Abstract
Plasma-derived microRNAs (miRNAs) are being used as biomarkers, and have been associated with human liver disease and function including fibrosis, inflammation, and drug-induced liver injury. They may also be biomarkers of the drug metabolism function of the liver. In order for plasma miRNA to function as a clinical biomarker, predictable variability is necessary during processing from whole blood to plasma. The current study evaluated the variability of miRNA in whole blood stored for 0.5, 1, 2, 4, 8, and 12 hours following the blood draw under clinical conditions (room temperature) prior to the separation of the plasma. Four healthy volunteers were recruited. Blood from all subjects was collected twice. MicroRNA-16 (miR-16) and miR-223 were evaluated because many studies have shown them to be reliably present in plasma and useful for normalization. miRNA concentrations were measured by real-time polymerase chain reaction. The coefficient of variability of the cycle threshold values for subjects for miR-223 and miR-16 ranged from ∼3.6 to 6.8% and ∼1.48 to 4.1%, respectively, over the 12-hour incubation. A second blood collection was performed to determine interday variability. The coefficient of variance from the initial blood draw compared with the final blood draw for each subject ranged from 0.42 to 7.9% for miR-16 and 1.7 to 8.3% for miR-223, indicating that these miRNAs have limited interday variability. We conclude that plasma miR-16 or miR-223 concentrations are stable in whole blood at room temperature for up to 12 hours.
Introduction
MicroRNAs (miRNAs) are short (19–25 nucleotide) noncoding endogenous RNAs that regulate gene expression within cells at the post-transcriptional level. They are also found in many body fluids, including blood, saliva, and urine (Ramachandran and Palanisamy, 2012). Plasma miRNA patterns have been associated with human and rodent liver disease and function. These include hepatitis C infection, fibrosis, inflammation, and drug-induced liver injury (Laterza et al., 2009; Wang et al., 2009; Zhang et al., 2010; Starkey Lewis et al., 2011; Bala et al., 2012; Murakami et al., 2012; Ward et al., 2012; Yamaura et al., 2012). In addition, reciprocal changes in liver and plasma miRNA concentrations occurred in models of liver injury (Wang et al., 2009). Thus, it is highly plausible that they are also associated with the drug metabolism function of the liver. Uptake of exosomes has also been demonstrated in liver-derived cells (Zhang et al., 2012a). Further, the miRNAs that are taken up appeared to be functional and altered gene expression patterns. Also, the transcription factor, hepatic nuclear factor 4α, is involved in the regulation of genes of drug disposition, and is itself regulated by miRNA (Takagi et al., 2010; Ramamoorthy et al., 2012). Furthermore, regulators of liver metabolism, CYP2C8 and CYP1B1, are also targets of miRNAs (Tsuchiya et al., 2006; Zhang et al., 2012b). Unlike the extraction of miRNA from cells, many methodological obstacles in measuring plasma miRNA are still being elucidated. The fundamental basis for the analysis of miRNA found in plasma was the work done by Mitchell et al. (2008) that showed that miRNAs in plasma were stable under various conditions. It was shown that plasma miRNA had very small variability over a 24-hour period when incubated at room temperature. Additionally, plasma miRNAs were stable after repeat freeze/thaw cycles (Mitchell et al., 2008). However, in order for plasma miRNA to function as a clinical biomarker, predictable variability is also necessary during the processing of whole blood to plasma, due to the potential of miRNAs to be released from blood cells and platelets. One factor of clinical importance that has not yet been addressed is the transport time required to get the whole blood from the phlebotomist to the laboratory and initiate the start of processing to plasma.
In mice, miRNAs have been evaluated directly from the whole blood (Hsieh et al., 2012). However, due to in vitro evidence showing concern that lysed human red blood cells (RBCs), which contain miRNA, added to human plasma changed the level of microRNA-16 (miR-16), it was thought that whole blood would need to be processed immediately into plasma to prevent highly varying miRNA levels (Kirschner et al., 2011). This is potentially problematic because miR-16, along with miR-223, has been considered to be consistently released into the blood and thought to have similar levels within a single individual and possibly across individuals with the same disease or health status (Mitchell et al., 2008; Kroh et al., 2010; Kim et al., 2012). However, there is no consensus of constitutively released miRNAs, and these miRNAs need to be evaluated with each experimental condition. Also, messenger RNA (coding RNA) in whole blood has been shown to change greater than 1000-fold during storage and transport of whole blood at room temperature (Rainen et al., 2002). These findings have potentially important implications for limiting the practical usefulness of plasma miRNA as a biomarker of drug metabolism and disposition in the clinic, due to variability in blood transport and processing times. However, since RBCs have a ∼120-day life span (Pietrzik et al., 2007), which makes RBC lysis unlikely, and miRNAs in isolated plasma are relatively stable, we hypothesized that the storage of whole blood at room temperature for extended periods of time would not affect the plasma miRNA concentrations. Therefore, the current study was designed to evaluate the variability in plasma miRNA concentrations obtained from whole blood that was stored under clinical conditions prior to the separation of the plasma.
Materials and Methods
Human Subjects.
Four healthy volunteers (3 men and 1 woman) were recruited for minimally invasive venous punctures for whole-blood specimens. Written informed consent was obtained from all subjects in the study. This study was approved by the Indiana University Institutional Review Board.
Blood Sample Collection.
Blood from all subjects was collected twice. The second sample was taken at variable times after the initial phlebotomy: subject 1 was sampled 49 days after the initial draw, subject 2 was 39 days, subject 3 was 16 days, and subject 4 was only 7 days after the first draw. Three to four milliliters of whole blood was collected at each time point by venous puncture with 21-gauge butterfly needles directly into 8-ml BD Bioscience EDTA Vacutainers (BD, Franklin Lakes, NJ). EDTA has been shown to be a suitable anticoagulant for collection of plasma for miRNA measurements (Hastings et al., 2012). Also, we have confirmed the findings that heparin interferes with miRNA measurements and can be mostly eliminated by treating the plasma with heparinase (Supplemental Fig. 1). Each tube was inverted eight times per BD protocol. Whole blood was incubated at room temperature under normal laboratory lighting. At each experimental time point, whole blood was processed to plasma by centrifugation at 1500 relative centrifugal force on a Beckman GH3.8 Rotor (Beckman Coulter, Pasadena, CA) at 4°C for 15 minutes, then the supernatant was transferred to a RNase-free 15-ml tube, inverted twice, and transferred to 1.5-ml RNase-free microcentrifuge tubes and immediately frozen at −20°C.
miRNA Preparation.
Following the Qiagen miRNeasy plasma protocol, plasma samples were thawed on ice, centrifuged at 1500 relative centrifugal force, 4°C for 10 minutes (because protein and/or lipid precipitates are present in the plasma) (Qiagen, Valencia, CA). An equal volume of plasma (50 µl) was used, since total miRNA concentrations in plasma are below the level of quantification by spectrometry. The centrifuged plasma (50 µl) was transferred to fresh 1.5-ml RNase-free microcentrifuge tubes, and the extraction followed the Qiagen protocol using 250 µl of QIAzol lysis reagent, according to the manufacturer’s instructions. Optional Caenorhabditis elegans miR-39 was added (5 µl of 5 nM stock) as a positive control for extraction efficiency immediately after the denaturation of the plasma. Additionally, 0.3 µl (0.8 µg/µl) of bacteriophage M2 RNA per 200 µl of Qiazol buffer was added (similar to the Exiqon protocol; Exiqon, Woburn, MA) to the denatured plasma at the same step as the addition of C. elegans miR-39. Bacteriophage M2 RNA acts as a carrier RNA to improve miRNA yield by preventing miRNA from sticking to the sides of tubes and columns. Cycle threshold (CT) values from controls (bacteriophage RNA only) were ∼15 cycles higher than reactions with template (data not shown). All samples were eluted twice in 30 µl of RNase/DNase-free water (total 60 µl) and stored at −80°C.
miRNA Analysis.
miRNA samples were analyzed by real-time polymerase chain reaction (PCR) using an iCycler (Bio-Rad, Hercules, CA) using Taqman Small RNA Assays (Applied Biosystems, Foster City, CA) for human miRNAs miR-223 and miR-16 along with C. elegans miR-39, and following the Taqman Small RNA Assay protocol. Technical triplicate PCR assays were run, and the four different subjects provided the biological replicates. Standard deviations and coefficients of variation of the CT values were calculated using Microsoft Excel (Microsoft, Redmond, WA). The effect of incubation time on miRNA concentrations was tested statistically using a repeated-measures general linear model in SPSS (SPSS Software, Chicago, IL).
Results
Subject Population.
To include subjects of both sexes and multiple ethnicities, 3 male subjects and 1 female subject of varying ethnicities, including Caucasian American, African American, South American, and Asian, were enrolled in the study. Subjects’ diets were also recorded from dinner the night before and any breakfast the morning of the phlebotomy.
miRNA Intrasubject Variability in Whole Blood.
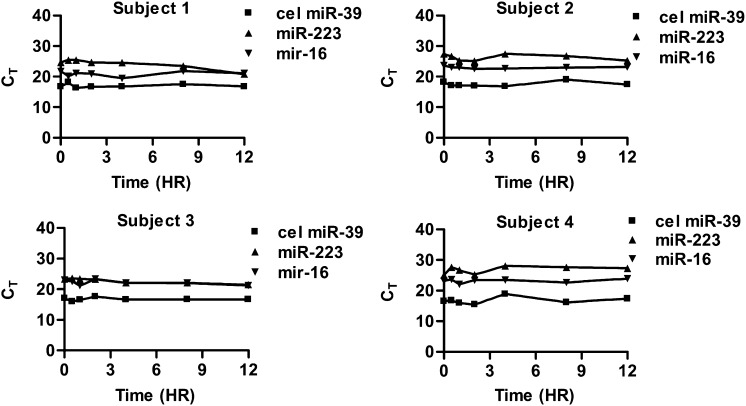
Whole blood was collected in EDTA (anticoagulant) tubes and incubated at room temperature under normal lighting similar to real-world clinical environments prior to conversion to plasma. Homo sapiens miR-16 and miR-223 were selected because many studies have shown them to be reliably present in the plasma and useful for normalization, although there is no gold standard for constitutively expressed plasma miRNAs. However, we do acknowledge that one limitation of our study and others that have used only miR-16 and miR-223 (Kim et al., 2012) is that the stability of these miRNAs may not be representative of all miRNAs in plasma. The plasma was stored at −20°C prior to analysis by Taqman real-time PCR. The miRNA concentrations did not consistently change over a 12-hour incubation in whole blood (Fig. 1; P > 0.10). In subject 1, the average CT values (± S.D.) over the 12-hour period for miR-223 and miR-16 were 24.1 ± 1.63 and 20.8 ± 0.85, respectively. In subject 2, the average CT values for miR-223 and miR-16 were 26.4 ± 1.05 and 23.0 ± 0.34, respectively. In subject 3, the average CT values for miR-223 and miR-16 were 22.7 ± 0.83 and 22.2 ± 0.81, respectively, and in subject 4, the average CT values over the 12 hours were 26.8 ± 1.19 and 23.2 ± 0.64, respectively. Although the absolute averages between subjects were different, the variability within subjects was relatively consistent over the incubation time. The coefficients of variability of the CT values for subjects 1, 2, 3, and 4 for miR-223 were 6.75, 4.07, 3.67, and 4.47% over the 12-hour incubation, respectively, whereas for miR-16, the CT values were 4.09, 1.48, 3.64, and 2.7%, respectively. There was no statistically significant change in miRNA concentration over time for either miR-16 or miR-223.
Fig. 1.
Analysis of stability of miRNA in whole blood. Whole blood was incubated for 0, 0.5, 2, 4, 8, and 12 hours at room temperature and then processed into plasma. Expression of miRNAs by real-time PCR of each subject showing limited variability is shown to the left. P values >0.10 for repeated-measures analysis over time for all three miRNAs revealed no significant changes. cel miR-39, C. elegans miR-39.
In addition, to evaluate the interday variability in miRNA concentrations, each subject had a repeat collection of whole blood (Fig. 2). Blood from all subjects was collected at variable times from the initial phlebotomy (7–49 days). The whole blood was incubated for 30 minutes at room temperature prior to separation of the plasma. The average CT values for subjects 1, 2, 3, and 4 were 25.8 ± 0.47, 26.3 ± 0.48, 24.2 ± 1.06, and 26.1 ± 2.17, respectively, for miR-223, and 20.7 ± 0.85, 22.2 ± 1.20, 22.5 ± 0.09, and 22.4 ± 1.77, respectively, for miR-16. The coefficient of variance from the initial blood draw at the 30-minute incubation compared with the final blood draw for each subject ranged from 0.42 to 7.9% for miR-16 and 1.7 to 8.3% for miR-223, indicating miRNA has limited interday variability within these healthy subjects.
Fig. 2.
Variability of whole blood miRNA compared with initial blood draw. Repeat collection of venous whole blood at various times (subject 1, 49 days; subject 2, 39 days; subject 3, 16 days; subject 4, 7 days) from initial collection for miR-223 and miR-16.
Discussion
miRNAs have been used as a biomarker for many disease states and are now being used to understand drug metabolism and disposition, but their practicality in the clinical setting has not been thoroughly explored. The study by Mitchell et al. (2008) suggested that further investigation into miRNA variability in whole blood was needed, and although Kirschner et al. (2011) have spiked lysed red blood cells into their plasma preparations, no whole blood variability study in subjects has been reported. Here, we show that incubating whole blood from healthy volunteers at room temperature causes only limited variability. This variability is small compared with the 1000-fold changes seen in messenger RNAs after incubation of whole blood at room temperature for 24 hours (Rainen et al., 2002). In fact, there is not any obvious sign of miRNA degradation occurring. Upon visual inspection of the samples, there was no pink hue to any of the samples even with time, indicating that the contribution of RBC lysis is likely minimal. The stability of endogenous miRNA is likely secondary to the finding that miRNA released from cells is usually packaged in some form, including high-density lipoprotein, exosomes, vesicles, and apoptotic bodies. In contrast, free miRNAs that are spiked into nondenatured plasma are rapidly degraded (Mitchell et al., 2008). Our studies show that the incubation of whole blood at room temperature has a minimal effect on miRNA concentrations. The red blood cells remain stable in the EDTA anticoagulant, and miRNA does not appear to be changing due to factors such as degradation or release from blood cells. Although it is possible that the stability of other miRNAs may be different, or that concurrent medications may cause altered stability, the results of this study reflect the stability of miR-16 and miR-223 in human blood and plasma. The extent to which these results can be extrapolated to future miRNAs of interest will require additional study. This study indicates that the storage of whole blood at room temperature prior to the isolation of plasma does not appear to affect plasma miRNA concentrations.
Supplementary Material
Abbreviations
- CT
cycle threshold
- miR-16
microRNA-16
- miR-223
microRNA-223
- miRNA
microRNA
- PCR
polymerase chain reaction
- RBC
red blood cell
Authorship Contributions
Participated in research design: Benson, Skaar.
Conducted experiments: Benson.
Performed data analysis: Benson, Skaar.
Wrote or contributed to the writing of the manuscript: Benson, Skaar.
Footnotes
This work was supported by grants from the National Institutes of Health Institute of General Medical Sciences [Grants T32GM008425-19 and RO1-GM088076]; and the Indiana University Institute of Personalized Medicine Brater Scholar award.
Part of this work was previously presented as: Benson EA and Skaar TC (2013) Analysis of the stability of MiRNA in whole blood. American Society for Clinical Pharmacology and Therapeutics; 2013 Mar 5–9; Indianapolis, IN. Abstract PI-1.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. (2012) Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 56:1946–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Palma J, Duelli DM. (2012) Sensitive PCR-based quantitation of cell-free circulating microRNAs. Methods 58:144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CH, Rau CS, Jeng JC, Chen YC, Lu TH, Wu CJ, Wu YC, Tzeng SL, Yang JC. (2012) Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J Biomed Sci 19:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JY, Gilman-Sachs A, Beaman K, Hastings ML, Martin JN, et al. (2012) Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn 14:71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G. (2011) Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 6:e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroh EM, Parkin RK, Mitchell PS, Tewari M. (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50:298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, et al. (2009) Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem 55:1977–1983 [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105:10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. (2012) Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS ONE 7:e48366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzik K, Lamers Y, Brämswig S, Prinz-Langenohl R. (2007) Calculation of red blood cell folate steady state conditions and elimination kinetics after daily supplementation with various folate forms and doses in women of childbearing age. Am J Clin Nutr 86:1414–1419 [DOI] [PubMed] [Google Scholar]
- Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, et al. (2002) Stabilization of mRNA expression in whole blood samples. Clin Chem 48:1883–1890 [PubMed] [Google Scholar]
- Ramachandran S, Palanisamy V. (2012) Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdiscip Rev RNA 3:286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy A, Li L, Gaedigk A, Bradford LD, Benson EA, Flockhart DA, Skaar TC. (2012) In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4α expression. Drug Metab Dispos 40:726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, French NS, Dhaun N, Webb DJ, Costello EM, et al. (2011) Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 54:1767–1776 [DOI] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. (2010) MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 285:4415–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. (2006) MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66:9090–9098 [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. (2009) Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 106:4402–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J, Bala S, Petrasek J, Szabo G. (2012) Plasma microRNA profiles distinguish lethal injury in acetaminophen toxicity: a research study. World J Gastroenterol 18:2798–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaura Y, Nakajima M, Takagi S, Fukami T, Tsuneyama K, Yokoi T. (2012) Plasma microRNA profiles in rat models of hepatocellular injury, cholestasis, and steatosis. PLoS ONE 7:e30250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, et al. (2012a) Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22:107–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SY, Surapureddi S, Coulter S, Ferguson SS, Goldstein JA. (2012b) Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. Mol Pharmacol 82:529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. (2010) Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem 56:1830–1838 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.