Abstract
It was demonstrated previously that symmetric, homodimeric proteins are energetically favored, which explains their abundance in nature1. It has been proposed that such symmetric homodimers underwent gene duplication and fusion to evolve into protein topologies that have a symmetric arrangement of secondary structure elements2– “symmetric superfolds”3; 4.Here, the ROSETTA protein design software was used to computationally engineer a perfectly symmetric variant of imidazole glycerol phosphate synthase (HisF) and its corresponding symmetric homodimer. The new protein, termed FLR, adopts the symmetric (βα)8 TIM-barrel superfold. The protein is soluble, monomeric, and exhibits two-fold symmetry not only in the arrangement of secondary structure elements, but in sequence and at atomic detail as verified by crystallography. When cut in half, FLR dimerizes readily to form the symmetric homodimer. The successful computational design of FLR demonstrates progress in our understanding of the underlying principles of protein stability and presents an attractive strategy for the in silico construction of larger protein domains from smaller pieces.
Structural studies of globular proteins have demonstrated that despite thousands of uniquely different proteins within living organisms, almost all tertiary structures can be categorized into one of ten fundamental protein folds3 Six of these fundamental “superfolds” exhibit symmetry at the level of the tertiary fold: A set secondary structure element is repeated at least twice in a defined sequential order and internally symmetric spatial arrangement4. It has been postulated that these symmetric superfolds have evolved via gene duplication and fusion events from homooligomeric proteins (Figure 1): Fusion of monomer units into a single domain removes the entropic cost of assembling the oligomer, thereby increasing thermodynamic stability and kinetic foldability2. Diversification on the sequence level achieves more complex biological functions and removes evidence of symmetry at the level of the primary sequence5. However, the overall fold remains symmetric.
Figure 1.

The self-attraction of a monomeric protein (A) yields a homodimeric complex with N and C termini close in space (B), and thereby a symmetric interface. If N- and C-termini are spatially proximal, gene duplication (C) and fusion (D) preserve the energetically favorable interaction across the interface. Diversification on the sequence level (E) allows for more complex function to be achieved (the introduction of mutations is represented by gray shading). The circles represent the alpha helices, while the triangles represent the beta strands.
Interestingly, the vast majority of homodimeric complexes in the Protein Data Bank (PDB) exhibit a symmetric arrangement of the two monomer units6. Andre et al. use explicit energy docking calculations with ROSETTA to investigate the bias toward very-low-energy complexes in symmetric homodimeric complexes1. The study finds a two-fold greater variance in the interaction energy of random symmetric protein-protein docking arrangements leading to an increased chance of observing highly attractive interactions. It is concluded that symmetric homodimers are select ed for in evolution, thus explaining their abundance in nature.
This bias towards symmetry in homodimers would be preserved on the level of folds that arose through gene duplication and fusion. A boundary condition for this evolutionary strategy is that the C-terminus of one domain is spatially close to the N-terminus of the other domain in the tertiary structure of the homodimer so that after the gene duplication and fusion event, the new protein domain can fold without disrupting the structure of the symmetric subunits. Figure S-1 demonstrates that 6.5% of 461 representative symmetric homodimers in the PDB have N- and C-termini closer than 20Å. The twelve homodimers with the shortest distance between their N and C termini are also displayed in Figure S-1.
The (βα)8-barrel superfold is one of the most frequently observed folds in nature, comprising 10% of proteins with known structures7. The domain is composed of eight (βα) units, linked together by loops which wrap around to form a cylinder of parallel β-strands (β-barrel, Figure S-2) surrounded by a layer of parallel α-helices. The wide variety of amino acid sequences that adopt the fold makes it difficult to determine an evolutionary history. It is possible that the (βα)8-barrel fold was created several times independently and via different evolutionary routes8. However, one of the most popular hypotheses is that it arose through gene duplication and fusion of (βα)2n units (Figure 1). Wierenga suggests that the (βα)4-half-barrel might be the smallest evolutionary unit because of its prominence in two-fold symmetric (βα)8-barrel proteins9. However, structure-based multiple sequence alignments reveal a common GXD motif in the loops that precede even-numbered β-strands suggesting evolution from (βα)2 quarter barrel units. Soding et al.10 detected a distinct two- and four-fold internal symmetry in members from several different SCOP superfamilies of the (βα)8-fold.
More evidence indicating the evolution of (βα)8 barrels from gene duplication and fusion comes from imidazole glycerol phosphate synthase (HisF, Figure 2A). The HisF (βα)4 half barrel structures have a sequence identity of only 16% but superimpose with root mean square distance (rmsd) deviations of 2.1 Å. In addition, the N- and C-terminal halves of HisF can be expressed separately and self-associate to form inactive homodimers11. When co-expressed in vivo or refolded together in vitro, the two half barrels combine to form an active heterodimer. In an attempt to reconstruct the evolutionary events that gave rise to HisF, Sterner et al. fused two copies of the gene encoding the C-terminal HisF (βα)4-half-barrel. Although the resulting protein “CC” was poorly soluble and unfolded with low cooperativity12, an iterative process combining rational redesign followed by random mutagenesis and selection generated a stable protein “C***C” with native-like properties13. However, while obtaining impressive results, this strategy has disadvantages: a) the resulting protein C***C is no longer perfectly symmetric on the sequence level as rational redesign and random mutagenesis introduced different mutations in both subunits; b) the approach assumes that the C-terminal half of the barrel was duplicated and all mutations accumulated in the N-terminal half during evolution which is highly unlikely; and c) the process involves an iterative improvement of the designed protein through trial-and-error, offering limited insight in the fundamental forces that determine protein stability and limiting its application to other proteins.
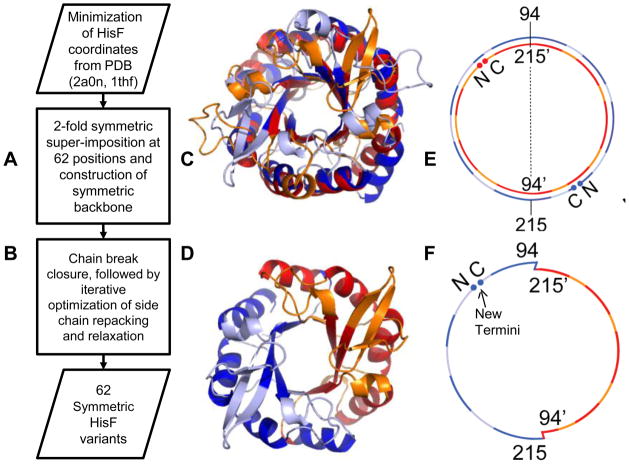
Figure 2.
The left panel shows the steps taken to computationally create the symmetric variants. Panel C is the superimposition of HisF, where the 62 cut sites are shown in dark blue on one copy, and red on another. The non-cut sites are shown in light blue and orange, respectively. Panel D is the symmetric variant created from duplicating cut sites 94–215 on each of the superimposed halves, which is termed FLR. Panels E and F are a schematic representation of the same process, showing the cut sites of FLR at 94 and 215. Panel F shows the location of the new termini. A larger copy of Figure 2 is also found in the supplement (Figure S-3).
Assembly of larger proteins from symmetric subunits presents not only an attractive strategy in evolution; it could also facilitate the computational design of large proteins, as the symmetry constraint reduces the sequence and conformational search space. Further, it enables a stepwise protocol that first designs and characterizes stable subunits before optimizing interfaces between these subunits for self-assembly. Both strategies will reduce the computational resources needed thereby enabling the design of larger proteins.
The present study reverse-engineers a perfectly two-fold symmetric (βα)8-barrel on the basis of a well-defined energy potential and with a reproducible in silico protocol (Figures 2, S-4 and S-5). It thereby overcomes above-mentioned limitations of previous studies and explores the potential to exploit protein symmetry for the design of larger protein domains. The promising results obtained by Seitz et al. when fusing the C-terminal half of HisF to form CC12 inspired this research to systematically test 62 symmetrized HisF-variants in silico. We expect to identify energetic hotspots in the CC protein and determine a low-energy symmetric version of HisF. Note, that this study was completed independently and before the experimental structure of the asymmetric C***C became available13. While the resulting protocol is based on the HisF structure as a template, the general strategy can be applied for de novo design of larger proteins. Specifically, the symmetry constraint reduces the sequence and conformational search space by a factor of two, making the respective computer simulations feasible.
HisF was first superimposed on itself with a 180° rotation around the main β-barrel axis using a structure-structure alignment algorithm14 (Figure 2A, C and E). As a result of the two-fold symmetry on the topology level, 62 sequence position pairs superimpose at 2.1Å in the protein backbone. These 62 positions reside in parts of the structure that follow the two-fold symmetry most closely; i.e. α-helices and β-strands. At each of these 62 positions it is possible to cross-over from one HisF copy to the other. For example, looking at the position pair (94::215) – starting at amino acid 94 of copy 1 follow the HisF backbone trace to amino acid 215 of copy 1. It superimposes with amino acid 94 of the 180° rotated copy 2 of HisF. Then, continue tracing on copy 2 until residue 215 in copy 2 is reached and it is possible to jump back to amino acid 94 of copy 1 (Figure 2D and F). In result, 62 cyclic symmetric HisF variants were created, each duplicating a different half of the original protein. Note, that depending on the cut point, a different set of HisF loops is kept and duplicated resulting in symmetric variants of different length. In essence this protocol is a protein design experiment with a constraint on the sequence and conformational space.
Cyclic coordinate decent (CCD)15 was used to rectify the slight geometry imperfections at the jump points. N- and C-termini are reintroduced into the cyclic proteins at positions equivalent to the termini positions in HisF between β1 and α8. Iterative energy optimization including backbone perturbation, side chain repacking, and gradient-based energy minimization16; 17 were applied to optimize the structure. For each of the 62 symmetric HisF variants, this energy minimization protocol was repeated 40 times in independent runs that started from either one of two experimental structures (1thf, 2a0n)18 and one of ten backbone conformations created in the CCD loop closure protocol. Repeating the protocol from slightly different backbone conformations ensures dense sampling of the local conformational space thereby providing a more accurate determination of the minimum energy.
To prioritize symmetric HisF variants for experimental validation, the 62 variants were ranked by energy. Depending on the length of the loops the variants had between 238 and 248 residues. To remove a bias towards larger proteins, the energy was normalized by the number of amino acids prior to ranking and is hence reported as ROSETTA Energy Units per Amino Acid (REU/AA). The top panel of Figure S-4 graphically depicts the lowest energy for each of the 62 backbones ranging between −2.80 REU/AA (68::189) and −3.16 REU/AA (94::215). This most stable variant 94::215 was termed FLR based on the amino acid sequence at the cut point. To obtain a baseline for comparison, the experimental structures of HisF were (1thf, 2a0n) minimized using the identical protocol and yielding −3.06 REU/AA.
All designs with energies better than −3.10 REU/AA were located in regions between sequence position pairs 93::214 (last turn of α3 and throughout β4) and 102::223 (last turn of α7 and throughout β8). Consistently low energies throughout these regions of secondary structure suggest that the half-barrel that contains β4-α4-β5-α5-β6-α6-β7-α7 of HisF yields the most stable two-fold symmetric variants largely independent from the precise position of the cut points. This region contains the elongated β5-α5 loop which consists of a three-stranded β-sheet. As this region is duplicated, the β-strand content of these symmetric variants increased from 24% in HisF to 30% in the symmetric HisF variants. The α-helical content remained constant with 35%.
Interestingly, the variant that is most similar to the fusion of the C-terminal half CC described by the Sterner laboratory (120::244) scored among the best (−3.06 REU/AA) giving an indication of why the experiments by the Sterner group were successful. The Sterner group further noted a salt-bridge cluster in HisF which contained R5 (β1), E46 (β2), K99 (β4), and E167 (β6). The cluster is irregular in the sense that not all four amino acids originate from β-strands with even numbers, i.e. they fail to form a single layer. The uncharged amino acid A220 in β8 cannot contribute to the salt-bridge cluster and is replaced with R5 (β1). This irregularity is responsible for the absence of the salt-bridge cluster in CC. Reintroduction of this salt-bridge cluster into the fusion of the C-terminal half of HisF greatly improved the proteins stability experimentally19; 20 and also in our simulations from −3.06 REU/AA to −3.10 REU/AA. The lowest energy symmetric HisF variants of the present study, including FLR, duplicate β4 instead of β8 when compared to CC. Thereby these proteins contain the salt-bridge cluster at the base of the β-barrel consisting of E46 (β2), K99 (β4), E167 (β2), and K220 (β4).
The active site of HisF is located at the C-terminal face of the barrel. The conserved and catalytically essential residues in HisF are located in positions D11 (β1) and D130 (β5). In FLR D130 is duplicated and D130′ is placed in an equivalent position to D11. Further, HisF binds two phosphate groups of the substrate through residues G82, N103, T104 in site 1 and D176, G177, G203, A224, S225 in site 2. FLR duplicates N103, T104, D176, G177, G203 forming two intact phosphate binding sites.
A truncated variant consisting of amino acids 1::121 of FLR was constructed and termed halfFLR (see supplement for sequence details). The ROSETTA energy of the monomer is substantially reduced when compared to FLR (−2.82REU/AA). A symmetric homodimer of halfFLR mimicking the structure of FLR is predicted to regain full stability (−3.16 REU/AA). The dimer interface is around 1700Å2. Dimerization therefore stabilizes the protein by around 11% in REU/AA and is predicted to occur spontaneously. This property of halfFLR further validates the hypothesis of the creation of symmetric superfold from symmetric homodimers through the generation of a hypothetical, ancestral homodimer for HisF (Figure 1C).
In an additional step, the sequence of all 62 variants was optimized enforcing a symmetry constraint to test if additional mutations can further stabilize the protein. While mutations were introduced in many of the 62 variants, FLR remained unaltered, indicating that its sequence is optimal. Even after optimizing the sequence of all 62 variants, FLR maintains the best overall energy and was therefore selected for experimental verification.
Details on the construction of the genes for FLR and halfFLR are given in the supplement. The plasmids were transformed into E. coli host strain BL21 (de3) pLysS for expression. The designed proteins expressed at greater than 75% within the soluble fraction. Purification by metal affinity resin yielded approximately 20 mg/ml protein per liter induction at greater than 95% purity.
We observed a mono-dispersed particle size distribution with an average hydrodynamic radius of 50±20Å for FLR and 60±20 Å for halfFLR. Both are within error of the expected value for the FLR monomer and the halfFLR homodimer. Analytical size-exclusion chromatography (SEC) indicated a single symmetric peak at a volume that corresponds to a 30kDa species for both proteins, further suggesting homodimeric halfFLR (Figure S-6). Secondary structure element percentages were calculated based on far-UV circular dichroism (CD) spectra and confirmed the predicted constant α-helical and increased β-strand content relative to HisF (Figure S-7). The stability of halfFLR and FLR were assessed by guanidine-induced denaturation and indicated a slightly decreased stability (2.6 and 2.8M guanidine, respectively) than HisF (3.5M guanidine), but showed cooperative unfolding (Figure S-8). Differential scanning calorimetry indicates the protein aggregates at high temperatures. Two-dimensional NMR (1H-15N HSQC) indicated compactly folded proteins with approximately half the number of peaks as HisF (140 vs. 252 peaks, Figure S-9). The number can be slightly larger than precisely half of the 252 signals for HisF as the perfect 2-fold symmetry is broken at the N-terminus (see supplement).
The expressed protein variants (FLR and half-FLR) are less stable than the wild-type protein HisF, even though at least FLR should be more stable according to the computational calculation. This is not surprising for two reasons: Firstly, FLR has a slightly different sequence composition than HisF which biases the energy function. When computing and expected energy of FLR from HisF through summation of the ROSETTA energies of the duplicated residues 94::215 in HisF we predict −3.17 REU/AA matching closely our actual findings. Secondly, the ROSETTA energy function is inaccurate and its correlation with free energies is nonlinear. A conversion of REU to predicted ΔG is therefore questionable for a variety of reasons including that entropic contributions to the energy function are largely ignored and that some interactions are double-counted. However, the ROSETTA energy function is generally accurate when ranking variants and mutants making it a successful tool in protein design. The standard deviation of the REU/AA values as computed from multiple minimizations of the same design variants is 0.02 REU/AA.
Analytical ultracentrifugation (AUC) of the halfFLR species was performed to assess the percent dimerization of the protein. Sedimentation velocity AUC experiments indicate a single dimeric species. Similarly SEC and dynamic light scattering experiments display a single dimeric species. No monomer or other oligomeric state can be observed under any conditions, thereby preventing the determination of the dissociation constant. Using the protein concentration of 160μM in the AUC experiment and assuming the fraction of the monomeric version at less than 1%, we determine a conservative upper limit for Kd of 20nmol, thus confirming the tight interaction predicted computationally.
The experimental structure of FLR was determined through X-ray crystallography to a resolution of 1.4 Å using the computational model for molecular replacement (PDB-code 3DTN). The experimental structure shows the protein is folded into the predicted (βα)8 barrel structure with 0.87 Å RMSD between the computational and experimental model backbones. Amino acid side chain conformations agree in 87% between model and experiment. Interestingly, 1.5 copies of FLR reside in each unit cell which is diagnostic of the structural symmetry. The distances between equivalent positions agree to a RMSD of 0.29 Å The two halves superimpose to a RMSD of 0.34 Å for Cα positions making FLR perfectly symmetric within the resolution of the experiment. The two halves superimpose for Cα positions to an RMSD of 0.339Å. The FLR structure also indicates the predicted salt-bridge cluster at the base of the β-barrel consisting of residues E46 (β2), K99 (β4), E167 (β2), and K220 (β4) is intact (shown in Figure 3-A). The catalytic aspartate residues D9/130 and the phosphate binding sites N103/224, T104/225, D55/176, G56/177, G82/203 are largely unperturbed (Figure 3-C).
Figure 3.
The computationally predicted models are shown in blue, while the experimental structures are shown in green in all panels. (A) The density of the salt bridge cluster (grey) of FLR is shown superimposed with the computational side chains in red. These are residues E46, K99, E167, K220. (B) The density of the contacts between helix 1 and strand 1 is shown superimposed with the computational side chains in red revealing an excellent side chain recovery. (C) The catalytically important residues are shown superimposed with the predicted model and appear unperturbed. However, the missing density in the loops could explain the loss of activity in FLR. (D) There is an overall agreement between the model and the experimental structure of halfFLR. (E) The interface between the two halves of dimeric halfFLR shows slight deviations from the computational model, which is likely due to the model being a monomeric half. (F) The catalytically important residues of halfFLR show the same flexibility as the FLR protein, again possibly explaining the lack of activity.
The experimental structure of halfFLR was determined with a resolution 2.3Å (PDB-code 3DTM). The computational model of halfFLR was used for phasing and all comparisons. The experimental structure shows the protein is folded into the predicted (βα)8 barrel structure with 0.49 Å difference between the computational and experimental backbone coordinates. HalfFLR clearly shows the two monomeric halves of the protein are assembled as a symmetric dimer (Figure 3-D). Important structural features such as interface contacts and catalytically important residues are represented in Figure 3-E and F, and show agreement with the predicted model. HalfFLR’s phosphate binding sites are occupied with phosphate ions which were present in the crystallization buffer at a concentration of 12.5 mmol·L−1. All crystallography data collection and refinement statistics are listed in Table 1 of the supplement.
Wild-type HisF converts PRFAR (N1-[(5′-phosphoribulosyl) formimino]-5-aminoimidazole-4-carboxamide ribonucleotide) to AICAR (5-aminoimidazole-4-carboxamide ribonucleotide) and IGP (Imidazole glycerol phosphate). This reaction is monitored by a decrease in adduct absorbance at 300nm. FLR and halfFLR did not yield measureable activity. Although the catalytic residues were largely unperturbed, missing density caused by the flexibility of the loop containing D176 and G177 could explain the loss of activity (Figure 3-C). These results will be investigated in ongoing studies.
This study presents the first structure of a sequence and structurally symmetric (βα)8 barrel protein which is soluble, monomeric, and folds cooperatively. Primary structure can be constrained to conform to the symmetry of the tertiary structure and the protein still folds properly. The results of the present study are consistent with the gene duplication and fusion hypothesis of symmetric superfolds. Moreover, it creates two hypothetical ancestral variants of HisF: a sequence-symmetric variant of HisF and a related half-barrel protein that spontaneously dimerizes to a symmetric homodimeric (βα)8 barrel. Conserved structural traits such as salt-bridges and core packing are noted in these symmetric designs. The computational design protocol was highly accurate as the x-ray structures agreed within 0.87 Å and 0.49 Å with the predicted models. To date, the largest de novo designed protein consists of 106 amino acids. By taking advantage of the inherent symmetry of the (βα)8 barrel fold in the protein HisF, a protein of 242 amino acids was computationally designed; although arguably not de novo. However, the strategy to connect identical small proteins to larger architectures can be extended to the de novo design of larger domains.
Supplementary Material
Acknowledgments
This work was supported by Defense Advanced Research Projects Agency, Protein Design Project.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Experimental procedures and crystallographic data are included in the supplementary information. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Andre I, Strauss CE, Kaplan DB, Bradley P, Baker D. Proc Natl Acad Sci U S A. 2008;105(42):16148–16152. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolynes PG. Proc Natl Acad Sci U S A. 1996;93(25):14249–14255. doi: 10.1073/pnas.93.25.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orengo CA, Jones DT, Thornton JM. Nature. 1994;372(6507):631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 4.Brych SR, Kim JW, Logan TM, Blaber M. Protein Science. 2003;12(12):2704–2718. doi: 10.1110/ps.03374903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Genome Biol. 2007;8(4):R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Nature. 2008;453(7199):1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlt JA. Nat Struct Biol. 2000;7(3):171–173. doi: 10.1038/73249. [DOI] [PubMed] [Google Scholar]
- 8.Nagano N, Orengo CA, Thornton JM. J Mol Biol. 2002;321(5):741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 9.Wierenga RK. FEBS Lett. 2001;492(3):193–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- 10.Soding J, Remmert M, Biegert A. Nucleic Acids Res. 2006;34(Web Server issue):W137–142. doi: 10.1093/nar/gkl130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hocker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R. Nat Struct Biol. 2001;8(1):32–36. doi: 10.1038/83021. [DOI] [PubMed] [Google Scholar]
- 12.Seitz T, Bocola M, Claren J, Sterner R. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 13.Hocker B, Lochner A, Seitz T, Claren J, Sterner R. Biochemistry. 2009 doi: 10.1021/bi802125b. [DOI] [PubMed] [Google Scholar]
- 14.Ortiz AR, Strauss CEM, Olmea O. Protein Sci. 2002;11:2606–2611. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canutescu AA, Dunbrack RL. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley P, Misura KM, Baker D. Science. 2005;309(5742):1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 17.Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. Nature. 2007 doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Science. 2000;289(5484):1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 19.Hocker B, Claren J, Sterner R. Proc Natl Acad Sci U S A. 2004;101(47):16448–16453. doi: 10.1073/pnas.0405832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sterner R, Hocker B. Chem Rev. 2005;105(11):4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.