Abstract
Most solid tumors are aneuploid, having a chromosome number that is not a multiple of the haploid number, and many frequently mis-segregate whole chromosomes in a phenomenon called chromosomal instability (CIN). CIN positively correlates with poor patient prognosis, indicating that reduced mitotic fidelity contributes to cancer progression by increasing genetic diversity among tumor cells. Here, we review the mechanisms underlying CIN, which include defects in chromosome cohesion, mitotic checkpoint function, centrosome copy number, kinetochore–microtubule attachment dynamics, and cell-cycle regulation. Understanding these mechanisms provides insight into the cellular consequences of CIN and reveals the possibility of exploiting CIN in cancer therapy.
Introduction
Most solid tumors are aneuploid and many mis-segregate chromosomes at very high rates in a phenomenon termed chromosomal instability (CIN). Aneuploidy is a state in which the number of chromosomes in a cell or organism deviates from multiples of the haploid number of chromosomes. Chromosomal instability (CIN) is defined as a persistently high rate of loss and gain of whole chromosomes. For the purpose of this review, we adhere to the strict definition of CIN as whole chromosome mis-segregation and do not include structural rearrangements of chromosomes (translocations, deletions, inversions), although these structural rearrangements may also be linked to mis-segregation.
Aneuploidy was first associated with tumors in the late 19th century. In 1890, David von Hansemann examined tissue sections from epithelial tumors and discovered cells that were going through multipolar divisions as well as bipolar yet asymmetric divisions of chromosomes [1]. Subsequently, Theodor Boveri compared defects in sea urchin embryos that had gone through multipolar divisions and proposed that a “certain abnormal chromatin constitution”, regardless of how it originated, “would result in the origin of a malignant tumor” [2]. The consequence of CIN is aneuploidy but the line between aneuploidy and CIN was blurred in these early studies because tools were not available to discriminate between aneuploidy (a state that describes the cellular karyotype) and CIN (increased rates of chromosome mis-segregation). This distinction is important because aneuploidy can arise in different ways; however, the fact that the majority of aneuploid tumors have chromosome numbers within the range of diploid cells — i.e. 40–60 chromosomes (http://cgap.nci.nih.gov/Chromosomes/Mitelman; also see [3]) — indicates that the accumulation of chromosome imbalances generated by the sequential loss and gain of single chromosomes through CIN may be the most common pathway to aneuploidy. Because aneuploidy represents a state of having an abnormal number of chromosomes and CIN is a condition of an increased rate of chromosome mis-segregation, the criteria needed to establish each condition are different. Aneuploidy can be detected by any method that quantifies chromosome numbers, including karyotype analysis, fluorescence in situ hybridization, spectral karyotyping, or array-based comparative genomic hybridization analyses. However, by themselves, these techniques are not sufficient to yield quantitative measures of CIN. Detection of CIN requires the determination of chromosome mis-segregation rates [4], which can be achieved by coupling tools for counting chromosomes with clonal cell assays that allow the analysis of chromosomal variation in the resulting clonal population. In these assays, populations of cells derived from chromosomally stable precursors will show little variation in chromosome content (regardless of whether or not they are aneuploid); in contrast, cells in a population derived from a CIN precursor cell will show high levels of deviance in chromosome content.
Using this single-cell colony assay, Vogelstein and colleagues [5] ignited research into the mechanisms underlying CIN when they demonstrated two key properties of colon cancer cell lines. First, they showed that colon cancer cells with microsatellite instability (MIN) maintain a stable chromosome content, but aneuploid colon carcinoma cells exhibited deviations from the modal chromosome number that ranged from 16% to 66%, indicating the presence of CIN. High deviations in chromosome content in clonal populations were subsequently reported in cells derived from many other tumor types, including breast and lung [6,7], indicating that CIN is a general property of aneuploid cancer cells. Direct measurement of chromosome mis-segregation rates in CIN cancer cell lines has recently shown that these cells mis-segregate a chromosome, on average, once every one to five cell divisions [8]. This may represent the upper limit of tolerable chromosome changes because massive chromosome mis-segregation caused by checkpoint failure [9,10] or multipolar anaphase [11] is lethal. Secondly, Vogelstein and colleagues [5] showed that fusion of MIN and CIN cells resulted in hybrid cells that retained the CIN phenotype, suggesting that the underlying mechanisms that cause CIN behave as dominant traits.
Here, we discuss recent advances that illuminate the underlying mechanisms causing CIN in human tumor cells. These mechanisms reduce mitotic fidelity and include defects in chromosome cohesion, the spindle assembly checkpoint (SAC), centrosome copy number, kinetochore– microtubule attachment dynamics, and cell-cycle regulation. We further discuss how the knowledge gained by uncovering these mechanisms unveils strategies to exploit CIN to improve cancer therapy.
Chromosome Segregation in Mitosis
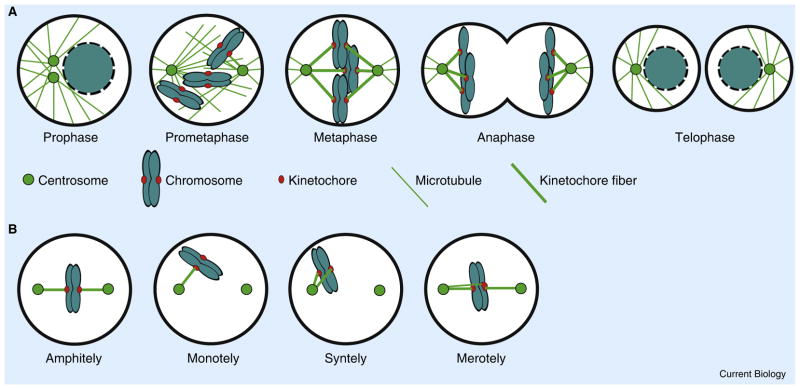
CIN represents the loss of chromosome segregation fidelity in mitosis, so it is relevant to review the salient features of mitosis that support faithful chromosome segregation. Mitosis is carefully choreographed to ensure that all sister chromatids segregate to opposite daughter cells (Figure 1A). Central to this choreography is sister chromatid cohesion, which is established at the time of DNA replication by the deposition of the cohesin complex. Proper chromosome segregation requires that cohesion be maintained through the G2 and M phases of the cell cycle and then abruptly disrupted at the onset of anaphase. The precise timing of the destruction of sister chromatid cohesion during mitosis is determined by the coordinated activities of cyclin-dependent kinases and the SAC. Chromosomes that are not properly attached to spindle microtubules emit a signal to maintain cyclin-dependent kinase activity, which inhibits anaphase onset and preserves sister chromatid cohesion. For example, mono-oriented chromosomes that lack attachment at one kinetochore (monotely) emit the ‘wait anaphase’ signal, and a single unattached kinetochore is sufficient to prevent anaphase onset [12,13]. Once all chromosomes achieve proper bi-oriented attachments to spindle microtubules, the SAC is satisfied and sister chromatid separation is abruptly and synchronously induced on all chromosomes by the cell-cycle machinery that proteolytically cleaves the cohesin complex. Finally, in human cells each kinetochore attaches to an average of ~20 microtubules and all of these microtubules must orient toward the same spindle pole to support faithful chromosome segregation. The back-to-back geometry of sister kinetochores favors chromosome bi-orientation on spindles [14–16], but the stochastic nature of kinetochore–microtubule attachments results in frequent misattachments in which single kinetochores simultaneously bind to microtubules emanating from both spindle poles — known as merotely (for a schematic representation of types of kinetochore–microtubule attachment, see Figure 1B). These merotelic attachments occur naturally in early mitosis and are corrected prior to anaphase onset to ensure faithful chromosome segregation [17]. It is noteworthy that merotely is not sensed by the SAC and, if left uncorrected, can cause sister chromatids to migrate to the same spindle pole in anaphase leading to chromosome mis-segregation [18].
Figure 1. Chromosome segregation in mitosis.
(A) Chromosomes associate with spindle microtubules following nuclear envelope breakdown. The spindle assembly checkpoint is responsible for preventing anaphase onset until all chromosomes form proper bi-oriented attachments to spindle microtubules. Destruction of cohesins permits sister chromatid separation to define anaphase. To ensure faithful chromosome segregation all sister chromatids must segregate to the same daughter cell. (B) Types of microtubule attachment. Correctly attached sister kinetochores orient toward opposite spindle poles (amphitely). Erroneous attachments include situations where only one kinetochore is attached to a spindle pole (monotely), both sister kinetochores are attached to the same pole (syntely), or one sister kinetochore is attached to both poles (merotely).
Causes of Chromosomal Instability
Since the time that Vogelstein and colleagues [5] clearly established the presence of CIN in aneuploid human tumor cells, many research groups have pursued its underlying cause. These efforts have revealed various mechanisms through which tumor cells lose mitotic fidelity and proteins involved in CIN are summarized in Table 1.
Table 1.
Proteins associated with CIN.
| Protein | Alteration | Putative mechanism(s) | Reference |
|---|---|---|---|
| APC | Depletion, mutation | Checkpoint defects, merotely | [49,83,92–94,127,128] |
| Aurora A | Overexpression | Centrosome amplification, cytokinesis failure | [58,59] |
| Aurora B | Depletion, drug inhibition | Checkpoint defects, merotely | [55,69,70] |
| β-catenin | Mutation | Dysregulation of cell-cycle proteins, merotely | [127] |
| BRCA1 | Mutation | Dysregulation of cell-cycle proteins, merotely | [102] |
| BRCA2 | Mutation | Dysregulation of cell-cycle proteins | [102] |
| Bub1 | Heterozygous knockout, hypomorph, mutation | Checkpoint defects | [32,41] |
| Bub3 | Heterozygous knockout | Checkpoint defects | [42,46] |
| BubR1 | Knockout, mutation | Checkpoint defect | [30,31,43,45] |
| CAML | Knockout | Cytokinesis failure, merotely | [129] |
| hCdc4/FBXW7 | Depletion, knockout | Dysregulation of cell-cycle proteins, merotely | [104] |
| Cdc20 | Mutation | Checkpoint defects | [44] |
| CENP-E | Depletion, knockout | Checkpoint defects, merotely | [47,84,85,130] |
| CENP-F | Depletion | Checkpoint defects, merotely | [86,106] |
| CENP-H | Overexpression | Cytokinesis failure, merotely | [131] |
| CLASP | Depletion | Merotely | [84,90] |
| Conductin/AXIN2 | Overexpression | Checkpoint defects, dysregulation of cell-cycle proteins | [128] |
| Cyclin E | Overexpression | Centrosome amplification, dysregulation of cell-cycle proteins, merotely | [104,105] |
| EB1 | Depletion | Merotely | [94] |
| ECRG2 | Depletion | Centrosome amplification, checkpoint defects, dysregulation of cell-cycle proteins | [132] |
| Eg5 | Overexpression | Cytokinesis failure | [62] |
| FoxM1 | Depletion, knockout | Dysregulation of cell-cycle proteins | [106] |
| Hec1–NDC80 complex | Antibody inhibition, mutation, overexpression | Cytokinesis failure, merotely | [73,87,133] |
| Hice-1 | Depletion | Cytokinesis failure, merotely | [134] |
| Id1 | Overexpression | Cytokinesis failure | [135] |
| Kif2a | Depletion, with MCAK depletion | Merotely | [78] |
| Kif2b | Depletion | Merotely | [67] |
| Kif4 | Knockout | Centrosome amplification, merotely | [136] |
| Kruppel-like factor 4 | Knockout | Centrosome amplification, chromosome breakage | [111] |
| Mad1 | Heterozygous knockout | Checkpoint defects | [40] |
| Mad2 | Depletion, heterozygous knockout, knockout, overexpression | Checkpoint defects, merotely | [38,39,100,103,108] |
| MCAK | Depletion | Merotely | [67,76,77] |
| MCT-1 | Overexpression | Merotely | [137] |
| Mdm2 | Overexpression | Dysregulation of cell-cycle proteins | [112] |
| MdmX | Knockout | Centrosome amplification, cytokinesis failure multipolar anaphases | [113] |
| Mps1 | Mutation | Checkpoint defects, merotely | [89] |
| p53 | Knockout | Dysregulation of cell-cycle proteins | [101,108,137] |
| PRP4 | Depletion | Checkpoint defects, merotely | [138] |
| Rad21/SCC1 (cohesin subunit) | Mutation | Cohesion defects | [19] |
| Rae1 | Heterozygous knockout | Checkpoint defects | [46] |
| RanBP1 | Depletion | Merotely | [114] |
| Rb | Depletion | Centrosome amplification, dysregulation of mitosis proteins causing overactivation of checkpoint | [103] |
| REST | Mutation | Dysregulation of mitosis proteins causing checkpoint defects | [109] |
| SCC3 (cohesin subunit) | Mutation | Cohesion defects | [19] |
| Securin | Knockout, overexpression | Cohesion defects | [22,24] |
| Separase | Knockout, overexpression | Cohesion defects, cytokinesis failure | [21,23] |
| SMC1 (cohesin subunit) | Depletion, mutation | Cohesion defects, cytokinesis failure | [19] |
| SMC3 (cohesin subunit) | Mutation | Cohesion defects | [19] |
| Sgo1 | Depletion | Cohesion defects, cytokinesis failure | [20] |
| Sgo2/tripin | Depletion | Cohesion defects, merotely | [88] |
| TMAP/CKAP2 | Depletion | Merotely | [139] |
| Topoisomerase II | Drug inhibition | Catenation, merotely | [140] |
| Von Hippel Lindau | Depletion | Checkpoint defects | [110] |
Cohesion Defects
Proteins that participate in chromosome cohesion were identified through genetic strategies using assays that measure the efficiency of transmission of minichromosomes in budding yeast. Interestingly, somatic mutations in genes whose products regulate sister chromatid cohesion, including subunits of the cohesin complex, were recently identified in human tumors in an expansive sequencing strategy designed to identify mutations of the human homologues of each gene known to induce CIN in budding yeast [19]. No direct analysis was performed to test how these mutations affect chromosome cohesion. Thus, it is unknown whether cells carrying these mutations are disposed to premature sister chromatid separation or to failure of chromosome disjunction at anaphase onset, making it impossible to evaluate their role in CIN. Nevertheless, the acute depletion of cohesins or of the cohesion regulators Sgo1 and separase (the enzyme that cleaves cohesins to initiate anaphase) using RNA interference elevates the numbers of tetraploid cells [19–21]. Tetraploid cells also arise in cells overexpressing separase or a non-degradable form of securin (the protein that prevents separase activity prior to anaphase) [20,22–24], indicating that disturbances of cohesion tend to cause global effects on chromosome segregation rather than elevating the incidence of single chromosome mis-segregation. In addition, large-scale sequencing of tumor genomes reveals that cohesin genes are rarely mutated [25–29]. Nevertheless, the possibility that dominant mutations in cohesin subunits increase chromosome mis-segregation by preventing timely chromosome separation remains open, and this scenario would fit with the dominant behavior of CIN in cell fusion experiments. An alternative view worth exploring is whether mutations in cohesin subunits impair the orderly packing of centromeric chromatin leading to disruption of the typical back-to-back orientation of sister kinetochores. Beyond these specific mutations, there are other hints that disruption of cohesion may contribute to CIN. For example, high levels of separase are seen in breast cancer tumor samples compared with normal tissue controls [23]. Excessive separase could induce premature sister chromatid disjunction leading to chromosome mis-segregation. Yet the interpretation of altered mRNA levels in tumors is unclear because of differences in mitotic indices between tumor and normal tissue. This discrepancy is widespread and applies to many other areas comparing normal and tumor tissues.
SAC Defects
Molecular components of the SAC were identified in budding yeast using genetic strategies that led to the isolation of mutant strains that failed to arrest cell-cycle progression in the presence of spindle poisons. Homologues of most of these yeast genes have been identified in other eukaryotic species, indicating that the molecular underpinnings of this checkpoint are highly conserved. These components generate the checkpoint signal emanating from kinetochores to prevent anaphase onset until all chromosomes form proper bipolar attachments to spindle microtubules. Impairment of the SAC permits precocious anaphase onset and significantly increases the likelihood for chromosome mis-segregation, meaning that mutations in SAC genes can cause CIN. Support for this view comes from mosaic variegated aneuploidy, an extremely rare disease linked to mutations in the gene encoding the SAC protein Bub1-related kinase (BubR1). Mosaic variegated aneuploidy causes growth retardation, microcephaly, childhood cancer, and often results in death at a young age [30,31]. Premature sister chromatid separation is often seen in >50% of lymphocytes from patients with mosaic variegated aneuploidy, and in many tissues more than 25% of cells are aneuploid; presumably, this high level of aneuploidy contributes to the elevation of cancer in these patients [30,31]. In addition to the clinical evidence, experimental data show that aneuploid tumor cells with CIN fail to maintain mitotic arrest when exposed to spindle poisons for extended times, resulting in a lower mitotic index between CIN cells and stable diploid cells [32]. This indicates a failure of the SAC to maintain arrest compared with what is seen in normal cells. Indeed, it was shown that some tumor cell lines have decreased levels of the SAC protein Mad2 [33–35], and mutations have been identified in a few colon cancer cell lines in the gene encoding Bub1 [32], another SAC protein, as well as in the Mad2 gene in a breast cancer cell line and in multiple gastric cancer tumor cell lines [36,37]. Finally, experimentally induced deletion of one copy of the Mad2 gene in otherwise stable diploid HCT116 colon carcinoma cells increased chromosome mis-segregation [38].
These initial reports linking CIN to defective SAC activity spawned a series of experiments using mouse models to test whether aneuploidy and CIN contribute to tumorigenesis. Because complete SAC ablation is lethal to cultured cells [9,10], it is no surprise that homozygous deletion of many SAC genes, including Mad2, Mad1, Bub1, BubR1, and Bub3, are embryonic lethal in mice [39–43]. However, mice heterozygous for mutations in specific SAC genes are viable. These animals develop normally but display elevated chromosome mis-segregation rates compared with control animals, consistent with attenuated checkpoint activity. Many of these heterozygous animals have an increased tumor incidence late in life, although this is highly dependent on the gene that was targeted and there is also variability in the tissue affected [38,40,41,44]. For example, mice heterozygous for mutations in Mad2 develop spontaneous lung tumors after 18 months [38], and mice with hypomorphic Bub1 alleles display an increase in hepatocellular carcinomas, lung adenocarcinomas, sarcomas, and lymphomas after 19 months [41]. In contrast, mice heterozygous for Bub1, BubR1, Bub3, or Rae1 do not form spontaneous tumors [41,45,46], despite having similar levels of aneuploidy in spleen cells and mouse embryonic fibroblasts when compared with other checkpoint-deficient mice that do get tumors. The fact that tumors are seen in many of these transgenic mouse models suggests that aneuploidy or CIN promotes tumorigenesis, although recent work shows that loss of mitotic fidelity can also suppress tumorigenesis depending upon the tissue context [47]. Assuming these animal models have an elevated frequency of chromosome mis-segregation throughout life, the delay between tumor appearance and the onset of aneuploidy and CIN (beginning at embryogenesis) — often 18 months or more — suggests that chromosome mis-segregation promotes tumorigenesis in cells that have acquired additional mutations. Chromosome loss is a known mechanism for revealing recessive mutations in tumor suppressor genes in human tumors [48]. It is important to emphasize that these animal studies fail to distinguish between the distinct contributions of aneuploidy (a state of improper numbers of chromosomes) and CIN (a high rate of chromosome mis-segregation) in the promotion or suppression of tumorigenesis because both CIN and aneuploidy are induced in these models.
Despite the aforementioned evidence, the role of checkpoint impairment in the widespread occurrence of CIN is still a matter of debate. It is becoming increasingly accepted that most cancer cells with CIN have a functional SAC. For example, no difference was reported between the abilities of diploid cells and cancer cells with CIN to arrest in mitosis in the presence of spindle poisons [49]. Moreover, direct observation of multiple CIN cell lines undergoing mitosis revealed that no cells entered anaphase prior to alignment of all chromosomes [8,50]. Collectively, these results provide robust evidence that the checkpoint in many CIN cell lines is functioning to prevent anaphase onset until kinetochores on all chromosomes attach to spindle microtubules. It was recently shown that cells with CIN have a higher rate of death during prolonged arrest in mitosis, providing a reasonable explanation for the previously observed decrease in mitotic index for CIN cell lines [51], although the differences in response to microtubule-targeted drugs between cells with CIN and MIN may not be universal [50,52]. Furthermore, complete ablation of the SAC is lethal due to massive chromosome mis-segregation [9,10], and the purported loss of function of the SAC in CIN cells does not fit with the dominant behavior of CIN in cell fusion studies [5]. Finally, no mutations in checkpoint genes were found when DNA was sequenced from over 100 cancer cell lines [19] or from hundreds of tumors from multiple tissues [25–29]. Taken together, it appears that mutations that erode SAC function are quite rare in human CIN tumors. Whereas some tumor cells may have a reduced capacity to maintain mitosis for extended periods when challenged with spindle poisons, the bulk of the evidence indicates that the SAC in most cancer cells with CIN is functional and prevents precocious anaphase under normal growth conditions.
Supernumerary Centrosomes
Direct analysis of human tumor cell lines with CIN shows that the most common mitotic defect is lagging chromosomes in anaphase [8]. Lagging chromosomes can also be observed during anaphase in human tumors, indicating that these errors are not unique to cultured cell lines (Figure 2). Examination of CIN cell lines using fixed cell analyses shows that this mitotic defect represents single chromatids that fail to segregate because the kinetochore is attached to spindle microtubules that are oriented toward opposite spindle poles in a merotelic conformation [8]. Merotely is only possible in eukaryotic cells where single kinetochores bind multiple microtubules and is usually disfavored by the back-to-back geometry of sister kinetochores [14–16]. However, many kinetochores become merotelic in early phases of mitosis due to the stochastic interaction between kinetochores and spindle microtubules [17]. The microtubule occupancy of merotelic kinetochores is equivalent to that of kinetochores on properly bioriented chromosomes, so the SAC is appropriately satisfied in the presence of these erroneous attachments [18]. Thus, it is critical that merotelic kinetochores be corrected prior to anaphase onset to ensure error-free chromosome segregation. Indeed, the incidence of lagging chromosomes during anaphase increases in cells with attenuated SAC activity, suggesting that those cells did not have sufficient time to correct all merotelic attachments prior to anaphase onset.
Figure 2. Lagging chromosomes in human melanoma.

Anaphase cells in a hematoxylin-and-eosin-stained section of a human melanoma show lagging chromosomes (arrow) consistent with mitotic defects observed in cultured tumor cell lines. Image provided by Vincent Memoli (Department of Pathology, Dartmouth-Hitchcock Medical Center and Norris Cotton Cancer Center).
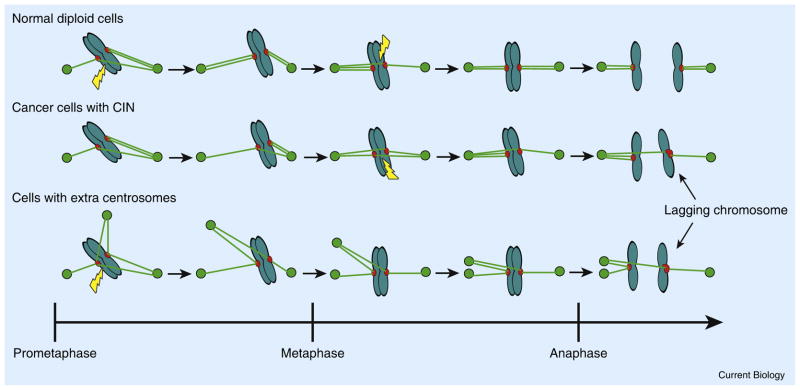
The prevalence of merotelic attachments is determined by two rates: the rate of their formation and the rate of their correction (Figure 3). Treatments that disrupt spindle geometry significantly increase the formation of merotelic attachments. For example, mitotic cells recovering from microtubule- depolymerizing agents, such as nocodazole, proceed through a multipolar spindle intermediate that induces high rates of merotely and subsequent lagging chromosomes [17,53,54]. Recovery from inhibitors of the kinesin-5 motor protein Eg5 proceed through monopolar spindle intermediates and similarly have elevated levels of merotely and lagging chromosomes at anaphase [55]. These drug recovery approaches have been used to show that CIN can be induced in otherwise stable cells by transiently disrupting spindle geometry to increase the prevalence of merotely [8]. Cells with more than two centrosomes often coalesce the extra centrosomes during mitosis to ensure that anaphase occurs with a bipolar spindle [56]. It was recently shown that extra centrosomes induce transient multipolar spindles prior to coalescence of centrosomes into bipolar spindles, and this event significantly increases the incidence of merotely and elevates chromosome mis-segregation rates (Figure 3) [11,57]. This provides a straightforward explanation for the long-standing correlation between extra centrosomes and CIN in many tumor cells. More direct analysis showed that induction of extra centrosomes in otherwise chromosomally stable cells was sufficient to elevate lagging chromosomes and induce chromosome mis-segregation through excessive merotely induced by transient multipolar spindles [11].
Figure 3. Formation and correction of merotelic kinetochore–microtubule attachments.
Merotely arises in early mitosis in normal cells, but is efficiently corrected (lightning bolts) to establish proper bi-orientation needed for error-free chromosome segregation. Kinetochore–microtubule attachments in cancer cells with CIN are hyperstable, which reduces their intrinsic ability to correct merotelic attachments. Spindle assembly in cells with extra centrosomes progresses through transient multipolar intermediates that greatly increase the formation rate of merotelic attachments.
Extra centrosomes are produced either by deregulation of the centrosome duplication cycle or as a by-product of cells that become tetraploid. For example, centrosome amplification has been shown to arise through overexpression of Aurora A kinase [58,59] or viral infection [60]. Tetraploidy can arise through viral-induced cell fusion [61], overexpression of Eg5 [62], or cytokinesis failure. Tetraploidy is a likely precursor to tumor cells with near-tetraploid karyotypes and has long been hypothesized to be a precursor of aneuploidy because it is found prior to gross aneuploidy in early tumors in Barrett’s esophagus and cervical cancer [63,64], and p53-deficient tetraploid cells (but not p53-deficient diploid cells) form tumors when injected into immune-compromised mice [65]. Since mechanisms generating tetraploidy also result in the presence of extra centrosomes, it is likely that tetraploid cells are highly prone to have CIN. Taken together, it appears that a common route to CIN can be through the acquisition of extra centrosomes that significantly increase the rate of formation of merotelic kinetochore attachments. Presumably, the levels of merotely in these situations exceed the capacity of the correction machinery to efficiently repair them prior to anaphase onset.
Defects in Kinetochore–Microtubule Attachment Dynamics
Microtubule attachment to kinetochores is reversible [66] and the rate-limiting step in the correction of erroneous kinetochore–microtubule attachments is the release of microtubules from kinetochores [67]. The repeated association and dissociation of individual microtubules from kinetochores generates a dynamic kinetochore–microtubule attachment that is necessary to promote error correction. The large number of microtubules bound to each kinetochore ensures that chromosomes remain tethered to spindle microtubules despite the continuous association/dissociation of individual microtubules. The dynamic turnover of kinetochore–microtubule attachments has been measured in various cell lines and is rapid in early mitosis (t1/2 = 2–3 minutes), slows as chromosomes align in metaphase (t1/2 = 5–7 minutes), and slows further when cells enter anaphase (t1/2 = 50 minutes) [67–69]. Thus, kinetochore– microtubule attachment stability is regulated in a very narrow range during different phases of mitosis, particularly at the crucial stage between prometaphase and metaphase when many erroneous attachments need to be corrected.
The molecular machinery involved in regulating kinetochore– microtubule attachment dynamics is beginning to emerge. The centromere-localized Aurora B kinase is a central controller of this process because inhibition of this kinase renders the attachment of microtubules to kinetochores virtually irreversible [69,70]. Among the many centromere and/or kinetochore targets of Aurora B kinase are the Ndc80 complex, which is required for kinetochore–microtubule attachment, and members of the kinesin-13 family of microtubule-depolymerizing enzymes [67,71–73]. Loss of function of the Ndc80 complex virtually eliminates stable kinetochore–microtubule attachments, indicating that it plays a key role in kinetochore–microtubule attachment [74,75]. Conversely, loss of function of kinesin-13 proteins, such as Kif2b and MCAK, results in higher rates of lagging chromosomes, showing that they are instrumental in correction of erroneous kinetochore-microtubule attachments [67,76–78]. Two of the three members of the kinesin-13 subfamily have overlapping responsibilities in this role in human cells. Kif2b localizes to outer kinetochores and destabilizes kinetochore–microtubule attachments in prometaphase [67,79]. This role appears to be critical to correct the numerous erroneous kinetochore–microtubule attachments that arise in prometaphase. Conversely, MCAK localizes to centromeres and destabilizes kinetochore– microtubule attachments predominantly during metaphase [67,79], which appears to be essential to prevent the formation of new merotelic attachments once chromosomes have aligned on the spindle.
Evidence indicates that tension generated between sister kinetochores regulates these correction mechanisms. Aurora B kinase generates a phosphorylation gradient emanating from the inner centromere [80,81] that influences both the localization and activities of MCAK and Kif2b. With few microtubule attachments in early mitosis, tension between sister kinetochores is low and the phosphorylation gradient extends to suppress MCAK activity and recruit Kif2b to outer kinetochores. As chromosomes biorient, tension builds between sister kinetochores and shifts the position of target proteins from the boundaries of the phosphorylation gradient. This results in the release of Kif2b from kinetochores and the activation of MCAK [67,71,82]. Accordingly, disruption of the spatial boundaries of this phosphorylation gradient by repositioning Aurora B kinase close to the kinetochore strongly destabilizes kinetochore–microtubule attachments and prevents the onset of anaphase [81]. These (and probably other) molecular changes are reflected in the stabilization of kinetochore–microtubule attachments as cells progress from prometaphase to metaphase and, presumably, into anaphase. A remaining question is whether indiscriminate changes in kinetochore–microtubule attachment dynamics that occur during these mitotic transitions are sufficient for efficient error correction [67] or whether erroneous kinetochore attachments are uniquely recognized by cellular machinery to promote their correction [55].
The narrow range for the regulation of kinetochore–microtubule dynamics as cells progress through mitosis makes it a sensitive target for the disruption of mitotic fidelity. Insults that destabilize kinetochore–microtubule attachments, such as the repositioning of Aurora B kinase described previously [81], mean that satisfaction of the checkpoint and progression into anaphase are impossible because kinetochores never achieve sufficient microtubule occupancy. In contrast, insults that stabilize kinetochore–microtubule attachments would prevent efficient correction of attachment errors (mediated by the release of inappropriately attached microtubules) and promote chromosome mis-segregation. Accordingly, it was recently shown that kinetochore–microtubule attachments are more stable in tumor cells with CIN than in a chromosomally stable diploid cell line [83]. CIN cancer cells displayed hyperstable kinetochore–microtubule attachments in prometaphase, or metaphase, or both and this was not dependent on the numbers of centrosomes present, indicating that the tested cell lines have an inherent defect in correcting erroneous kinetochore–microtubule attachments. Moreover, overexpression of the kinesin-13 microtubule depolymerases MCAK and Kif2b reduced kinetochore– microtubule attachment stability and restored faithful chromosome segregation to cancer cells that otherwise showed CIN [67]. This is the only example in which chromosome segregation fidelity has been restored to cancer cells that have CIN and provides compelling evidence that excessively stable kinetochore–microtubule attachments are the root cause of CIN in many cancer cells.
The list of centromere/kinetochore proteins whose perturbation reduces segregation fidelity due to elevation of kinetochore– microtubule mal-attachment is growing rapidly. Perturbation of many of these (Kif2b [67], MCAK [76,77], CENP-E [84,85], CENP-F [86], the Ndc80 complex [73,87], Aurora B [55,69,70], Sgo2 [88], Mps1 [89], CLASPs [84,90], Ndel1 [91], and APC [83,92–94]) has been shown to elevate the frequency of merotely and lagging chromosomes in anaphase. This infers that kinetochore–microtubule attachments become excessively stable upon perturbation of these proteins and this in turn reduces the efficiency of correction of erroneous kinetochore–microtubule attachments leading to CIN. This inference has been verified in several instances. As mentioned previously, perturbation of either Aurora B or MCAK activity stabilizes kinetochore– microtubule attachments and increases the numbers of lagging chromosomes in anaphase [69,70,95]. Furthermore, APC is frequently mutated in colon cancer and cells with mutations in APC show high rates of lagging chromosomes in anaphase [92–94]. It was recently shown that depletion of APC increased kinetochore–microtubule stability and induced lagging chromosomes in otherwise stable diploid cells, and this defect was rescued by concomitant overexpression of proteins that promoted kinetochore–microtubule dynamics [83]. Moreover, depletion of CENP-E stabilizes kinetochore–microtubule attachments [84] and we have observed a consequent increase in lagging chromosomes in anaphase in human HCT116 cells lacking CENP-E (S.L.T. and D.A.C., unpublished observations).
Despite this cellular evidence, the only gene from the above list for which mutations have been directly observed at high frequencies in tumors is APC and mutations in others are quite rare (Aurora B [28] and CLASPs [26]) or not detected at all [25–29]. However, loss-of-function mutations would not be expected based on the dominant behavior of CIN in cell fusion experiments, and there is evidence for a dominant effect of APC mutations [5,96,97]. Thus, a more likely scenario is that imbalances in the levels of these proteins act dominantly to stabilize kinetochore–microtubule attachments beyond a certain threshold, leading to deficiencies in error correction and chromosome mis-segregation. These imbalances in protein levels could be generated by an initial chromosome mis-segregation event or by epigenetic mechanisms. The idea that protein imbalances induce CIN has been proposed previously based on the consistency with which chemically induced transformation associates with aneuploidy and CIN in cultured hamster cells [98,99]. This view is further supported by experiments showing that overexpression of Hec1, a component of the Ndc80 complex, causes high levels of lagging chromosomes and chromatin bridges in mouse cells and can induce tumors [87]. Mad2 overexpression also increases the frequency of lagging chromosomes in anaphase and can increase the rate of spontaneous tumor formation [100]. Moreover, recent data shows substantial variability in the levels of mitotic proteins that participate in kinetochore–microtubule dynamics in tumor cells compared with stable diploid cells [83]. Accordingly, when diploid cells are rendered tolerant for a non-diploid genome through loss of p53, they display CIN following initial chromosome mis-segregation events [101], showing that perturbation of chromosomal content (and hence a plethora of transcript levels) may have a cyclical effect in driving a perpetual state of CIN. This view is counter to initial data showing that trisomy 3 was insufficient to induce CIN [5], but this particular trisomy may not imbalance kinetochore–microtubule attachment dynamics and other compensatory changes may have arisen. It remains unknown whether kinetochore–microtubule attachments are indeed stabilized by protein imbalances that arise from spontaneous chromosome mis-segregation. Nevertheless, a clear picture has emerged that can explain why lagging chromosomes are the most commonly observed cause of CIN in cancer cells: the relatively narrow acceptable range of kinetochore–microtubule dynamics needed for error-free mitosis, which is coupled to the large number of available protein targets that could disrupt these dynamics, makes CIN-causing alterations highly likely.
Defects in Cell-Cycle Regulation
In addition to proteins that function directly in the process of chromosome segregation during mitosis, there are numerous cell-cycle regulators that have been associated with CIN (including BRCA1 and BRCA2 [102], Rb [103], hCDC4 [104], cyclin E [105], FoxM1 [106,107], p53 [101,108], REST [109], Von Hippel Lindau [110], Kruppel-like factor [111], Mdm2 [112], MdmX [113], and RanBP1 [114]). The precise role that many of these cell-cycle regulators play in inducing CIN is not currently clear, but three general pathways are possible. First, these well-known cell-cycle regulators may directly participate in supporting chromosome segregation fidelity during mitosis in roles that had not been previously recognized. Second, deregulation of cell-cycle events outside of M phase (e.g. centrosome duplication during S phase) may indirectly promote chromosome mis-segregation during the ensuing mitosis. Finally, disruption of cell-cycle checkpoints may permit cells that have mis-segregated chromosomes to continue cell-cycle progression. In the following paragraphs we summarize data highlighting how a few of these cell-cycle regulatory proteins participate in generating CIN. We have selected these particular examples simply to emphasize the general principles for how deregulation of the cell cycle can lead to CIN.
hCDC4 (also called Fbw7) is an E3 ubiquitin ligase that targets cyclin E for degradation and thereby regulates the G1–S cell cycle transition [104]. hCDC4 is mutated in many cancers, including ~12% of colorectal cancers [104]. Homozygous knockout of hCDC4 in otherwise chromosomally stable HCT116 cells generates CIN, which can be ameliorated by depletion of cyclin E, indicating that mitotic defects are created through exaggerated cyclin E–Cdk activity [104]. Cyclin E regulates centrosome duplication during the cell cycle [115,116] and extra centrosomes promote CIN by increasing the frequency of merotelic kinetochore attachments, providing a mechanistic link between cyclin E levels and CIN. However, the role of hCDC4 mutations in CIN is still under debate [117], and mutations in hCDC4 do not seem to occur more frequently in CIN tumors compared with chromosomally stable, MIN tumors [118].
Cell-cycle regulators also control the levels of components of the SAC. For example, repressor-element-1-silencing transcription factor (REST) is a transcriptional repressor that must be degraded to permit transcription of the Mad2 gene [109]. Timely destruction of REST in the G2 phase of the cell cycle is mediated by the E3 ligase β-TrCP. Expression of a REST mutant that does not interact with β-TrCP results in low levels of Mad2 that lead to chromosome segregation defects in mitosis akin to those observed in Mad2 heterozygotes [109]. Another example is the Rb tumor suppressor protein. Rb regulates the transcriptional activator E2F [103]. One target of E2F transcriptional activation is the Mad2 gene and loss of Rb results in overexpression of Mad2, which has been shown to induce CIN [100,103].
Finally, FoxM1 is a cell-cycle regulator that has been associated with CIN: 80% of mouse embryo fibroblasts lacking FoxM1 are aneuploid compared with 20% of wild-type controls [106]. Human U2OS cells depleted of FoxM1 have multiple problems because they enter anaphase with unaligned chromosomes, have high numbers of lagging chromosomes, and often fail in cytokinesis [106]. Additionally, FoxM1 overexpression has been reported in tumor cell lines with CIN [107]. FoxM1 activates the transcription of many mitotic proteins, including cyclin B, CENP-F, and Plk-1 [106]. Thus, loss of FoxM1 may cause CIN by altering the expression of one or more of these mitotic proteins or by creating imbalances in mitotic proteins that disrupt normal mitotic progression.
CIN as a Target for Cancer Therapy
CIN in tumors is associated with poor patient prognosis [119–121] and contributes to tumor evolution through the acquisition of metastatic potential and resistance to some chemotherapeutic agents [52,122,123]. Through persistent chromosome shuffling, tumor cells with CIN continuously sample the genetic landscape with an almost limitless number of combinations, based on evidence in vertebrate cells that all chromosomes have an equal probability for mis-segregation [124]. Whereas some chromosome combinations may provide no selective advantage or a disadvantage, others could provide growth advantages to cells in the tumor microenvironment, particularly under the selective pressure applied by chemotherapy. In this context, CIN provides tumor cells with adaptability that permits them to acquire new phenotypes, such as drug resistance. It may therefore prove therapeutically beneficial to suppress CIN and thereby eliminate tumor cell adaptability. Suppression of CIN through the overexpression of MCAK and Kif2b provides proof of principle that CIN can be suppressed in tumor cells [67], and this avenue could be exploited to prevent acquisition of resistance to conventional chemotherapy.
An alternative strategy would be to intentionally promote chromosome mis-segregation. Unlike aneuploid tumor cells, diploid cells fail to propagate efficiently following chromosome mis-segregation [8]. One molecular pathway that limits the growth of diploid cells following chromosome mis-segregation involves p53 [101] and serves to highlight that aneuploid tumor cells have acquired tolerance for non-diploid genomes. This tolerance permits them to propagate efficiently as they constantly mis-segregate single chromosomes at high rates in association with CIN. However, tolerance is limited because aneuploid tumor cells die when large numbers of chromosomes mis-segregate in a single cell division cycle. Such massive chromosome changes arise when the SAC is abrogated [9,10] or when cells progress through a multipolar anaphase induced by extra centrosomes [11]. Thus, strategies that elevate chromosome mis-segregation beyond tolerable levels could be therapeutically beneficial. An obvious approach for this would be to abrogate the SAC in tumor cells, and a variation on this theme includes partial checkpoint inhibition coupled with treatments that disrupt proper chromosome segregation [125]. Another approach would involve induction of lethal multipolar anaphase in tumor cells carrying extra centrosomes by targeting proteins involved in centrosome clustering (e.g. the kinesin-14 motor HSET). Proof of principle for this strategy comes from recent work showing that multipolar anaphase is induced by depletion of HSET [11] and that Cdk2 inhibitors suppress the growth of lung tumors that overexpress cyclin E by inducing multipolar anaphases [126].
Conclusions
In summary, investigations into the basic mechanisms of chromosome segregation have revealed many defects that increase chromosome mis-segregation rates and cause CIN. These insights provide the tools to directly manipulate the frequency of chromosome mis-segregation in tumor cells to induce or suppress CIN. Importantly, these tools will prove invaluable in determining how CIN, independently of aneuploidy, contributes to tumorigenesis and could be used therapeutically to limit tumor growth.
Acknowledgments
We apologize to those investigators whose work we could not cite due to length constraints. Work in the authors’ lab is supported by the National Institutes of Health (GM51542).
References
- 1.von Hansemann D. Ueber asymmetrische zelltheilung in epithelkrebsen und deren biologische bedeutung. Virschows Arch Pathol Anat. 1890;119:299–326. [Google Scholar]
- 2.Wolf U. Theodor Boveri, and his book on the problem of the origin of malignant tumors. In: German J, editor. Chromosomes and Cancer. New York: Wiley; 1974. pp. 3–20. [Google Scholar]
- 3.Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 6.Yoon DS, Wersto RP, Zhou W, Chrest FJ, Garrett ES, Kwon TK, Gabrielson E. Variable levels of chromosomal instability and mitotic spindle checkpoint defects in breast cancer. Am J Pathol. 2002;161:391–397. doi: 10.1016/S0002-9440(10)64194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haruki N, Harano T, Masuda A, Kiyono T, Takahashi T, Tatematsu Y, Shimizu S, Mitsudomi T, Konishi H, Osada H, et al. Persistent increase in chromosome instability in lung cancer: possible indirect involvement of p53 activation. Am J Path. 2001;159:1345–1352. doi: 10.1016/S0002-9440(10)62521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VV, Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci. 2004;101:4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301– 1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produce by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loncarek J, Kisurina-Evgenieva O, Vinogradova T, Hergert P, La Terra S, Kapoor TM, Khodjakov A. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 16.Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner A. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc Natl Acad Sci. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimini D, Moree B, Canman J, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 18.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barber T, McManus K, Yuen KWY, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, Igarashi H, Naramura T, Suzuki H, Watanabe Y, et al. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- 21.Wirth KG, Wutz G, Kudo NR, Desdouets C, Zetterberg A, Taghybeeglu S, Seznec J, Ducos GM, Ricci R, Firnberg N, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, Zhao YJ, Li XN, Cai WW, El-Naggar AK, et al. Overexpression of separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci USA. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu R, Lu W, Chen J, McCabe CJ, Melmed S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology. 2003;144:4991–4998. doi: 10.1210/en.2003-0305. [DOI] [PubMed] [Google Scholar]
- 25.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807– 1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 28.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 30.Hanks S, Coleman K, Reid S, Plaja A, Firth H, FitzPatrick D, Kidd A, Mehes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 31.Matsuura S, Matsumoto Y, Morishima KI, Izumi H, Matsumoto H, Ito E, Tsutsui K, Kobayashi J, Tauchi H, Kajiwara Y, et al. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am J Med Genet A. 2006;140:358–367. doi: 10.1002/ajmg.a.31069. [DOI] [PubMed] [Google Scholar]
- 32.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JKV, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Benezra R. Identification of a human checkpoint gene: hsMad2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Jin DY, Wong YC, Cheung ALM, Chun ACS, Lo AKF, Liu Y, Tsao SW. Correlation of defective mitotic checkpoint with aberrantly reduced expression of Mad2 protein in nasopharyngeal carcinoma cells. Carcinogenesis. 2000;21:2293–2297. doi: 10.1093/carcin/21.12.2293. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Jin DY, Ng RWM, Feng H, Wong YC, Cheung AL, Tsao SW. Significance of Mad2 expression to mitotic checkpoint control in ovarian cancer cells. Cancer Res. 2002;62:1662–1668. [PubMed] [Google Scholar]
- 36.Percy MJ, Kenute MA, Neeley CK, Azim JN, Ethier SP, Petty EM. Expression and mutational analysis of the human MAD2L1 gene in breast cancer cells. Genes Chromosomes Cancer. 2000;29:356–362. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1044>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 37.Kim HS, Park KH, Kim SA, Wen J, Park SW, Park B, Gham CW, Hyung WJ, Noh SH, Kim HK, et al. Frequent mutations of human Mad2, but not Bub1, in gastric cancers cause defective mitotic spindle checkpoint. Mut Res. 2005;578:187–201. doi: 10.1016/j.mrfmmm.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 38.Michel LS, Liberal V, Chatterjee A, Kirchweffer R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VVVS, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 39.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 40.Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Pelopenese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 41.Jeganathan K, Malureanu L, Baker DJ, Abaraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalitsis P, Earle E, Fowler KJ, Choo KH. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, Ramaswamy G, Sakamoto KM, Darzynkiewicz Z, Xu M, et al. BubR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 44.Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, deGroen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 46.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome mis-segregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 49.Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gascoigne KE, Taylor S. Cancer cells display profound intra-and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;12:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Brito DA, Rieder CL. The ability to survive mitosis in the presence of microtubule poisons differs significantly between human nontransformed (RPE-1) and cancer (U2OS, HeLa) cells. Cell Motil Cytoskeleton. 2009;8:437–447. doi: 10.1002/cm.20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci USA. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cimini D, Tanzarella C, Degrassi F. Differences in mis-segregation rates obtained by scoring ana-telophases or binucleate cells. Mutagenesis. 1999;14:563–568. doi: 10.1093/mutage/14.6.563. [DOI] [PubMed] [Google Scholar]
- 54.Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 56.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 57.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lentini L, Amato A, Schillaci T, Di Leonardo A. Simultaneous Aurora A/STK15 overexpression and centrosome amplification induce chromosomal instability in tumor cells with a MIN phenotype. BMC Cancer. 2007;7:212. doi: 10.1186/1471-2407-7-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishida N, Nagasaka T, Koshiwagi K, Boland CR, Goel A. High copy amplification of the Aurora-A gene is associated with chromosomal instability phenotype in human colorectal cancers. Cancer Biol Therapy. 2007;6:525–533. doi: 10.4161/cbt.6.4.3817. [DOI] [PubMed] [Google Scholar]
- 60.Duensing S, Munger K. Human papillomaviruses and centrosome duplication errors: modeling the origins of genomic instability. Oncogene. 2002;21:6241–6248. doi: 10.1038/sj.onc.1205709. [DOI] [PubMed] [Google Scholar]
- 61.Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, Lazebnik Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol. 2007;17:431–437. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 62.Castillo A, Morse HC, III, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 63.Barrett MT, Pritchard D, Palanca-Wessels C, Anderson J, Reid BJ, Rabinovitch PS. Molecular phenotype of spontaneously arising 4N (G2-Tetraploid) intermediates of neoplastic progression in Barrett’s esophagus. Cancer Res. 2003;63:4211–4217. [PubMed] [Google Scholar]
- 64.Olaharski AJ, Sotelo R, Solorza-Luna G, Gonsebatt ME, Guzman P, Mohar A, Eastmond DA. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006;27:337–343. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- 65.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1046. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 66.Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cimini D, Wan XH, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 70.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore- microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow J. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 72.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 73.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 74.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 76.Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1145–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 79.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distict roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of Aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere- associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maffini S, Maia ARR, Manning AL, Maliga Z, Pereira AL, Junqueria M, Shevchenko A, Hyman A, Yates JR, III, Galjart N, et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 86.Holt SV, Vernolle MAS, Hussein D, Wozniak MJ, Allan VJ, Taylor SS. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J Cell Sci. 2005;118:4889–4900. doi: 10.1242/jcs.02614. [DOI] [PubMed] [Google Scholar]
- 87.Diaz-Rodriguez E, Sotillo R, Scvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GKT, Yen TJ. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, Medema RH, Kops GJ. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 90.Pereira AL, Pereira AJ, Maia ARR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Strapnaova T, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vergnolle MA, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubules motor complexes. Curr Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- 92.Green RA, Kaplan KB. Chromosome instability in colorectal tumor cells is associated with defects in microtubule plus-end attachments caused by dominant mutation in APC. J Cell Biol. 2003;163:949–961. doi: 10.1083/jcb.200307070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, et al. Mutations in the APC tumor suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 94.Draviam VM, Shapiro I, Aldridge B, Sorger PK. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 2006;25:2814–2827. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takacs CM, Baird JR, Hughes EG, Kent SS, Benchabane H, Paik R, Ahmed Y. Dual positive and negative regulation of wingless signaling by adenomatous polyposis coli. Science. 2008;319:333–336. doi: 10.1126/science.1151232. [DOI] [PubMed] [Google Scholar]
- 97.Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominat effects on proliferation, spindle checkpoint control, survival chromosome stability. J Cell Sci. 2004;117:6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- 98.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- 100.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. The BRCA1/BARD1 heterodimer modulates Ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 103.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 104.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosome instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 105.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 106.Laoukili J, Kooistra MRH, Bras A, Kauw J, Kerkhoven RM, Morrision A, Clevers, Medema RH. FoxM1 is required for execution of the mitotic program and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 107.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 108.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci USA. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Change S, Hernando E, et al. Control of chromosome stability by the β-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, Hergovich A, Moch H, Meraldi P, Krek W. VHL loss causes spindle misoreintation and chromosome instability. Nat Cell Biol. 2009;11:994– 1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 111.Hagos EG, Ghaleb AM, Dalton WB, Bialkowska AB, Yang VW. Mouse embryonic fibroblasts null for the Kruppel-like factor 4 gene are genetically unstable. Oncogene. 2009;28:1197–1205. doi: 10.1038/onc.2008.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2007;27:1590–1598. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- 113.Matijasevic Z, Krzywicka-Racka A, Sluder G, Jones SJ. MdmX regulates transformation and chromosomal stability in p53-deficient cells. Cell Cycle. 2008;7:1–7. doi: 10.4161/cc.7.19.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tedeschi A, Ciciarello M, Mangiacasale R, Roscioli E, Rensen WM, Lavia P. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J Cell Sci. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- 115.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 116.Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase controls of centrosome duplication. Proc Natl Acad Sci USA. 1999;96:2817– 2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Byrd KN, Huey B, Roydasgupta R, Fridlyand J, Snijdres AM, Albertson DG. FBXW7 and DNA copy number instability. Breast Cancer Res Treat. 2007;109:47–54. doi: 10.1007/s10549-007-9623-7. [DOI] [PubMed] [Google Scholar]
- 118.Kemp Z, Rowan A, Chambers W, Wortham N, Halford S, Sieber O, Mortensen N, von Herbay A, Gunther T, Ilyas M, et al. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 2005;65:11361–11366. doi: 10.1158/0008-5472.CAN-05-2565. [DOI] [PubMed] [Google Scholar]
- 119.Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, Werts A, Haak P, Vande Woude GF. Chromosomal instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci USA. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heilig CE, Loffler H, Mahlknecht U, Janssen JW, Ho AD, Jauch A, Kramer A. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00905.x. 10.1111/j.1582-4934. 2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Choi CM, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, Lee DS, Lee SD. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer. 2009;64:66–70. doi: 10.1016/j.lungcan.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 122.Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi OP, Isola J. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597– 1604. [PubMed] [Google Scholar]
- 123.McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–3266. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- 124.Torosantucci L, de Santis Puzzonia M, Cenciarelli C, Rens W, Degrassi F. Aneuploidy in mitosis of PtK1 cells is generated by random loss and nondisjunction of individual chromosomes. J Cell Sci. 2009;122:3455–3461. doi: 10.1242/jcs.047944. [DOI] [PubMed] [Google Scholar]
- 125.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Galimberti F, Thompson SL, Liu X, Li H, Memoli V, Green SR, DiRenzo J, Greninger P, Sharma SV, Settleman J, et al. Targeting the cyclin E-Cdk-2 complex represses lung cancer growth by triggering anaphase catastrophe. Clin Cancer Res. 2010;16:109–120. doi: 10.1158/1078-0432.CCR-09-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aoki K, Aoki M, Sugai M, Harada N, Miyoshi H, Tsukamoto T, Mizoshita T, Tatematsu M, Seno H, Chiba T, et al. Chromosomal instability by β-catenin/TCF transcription in APC or β-catenin mutant cells. Oncogene. 2007;26:3511–3520. doi: 10.1038/sj.onc.1210141. [DOI] [PubMed] [Google Scholar]
- 128.Hadjihannas MV, Bruckner M, Jerchow B, Birchmeier W, Dietmaier W, Behrens J. Aberrant Wnt/B-catenin signaling can induce chromosomal instability in colon cancer. Proc Natl Acad Sci USA. 2006;103:10747–10752. doi: 10.1073/pnas.0604206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Y, Malureanu L, Jeganathan KB, Tran DD, Lindquist LD, van Deursen JM, Bram RJ. CAML loss causes anaphase failure and chromosome missegregation. Cell Cycle. 2009;8:940–949. doi: 10.4161/cc.8.6.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weaver BAA, Bonday ZQ, Putkey FR, Kops GJPL, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tomonaga T, Matushita K, Ishibashi M, Nezu M, Shimada H, Ochiai T, Yoda K, Nomura F. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 2005;65:4683–4689. doi: 10.1158/0008-5472.CAN-04-3613. [DOI] [PubMed] [Google Scholar]
- 132.Cheng X, Shen Z, Yang J, Lu SH, Cui Y. ECRG2 disruption leads to centrosome amplification and spindle checkpoint defects contributing chromosome instability. J Biol Chem. 2008;283:5888–5898. doi: 10.1074/jbc.M708145200. [DOI] [PubMed] [Google Scholar]
- 133.Du J, Cai X, Yao J, Ding X, Wu Q, Pei S, Jiang K, Zhang Y, Wang W, Shi Y, et al. The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene. 2008;27:4107–4114. doi: 10.1038/onc.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu G, Lin YT, Wei R, Chen Y, Shan Z, Lee WH. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol Cell Biol. 2008;28:3652– 3662. doi: 10.1128/MCB.01923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang X, Di K, Zhang X, Han HY, Wong YC, Leung SC, Ling MT. Id-1 promotes chromosomal instability through modification of APC/C activity during mitosis in response to microtubule disruption. Oncogene. 2008;27:4456–4466. doi: 10.1038/onc.2008.87. [DOI] [PubMed] [Google Scholar]
- 136.Mazumdar M, Lee JH, Sengupta K, Reid T, Rane S, Misteli T. Tumor formation via loss of a molecular motor protein. Curr Biol. 2006;16:1559–1564. doi: 10.1016/j.cub.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kasiappan R, Shih HJ, Chu KL, Chen WT, Liu HP, Huang SF, Choy CO, Shu CL, Din R, Chu JS, et al. Loss of p53 and MCT-1 overexpression synergistically promote chromosome instability and tumorigenicity. Mol Cancer Res. 2009;7:536–548. doi: 10.1158/1541-7786.MCR-08-0422. [DOI] [PubMed] [Google Scholar]
- 138.Montembault E, Dutertre S, Prigent C, Giet R. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J Cell Biol. 2007;179:601–609. doi: 10.1083/jcb.200703133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hong KU, Kim E, Bae CD, Park J. TMAP/CKAP2 is essential for proper chromosome segregation. Cell Cycle. 2009;8:314–324. doi: 10.4161/cc.8.2.7597. [DOI] [PubMed] [Google Scholar]
- 140.Cimini D, Antoccia A, Tanzarella C, Degrassi F. Topoisomerase II inhibition in mitosis produces numerical and structural chromosomal aberrations in human fibroblasts. Cytogenet Cell Genet. 1997;76:61–67. doi: 10.1159/000134517. [DOI] [PubMed] [Google Scholar]