB mutagenesis. Whereas in the mitochondrial assay, the effect of PUMA SAHBA1 on BAX activity can be amplified by simultaneous inhibition of membrane-embedded anti-apoptotic proteins, the requirement of PUMA SAHBA1 to initiate BAX activation in both assays underscores the functional role of the direct PUMA SAHB/BAX binding interaction in triggering the membrane release activity of BAX.
Figure 5. PUMA SAHBA1 Triggers BAX-mediated Liposomal and Cytochrome c Release.
(A) PUMA SAHBA1 triggered dose-responsive BAX-mediated liposomal release of entrapped fluorophore (left), whereas BAX or PUMA SAHBs alone, or the combination of PUMA SAHBA1mut with BAX (right), had little to no effect. Data are mean ± S.D. for experiments performed in triplicate and repeated twice.
(B) BAX/BAK-deficient mitochondria dose-responsively released cytochrome c upon incubation with recombinant BAX and increasing amounts of PUMA SAHBA1, whereas BAX or PUMA SAHBs alone, or the combination of PUMA SAHBA1mut with BAX, had little to no effect. Data are mean ± S.D. for experiments performed in sextuplicate and repeated three times.
PUMA SAHB Induces Caspase 3/7 Activation and Cell Death by Engagement of Anti- and Pro-apoptotic Targets
We next aimed to correlate our in vitro biochemical and structural findings to PUMA BH3 activity in cells. Among the BH3-only proteins, PUMA has been implicated as a major driver of apoptosis induction in neurons and neuronal precursor cells(Akhtar et al., 2006; Reimertz et al., 2003; Steckley et al., 2007; Wyttenbach and Tolkovsky, 2006). Therefore, we examined the pro-apoptotic activity of PUMA SAHBA1 in neuroblastoma cells, which can mount a substantial and clinically-relevant apoptotic blockade(Goldsmith et al., 2012). We first screened PUMA SAHBA1 for cellular uptake by treating SH SY5Y neuroblastoma cells with its FITC-derivatized analog for six hours, followed by trypsinization, repeated washing, cellular lysis, electrophoresis, and fluorescence scan detection. Of note, we initially test for cell permeability in serum-free media to avoid potential effects of serum binding on peptide exposure(Bird et al., 2011; Braun et al., 2010; Pitter et al., 2008). A comparison of lysates from cells treated with the unmodified FITC-PUMA BH3 peptide and the corresponding A-stapled analog revealed no significant intracellular peptide fluorescence (Figure 6A), consistent with our prior observations that stapling of peptides alone does not uniformly confer cellular penetrance, particularly when the overall charge of the construct is less than 0 (Bernal et al., 2007; Bird et al., 2011; Gavathiotis et al., 2008; Stewart et al., 2010), as is the case with PUMA SAHBA1. To facilitate cellular penetrance, we previously adjusted the overall charge of α-helical stapled peptides from <0 to the 0 to +2 range either by converting aspartic and glutamic acid residues to asparagines and glutamines(Bernal et al., 2007), or by shifting the sequence composition to eliminate N- or C-terminal negatively charged residues and/or include N- or C-terminal positively charged residues(Gavathiotis et al., 2008; Stewart et al., 2010; Walensky et al., 2004). In this case, we eliminated the N-terminal glutamic acid of PUMA SAHBA1 and appended two native C-terminal arginines, converting the overall peptide charge from −1 to +2, while maintaining high α-helical content (Figure S3A) and analogous binding activity (Figure 1A). When the FITC-derivatized analog of this alternate construct, PUMA SAHBA2, was subjected to cellular uptake analysis, a robust fluorescent band was detected in the lysate of treated cells, whereas the corresponding unmodified peptide again showed no uptake (Figure 6B). As previously observed for a variety of cell-permeable stapled peptides(Bernal et al., 2007; Takada et al., 2012; Walensky et al., 2004), the uptake was time-dependent and the intracellular peptide proteolytically stable (Figure 6C). Live cell confocal microscopy of FITC-PUMA SAHBA2-treated SH SY5Y cells further confirmed peptide uptake (Figure 6D).
Figure 6. Development and Evaluation of a Cell-Permeable PUMA SAHB.
(A, B) SH SY5Y cells were treated with the indicated FITC-PUMA BH3 peptides (5 μM) in serum-free media for 6 hours, followed by washing, trypsinization, cellular lysis, electrophoresis of the isolated supernatant, and fluorescence detection. Whereas little to no unmodified PUMA BH3 peptide or PUMA SAHBA1 fluorescence was detected in the corresponding cellular lysates, PUMA SAHBA2 demonstrated robust fluorescence.
(C) The cellular uptake of FITC-PUMA SAHBA2 was time-dependent, as reflected by a progressive increase in intracellular peptide that co-migrated with FITC-PUMA SAHBA2 marker peptide.
(D) Live cell confocal microscopy of SH SY5Y cells treated with FITC- PUMA SAHBA2 demonstrated its intracellular distribution. Prior to imaging, the cells were treated with Hoechst dye (blue) to label nuclei and CellMask Orange (orange-red) to delineate the plasma membrane. Original magnification, 100×.
(E) Cellular uptake of FITC-PUMA SAHBA2 was dose-responsively reduced by the addition of fetal bovine serum (FBS; 0%, 2.5%, 5%, and 10% [v/v]) to the tissue culture medium.
(F) Addition of the indicated amounts of serum to PUMA SAHBA2-treated SH SY5Y cells during the initial 6 hour incubation period, dose-responsively reduced cytotoxicity, as measured at 24 hours (18 hours after serum replacement to 10% [v/v]) by CellTiter-Glo assay. Incubating SH SY5Y cells with vehicle and media containing 0%, 2.5%, 5%, or 10% serum for 6 hours, followed by serum replacement to 10% (v/v), had no independent effect on cell viability, as measured by CellTiter-Glo at 24 hours. Data are mean ± S.D. for experiments performed in triplicate.
See also Figure S3.
Whereas some stapled peptides manifest robust cellular uptake and bioactivity in serum-containing tissue culture media(Brown et al., 2013; Danial et al., 2008; Takada et al., 2012), others demonstrate reduced uptake and activity in the presence of serum, necessitating a serum-free window to facilitate cellular exposure(Bernal et al., 2007; LaBelle et al., 2012). Here, we find that the addition of serum dose-responsively impaired the uptake of PUMA SAHBA2 by SH SY5Y cells (Figure 6E), with optimal peptide loading achieved in serum-free conditions by 6 hours (Figure 6C). The level of intracellular peptide detected at 6 hours correlated with the observed cytotoxicity, as measured after an additional 18 hour incubation in the presence of 10% serum (Figure 6F). Importantly, varying the serum concentration from 0% to 10% during the first 6 hours followed by full serum replacement had no independent effect on cell viability (Figure 6F). Although this does not rule out the possibility that viability impairment is enhanced by the combination of serum withdrawal and PUMA SAHBA2 treatment, the dose-responsive decrease in PUMA SAHBA2 uptake appears to be the predominant mechanistic effect of serum in shifting the cellular response curves to the right (Figures 6E and 6F).
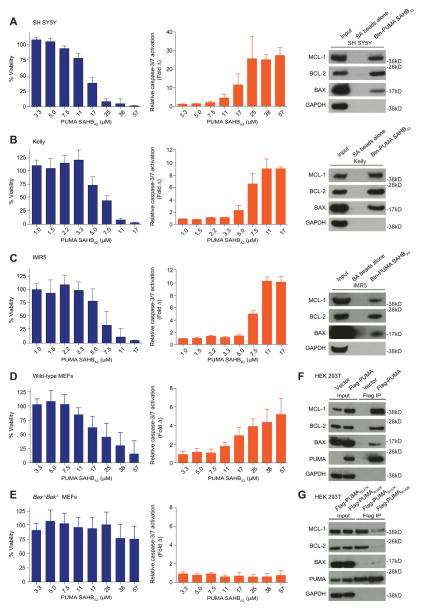
With a cell permeable version of PUMA SAHB in hand and practical treatment conditions defined, we exposed a series of neuroblastoma cells, including SH SY5Y, Kelly, and IMR5, to PUMA SAHBA2 in the absence of serum, followed by full serum replacement at 6 hours and cell viability measurement at 24 hours. In each case, a dose-responsive impairment in cell viability was observed (Figure 7A–C, left), which correlated with dose-responsive caspase 3/7 activation, as measured at 8 hours (Figure 7A–C, middle). To investigate whether the observed caspase 3/7 activation and cell death correlated with PUMA SAHB engagement of BCL-2 family targets, we incubated cellular lysates from each of the three cell lines with an N-terminal biotinylated analog of PUMA SAHBA, followed by streptavidin pull-down, and western analysis. In each case, a diverse spectrum of BCL-2 family targets was engaged, as reflected by PUMA SAHB pull-down of anti-apoptotic BCL-2 and MCL-1, and pro-apoptotic BAX (Figure 7A–C, right). Taken together, these data demonstrate that cell permeable PUMA SAHBA2 can activate the death pathway in neuroblastoma cells, and this biological activity correlates with the capacity of a biotinylated PUMA BH3 helix to bind both anti- and pro-apoptotic BCL-2 family members.
Figure 7. Pro-apoptotic Activity and Multimodal Interactions of PUMA BH3.
(A–C) PUMA SAHBA2 dose-responsively impaired the viability of a series of neuroblastoma cell lines including, SH SY5Y, Kelly, and IMR5, as measured by CellTiter-Glo at 24 hours after treatment (left column). The observed decrease in cell viability corresponded to dose-responsive caspase 3/7 activation, as measured by Caspase-Glo at eight 8 hours after treatment (middle column). Data are mean ± S.D. for experiments performed in triplicate. Incubation of cellular lystates with biotinylated PUMA SAHBA3, followed by streptavidin pull-down, washing, electrophoresis of eluates, and BCL-2 family western analysis, documented engagement of native MCL-1, BCL-2, and BAX in all three neuroblastoma lines (right column). (D, E) PUMA SAHBA2 dose-responsively impaired the viability of wild-type mouse embryonic fibroblasts (MEFs), but had little to no effect on Bax−/−Bak−/− MEFs, as measured by CellTiter-Glo at 24 hours after treatment (left column). Correspondingly, dose-responsive caspase 3/7 was observed in wild-type MEFs but not in Bax−/−Bak−/− MEFs, as measured by Caspase-Glo at 8 hours after treatment (right column). Data are mean ± S.D. for experiments performed in triplicate and repeated four times.
(F, G) Flag-Pumaα, its R142E or D147K mutants, or vector control was transiently transfected into human embryonic kidney 293T (HEK 293T) cells and lysates subjected to immunoprecipitation using Flag affinity beads, followed by washing, electrophoresis of eluates, and anti-MCL-1, BCL-2, BAX, PUMA, and GAPDH western analysis.
See also Figure S4.
Specificity and Physiologic Relevance of PUMA SAHB Activity
An important measure of pro-apoptotic specificity for ligands designed to operate via the BCL-2 family pathway is whether death induction occurs in a BAX/BAK-dependent fashion. Thus, we treated wild-type and Bax−/−Bak−/− mouse embryonic fibroblasts (MEFs) with PUMA SAHBA2 and measured cell viability and caspase 3/7 activation, as performed for the neuroblastoma cell lines. Albeit at a higher dosing range, PUMA SAHBA2 dose-responsively impaired cell viability of wild-type MEFs (Figure 7D, left), with caspase 3/7 activation observed at the corresponding doses that induced cell death (Figure 7D, right). In contrast, little to no effect of PUMA SAHBA2 was observed in Bax−/−Bak−/− MEFs, with cell viability maintained in the 80–100% range (Figure 7E, left) and no activation of caspase 3/7 observed (Figure 7E, right).
We next explored whether or not the protein interactions of stapled PUMA BH3 helices, and their documented functional consequences, bear on the physiologic interactions of full-length PUMA protein. Flag-Pumaα was transiently transfected into human embryonic kidney (HEK) 293T cells and immunoprecipitated using anti-Flag resin. As observed for biotinylated PUMA SAHBA3, Flag-PUMA engaged a diversity of anti- and pro-apoptotic BCL-2 family targets, as reflected by western analysis detection of co-precipitated BCL-2, MCL-1, and BAX (Figure 7F). As for the PUMA SAHB pull down (Figure 7A–C, right), the relative ratio of Flag-PUMA pull down to protein input was greater for anti-apoptotic BCL-2 and MCL-1 than pro-apoptotic BAX, reflecting either (1) the more stable nature of BH3/anti-apoptotic interactions compared to BH3/pro-apoptotic interactions or (2) a frank preference for PUMA BH3 engagement of anti-apoptotic targets, and once this reservoir is saturated, direct activation of pro-apoptotic targets can ensue.
Finally, we investigated whether disrupting key charge-charge interaction pairs between the PUMA BH3 domain and its binding sites on multidomain BCL-2 family proteins could be harnessed to probe the selectivity of PUMA interactions. PUMA shares striking sequence homology with BIM and BAX in the core BH3 domain sequence, as reflected by the hydrophobic and electrostatic distribution in the consensus L(K/R)R(I/M)(A/G)D(D/E) (Figure S4A). The first positively charged residue of the consensus consistently engages in a conserved electrostatic interaction with a negatively charged residue at both canonical and BAX α1/α6 BH3-binding sites, as exemplified by PUMA R142/MCL-1 D237, BAX K64/BCL-2 D140, BAX K64/BAX D102, and BIM R153/BAX E131 in the corresponding structures (Figure S4B–E). However, the second negatively charged residue of the consensus is less commonly engaged in a complementary electrostatic pairing at canonical sites (e.g. PUMA R142 in the PUMA BH3/MCL-1 complex, BAX E69 in the BAX BH3/BAX complex) (Figure S4B,D), but explicitly engages K21 at the BAX α1/α6 trigger site (e.g. BIM E158/BAX K21 in the BIM BH3/BAX complex) (Figure S4E). Indeed, we previously found that BAX K21E mutagenesis slows (but does not wholly abrogate) the kinetics of direct BAX activation by BIM and BAX BH3 helices in vitro, and in Bax−/−Bak−/− mouse embryonic fibroblasts (MEFs) reconstituted with BAX K21E and exposed to staurosporine(Gavathiotis et al., 2010; Gavathiotis et al., 2008). In addition, recombinant BAX is unable to undergo heat-induced oligomerization in solution upon K21E (trigger site) or E69K (BH3 domain) mutagenesis, but activity is fully restored in the double K21E, E69K mutant, further underscoring the functional relevance of activator BH3 domain interaction at the α1/α6 trigger site in initiating and propagating BH3-mediated direct activation of cytosolic BAX.
Here we find that PUMA R142E mutagenesis eliminates co-immunoprecipitation with MCL-1, BCL-2, and BAX, whereas PUMA D147K mutagenesis only disrupts the BAX interaction (Figure 7G). These data reinforce that PUMA engages BCL-2 family multidomain canonical pockets and the BAX α1/α6 site, as reflected by binding sensitivity to PUMA R142E mutagenesis, which establishes a charge-charge repulsion with MCL-1 D237 and, by analogy, with BCL-2 D140 and BAX D102 at the canonical sites, and BAX E131 at the α1/α6 site (Figure S4B–E). In contrast, PUMA D147 projects away from the canonical pocket of MCL-1 (Figure S4B), and also has no electrostatic pairing at the BAX canonical site (Figure S4D), yet would be predicted to interact with the functionally relevant K21 residue at the BAX α1/α6 site based on the calculated model structure of a BIM BH3 helix bound to full-length BAX (Figure S4E). Indeed, we find that PUMA D147K mutagenesis selectively impairs co-immunoprecipitation with native BAX but spares the interactions with anti-apoptotic BCL-2 and MCL-1 (Figure 7G). Further, the finding that PUMA D147K mutagenesis alone disrupts BAX binding is consistent with our NMR, crosslinking, and FP analyses, all of which indicate that the site of first contact between PUMA BH3 and full-length BAX is at the α1/α6 trigger site. Taken together, our data demonstrate that PUMA BH3 harbors the biochemical capacity to engage both anti- and pro-apoptotic BCL-2 family proteins in a sequence-specific manner and, depending on the cellular context, may harness this multimodal interaction potential to activate the apoptotic pathway.
DISCUSSION
PUMA is a potent pro-apoptotic BH3-only protein implicated in a variety of apoptotic signaling pathways and subject to transcriptional and post-translational regulation(Jeffers et al., 2003). Like BIM, PUMA broadly interacts with BCL-2 family anti-apoptotic proteins, and the two proteins have been shown to compensate for and cooperate with one another in a variety of physiologic death pathways(Erlacher et al., 2006; Erlacher et al., 2005; Garrison et al., 2012; Gray et al., 2012). In addition, PUMA has recently been shown to promote apoptosis by selectively displacing p53 from anti-apoptotic BCL-XL via an allosteric mechanism(Follis et al., 2013). Published biochemical, cellular, and in vivo studies differ, however, on whether a component of PUMA’s pro-apoptotic potency also derives from its capacity to directly engage pro-apoptotic proteins, as demonstrated for BID and BIM. Notably, alignment of BCL-2 family BH3 domains reveals that PUMA BH3 is most similar to the BH3 domains of BIM and BAX in the very region implicated in direct interaction at the recently identified α1/α6 trigger site of BAX (Figure S4A). Thus, based on the defined BIM BH3 triggering and BAX BH3 auto-propagating interactions at the α1/α6 site(Gavathiotis et al., 2010; Gavathiotis et al., 2008), it seemed likely that PUMA BH3 could likewise bind BAX at this same location. Indeed, by a series of distinct methods, including NMR spectroscopy, mass spectrometry, fluorescence polarization binding, and co-immunoprecipitation analyses, we identified PUMA BH3 interaction at the α1/α6 interface of full-length BAX. We linked this binding event to direct and functional activation of BAX in both liposomal and mitochondrial assay systems, in which PUMA SAHB dose-responsively triggered BAX-mediated release activity in a BH3 sequence-dependent manner. The physiologic relevance of these structural and biochemical findings are supported by the capacity of PUMA SAHB and PUMA protein to likewise engage native anti-apoptotic BCL-2 family proteins and pro-apoptotic BAX from cells. Taken together, these data support a model in which PUMA can function as a direct activator of BAX, consistent with a recent study documenting that in the absence of the direct activators BID, BIM, and p53, PUMA was sufficient to activate BAX(Garrison et al., 2012).
In mapping the PUMA BH3 interaction site by pSAHB-based photoaffinity labeling, the majority of crosslinked residues in full-length BAX localized to the α1/α6 trigger site, yet a small subset included residues of the canonical BH3-binding pocket. As the initial chemical shift changes observed by NMR upon PUMA SAHBA1 binding localize to surface residues of the α1/α6 trigger site, we conclude that allosteric release of α9 upon triggering at the N-terminal site exposes the canonical pocket, which is likewise compatible with PUMA BH3 binding. Indeed, when we performed the MS-based site identification analyses with C-terminally deleted BAX, the PUMA pSAHB crosslinking pattern shifted to predominantly involve residues of the canonical pocket. This dual but sequential compatibility of PUMA BH3 binding at distinct sites on BAX was further supported by FP analyses using two constructs of BAX that favor interaction either at its N-terminal trigger site or canonical pocket. A similar compatibility for activator BH3 domains at the traditional BH3-binding pocket was also observed for C-terminally deleted BAX by X-ray crystallography(Czabotar et al., 2013). It is noteworthy that these structural anaylses of select activator BH3 domains in stable complex with BAXΔC only captured the canonical pocket interaction, just as our MS analyses with BAXΔC markedly favored crosslinking to canonical site residues. However, in the context of the full-length BAX protein, our NMR, MS, and binding analyses demonstrate primary BH3 engagement of the α1/α6 trigger site, a key regulatory feature that is missed when studying C-terminally truncated BAX protein. Interestingly, a series of recent studies performed with full-length(Leshchiner et al., 2013) and C-terminally deleted(Dai et al., 2011; Moldoveanu et al., 2013) BAK protein, which in its native state constitutively resides in the outer mitochondrial membrane, identified direct BH3 interactions exclusively at the canonical site. Taken together, these data suggest that activator BH3-engagement at the canonical pocket may represent a common mechanism for propelling the activation of mitochondrial-localized BAK and BAX, with the α1/α6 triggering mechanism of BAX representing a unique afferent step required to regulate the activation and mitochondrial translocation of cytosolic BAX(Walensky, 2013a, b).
The stability and high affinity of BH3-only interactions with anti-apoptotic BCL-2 family proteins suggests that upon BH3-only deployment in response to cellular stress, the principal wrestling site between pro-life and pro-death forces may reside in these neutralizing complexes. However, with continued pro-apoptotic signaling, displacement of activator BH3-only proteins from anti-apoptotics or depletion of the anti-apoptotic reservoir, produces a BID/BIM/PUMA overflow that directly activates BAX and BAK at their respective BH3 trigger sites. Hydrocarbon-stapled peptides modeled after the BH3 domain helices of BCL-2 family proteins have proven to be high fidelity tools for dissecting physiologically-relevant binding interfaces and mechanisms (Gavathiotis et al., 2010; Gavathiotis et al., 2008; Stewart et al., 2010; Walensky et al., 2006), while also serving as prototype therapeutics for the modulation of apoptosis in vivo(LaBelle et al., 2012; Walensky et al., 2004). Whereas BAD and NOXA SAHBs recapitulate the finely-tuned selectivities of the corresponding native BH3-only proteins(Stewart et al., 2010; Walensky et al., 2006), BID, BIM, and PUMA SAHBs manifest the broad anti-apoptotic targeting capacity and direct BAX and BAK triggering activity of their cognate proteins(Gavathiotis et al., 2008; Leshchiner et al., 2013; Walensky et al., 2006). Importantly, the very potency of PUMA and the capacity of its BH3 helix to engage native anti- and pro-apoptotic BH3-binding pockets, suggests that pharmacologic mimicry of this dual functionality carries the potential to reactivate apoptosis in the context of resistant cancer, as observed here for neuroblastoma. Indeed, the BH3-binding pockets of both anti- and pro-apoptotic BCL-2 family proteins are bona fide drug targets. Whereas therapeutic targeting of the anti-apoptotic BCL-2/BCL-XL or BCL-2 canonical grooves in the form of ABT-737/ABT-263 and ABT-199, respectively, has led the way for “inhibiting the inhibitors” of apoptosis(Oltersdorf et al., 2005; Souers et al., 2013; Tse et al., 2008), new approaches to “activating the activators” by simulating BID, BIM and PUMA engagement of BH3 trigger sites on BAX/BAK are underway(Gavathiotis et al., 2012; LaBelle et al., 2012) and may add a new dimension to the growing arsenal of BCL-2 family modulators.
SIGNIFICANCE
BH3-only proteins are critical modulators of mitochondrial apoptosis and their array of protein interactions are providing blueprints for the development of novel therapeutics to manipulate the death pathway for clinical benefit. The BH3-only proteins BID and BIM have been implicated in direct triggering interactions with multidomain pro-apoptotic BCL-2 family members such as BAX and BAK, but whether PUMA is also a member of this direct activator group has been a longstanding controversy in the cell death field. We sought to address this issue using multidisciplinary chemical biology approaches, including NMR structural analysis, photoreactive stapled peptide crosslinking and MS-based binding site identification, and correlative biochemical and cellular assays. We report that a PUMA BH3 helix directly binds to BAX at the novel α1/α6 trigger site, resulting in its functional activation. Once the α9 helix of BAX is allosterically released from its binding pocket, the canonical BH3-binding groove of BAX is also compatible with PUMA BH3 binding. These data provide direct evidence of a sequential binding mode for activator BH3 domains in (1) triggering the activation and mitochondrial translocation of BAX, and then (2) propelling the activated state at the mitochondrial membrane to facilitate oligomerization. We validate our in vitro findings in the cellular context, demonstrating that (1) a cell permeable PUMA BH3 helix induces caspase 3/7 activation and cell death of neuroblastoma cell lines, and (2) both biotinylated PUMA BH3 helix and expressed full-length PUMA protein engage native anti-apoptotic and pro-apoptotic proteins in cells. Thus, we provide structural, biochemical and cellular evidence for how PUMA, a critical pro-apoptotic mediator of p53-dependent and -independent apoptotic responses, transmits its death message through multimodal protein interaction. PUMA mimetics with dual capacity to “inhibit the inhibitors” and directly “activate the activators” of cell death hold promise to overcome the apoptotic resistance of human cancers.
EXPERIMENTAL PROCEDURES
Stapled Peptide Synthesis and Characterization
SAHBs were synthesized, derivatized, purified to >95% homogeneity by LC/MS, quantified by amino acid analysis, and subjected to circular dichroism (Aviv Biomedical spectrophotometer) in 5 mM potassium phosphate buffer, pH 7.4, as previously described(Bird et al., 2008; Bird et al., 2011; Braun et al., 2010). All BH3 peptides generated and applied in the biochemical, structural, and cellular analyses are listed in Table S1.
Recombinant Protein Production
BCL-2ΔC, BCL-XLΔC, BCL-wΔC, MCL-1ΔNΔC, BFL1ΔC, BAXΔC and full-length BAX proteins were produced and purified as described(Gavathiotis et al., 2008; LaBelle et al., 2012). Briefly, BCL-2ΔC, BCL-XLΔC, BCL-wΔC, and MCL-1ΔNΔC were expressed as glutathione-S-transferase (GST) fusion proteins in Escherichia coli BL21 (DE3) from the pGEX2T vector (Pharmacia Biotech) and purified by affinity chromatography using glutathione sepharose beads (GE Healthcare), followed by thrombin cleavage of the GST tag. GB1-BFL1ΔC-His was similarly expressed in Escherichia coli BL21 (DE3) from the pGEV2 vector, purified by affinity chromatography using nickel-NTA agarose beads (Qiagen), and eluted according to the manufacturer’s instructions. BAX, BAXΔC, and BAX A112C/V177C were expressed in Escherichia coli BL21 (DE3) from the pTYB1 vector and purified by affinity chromatography using chitin beads (New England BioLabs), and the chitin tag cleaved by overnight incubation in 50 mM dithiothreitol. In each case, pure, monomeric protein was isolated by gel filtration FPLC.
Fluorescence Polarization Binding Assay
FITC-PUMA SAHBs (50 nM) were added to serial dilutions of recombinant protein in binding buffer (anti-apoptotic binding: 100 mM NaCl, 50 mM Tris, pH 8.0; BAX binding: 140 mM NaCl, 50 mM Tris, pH 7.4) in 96-well black opaque plates. For binding assays employing BAX A112C/V177C, the protein stock and binding buffer was supplemented with GSSG (0.5 mM). The plates were incubated in the dark at room temperature and then fluorescence polarization measured at 20 minutes on a microplate reader (Spectramax M5 Microplate Reader, Molecular Devices). Dissociation constants (KD) were calculated by nonlinear regression analysis of dose-response curves with Prism software 5.0 (GraphPad).
NMR analysis
Uniformly 15N-labeled full-length human BAX was generated as previously described(Gavathiotis et al., 2008; Suzuki et al., 2000). Protein samples were prepared in 25 mM sodium acetate, 50 mM NaCl solution at pH 6.0 in 5% D2O. PUMA SAHBA1 (6 mM stock) was titrated into a solution of 50 μM BAX to achieve the indicated molar ratios. Correlation 1H-15N HSQC spectra(Grzesiek and Bax, 1993) were acquired at 25°C on a Bruker 800 MHz NMR spectrometer equipped with a cryogenic probe, processed using NMRPipe(Delaglio et al., 1995), and analyzed with NMRView(Johnson, 2004). The weighted average chemical shift difference Δ at the indicated molar ratio was calculated as [{(ΔH)2 + (ΔN/5)2}/2]1/2 in p.p.m. The absence of a bar indicates no chemical shift difference, or the presence of a proline or residue that is overlapped or not assigned. BAX cross-peak assignments were applied as previously reported(Suzuki et al., 2000). The significance threshold for backbone amide chemical shift changes was calculated based on the average chemical shift across all residues plus the standard deviation, in accordance with standard methods(Marintchev et al., 2007).
Binding Site Identification by pSAHB Photoaffinity Labeling and Mass Spectrometry
Recombinant protein (10 μM) and biotinylated pSAHB (30 μM) were mixed and then irradiated (365 nm, Spectroline Handheld UV Lamp Model En280L, Spectronics Corp.) for 1.5 hours on ice. Unreacted peptide was removed from the irradiated samples by overnight dialysis at 4°C in dialysis buffer (200 mM NaCl, 50 mM Tris, pH 7.5) using 6–8 kD molecular weight cutoff D-Tube dialyzers (EMD Biosciences). Biotinylated species were captured by incubation with high-capacity streptavidin agarose (Thermo) for 2 hours at 4°C. Streptavidin beads were sequentially washed at room temperature three times each in 1% SDS in PBS, 1 M NaCl in PBS, and 10% ethanol in PBS, boiled 2 × 10 minutes in a 10% SDS solution (Promega) containing D-biotin (10 mg/ml), the eluates electrophoresed using 4%–12% gradient Bis-Tris gels (Invitrogen), and then stained for protein content by Coomassie Blue. The crosslinked bands were excised and prepared for mass spectrometry analysis as previously described(Braun et al., 2010). Samples were subjected to LC-MS/MS in an LTQ Orbitrap XL hybrid mass spectrometer (ThermoFisher, San Jose, CA). MS/MS spectra were searched using the SEQUEST algorithm(Yates et al., 1995) against a partially tryptic database of the relevant protein. Benzophenone-crosslinked tryptic fragments were identified by using the mass of the tryptic fragment of the pSAHB peptide as a required modification. Reversed protein sequences were used to generate an estimate of the false discovery rate. Identified crosslinked peptides were filtered based on tryptic state, charge state, mass accuracy, and number of crosslinks per peptide.
Liposomal Release Assay
Liposomes were generated and applied in BAX-mediated release assays as described(Gavathiotis et al., 2010; Yethon et al., 2003). Briefly, a lipid mixture composed of a 48:28:10:10:4 molar ratio of phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, dioleoyl phosphatidylserine, and tetraoleolyl cardiolipin (Avanti Polar Lipids) in chloroform was generated to mimic the lipid composition of the outer mitochondrial membrane. Solvent was removed by evaporation under nitrogen gas and then by vacuum for two hours. Lipid films were hydrated in liposome buffer (150 mM KCl, 20 mM HEPES, pH 7.0) and mixed with 12.5 mM ANTS fluorophore and 45 mM DPX quencher. After five freeze-thaw cycles, the mixture was extruded through 100 nm nucleopore polycarbonate membranes (Avestin) and intact liposomes purified by gel filtration chromatography using a 10 mL Sepharose CL2B column (GE Healtcare). To monitor ligand-triggered BAX-mediated release, liposomes were incubated in liposome buffer with the indicated amounts of recombinant monomeric BAX and PUMA SAHBs in black opaque 96 well plates (total reaction volume, 100 μL). ANTS/DPX release was monitored over a 120 minute period in a spectrofluorometer (Tecan Infinite M1000) using an excitation wavelength of 355 nm, an emission wavelength of 540 nm, and a bandwidth of 20 nm. Maximal release was determined by addition of Triton X-100 to a final concentration of 0.2% (v/v). Percent release was calculated as [(F−F0)/(FTriton−F0)] ×100, where F is the observed release, and F0 and F100 are baseline and maximal fluorescence, respectively.
Cytochrome c Release Assay
Mouse liver mitochondria (1.0 mg/mL) were isolated from Alb-creposBaxfl/− Bak−/− mice as described(Pitter et al., 2008), and incubated with the indicated combinations of recombinant BAX and PUMA SAHBs at 37°C for 40 minutes. The pellet and supernatant fractions were isolated by centrifugation, and released cytochrome c was quantitated using a colorimetric ELISA assay (R&D Systems). Percent cytochrome c released into the supernatant (%cytocsup) from releasable mitochondrial pools was calculated according to the following equation: %cytoc=[(cytocsup−cytocbackgr)/(cytoctotal−cytocbackgr)]×100, where background release represents cytochrome c detected in the supernatant of vehicle-treated samples and total release represents cytochrome c measured in 1% Triton X-100-treated samples.
Cell Culture
Wild-type (WT) and Bax−/−Bak−/− (DKO) mouse embryonic fibroblasts (MEFs) and human embryonic kidney (HEK) 293T cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS, 100 U/mL penicillin/streptomycin, 2 mM L-glutamine, 0.1 mM nonessential amino acids, and 50 μM β-mercaptoethanol. Neuroblastoma cell lines SH SY5Y, Kelly, and IMR5 were maintained in RPMI 1640 GlutaMAX (Invitrogen) supplemented with 10% FBS and 100 U/mL penicillin/streptomycin.
Cellular Uptake Analysis
SH SY5Y cells were plated in 6-well plates at 5×105 cells per well and, after overnight incubation, full media was removed and replaced with OptiMEM (Invitrogen) containing 100 U/mL penicillin/streptomycin and either no or the indicated amount of added serum. FITC-PUMA peptide (1 mM stock in DMSO) or vehicle was diluted into 1 mL cultures for a final treatment concentration of 5 μM and incubated at 37°C for 6 hours, followed by washing twice in PBS, trypsinizing for 10 minutes to remove any surface-bound peptide, and washing in PBS twice more. Cells were then lysed in 1% CHAPS buffer on ice and incubated for 20 minutes. Supernatants were collected after table top centrifugation, electrophoresed, and intracellular FITC peptide detected by fluorescence imaging using a Typhoon 9400 (GE Healthcare Life Sciences). For live cell confocal microscopy, SH SY5Y cells were plated in poly-D-lysine-coated culture plates (MatTek Corporation) at 2×105 cells per well and, after overnight incubation, full media was replaced with OptiMEM (Invitrogen) containing 100 U/mL penicillin/streptomycin. FITC-PUMA peptide (1 mM stock in DMSO) or vehicle was diluted into 1 mL cultures for a final treatment concentration of 10 μM and incubated at 37°C. After 6 hours, cells were incubated with CellMask Orange (5 μg/mL) and Hoeschst 33342 (5 μg/mL, Invitrogen) for 10 minutes, followed by washing three times with PBS and incubation with DMEM media lacking phenol red. Confocal images were acquired on a Yokogawa CSU-X1 spinning disk confocal system (Andor Technology) attached to a Nikon Ti-E inverted microscope (Nikon Instruments). Excitation of the three fluorophores was performed sequentially using 405 nm, 488 nm, and 561 nm lasers. Images were acquired using a 100× Plan Apo objective lens with a Hamamatsu OrcaER camera (Hamamatsu Photonics). Acquisition parameters, shutters, filter positions, and focus were controlled by Andor iQ software (Andor Technology).
Cell Viability and Caspase-3/7 Activation Assays
Cells were plated in 96-well plates at 2.5×103 cells per well and, after overnight incubation, full media was replaced with OptiMEM containing 100 U/mL penicillin/streptomycin and no added serum. A serial dilution of PUMA SAHBA2 from a 1 mM DMSO stock or vehicle was added to the cells in a final volume of 100 μL and incubated at 37°C for 6 hours, followed by addition of 10 μL FBS (serum replacement to 10% [v/v]). For the correlative cellular uptake and viability analyses (Figure 6E–F), the culture media during the initial 6 hour treatment period contained either no added serum or 2.5%, 5%, or 10% [v/v] FBS. To measure caspase-3/7 activation, cells were analyzed after eight hours of total treatment using the Caspase-Glo 3/7 chemiluminescence reagent (Promega). For cell viability analysis, cells were analyzed after 24 hours of total treatment using the CellTiter-Glo chemiluminescence reagent (Promega). Luminescence was detected by a microplate reader (Spectramax M5 Microplate Reader, Molecular Devices).
PUMA SAHB Co-precipitation from Cellular Lysates
Neuroblastoma cells were trypsinized, collected by centrifugation, washed twice in cold PBS, and lysed in a 1% CHAPS buffer (50 mM Tris [pH 7.5], 200 mM NaCl, 1% [w/v] CHAPS, 1 mM EDTA, 1.5 mM MgCl2, complete protease inhibitor tablet [Roche]), on ice for 20 minutes. Lysates were isolated after table top centrifugation and then incubated (1.25 mg) with 50 nmol biotinylated-PUMA SAHBA3 in lysis buffer overnight at 4°C. Biotin pull down was accomplished by incubation with high-capacity streptavidin agarose (Thermo) for 2 hours at 4°C, followed by washing the beads with 3 x 500 μL lysis buffer. Precipitated proteins were eluted by treating the beads with 4× LDS buffer containing 50 mM DTT for 10 minutes at 70°C, and then subjected to electrophoresis (25 μg protein/lane) and western blotting using the following antibodies: BAX N-20 (1:2000) (sc-493, Santa Cruz Biotechnology), BCL-2 (1:200) (sc-509, Santa Cruz Biotechnology), MCL-1 s-19 (1:200) (sc-819, Santa Cruz Biotechnology).
Cell Transfection and Flag-PUMA Co-immunoprecipitation
HEK 293T cells were transiently transfected with pcDNA3 plasmid containing Flag-PUMAα or its R142E or D147K mutants, using polyethylenimine(Yethon et al., 2003) (PEI) at a 4:1 PEI:DNA ratio. Two days following transfection, cells were lysed in 1% CHAPS buffer (as above) and isolated lysate (2 mg) was incubated with 30 μL of anti-Flag magnetic affinity beads (Sigma) in lysis buffer overnight at 4°C. The beads were washed with 3 × 500 μL lysis buffer, and immunoprecipitated proteins eluted by incubation with 4× LDS containing 50 mM DTT for 10 minutes at 70°C. The immunoprecipitates were subjected to electrophoresis and analyzed by western blot using the above-described BAX, BCL-2, and MCL-1 antibodies.
Supplementary Material
HIGHLIGHTS.
PUMA BH3 directly activates BAX by engaging its α1/α6 trigger site
Upon allosteric release of BAX α9, PUMA BH3 can also target BAX’s C-terminal pocket
PUMA BH3 triggers neuroblastoma cell death by multimodal BCL-2 family interaction
PUMA mimetics targeting anti- and pro-apoptotic BCL-2 proteins have therapeutic potential
Acknowledgments
We thank E. Smith for editorial and graphics assistance, R. George and T. Look for neuroblastoma cell lines, and D. Andrews for guidance on the liposomal assay system. This work was supported by NIH grants 5R01CA050239 and 5R01GM090299, and a Stand Up to Cancer Innovative Research Grant to L.D.W. and a National Science Foundation Graduate Research Fellowship to A.L.E. L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar RS, Geng Y, Klocke BJ, Latham CB, Villunger A, Michalak EM, Strasser A, Carroll SL, Roth KA. BH3-only proapoptotic Bcl-2 family members Noxa and Puma mediate neural precursor cell death. J Neurosci. 2006;26:7257–7264. doi: 10.1523/JNEUROSCI.0196-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Crannell CW, Walensky LD. Chemical Synthesis of Hydrocarbon-Stapled Peptides for Protein Interaction Research and Therapeutic Targeting. Curr Protoc Chem Biol. 2011;3:99–117. doi: 10.1002/9780470559277.ch110042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun CR, Mintseris J, Gavathiotis E, Bird GH, Gygi SP, Walensky LD. Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem Biol. 2010;17:1325–1333. doi: 10.1016/j.chembiol.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Quah ST, Jong J, Goh AM, Chiam PC, Khoo KH, Choong ML, Lee MA, Yurlova L, Zolghadr K, et al. Stapled peptides with improved potency and specificity that activate p53. ACS Chem Biol. 2013;8:506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- Callus BA, Moujallad DM, Silke J, Gerl R, Jabbour AM, Ekert PG, Vaux DL. Triggering of Apoptosis by Puma Is Determined by the Threshold Set by Prosurvival Bcl-2 Family Proteins. J Mol Biol. 2008;384:313–323. doi: 10.1016/j.jmb.2008.09.041. [DOI] [PubMed] [Google Scholar]
- Cartron PF, Gallenne T, Bougras G, Gautier F, Manero F, Vusio P, Meflah K, Vallette FM, Juin P. The first alpha helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol Cell. 2004;16:807–818. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- Dai H, Smith A, Meng XW, Schneider PA, Pang YP, Kaufmann SH. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol. 2011;194:39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, Smits C, Fan FC, Lee EF, Fairlie WD, Hinds MG. Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J Mol Biol. 2008;380:958–971. doi: 10.1016/j.jmb.2008.05.071. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Du H, Wolf J, Schafer B, Moldoveanu T, Chipuk JE, Kuwana T. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 2011;286:491–501. doi: 10.1074/jbc.M110.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G, Michalak E, Strasser A, Villunger A. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med. 2006;203:2939–2951. doi: 10.1084/jem.20061552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, Adams JM, Strasser A, Villunger A. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005;106:4131–4138. doi: 10.1182/blood-2005-04-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis AV, Chipuk JE, Fisher JC, Yun MK, Grace CR, Nourse A, Baran K, Ou L, Min L, White SW, et al. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat Chem Biol. 2013;9:163–168. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallenne T, Gautier F, Oliver L, Hervouet E, Noel B, Hickman JA, Geneste O, Cartron PF, Vallette FM, Manon S, et al. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J Cell Biol. 2009;185:279–290. doi: 10.1083/jcb.200809153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Phillips DC, Jeffers JR, Chipuk JE, Parsons MJ, Rehg JE, Opferman JT, Green DR, Zambetti GP. Genetically defining the mechanism of Puma- and Bim-induced apoptosis. Cell Death Differ. 2012;19:642–649. doi: 10.1038/cdd.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna D, Davis M, Bird G, Walensky L. BH3-Triggered Structural Reorganization Drives the Activation of Proapoptotic BAX. Mol Cell. 2010;40:481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES, Walensky LD. Direct and selective small-molecule activation of proapoptotic BAX. Nat Chem Biol. 2012;8:639–645. doi: 10.1038/nchembio.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EHY, Tjandra N, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith KC, Gross M, Peirce S, Luyindula D, Liu X, Vu A, Sliozberg M, Guo R, Zhao H, Reynolds CP, et al. Mitochondrial Bcl-2 family dynamics define therapy response and resistance in neuroblastoma. Cancer Res. 2012;72:2565–2577. doi: 10.1158/0008-5472.CAN-11-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DH, Kupresanin F, Berzins SP, Herold MJ, O’Reilly LA, Bouillet P, Strasser A. The BH3-only proteins Bim and Puma cooperate to impose deletional tolerance of organ-specific antigens. Immunity. 2012;37:451–462. doi: 10.1016/j.immuni.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S, Bax A. The importance of not saturating water in protein NMR: application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ, Zhu L, Chittenden T. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci USA. 2001;98:11318–11323. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, Day CL. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- Jabbour AM, Heraud JE, Daunt CP, Kaufmann T, Sandow J, O'reilly LA, Callus BA, Lopez A, Strasser A, Vaux DL, et al. Puma indirectly activates Bax to cause apoptosis in the absence of Bid or Bim. Cell Death Differ. 2009;16:555–563. doi: 10.1038/cdd.2008.179. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kim H, Tu H, Ren D, Takeuchi O, Jeffers J, Zambetti G, Hsieh J, Cheng E. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- LaBelle JL, Katz SG, Bird GH, Gavathiotis E, Stewart ML, Lawrence C, Fisher JK, Godes M, Pitter K, Kung AL, et al. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest. 2012;122:2018–2031. doi: 10.1172/JCI46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci USA. 2013;110:E986–995. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, He Q, Huang Y, Sheikh MS. Transcriptional upregulation of PUMA modulates endoplasmic reticulum calcium pool depletion-induced apoptosis via Bax activation. Cell Death Differ. 2005;12:1310–1318. doi: 10.1038/sj.cdd.4401659. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- Moldoveanu T, Grace CR, Llambi F, Nourse A, Fitzgerald P, Gehring K, Kriwacki RW, Green DR. BID-induced structural changes in BAK promote apoptosis. Nat Struct Mol Biol. 2013;20:589–597. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24:677–688. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Mustata G, Li M, Zevola N, Bakan A, Zhang L, Epperly M, Greenberger JS, Yu J, Bahar I. Development of small-molecule PUMA inhibitors for mitigating radiation-induced cell death. Curr Top Med Chem. 2011;11:281–290. doi: 10.2174/156802611794072641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Pitter K, Bernal F, Labelle J, Walensky LD. Dissection of the BCL-2 family signaling network with stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimertz C, Kögel D, Rami A, Chittenden T, Prehn JHM. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J Cell Biol. 2003;162:587–597. doi: 10.1083/jcb.200305149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJD, Cheng EHY. BID, BIM, and PUMA Are Essential for Activation of the BAX- and BAK-Dependent Cell Death Program. Science. 2010;330:1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits C, Czabotar PE, Hinds MG, Day CL. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure. 2008;16:818–829. doi: 10.1016/j.str.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Steckley D, Karajgikar M, Dale LB, Fuerth B, Swan P, Drummond-Main C, Poulter MO, Ferguson SSG, Strasser A, Cregan SP. Puma is a dominant regulator of oxidative stress induced Bax activation and neuronal apoptosis. J Neurosci. 2007;27:12989–12999. doi: 10.1523/JNEUROSCI.3400-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Takada K, Zhu D, Bird GH, Sukhdeo K, Zhao JJ, Mani M, Lemieux M, Carrasco DE, Ryan J, Horst D, et al. Targeted disruption of the BCL9/beta-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med. 2012;4:148ra117. doi: 10.1126/scitranslmed.3003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- Uren RT, Dewson G, Chen L, Coyne SC, Huang DCS, Adams JM, Kluck RM. Mitochondrial permeabilization relies on BH3 ligands engaging multiple prosurvival Bcl-2 relatives, not Bak. J Cell Biol. 2007;177:277–287. doi: 10.1083/jcb.200606065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Labi V, Bouillet P, Adams J, Strasser A. Can the analysis of BH3-only protein knockout mice clarify the issue of ‘direct versus indirect’ activation of Bax and Bak? Cell Death Differ. 2011;18:1545–1546. doi: 10.1038/cdd.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, Adams JM, Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Walensky LD. Direct BAKtivation. Nat Struct Mol Biol. 2013a;20:536–538. doi: 10.1038/nsmb.2579. [DOI] [PubMed] [Google Scholar]
- Walensky LD. Playing FullBAK. Cell Cycle. 2013b;12:1333–1334. doi: 10.4161/cc.24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Tolkovsky AM. The BH3-only protein Puma is both necessary and sufficient for neuronal apoptosis induced by DNA damage in sympathetic neurons. J Neurochem. 2006;96:1213–1226. doi: 10.1111/j.1471-4159.2005.03676.x. [DOI] [PubMed] [Google Scholar]
- Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Yee KS, Vousden KH. Contribution of membrane localization to the apoptotic activity of PUMA. Apoptosis. 2008;13:87–95. doi: 10.1007/s10495-007-0140-2. [DOI] [PubMed] [Google Scholar]
- Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- Yu J, Yue W, Wu B, Zhang L. PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin Cancer Res. 2006;12:2928–2936. doi: 10.1158/1078-0432.CCR-05-2429. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xing D, Liu L. PUMA promotes Bax translocation by both directly interacting with Bax and by competitive binding to Bcl-X L during UV-induced apoptosis. Mol Biol Cell. 2009;20:3077–3087. doi: 10.1091/mbc.E08-11-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.