Abstract
Background
Autophagy is a highly conserved mechanism for degradation and recycling of long-lived proteins and damaged organelle to maintain cell homeostasis. Deregulation of autophagy has been associated with tumorigenesis. Beclin 1 is an essential autophagy protein and its upregulation has been observed in most colorectal cancer tissues. However, there is a small population of colorectal cancers with downregulation of Beclin 1.
Aim
The purpose of this study was to investigate the role autophagy plays in colorectal cancers with downregulaion of Beclin 1.
Methods
LC3 protein, an autophagosome marker, was assessed by ICH and WB in colorectal cancers tissues. An anti-tumor effect of Beclin 1 was examined by introducing exogenous Beclin 1 in vitro. Colony formation assay, growth curves and mouse xenograft were analysed.
Results
Our results showed that LC3 was suppressed in the colorectal cancers (9.86 %) with downregulation of Beclin 1. Moreover, overexpression of Beclin 1 inhibited colorectal cancer cell growth and enhanced the rapamycin-induced antitumor effect in vitro.
Conclusion
Downregulation of Beclin 1 and autophagy inhibition play an important role in a part of colorectal cancers. Activating autophagy or overexperssion of Beclin 1 may be an effective treatment for some colorectal cancers. Detection of expression profile of Beclin 1 in colorectal cancers could be a strategy for new diagnostic and therapeutic methods.
Electronic supplementary material
The online version of this article (doi:10.1007/s10620-013-2732-8) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, Beclin 1, Autophagy, Rapamycin
Introduction
Autophagy is a cellular degradation pathway for the clearance of cytoplasmic materials, damaged organelles and aggregate-prone proteins in lysosomes. The recycling of these intracellular constituents also serves as an alternative energy source during periods of metabolic stress to maintain homeostasis and viability. In tumour cells with defects in apoptosis, autophagy allows prolonged survival. Intriguingly, defective autophagy is also reported to play a critical role in tumorigenesis. But the mechanism has not been determined. Recent evidence shows that autophagy provides a protective function to limit tumour necrosis and inflammation, and to mitigate genome damage in tumour cells in response to metabolic stress [1–3]. As the first identified mammalian autophagy effecter, Beclin 1 is essential for the initiation of autophagy via its interaction with the class III phosphatidylinositol-3-kinase Vps34 [4]. The beclin 1 gene maps to a tumour susceptibility locus on human chromosome 17q21 that is mono-allelically deleted in 75, 50 and 40 % of ovarian, breast, and prostate cancers, respectively [5–7]. Beclin 1 has been characterized as a tumor suppressor, since heterozygous disruption of Beclin 1 mice display increased proliferation and increased frequency of spontaneous malignancy and mammary neoplasia [8, 9]. And Beclin 1 expression suppresses tumorigenicity of cancer cells [10]. Furthermore, the Beclin 1 gene is monoallelically deleted in most cases of sporadic human cancers [11, 12].
Compared to Beclin 1 level in normal adjacent breast tissue, low Beclin 1 protein expression was confirmed in a small series of human breast tumors, but any correlation between allelic beclin 1 loss, and thus defective autophagy and clinical outcome in breast cancer remains to be investigated. Decreased Beclin 1 mRNA and protein expression was also demonstrated in glioblastoma multiforme and other high-grade brain tumors. In contrast, higher expression of Beclin 1 was detected in the majority of colorectal (95 %) and gastric (83 %) carcinomas when compared to the normal stomach and colorectal mucosa, which showed very low or undetectable Beclin 1 levels. However, in those cases, Beclin 1 expression did not show any correlation with pathological or clinical characteristics, suggesting that it plays a role in cancer development but not in cancer progression [13]. Interestingly, there is a small part of colorectal cancers with low expression of Beclin 1. Moreover, low expression of Beclin 1, associated with high Bcl-xL predicts a malignant phenotype and poor prognosis of gastric cancer [14]. Therefore, it is conceivable that the tumor suppressive function of Beclin 1 may be individual-specific, which is certainly worthy of further investigation.
In this study, we investigated the role that autophagy plays in colorectal cancer cases with low expression of Beclin 1. Our data revealed that some colorectal cancers, which are characterized by reduced Beclin 1 level, displayed impaired autophagy, while introducing exogenous Beclin 1 to colorectal cancer cells restored impaired autophagy and inhibited tumorigenesis.
Methods
Tissue Samples and Cell Lines
Surgically resected specimens from colorectal cancer and matched adjacent normal tissues were collected from 175 colorectal cancer patients undergoing surgery at Hunan Provincial People’s Hospital during the period 2010–2012 in accordance with China law and ethical guidelines, and informed consent was obtained from patients prior to resection. The corresponding adjacent normal tissue samples were selected 12–15 cm away from the site at which the primary tumor was sampled. HT29 cell line was purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (GIBCO) supplemented with 10 % fetal bovine serum (GIBCO) and 4 mM glutamine.
Immunohistochemistry
Paraffin-embedded sections were deparaffinized and subjected to immunohistochemical staining for Beclin 1 and LC3 antibodies (Cell Signal Technology). The sections were microwaved in 10 mM sodium citrate buffer (pH 6.0) at 10 min intervals for a total of 20 min. Endogenous peroxidase activity was blocked by incubating the sections in a solution of 3.0 % hydrogen peroxide for 20 min at room temperature. After washing in PBS the sections were incubated with the primary Beclin 1 (1:200) and LC3 (1:50) antibody, overnight at 4 °C. The sections were washed with PBS and incubated with biotinylated secondary antibody for 30 min, followed by incubation with streptavidin–biotin–peroxidase complex, a solution 3–3′diaminobenzidine (Sigma) containing 1.0 % hydrogen peroxide, and lightly counterstained with Harris hematoxylin. All images were analyzed using Image Pro Plus software (Media Cybernetics, Bethesda, MD). The total intensity of staining and the area covered by whitespace (non-tissue) were recorded for each image (5–6 images/slide).
Western Blot
Tissues were homogenized with lysis buffer containing 50 mM Tris–HCl, 150 mM NaCl, 1 % sodium deoxycholate, 0.1 % SDS, 20 mM EDTA, 1 mM NaF, and 1 % Triton X-100 (pH 7.4) with protease inhibitors (Promega). The protein concentration was determined using the BCA protein assay Kit (Priece). Lysis was run in a 8–15 % sodium dodecyl sulfate–polyacrylamide electrophoresis (SDS-PAGE) gel, transferred to PVDF membranes (Millipore), and incubated with antibodies against Beclin 1, LC3 and actin (Sigma) and visualized by enhanced chemiluminescence (GE).
Retrovirus Production
For retrovirus production, Beclin 1 was cloned into the vector pQCXIP (Clontech). HEK 293T cells were seeded in 10-cm diameter dishes. The HEK 293T packaging cells (at 30–50 % confluence) were co-transfected using Lipofectamine (Invitrogen) with pE-ampho vector (Takara Bio) and pQCXIP-Beclin 1. Retroviral particles were collected, filtered through the 0.45-μm syringe filter and used in the presence of polybrene (8 μg/ml final concentration) to infect cells for 12 h.
Colony Formation Assay and Growth Curves
Forty-eight hours after infection, the HT-29 cells were replated in 10-cm diameter dishes and maintained in conditional medium containing 800 μg/ml of G418 (GIBCO). Cultures were replated in the densities of 1 × 103, 5 × 102, or 2.5 × 102 on 10 cm-diameter dishes in triplicates and maintained for 2 weeks. The neoresistant colonies were fixed with methanol, stained with Giemsa, and counted. The number of colonies on the control dishes was used as the 100 % in this assay.
The cells were infected with retrovirus. Growth curves were generated by plating 105 cells in the DMEM medium for 24, 48 and 72 h in triplicates. The cells were harvested with trypsin and counted at intervals.
Mouse Xenograft Studies
For HT-29 cancer cell xenograft experiments, 4–5 weeks aged female nu/nu nude mice (purchased from Shanghai Slac Laboratory Animal) were approved by the University of Hunan Normal University Animal Care and Use Committee. Xenograft tumors were established by s.c. injecting 1 × 106 HT-29 cells suspended in 50 % matrigel into the flanks of severe combined immunodeficiency mice. Tumor measurements were taken on day 30. Tumor growth was monitored by calipers, and tumor volumes were calculated according to the formula: 0.5 × length × width2. Seven mice were injected and 13 tumors were assessed for each condition.
Statistical Analyses
Statistical analysis was performed using the Mann–Whitney U test. All of the results were expressed as mean ± SD values; statistical analysis for the multiplicity was performed using ANOVA or Student’s t test, where appropriate. The results were considered to be statistically significant when P values were <0.05.
Results
LC3 was Suppressed in the Tumour Tissues with Low-Expression Level of Beclin 1
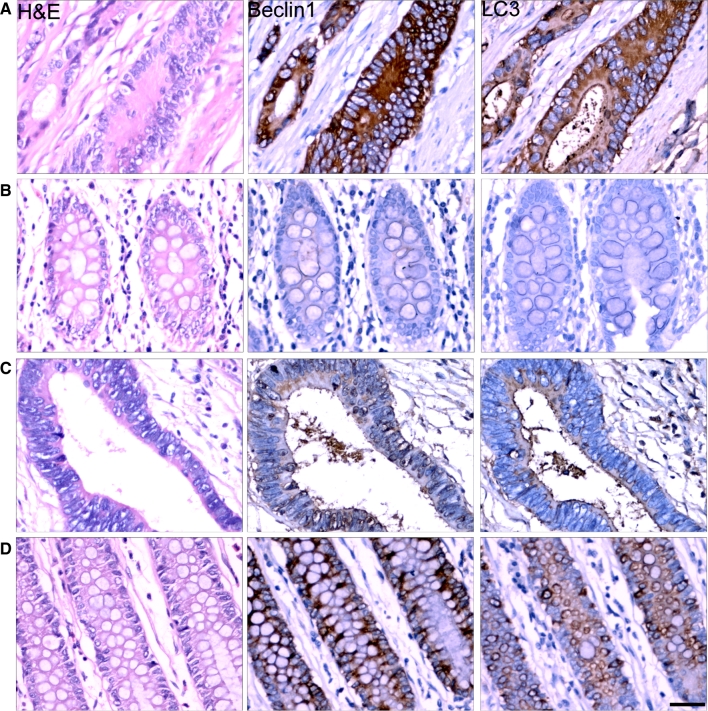
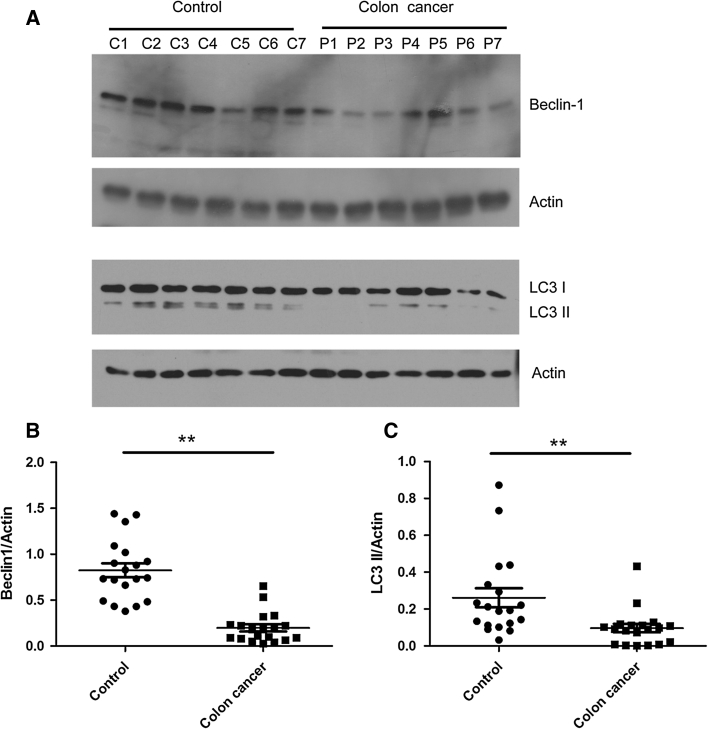
All of the tumors were categorized based on the histopathologic diagnosis. Tumor samples were reevaluated by a pathologist to confirm the diagnosis and were graded according to the World Health Organization criteria. Beclin 1, a haploinsufficient tumor suppressor gene [9], is essential for early embryonic development and may be involved in colorectal tumorigenesis [15]. To confirm the role of Beclin 1 in colorectal tumorigenesis, we detected the expression levels of Beclin 1 in 175 colorectal cancer tissues by immunohistochemstry. Most of them showed strong Beclin 1 staining (Fig. 1a) compared with paired normal adjacent tissues (Fig. 1b), but 19 colorectal cancer tissues present weak Beclin 1 and LC3 staining (Fig. 1c) in comparison to paired control (Fig. 1d). We further tested for Beclin 1 and LC3 II in these 19 samples by western blot (Fig. 2). Our results showed that the expression of Beclin 1 in cancer tissues were significantly lower than that in paired normal adjacent tissues. Meanwhile, we also found that expression of LC3 in these 19 samples was decreased (Fig. 2). It indicated that the expression of Beclin 1 was reduced at least in partial colorectal tissues, and decreased Beclin 1 may be involved in autophagy regulation in colorectal tumorigenesis.
Fig. 1.
Expression of Beclin 1 and LC3 in colorectal cancer tissues. Immunohistochemical staining for Beclin 1 and LC3 were performed in paraffin-embedded sections of colorectal cancer and normal adjacent tissues. Most tumors showed Beclin 1 and LC3 with strong staining (a) in comparison to paired normal adjacent tissues (b). But some colorectal cancer tissues showed weak staining (c) in comparison to paired normal adjacent tissues (d). Representative pictures were presented. Scale bars, 50 μm
Fig. 2.
The expression of LC3 II was decreased in colorectal cancer tissues with downregulation of Beclin 1. Western blot analysis of Beclin 1 and LC3 II expression in paired normal adjacent tissues (control) and colorectal cancer tissues (colorectal cancer). a Quantitative analysis of expression of Beclin 1 (b) and LC3 II (c) in colorectal cancer tissues and normal adjacent tissues. Actin is a loading control. Data were presented as a mean ± SD, n = 19, **P < 0.01
Overexpression of Beclin 1 Inhibits Colorectal Cancer Cell Growth
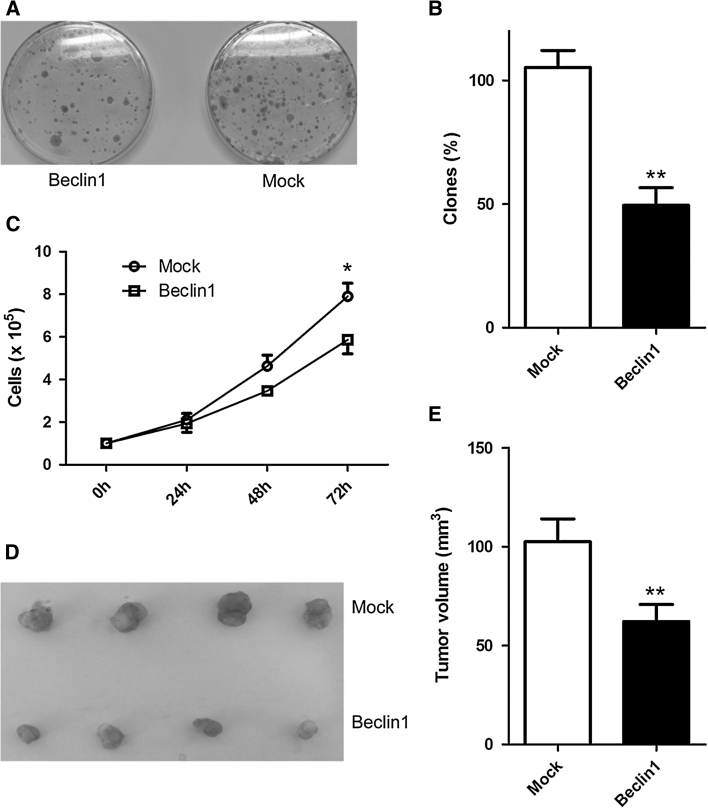
To investigate whether an anti-tumor effect of Beclin 1 was affected by exogenous Beclin 1, we expressed exogenous Beclin 1 by infection of retrovirus in HT29 cells in which the expression of Beclin 1 mRNA was lower than in the other colorectal cancer cell lines [16]. Overexpression of Beclin 1 potently inhibited colony-forming activity in the HT29 cell line (Fig. 3a, b) and resulted in a reduction in the rate of cell growth (Fig. 3c). Moreover, Beclin1 induced a decreased tumor growth in vivo when stably expressed Beclin1 cells were injected as xenografts (Fig. 3d, e). However, exogenous Beclin 1 did not show an affection on inhibition of cell growth in HCT-116 and SW48 cell lines with high-level Beclin 1 in comparison to HT-29 and wild type p53 [17] (Fig. S1).
Fig. 3.
Growth suppressive effects of Beclin 1 in HT-29 cells. a Colony formation assay demonstrating that wild-type Beclin 1 suppresses cell growth in HT29 cells. Representative dishes are shown for control virus infection (Mock) and Beclin 1 virus infection (Beclin 1). Quantization of results is shown in (b). Overexpression of Beclin 1 reduces growth rate in HT29 cells (c). HT29 cells were infected with control virus and Beclin 1 virus. All experiments were performed in triplicate. *P < 0.05 (t test) for Beclin 1 compared to control. Beclin 1 suppressed tumor growth in vivo (d, e). HT29 cells stably expressed Beclin 1 and control were injected as xenografts. Representative tumors are shown in d and tumor sizes are shown in e (n = 13). **P < 0.01 (t test) for Beclin 1 compared to control. Data presented as mean ± SD
Enhancement of the Rapamycin-Induced Antitumor Effect on HT29 Cells by Overexpression of Beclin 1
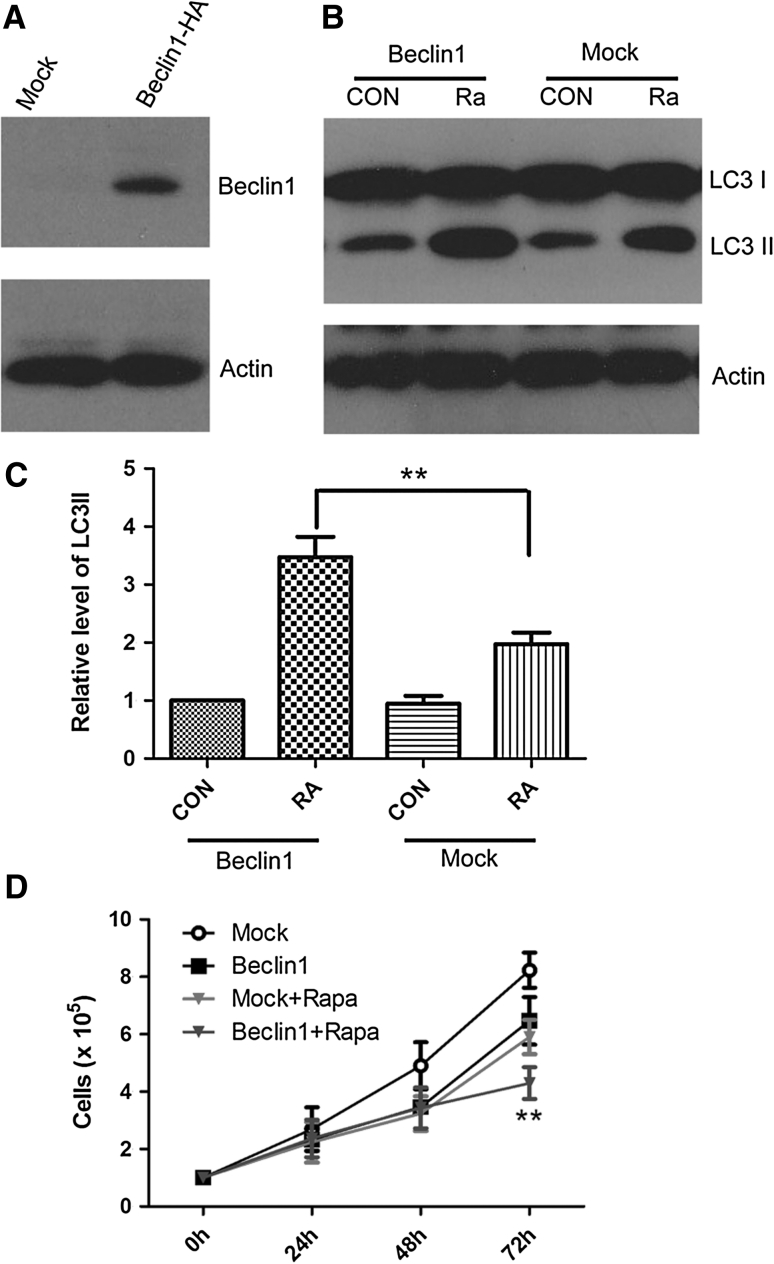
We proposed that mammalian autophagy gene beclin 1 negatively regulated tumorigenesis in the HT29 cell line. Therefore, we investigated the effects of beclin 1 gene transfer on the autophagic activity and growth properties of HT29 cells. HT29 cells have a low expression level of endogenous Beclin 1 [16]. To investigate whether impaired autophagy activity was affected by exogenous Beclin 1, we infected HT29 cells with a recombinant retrovirus expressed Beclin 1-HA. Exogenous Beclin 1 fused HA peptide tag was detected by western blot (Fig. 4a). Our results showed that Beclin 1 increased the rapamycin-induced level of LC3 II (Fig. 4b, c), and enhanced the rapamycin-induced antitumor activity on HT29 cells (Fig. 4d).
Fig. 4.
Effect of enhanced rapamycin-induced autophagy by overexpression of Beclin 1. Beclin 1 was overexpressed in HT29 cell by infection. HT29 cell was infected with virus containing Beclin 1-HA for 72 h and detected by immunoblotting with antibody against HA (a). Rapamycin-induced autophagy was enhanced of by infection of Beclin 1. HT29 cell was infected with Beclin 1 or mock, and treated with 100 nM rapamycine for 24 h. Cells were lysed and processed for immunoblotting with antibody against LC3 (b). LC3II/Actin was quantified (c). Data were presented as a mean ± SD of three independent experiments. **P < 0.01. Overexpression of Beclin 1 reduces growth rate in HT29 cells and enhances the rapamycin-induced anti-tumor effect on HT29 cells (d). ** Beclin 1 versus Beclin 1 + Rapa P < 0.01
Discussion
Decreased expression of Beclin 1 has been observed in human breast, ovarian and brain tumors [18, 19]. But Beclin 1 was rarely mutated in both colorectal and gastric cancers, suggesting that point mutations are not a main mechanism of Beclin 1 inactivation in colorectal cancers [20]. Surprisingly, increased expression of Beclin 1 was observed in most malignant colorectal and gastric epithelial cells [13, 15]. Consistent with our study, we also confirmed that most colorectal cancer showed strong Beclin 1 staining (156/175, 89.14 %). However, a small part of colorectal cancers showed weak Beclin 1 staining (19/175, 9.86 %), compared with paired normal adjacent tissues (Figs. 1 and 2). Although the mechanism by which overexpression of Beclin 1 contributes to colorectal tumorigenesis is under intense investigation, we focus on the mechanism of the colorectal cancer with weak Beclin 1 staining in this study. We found that some of the colorectal cancer tissues show downregulation of Beclin 1 and low autophagy activity. We also demonstrated that overexpression of Beclin 1 and activation of autophagy inhibit colorectal cancer cell proliferation in the cell with Beclin 1 downregulation.
Beclin 1, the mammalian orthologue of yeast Atg6, has an essential role in autophagy, a process of programmed cell survival, which is increased during periods of cell stress and extinguished during the cell cycle. Beclin 1 interacts with several cofactors to regulate the lipid kinase Vps-34 protein and promote formation of Beclin 1-Vps34-Vps15 core complexes, thereby inducing autophagy [21]. The biochemical and genetic properties of Beclin 1 suggest its role in the induction of autophagy and inhibition of tumorigenesis [10]. Beclin 1 maps to a tumor susceptibility locus that is monoallelically deleted in a high percentage of human breast, ovarian and prostate cancers [18]. The monoallelic deletions of Beclin 1 in human cancer likely contribute to tumorigenesis, as targeted mutant mice with heterozygous disruption of Beclin 1 have decreased autophagy, and spontaneously developed tumors including lymphomas, lung carcinomas, hepatocellular carcinomas and mammary precancerous lesions as they age and undergo accelerated hepatitis B virus-induced carcinogenesis [8, 9]. Furthermore, it is consistent with our results that exogenous Beclin 1 gene inhibits tumor growth in colorectal cancer cell lines [16].
Beclin 1 is involved in regulating autophagy and apoptosis [21]. Autophagy is reported to play a critical role in colorectal tumorigenesis. Autophagy is activated in colorectal cancer cells and tissues and contributes to the tolerance to nutrient deprivation [22, 23]. Inhibition of autophagy markedly reduced cell viability and enhanced anti-tumor effects of several drugs for colorectal cancer [24–26]. However, the role of autophagy in colorectal tumorigenesis is paradoxical. Evidence has indicated that activation of autophagy contributes to colorectal tumorigenesis, but it also provides some advantages to the tumor suppressor function. Autophagy has been proposed to play a tumor-suppressive role in the early stages of tumorigenesis. Human tumors were detected with mutations in the PI3-kinase pathway, leading to activation of mTOR and thus suppression of autophagy [27, 28]. More importantly, mice with impaired autophagy, such as Beclin1−/− and Atg4C−/−, are more prone to develop tumors [9]. Defection and down-regulated expression of autophagy-related genes were associated with colorectal tumorigenesis [29, 30]. Induction of autophagy contributes to proliferative arrest or apoptosis in human colorectal cancer cells [31, 32]. This evidence suggested autophagy had a dual role in colorectal tumorigenesis [33]. We also investigated the role of autophagy in the HT29 cell line, in which the expression of Beclin 1 mRNA was lower than that in the other colorectal cancer cell lines [16]. We also introduced exogenous Beclin 1 in HCT-116 and SW48 cell lines with high-level Beclin 1 comparison of HT-29 and wild type p53 [17]. But no inhibition of cell growth was found (Fig. S1). The HT-29 cell line has an overproduced p53 mutant [34]. It was reported that p53 plays a negative role in regulating HMGB1/Beclin 1 complex [35]. Meanwhile, Beclin 1 controlled the levels of p53 by regulating the deubiquitination activity of USP10 and USP13 [36]. Our results showed knockdown p53 expression can increase Beclin 1 level in HT-29 cell (Fig. S2). So, an overproduced p53 mutant has a potential inactive effect on Beclin 1 and impairs autophagy in the HT-29 cell. But the exact mechanism still needs further study.
We showed activation of autophagy significantly reduced cell viability and enhanced anti-tumor effects of rapamycin for colorectal cancer cell line HT29. Autophgy was inhibited in colorectal cancer tissues with low-expression Beclin 1 (Fig. 2). Importantly, we also found exogenous Beclin 1 increases the expression of LC3 II in HT-29 cells (Fig. 4b). Although the Bcl-2/Beclin 1 interaction was clearly an important checkpoint in autophagy induction of rapamycin (an mTOR inhibitor), less apparent effects were seen on the release of Beclin 1 from Bcl-XL [37]. mTORC1 was inhibited and autophagy was initiated by the ULK1 complex. During starvation, the phosphorylation of Beclin 1 by DAPK and JNK1 trigger the dissociation of Bcl-2 from Beclin 1 [21]. The functional relationship between autophagy initiation complexs and nucleation complexes remains to be determined. Unknown regulative pathways of Beclin 1 may act in parallel with the mTOR pathway to regulate autophagy. Additional studies to evaluate these possibilities should be informative.
Conclusion
Our results indicate that defective Beclin 1 and autophagy inhibition may play an important role in a part of colorectal tumorigenesis and activation of autophagy by overexperssion of Beclin 1, which may be an effective treatment for some colorectal tumor. Thus, detection of the expression profile of Beclin 1 in colorectal cancer might be a strategy for new diagnostic and therapeutic methods.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Growth suppressive effects of Beclin1 in HCT-116 and SW48 cells. HCT-116 and SW48 cells were infected with control virus (mock) and Beclin1 virus (Beclin 1-HA). A Exogenous Beclin 1 tagged HA peptide was detected by western blot using HA antibody. B Overexpression of Beclin 1 has no effect on cell growth in HCT-116 and SW48 cells. All experiments were performed in triplicate. Results shown are the mean ± SD. (TIFF 11550 kb)
Fig. S2
Knockdown of p53 induces an increased Beclin 1 in HT-29 cells. HT-29 cells were transfected with 10 nM p53-targeting siRNAs (siRNA target sequence: siRNA#1 GCACAGAGGAAGAGAAUCU, siRNA#2GAAGAAACCACUGGAUGGA, siRNA#3GAGGUUGGCUCUGACUGUA), or siRNA control for 48 h. p53 and Beclin 1 expression were detected by western blot. Actin was used as a loading control. (TIFF 3139 kb)
Acknowledgments
Conflict of interest
None.
Contributor Information
Zhihong Chen, Email: zhihong6662002@yahoo.com.cn.
Yanchun Li, Email: liyanchun99@yahoo.cn.
Chi Zhang, Email: zhangchi9966@163.com.
Hongmei Yi, Email: yihongmei2012@yahoo.cn.
Chang Wu, Email: wuchang_scs@163.com.
Junpu Wang, Email: junpuwang1212@yahoo.cn.
Yuwu Liu, Email: liuyuwu_8923@yahoo.cn.
Jieqiong Tan, Email: tanjieqiong@sklmg.edu.cn.
Jifang Wen, Phone: +86-731-82650410, FAX: +86-731-82650410, Email: jifang.wen@yahoo.com.cn.
References
- 1.Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009;1793:1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 4.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 5.Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 6.Eccles DM, Cranston G, Steel CM, Nakamura Y, Leonard RC. Allele losses on chromosome 17 in human epithelial ovarian carcinoma. Oncogene. 1990;5:1599–1601. [PubMed] [Google Scholar]
- 7.Gao X, Zacharek A, Salkowski A, et al. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55:1002–1005. [PubMed] [Google Scholar]
- 8.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 11.Gong C, Bauvy C, Tonelli G, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32:2261–2272. [DOI] [PMC free article] [PubMed]
- 12.Fu LL, Cheng Y, Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int J Biochem Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Ahn CH, Jeong EG, Lee JW, et al. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou WH, Tang F, Xu J, et al. Low expression of Beclin 1, associated with high Bcl-xL, predicts a malignant phenotype and poor prognosis of gastric cancer. Autophagy. 2012;8:389–400. doi: 10.4161/auto.18641. [DOI] [PubMed] [Google Scholar]
- 15.Li BX, Li CY, Peng RQ, et al. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009;5:303–306. doi: 10.4161/auto.5.3.7491. [DOI] [PubMed] [Google Scholar]
- 16.Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453–1457. [PubMed] [Google Scholar]
- 17.Liu Y, Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci USA. 2006;103:976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 19.Miracco C, Cosci E, Oliveri G, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–436. [PubMed] [Google Scholar]
- 20.Lee JW, Jeong EG, Lee SH, Yoo NJ. Somatic mutations of BECN1, an autophagy-related gene, in human cancers. APMIS. 2007;115:750–756. doi: 10.1111/j.1600-0463.2007.apm_640.x. [DOI] [PubMed] [Google Scholar]
- 21.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii S, Mitsunaga S, Yamazaki M, et al. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99:1813–1819. doi: 10.1111/j.1349-7006.2008.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato K, Tsuchihara K, Fujii S, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 24.Yang PM, Liu YL, Lin YC, Shun CT, Wu MS, Chen CC. Inhibition of autophagy enhances anticancer effects of atorvastatin in digestive malignancies. Cancer Res. 2010;70:7699–7709. doi: 10.1158/0008-5472.CAN-10-1626. [DOI] [PubMed] [Google Scholar]
- 25.Li DD, Sun T, Wu XQ, et al. The inhibition of autophagy sensitises colon cancer cells with wild-type p53 but not mutant p53 to topotecan treatment. PLoS ONE. 2012;7:e45058. doi: 10.1371/journal.pone.0045058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki K, Tsuno NH, Sunami E, et al. Chloroquine potentiates the anti-cancer effect of 5-fluorouracil on colon cancer cells. BMC Cancer. 2010;10:370. doi: 10.1186/1471-2407-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- 28.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 29.Marino G, Salvador-Montoliu N, Fueyo A, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. [DOI] [PubMed]
- 30.Cho DH, Jo YK, Kim SC, Park IJ, Kim JC. Down-regulated expression of ATG5 in colorectal cancer. Anticancer Res. 2012;32:4091–4096. [PubMed] [Google Scholar]
- 31.Xie CM, Chan WY, Yu S, Zhao J, Cheng CH. Bufalin induces autophagy-mediated cell death in human colon cancer cells through reactive oxygen species generation and JNK activation. Free Radic Biol Med. 2011;51:1365–1375. doi: 10.1016/j.freeradbiomed.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6:256–269. doi: 10.4161/auto.6.2.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang SY, Winslet MC. Dual role of autophagy in colon cancer cell survival. Ann Surg Oncol. 2011;18:S239. doi: 10.1245/s10434-011-1789-x. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues NR, Rowan A, Smith ME, et al. p53 mutations in colorectal cancer. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livesey KM, Kang R, Vernon P, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Xia H, Kim M, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1
Growth suppressive effects of Beclin1 in HCT-116 and SW48 cells. HCT-116 and SW48 cells were infected with control virus (mock) and Beclin1 virus (Beclin 1-HA). A Exogenous Beclin 1 tagged HA peptide was detected by western blot using HA antibody. B Overexpression of Beclin 1 has no effect on cell growth in HCT-116 and SW48 cells. All experiments were performed in triplicate. Results shown are the mean ± SD. (TIFF 11550 kb)
Fig. S2
Knockdown of p53 induces an increased Beclin 1 in HT-29 cells. HT-29 cells were transfected with 10 nM p53-targeting siRNAs (siRNA target sequence: siRNA#1 GCACAGAGGAAGAGAAUCU, siRNA#2GAAGAAACCACUGGAUGGA, siRNA#3GAGGUUGGCUCUGACUGUA), or siRNA control for 48 h. p53 and Beclin 1 expression were detected by western blot. Actin was used as a loading control. (TIFF 3139 kb)