Abstract
An increasing body of research on autophagy provides overwhelming evidence for its connection to various biological functions and human diseases. Beclin 1 is the first described mammalian autophagy protein and appears to act as a nexus point between autophagy, endosomal, and perhaps also cell death pathways. Beclin 1 performs these roles as part of a core complex that contains vacuolar sorting protein 34 (VPS34), a class III phosphatidylinositol-3 kinase. The precise mechanism of Beclin 1-mediated regulation of these cellular functions is unclear, but substantial progress has recently been made in identifying new players and their functions in Beclin 1-VSP34 complexes. Here, we review emerging studies, which are beginning to unveil physiological functions for Beclin 1-VPS34 in the central control of autophagic activity and other cellular trafficking events through the formation of distinct Beclin 1-VPS34 protein complexes.
Introduction
Autophagy is a catabolic pathway used to breakdown and recycle long-lived proteins and injured organelles (Box 1). At least thirty-three AuTophaGy-related (ATG) genes have been found thus far in yeast 1, 2, and homologs of many of these genes have been described in higher eukaryotes. The total group of ATG genes can be broken down into functional categories that follow the autophagic processes of induction, vesicle nucleation, vesicle expansion and completion, and recycling, as well as the cargo packaging that is specific to selective autophagic degradation of modified proteins or damaged organelles 3. One of the most-studied mammalian autophagy proteins that falls into the category of vesicle nucleation is Beclin 1, a 60 kDa coiled-coil protein that contains a Bcl-2-homology-3 (BH3) domain, a central coiled-coil domain (CCD), and an evolutionarily conserved domain (ECD). The function of Beclin 1 in autophagy was first suspected due to its 24.4% amino-acid sequence identity to the yeast autophagy protein Atg6. Beclin 1 was found to restore autophagic activity in Atg6-disrupted yeast, becoming one of the first identified mammalian genes to positively regulate autophagy 4 Interestingly, Beclin 1 was originally discovered not as an autophagy protein but rather as an interaction partner for the anti-apoptotic protein Bcl-25. Subsequent studies demonstrated Beclin 1 to be a haploinsufficient tumor suppressor gene that is either monoallelically deleted or shows reduced expression in several different cancers 6, 7. Several studies have also found Beclin 1 to be involved in other biological functions and human diseases including heart disease, pathogen infection, development, and neurodegeneration 8. However, the precise mechanism of Beclin 1 in these different physiological processes has yet to be elucidated. Clues to these mechanisms of Beclin 1 may be found through its interactions with other proteins. Since the initial finding that Beclin 1 binds Bcl-2 through its BH3 domain, several other partners have also been identified that interact through the CCD and ECD domains of Beclin 1. These interacting partners provide potential insight into Beclin 1’s different physiological roles, including autophagy and those outside of autophagy.
Box 1. Autophagy.
Autophagy is a “self-eating” catabolic process used to breakdown and recycle long-lived proteins and organelles in order to maintain a homeostatic environment within the cell. In yeast, the to-be-degraded cytoplasmic contents are delivered to the vacuole while, in mammals, the highly conserved autophagic process transports such molecules to the lysosome. There are three classes of autophagy (microautophagy, macroautophagy, and chaperone-mediated autophagy) that are defined by varying cellular mechanisms and functions 43. As the most prevalent form, macroautophagy is commonly referred to as autophagy and is so referenced in this review.
Autophagy follows a sequential course in the cell that begins with the formation of an isolation membrane, or phagophore, around a portion of cytosol and its inhabitants. The isolation membrane then elongates and seals on itself, trapping cytosolic components and resulting in the biosynthesis of a double-membrane vacuoles known as an autophagosome. The final step is the fusion of the autophagosome with the lysosome, allowing for the degradation of the entrapped components. Most autophagic activity in the cell is generally considered to be a nonspecific breakdown of the cytoplasm, occurring at basal and upregulated levels in response to environmental cues or stress conditions such as nutrient depletion. However, autophagy can also be selective, targeting modified proteins (e.g. ubiquitinated) as well as excess and damaged organelles (peroxisomes and mitochondria) 44-47. The cytoplasm-to-vacuole (Cvt) pathway in yeast is an example of selective autophagy that is required for the transport of specific hydrolases to the vacuole 48, 49. Although unique in its biosynthetic use, the Cvt pathway shares many key autophagy proteins with the nonspecific autophagy routes, suggesting the multiple roles played by these autophagy proteins.
Because the autophagic pathway is conserved among eukaryotes, many of the protein-protein interactions and protein complexes found in yeast are also identified in mammals. One primary example is the critical core complex formed from the yeast counterpart to Beclin 1, Atg6, and the class III phosphatidylinositol 3-kinase (PI(3) kinase) Vps34 (Box 2) that plays a role in vesicle nucleation in autophagy and hydrolase sorting through the vacuolar protein sorting (Vps) pathway 9. Similar to this yeast Atg6-Vps34 complex, a mammalian Beclin 1-VPS34 core autophagy complex also exists. Additionally, several different autophagy complexes consisting of Beclin 1-VPS34 and a selection of Beclin 1’s binding partners have recently been identified in mammalian cells 10-15. Recent studies suggest a central role for Beclin 1 complexes in controlling human VPS34-mediated vesicle trafficking pathways including autophagy. This review focuses on the newly identified Beclin 1 complexes and the regulation of the autophagic process by these distinct complexes and discusses the emerging questions that await future investigation in order to fully understand the molecular mechanism of autophagy control.
Box 2. PI(3)-kinases.
Yeast Vps34 and its human counterpart VPS34 belong to a family of kinases known as the phosphatidylinositol 3-kinases (PI(3)-kinases) that phosphorylate inositol lipids at the 3’ position of the inositol ring to produce the 3-phosphoinositides PI(3)P, PI(3,4) P2 and PI(3,4,5) P3. The PI(3)-kinases are divided into three distinct classes in mammalian cells with class I and class II enzymes that produce PI(3,4) P2 and PI(3,4,5) P3 as the most abundant. The PI(3)P producing VPS34 is the only enzyme in mammals designated as the class III type50. An increase in the amount of the PI(3)P class III product in mammalian cells is known stimulate autophagy whereas increases in the class I products have been shown to result in reduced autophagic activity51. Yeast lack the class I and class II PI(3) kinases and have only a single class III gene, vps34, that also generates the autophagy stimulating PI(3)P product50.
Multiple Autophagy Complexes with Distinct Functions
A central question for understanding the regulation of autophagy is whether Beclin 1-VPS34 shares a similar function as the yeast homolog in autophagy and whether it gains additional functions through evolution. While the yeast Atg-6-Vps34 complexes and functions have been well documented, the Beclin 1-VPS34 interactions with different proteins and the resulting changes in autophagic function are just beginning to be elucidated.
Two Atg6-Vps34 Complexes in Yeast
In yeast, Atg6 exists in two separate types of complexes that regulate either autophagy or the Vps pathway that sorts vacuolar proteins such as carboxypeptidase Y (CPY) at the trans-Golgi network and delivers them to the vacuole 16, 17 (Figure 1). Both complex types involve Vps34 and the serine-threonine kinase Vps15, as common factors. Vps15 acts to anchor the complex to the membrane whereas Vps34 is recruited and then activated by Vps15. The autophagy-related complex (complex I) contains Atg14, which associates with the peripheral membrane and specifically localizes to vaculoar membranes and the pre-autophagosomal structure (PAS) 18, 19. Its overexpression suppresses the autophagic deficiency in yeast expressing mutant Atg6 20. However, the Atg6 mutant also exhibits a deficit in the Vps pathway that Atg14 overexpression does not alleviate, demonstrating the specificity of complex I for autophagy. Vps38, which is the completing subunit for complex II, is the essential binding partner for Atg6 for proper functionality of the Vps pathway. Distinct from complex I, complex II localizes to the vacuolar membrane and endosomes; and Vps38 is necessary for this subcellular localization. It was shown that complex II is more abundant than complex I, suggesting that Atg6-Vps34-Vps15 is primarily involved in Vps pathways under normal conditions and that yeast autophagy is strictly regulated at a basal level 16.
Figure 1. Multiple Beclin 1-VPS34 complexes and their distinct functions.
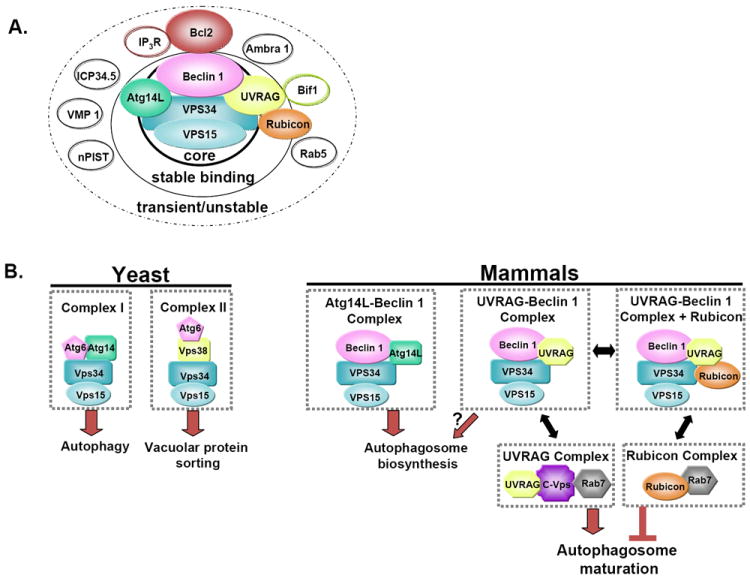
(A) An illustration of the multilayered Beclin 1 protein complexes: a core complex Beclin 1-VPS34-VPS15; stable binding partners Atg14L, UVRAG, and Rubicon; unstable or transient binding partners including Bcl-2 homologs, Bif-1, Ambra 1, VMP1, nPIST, Rab5, ICP 34.5, and inositol 1,4,5-triphosphate receptor (IP3R). Among the unstable binding partners, Bif-1 interacts with UVRAG, and IP3R interacts with Bcl-2.
(B) In yeast two Atg6-Vps34 complexes, I and II, regulate autophagy and vacuolar protein sorting, respectively. In mammals, multiple Beclin 1 complexes act in different stages of autophagy and perhaps also endocytic trafficking. Atg14L associates with the core Beclin 1-VPS34-VPS15 complex to function in the biosynthesis of autophagosomes, and UVRAG potentially associates with the same core complex to also influence autophagosome formation. However, UVRAG has an additional role in autophagosome maturation in a complex with C-Vps and the Rab7 GTPase that is independent of Beclin 1. Rubicon inhibits autophagosome maturation in a possibly Beclin 1-independent manner. Rubicon could sequester Rab proteins such as Rab 7 through its RUN domain to inhibit this stage of autophagy. The binding of Rubicon to the Beclin 1 core complex though UVRAG may aid in neutralizing such a negative function.
Beclin 1-VPS34 Core Complexes and Physiological Binding Partners in Mammals
For some time Beclin 1 has been known to form a protein complex consisting of VPS34 and p150/VPS15 and has been predicted to regulate autophagy in a similar manner to yeast 9. While the binding of Beclin 1 to VPS34/VPS15 suggests a conserved function of Beclin 1 in autophagy, the association of Beclin 1 with an additional set of proteins implicates Beclin 1 in diverse cellular functions, which have yet to be defined. These observations raise a question as to what proteins constitute the core complex of Beclin 1 and what functions are associated with the Beclin 1 complex under various conditions.
Affinity purification of a transgenic GFP-tagged Beclin 1 (functionally replacing endogenous Beclin 1) and its associated proteins from various mouse tissues has revealed VPS34/PI(3)K, p150/VPS15, and UVRAG (UV irradiation resistance-associated gene) binding to Beclin 1 in a nearly stoichiometric fashion and Atg14L and Rubicon (RUN domain, a cysteine-rich domain containing, Beclin 1-interacting protein) binding to Beclin 1 in a probably substoichiometric manner 11. Interestingly, other previously identified Beclin 1-binding proteins, including Bcl-2, Ambra 1 and Bif1, were not detected under the same experimental conditions, suggesting that they do not form a stable complex with Beclin 111. These results were confirmed independently by two other reports, which showed immuno-isolated Beclin 1 complexes from cultured cells 12, 21.
Atg14L, a putative mammalian homolog for yeast Atg14 was reported based on a sequence homology search 10. UVRAG, a previously found Beclin 1-binding protein15, was also predicted to be close to a homolog of yeast Vps3810, 22. The novel protein Rubicon, however, does not have a yeast homolog. These studies suggest a central core complex consisting of VPS34/PI(3)K, p150/VPS15, and Beclin 1, which are highly conserved from yeast to mammals. UVRAG, Atg14L, or Rubicon appear to form stable complexes with the core complex; but the UVRAG-containing complex is likely more stable or abundant than those with Atg14L or Rubicon under normal conditions. Furthermore, these studies also suggest that the binding of Beclin 1 to other previously identified proteins may be relatively unstable, transient, or condition specific (Figure 1A). However, these ‘peripheral’ binding proteins may play important roles in regulating core complex activity by responding to various physiological or pathophysiological conditions. Recent characterizations of UVRAG, Atg14L and Rubicon functions have started to reveal possible physiological functions of Beclin 1-VPS34 complexes. Moreover, they have provided important clues for how the ‘peripheral’ binding proteins are connected structurally and functionally to Beclin 1-VPS34 complex.
Atg14L
Atg14L, which is the likely mammalian homolog of yeast Atg14, contains two coiled-coil domains that are required for binding the CCD regions of Beclin 1 and VPS34. In general, Atg14L exists primarily in a Beclin 1-Atg14L-VPS34-VPS15 complex that is essential for the biosynthesis of autophagosomes (Figure 1B). This complex, likely equivalent to the yeast complex I, is therefore highly conserved through evolution. It is unclear where Atg14L normally localizes at physiological level, but it may translocate to the isolation membrane (IM)/phagophore during autophagy induction 10-12. The colocalization of Atg14L with several IM markers suggests that the Atg14L complex is critical in the early stages of autophagosome biosynthesis (Box 1). Furthermore, the presence of intracellular Atg14L is mainly dependent on Beclin 1 protein. For example, loss of Beclin 1 was found to greatly reduce Atg14L protein levels10, 11. Atg14L was also shown to enhance VPS34 lipid kinase activity, which is critical for autophagosome formation. However, this effect of Atg14L on VPS34 kinase activity was dependent on the simultaneous overexpression of Beclin 111. The Beclin 1-Atg14L stable complex may allow the two proteins to synergistically promote and positively regulate autophagy through VPS34-VPS15, whereas Atg14L expression alone is insufficient.
UVRAG
Similarly to Beclin 1, UVRAG is also a tumor suppressor and is monoallelically deleted in human colon cancer cells and tissues 15. Containing a central coiled-coil domain, UVRAG forms an α-helical heterodimer with the CCD of Beclin 1 via its own CCD 15. A portion of UVRAG is present in a Beclin 1-UVRAG-VPS34-VPS15 complex that normally excludes Atg14L (Figure 1B). This mutual exclusion of Atg14L and UVRAG for binding Beclin 1 is likely due to the overlap of their docking sites on Beclin 1 (e.g. CCD) 12. Our study and others suggest that more UVRAG than Atg14L is found in complex with Beclin 1 in different tissues and cell lines 11, 12. These results suggest that the Beclin 1-UVRAG containing complex is likely a more dominant species as compared to the Beclin 1-Atg14L complex under normal conditions. With respect to its function in autophagy, UVRAG was reported to act together with Beclin 1 to induce autophagosome biosynthesis (Figure 1B). The expression of UVRAG was also correlated with an increase in VPS34 enzymatic activity, suggesting a role for the Beclin 1-UVRAG-VPS34-VPS15 complex in autophagy regulation 15. However, a study by Itakura et al. questioned this particular autophagic role for UVRAG, reporting that suppressing UVRAG expression in cells did not affect autophagic flux or autophagosome formation 10.
Apart from the controversial role in the initial stages of autophagy, UVRAG has also been described to act in a separate complex that is independent of Beclin 1 to promote autophagosome maturation (Figure 1B) and endocytic trafficking. UVRAG was shown to associate with the class C Vps (C-Vps) complex, which is known in yeast to be required for vesicle tethering events that precede vesicle fusion with lysosomes. After recruiting C-Vps to the autophagosome, UVRAG reportedly stimulates Rab 7 GTPase activity, which is required for complete autophagosome maturation. This interaction of UVRAG with C-Vps was also shown to affect the endocytic pathway by promoting endosome-endosome fusion23. Further evidence for the involvement of UVRAG in endocytic trafficking is its localization to early/late endosomes. Specifically, UVRAG was found to colocalize with late endosomal marker Rab9 as well as partially with Rab5 and Rab710, 23. Due to a partial homology to yeast Vps38, it has been suggested that UVRAG could be the mammalian homolog of Vps3810, whose function is limited to the Vps pathway16. While it is clear that UVRAG plays a role in the endocytic pathway, the extent to which UVRAG (particularly in the complex with Beclin 1-VPS34) functions in autophagy regulation seems to be uncertain given the conflicting reports of cell culture data. Although different experimental conditions may contribute to the drastic difference in the observation, further investigation of possibly distinct UVRAG protein complexes should assist to resolve this conflict.
Rubicon
Rubicon contains a conserved RUN domain near the amino terminus, a cysteine-rich domain near the carboxy terminus, and a central CCD region. The central region containing the CCD is critical for the binding of Rubicon with both Beclin 1 and VPS34. Two reports demonstrated that at least a portion of Rubicon population is present in a Beclin 1 complex exclusive of Atg14L. However, Rubicon is found in the same complex with UVRAG when associated with Beclin 1 11, 12. The available evidence suggests that Rubicon and UVRAG are involved together with the core complex, forming a “Beclin 1-VPS34-VPS15-UVRAG-Rubicon” complex (Figure 1B). As aforementioned, UVRAG can interact with the core complex independently of Rubicon. However, Rubicon seems only to bind the core complex when UVRAG is also bound, suggesting the possibility that Rubicon interacts with Beclin 1 through UVRAG 11, 12. Although no Rubicon homolog is found in yeast, it is conserved among nematodes, slime mold, and mammals. This suggests that perhaps Rubicon could provide an extra layer of autophagic regulation that is unique to higher eukaryotes.
In opposition to the role of Atg14L, Rubicon was found to negatively regulate autophagy 11, 12. Available evidence revealed that Rubicon acts on a later stage of the autophagic process. For example, overexpression of Rubicon resulted in an abundance of immature autophagosomes suggesting that Rubicon may hinder autophagosome maturation 11, 12. Furthermore, Rubicon was found to decrease VPS34 kinase activity, and this inhibitory effect seems to be aborted with an excess of Beclin 1 11. This result raises the possibility that the negative regulation of autophagy by Rubicon does not require Beclin 1. Additionally, Beclin 1 does not appear to be involved in the formation of aberrant endosomal structures resulted from Rubicon overexpression, which apparently inhibits autophagosome maturation 11.
Therefore, based on the aforementioned results, Rubicon-mediated downregulation of autophagy is likely Beclin 1-independent, occurring when Rubicon is not involved with the Beclin 1-VPS34 complex (Figure 1B), while binding of Rubicon with Beclin 1 may suppress the downregulation of autophagy. Then how does Rubicon inhibit autophagosome maturation? Since Rubicon contains a RUN domain, a conserved motif that is known to interact with and regulate small GTPases 11, 12, 24, one possibility is that Rubicon interacts with specific Rabs that were previously shown to regulate the autophagic process 25-27. A likely candidate would be Rab 7, which has been shown to participate in the maturation of late autophagic vacuoles27 upon stimulation by UVRAG that has recruited C-Vps 23. However, this prediction awaits verification. Furthermore, the function of Rubicon in the larger Beclin 1-VPS34-VPS15-UVRAG-Rubicon also remains to be shown. Consistent with a conserved function of Beclin 1 in promoting autophagic activity, association of Beclin 1 with Rubicon included in such a complex perhaps neutralize the inhibitory effect of Rubicon in autophagy.
Moreover, like UVRAG, Rubicon may also participate in the endocytic pathway. Rubicon was found to colocalize with the early endosomal marker EEA1, multivesicular body (MVB) marker lysobisphosphatidic acid (LBPA), and late endosomal markers Rab9 and LAMP1 11, 12. In addition, Rubicon-associated endosomes are enriched in PI(3)-P, suggesting that endocytic functions of Rubicon may be associated with VPS34-mediated activity11. However, it is not yet clear if this “extra” role of Rubicon in the endosomal pathway involves Beclin 1.
Antiapoptotic Bcl-2 family in Regulation of Beclin 1 complex and Autophagy activity
Previous studies have shown that a few antiapoptotic members of the Bcl-2 family (Box 3), such as Bcl-2, Bcl-Xl, and viral Bcl-2 homologs vBcl-2 (Kaposi’s sarcoma associated herpes virus) and γHV68M11, interact with Beclin 1 5, 28-30. A series of investigations have shown that antiapoptotic Bcl-2 proteins inhibit autophagic function of Beclin 1 through binding to its BH3 domain 28, 31, thereby suggesting that Bcl-2 family members also function as inhibitors for autophagic pathways.
Box 3. Bcl-2 Family.
Bcl-2 is an antiapoptotic protein that suppresses the release of cytochrome c from mitochondria, preventing caspase activation in the cell death pathway 52. It is actually part of a growing family of proteins that are subdivided into three groups (Table 1). The first group consists of proteins such as Bcl-xL that have three to four Bcl-2 homolgy (BH) domains, which are important in regulating interactions with other Bcl-2 family members. The second group contains pro-apoptotic proteins whose function can be inhibited by Bcl-2, and the third group includes proteins that have only one short BH3 domain 53. Beclin 1 has an N-terminal BH3 domain and is considered to fall into this third category of the Bcl-2 family 54, 55. The BH3 domain of Beclin1 is critical for binding Bcl-2 as well as the Bcl-2 family member Bcl-xL 55.
A subsequent study suggests that endogenous Bcl-2 likely regulates Beclin 1-mediated autophagy, and this inhibition of autophagy by Bcl-2 may occur specifically at the ER rather than mitochondria 32. A model was proposed that the Beclin 1-Bcl-2 complex exists constitutively and functions as a rheostat, which maintains homeostasis of basal levels of autophagy 28. This model, however, does not appear to explain the aforementioned results from several groups showing the lack of endogenous Bcl-2 homologues in the Beclin 1-VPS34 stable complexes 10-12. It could be that a transient Bcl-2-Beclin 1 interaction occurring in vivo escapes the capture by the described purification procedures; future studies with advanced tools will be needed to address this possibility.
Interestingly, Liang et al reported that viral γHV68 Bcl-2 (vBcl-2), Beclin 1, UVRAG and VPS34 are present in the same protein complex obtained by affinity purification 15. Subsequent biochemical analysis showed that vBcl-2 displays markedly higher affinity than Bcl-2 or Bcl-XL to Beclin 1 (KD of 40nM vs. 1.7μM). This tighter binding of vBcl-2 to Beclin 1 is correlated with stronger inhibition of autophagy as compared to cellular Bcl-2 homologues 33. The evidence reveals a pathogenic mechanism whereby the virus targets Beclin 1-VPS34 complex to shut down autophagy. However, it is unclear how a relatively weak binding of cellular Bcl-2 homologs to the Beclin 1 BH3 domain would contribute to efficient inhibition of autophagy at constitutive levels in vivo.
Although the above studies provide strong evidence that the binding of Bcl-2 homologues, in particular vBcl-2, to the Beclin 1 BH3 domain blocks autophagy, the mechanism underlying this inhibition remains to be defined. It was shown that binding of Bcl-2 causes less Beclin 1-VPS34 interaction and reduced autophagic activity 28. However, it is unclear whether the Bcl 2-Beclin 1 binding interferes with the interaction of other physiological components in the VPS34 complex including Atg14L, UVRAG, Rubicon and VPS15. In one study, Liang et al did not find that vBcl-2 and UVRAG were in direct competition for binding of Beclin 1; however, Atg14L was not found in the isolated complex containing vBcl-2, UVRAG, Beclin 1 and VPS34 15. Interestingly, a more recent analysis demonstrated that vBcl-2 and Bcl-xL were indeed able to reduce the affinity of UVRAG for Beclin 1 34. This study also suggested that Beclin 1 can be found in a homodimeric form that is capable of interacting with Bcl-2 proteins; but binding of UVRAG disrupts this homodimerization, freeing Beclin 1 to bind other core complex members 34 (Figure 2). The potential role for Beclin 1 self-interacting in regulating the binding of other essential components in the Beclin 1-VPS34 complex was also proposed in a separate study, in which Beclin 1 was shown to form a large homo-oligomer. This study proposed that the oligomerized Beclin 1, which can be disrupted by vBcl-2 (M11), provides a platform for the assembly of other Beclin 1-binding partners and the coordination of their functions in autophagy 35.
Figure 2. Dynamic regulation of Beclin 1-VPS34 complex formation and autophagy by Bcl-2 family members and Beclin 1 dimerization.
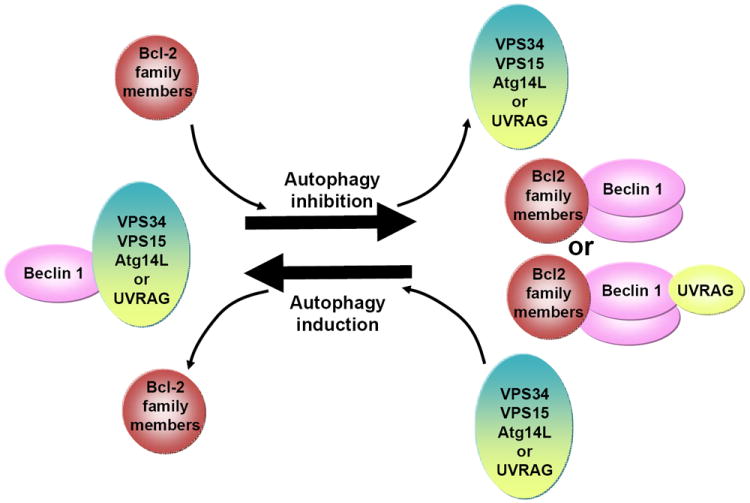
Beclin 1-VPS34-VPS15 forms pro-autophagic complexes with Atg14L or UVRAG. Binding of Bcl-2 and Beclin 1 is known to inhibit autophagy by possibly disrupting VPS34 binding; but Bcl-2 binding may also displace other stable interacting partners such as Atg14L (or UVRAG), resulting in autophagy inhibition. Additionally, the anti-autophagic binding of Bcl-2 promotes Beclin 1 homodimerization, which prevents heterodimerization with Atg14L or UVRAG.
Finally, protein phosphorylation was also shown to regulate Bcl-2-Beclin 1-mediated autophagic activity. The phosphorylation of the BH3 domain of Beclin 1 by death-associated protein kinase (DAPK) may cause disruption of the association between Beclin 1 and Bcl-xL, thereby enhancing autophagic activity 36. In addition, a previous study demonstrated that JNK1 activation through cell starvation phosphorylates Bcl-2 at three different sites in its BH3 and BH4 domains, resulting in the dissociation of Bcl-2 from Beclin 1 37. Future studies defining the upstream kinase and phosphatase activities for the above-mentioned phosphorylation/dephosphorylation events will be imperative for further understanding of the precise cellular signaling and control of autophagy.
Additional Beclin 1 binding proteins
Other proteins, including Ambra 1, Bif-1, nPIST, inositol 1,4,5-trisphosphate receptor (IP(3)R), and VMP1, have also been described previously as Beclin 1-binding proteins 13, 14, 38-40. As they were not found in the stable Beclin 1-VPS34 complexes as shown by affinity purification 10-12, 21, their association with this complex is probably “loose” or transient in nature. In some cases, the binding is indirect as shown for Endophilin B1 (Bif-1), which interacts with Beclin 1 through UVRAG 13. It was shown that Bif-1 levels critically affect the synthesis of autophagosomes likely through regulating the membrane curvature13, 41. Little is known about how others interact with the Beclin 1-VPS34-VPS15 core complex or relate to Atg14L, UVRAG or Rubicon, although current studies suggest that some may play roles in regulating Beclin 1-mediated autophagy or even autophagy-independent pathways in a cell/tissue or development stage-specific manner. Finally, endosomal protein Rab5 was also shown to bind Beclin 1, and this binding seems to require VPS34 25. This result is consistent with an earlier study that Rab 5 interacts with VPS34-VPS15 and activates its kinase activity 37. Furthermore, Rab 5 was shown to partially colocalize with UVRAG, but it is unknown whether Rab5 can also directly bind Rubicon through the RUN domain. Therefore, investigation of the molecular mechanisms whereby these additional binding proteins are involved with the Beclin 1-VPS34-VPS15 core complex will provide further information regarding how the Beclin 1-VPS34 complex is directed to different cellular trafficking pathways, including autophagy and endocytosis.
Concluding Remarks
Current studies demonstrate pleiotropic functions for Beclin 1-VPS34 complexes in autophagy control and the complexity of the autophagy process. The evidence reveals a core Beclin 1-VPS34-VPS15 complex that can be tightly associated with UVRAG, Atg14L or Rubicon under normal conditions. It remains possible that many other previously known Beclin 1-binding proteins are periphery to these proteins or function to regulate VPS34 kinase activity through these proteins. Therefore, a centralized function of Beclin 1 and its binding partners is the control of cellular VPS34 lipid kinase activity that is essential for autophagy and other membrane trafficking processes. By forming distinct protein complexes, Beclin 1 orchestrates the concerted action of VPS34/PI(3) kinase activity and specifically targets it at different steps of the autophagic process, such as autophagosome biogenesis and maturation.
While the precise role of each Beclin 1-VPS34 complex in regulating autophagy needs to be further examined (Box 4), the binding of Beclin 1 to VPS34, UVRAG or Rubicon, whose roles were shown in endosomal pathways, seems to link Beclin 1 to functions beyond the regulation of autophagy. This is likely, as yeast Atg6 has additional roles in the Vps pathway 16. Moreover, emerging evidence suggests the involvement of previously known endocytic proteins in mammalian autophagy. Salient examples are the small GTPase Rab5 (early endosome) and Rab7 (late endosome), which are also known to regulate autophagy 25-27. Given the connection of Beclin 1 to both Rab5 and Rab7 (possibly through Rubicon – QJW unpublished observation), it could be that Beclin 1 affects Rab GTPase activity and thereby the endocytic pathway. Despite these potential links, however, unequivocal evidence of a Beclin 1 endocytic function has yet to be formally demonstrated. A previous report refuted the idea that Beclin 1 cooperates with VPS34 in endosomal trafficking since cells in the study with Beclin 1 knocked down maintained full function of endocytic trafficking and protein trafficking from the TGN to the lysosomes42. Furthermore, Beclin 1 has not been found at endosomes, and the UVRAG-associated endosomal trafficking does not involve Beclin 1 23. Additionally, block of autophagosome maturation by Rubicon does not seem to depend on Beclin 1 11. Therefore, future investigation is needed to clarify if Beclin 1 does, indeed, collaborate with VPS34, UVRAG, or Rubicon to regulate the endosomal pathway.
Box 4. Future questions.
Numerous key binding partners for Beclin 1 are involved in endocytic pathways; however, the direct evidence for the participation of Beclin 1 in endocytosis remains to be demonstrated. Does Beclin 1 conduct the autophagy-specific action only, albeit binding to proteins whose functions are connected to endocytosis, or coordinate both autophagic and endocytic pathways depending on the cues of cellular context?
What is the exact function and mechanism of each newly identified Beclin 1 complex? Where are they localized under normal conditions and activated autophagy? For example, how does the Atg14L-Beclin 1 interaction exactly direct the VPS34-VPS15 lipid kinase to the biosynthesis of autophagosomes? What is the precise role of UVRAG-Beclin 1 or UVRAG-Rubicon-Beclin 1 in the autophagic process, e.g. pro-autophagy vs. anti-autophagy, biosynthesis vs. maturation? Does each of these complexes have roles outside of autophagy?
What is the sequence information for the binding between Beclin 1 and each partner? What is the dynamics of the composition and decomposition of each complex in response to autophagy signaling? Furthermore, what is the architecture and composition of each Beclin 1 protein complex at detailed structural levels?
What are the consequences of the binding of Beclin 1 and Bcl-2 family members on the interaction between Beclin 1 and Atg14L, UVRAG, or Rubicon? What are the molecular signaling and mechanism that induces the switch of Beclin 1 binding to different partners and control autophagy levels?
What are the functional associations of small GTPase protein (Rab5 and Rab7) with UVRAG or Rubicon-Beclin 1 complex in autophagy and endocytic pathway? Do Beclin 1, UVRAG and Rubicon affect GTPase activity of Rab5 or Rab7? Does the RUN domain of Rubicon provide a sequence motif for a direct binding of Rab5 or Rab7 and control Rab5 or 7-mediated autophagy activity?
Current knowledge also suggests that Bcl-2 family proteins play an important role in regulating autophagy through Beclin 1, therefore broadening our understanding of Bcl-2 cellular functions 28, 33, 35. However, the mechanism whereby Bcl-2 homologs, in particular endogenous Bcl-2, interferes with binding of Beclin 1 to Atg14L, UVRAG, or Rubicon has not yet been examined. In considering the Bcl-2-Beclin 1 complex being explored as a drug target, it is of particular interest to understand the exact mechanism of autophagy inhibition mediated by the Bcl-2 and Beclin 1 interaction.
Finally, the recent discovery of new Beclin 1-interacting partners and how they associate in various ways to form different complexes has the potential to be a significant gateway to elucidating the specific autophagic mechanisms that are triggered in various physiological and pathophysiological conditions. While studies involving cancer, heart disease, infectious disease, autophagic cell death, or neurodegeneration have focused on Beclin 1 expression in the past, the possibility now to examine different positive and negative regulators of a Beclin 1 complex could add another level of detail and clarity. The three newest additions to the mammalian core complex, UVRAG, Atg14L and Rubicon, especially have the potential to yield insight due to their distinct natures in promoting or inhibiting autophagy.
Table 1.
Three Groups of Bcl-2 Family Members
| Bcl-2 Family Groups | Members |
|---|---|
| Group 1: (Bcl-2) anti-apoptotic | Bcl-2, Bcl-XL, Bcl-w, Mcl-1, Bcl2A1, and Bcl-B |
| Group 2: (Bax) pro-apoptotic | Bax, Bak and Bok |
| Group 3: (BH3) pro-apoptotic | Bad, Bid, Bik, Hrk, BimL, PUMA, NOXA, and BMF |
Footnotes
Note: The authors declare No conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawamata T, et al. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 5.Liang XH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 7.Yue Z, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kihara A, et al. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Molecular biology of the cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nature cell biology. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 15.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 16.Kihara A, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schu PV, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 18.Obara K, et al. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. Embo J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kametaka S, et al. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 21.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 23.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callebaut I, et al. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends in biochemical sciences. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- 25.Ravikumar B, et al. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. Journal of cell science. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez MG, et al. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. Journal of cell science. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 27.Jager S, et al. Role for Rab7 in maturation of late autophagic vacuoles. Journal of cell science. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 28.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Maiuri MC, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. The EMBO journal. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinha S, et al. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattingre S, Levine B. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer research. 2006;66:2885–2888. doi: 10.1158/0008-5472.CAN-05-4412. [DOI] [PubMed] [Google Scholar]
- 32.Maiuri MC, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 33.Ku B, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS pathogens. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noble CG, et al. BCL-XL and UVRAG cause a monomer-dimer switch in beclin1. The Journal of biological chemistry. 2008 doi: 10.1074/jbc.M804723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ku B, et al. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy. 2008;4:519–520. doi: 10.4161/auto.5846. [DOI] [PubMed] [Google Scholar]
- 36.Zalckvar E, et al. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- 37.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nature cell biology. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 38.Yue Z, et al. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35:921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 39.Ropolo A, et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. The Journal of biological chemistry. 2007;282:37124–37133. doi: 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- 40.Vicencio JM, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell death and differentiation. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi Y, et al. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell death and differentiation. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng X, et al. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. Journal of cell science. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 43.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annual review of cell and developmental biology. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 44.Xue L, et al. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 45.Iwata J, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 46.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 47.Cavallini G, et al. Evidence for selective mitochondrial autophagy and failure in aging. Autophagy. 2007;3:26–27. doi: 10.4161/auto.3268. [DOI] [PubMed] [Google Scholar]
- 48.Klionsky DJ, et al. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takegawa K, et al. Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J Cell Sci. 1995;108(Pt 12):3745–3756. doi: 10.1242/jcs.108.12.3745. [DOI] [PubMed] [Google Scholar]
- 51.Petiot A, et al. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, et al. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 53.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat Rev Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 54.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oberstein A, et al. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]


