Abstract
Phospholipase C (PLC) enzymes convert phosphatidylinositol-4,5-bisphosphate into the second messengers diacylglycerol and inositol-1,4,5-triphosphate. The production of these molecules promotes the release of intracellular calcium and activation of protein kinase C, which results in profound cellular changes. The PLCβ subfamily is of particular interest given its prominent role in cardiovascular and neuronal signaling and its regulation by G protein–coupled receptors, as PLCβ is the canonical downstream target of the heterotrimeric G protein Gαq. However, this is not the only mechanism regulating PLCβ activity. Extensive structural and biochemical evidence has revealed regulatory roles for autoinhibitory elements within PLCβ, Gβγ, small molecular weight G proteins, and the lipid membrane itself. Such complex regulation highlights the central role that this enzyme plays in cell signaling. A better understanding of the molecular mechanisms underlying the control of its activity will greatly facilitate the search for selective small molecule modulators of PLCβ.
Introduction
Phospholipase C (PLC) enzymes are responsible for the hydrolysis of the inner membrane component phosphatidylinositol-4,5-bisphosphate (PIP2), generating the second messengers inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 is freely diffusible and binds to IP3-specific receptors, leading to the release of intracellular Ca2+. DAG remains membrane associated and, together with increasing Ca2+, activates protein kinase C. These events are associated with the regulation of numerous physiological processes, including muscle contraction (Berridge, 2003; Woodcock et al., 2009a), chemotaxis (Jiang et al., 1997; Li et al., 2000), opioid sensitivity (Murthy and Makhlouf, 1996; Wu et al., 1998; Mathews et al., 2008), and cell proliferation and survival (Braz et al., 2004; Palaniyandi et al., 2009; Newton, 2010).
There are six subfamilies of PLC in higher eukaryotes (Gresset et al., 2012; Kadamur and Ross, 2013). Of these, the PLCβ subfamily is among the most intensively studied. These enzymes are the canonical downstream targets of the Gq subfamily of G protein–coupled receptors (GPCRs) and play prominent roles in cardiovascular function, chemotaxis, and neuronal signaling. In the absence of extracellular stimuli, PLCβ exhibits very low intrinsic PIP2 hydrolysis, but is robustly activated upon direct interactions with Gαq. GPCR-mediated activation of PLCβ also occurs through release of the Gβγ heterodimer, which is thought to be mediated by activation of Gi-coupled GPCRs (Camps et al., 1992; Katz et al., 1992; Wu et al., 1998; Xie et al., 1999). Members of the Rho family of small molecular weight G proteins, such as the Rac isoforms, also directly bind and activate PLCβ, linking PLCβ activity to GPCR-independent signaling cascades (Gresset et al., 2012; Kadamur and Ross, 2013). It is also increasingly recognized that the membrane itself plays a role in the regulation of PLCβ, as may interactions with scaffolding proteins (Cartier et al., 2011; Grubb et al., 2011, 2012; Sun et al., 2013). In this review, we highlight the current understanding of the molecular basis of regulation of mammalian PLCβ enzymes and their modulation by small molecules, with an emphasis on recent structural discoveries.
Structure of the PLCβ Catalytic Core and Its C-Terminal Extension
As in most other PLC enzymes, PLCβ proteins share a highly conserved catalytic core composed of an N-terminal pleckstrin homology (PH) domain, four tandem EF hand repeats, a triose phosphate isomerase (TIM)–like barrel domain split into X and Y halves and which houses the active site, and a C2 domain (Figs. 1 and 2). With the exception of the TIM barrel, the domains have somewhat unconventional roles. Unlike the PLCδ PH domain, which binds PIP2 with high specificity and affinity, the PLCβ PH domain only weakly contributes to membrane association (Tall et al., 1997; Wang et al., 1999b) and is intimately associated with the rest of the catalytic core. Instead, its most significant role is arguably its contribution to regulatory protein–protein interactions. The EF hands, contrary to their role in other well-known proteins such as calmodulin, do not bind Ca2+. In PLCβ, they serve as a scaffold and support the loop responsible for stimulating GTP hydrolysis when Gαq is bound. Finally, unlike many other C2 domains, the PLCβ C2 domain does not participate in Ca2+-mediated interactions with the membrane, but instead contributes to intra- and intermolecular regulatory binding sites.
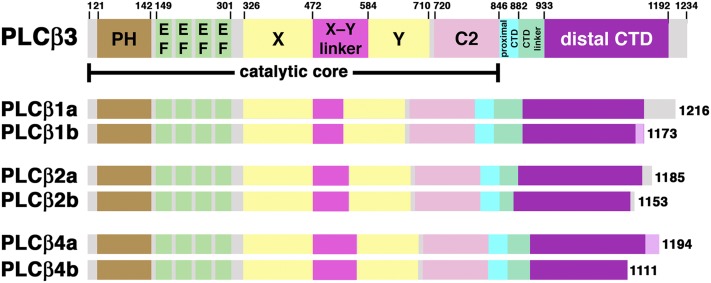
Fig. 1.
Primary structure of PLCβ isozymes and splice variants. Numbers above the diagram correspond to domain boundaries in human PLCβ3, and all domain diagrams correspond to human isoforms, with the exception of PLCβ4b, which is from Rattus norvegicus. All identified PLCβ variants share the same catalytic core, which is the minimal fragment of PLCβ that hydrolyzes PIP2, defined as the N terminus through the end of the C2 domain. The PLCβ isoforms differ most significantly in the length of the X–Y linker, whereas the splice variants reported for each isoform primarily vary the length and sequence of the CTD linker and extreme C terminus. Regions with sequences unique to the PLCβ1b and PLCβ4a splice variants are shown in pink.
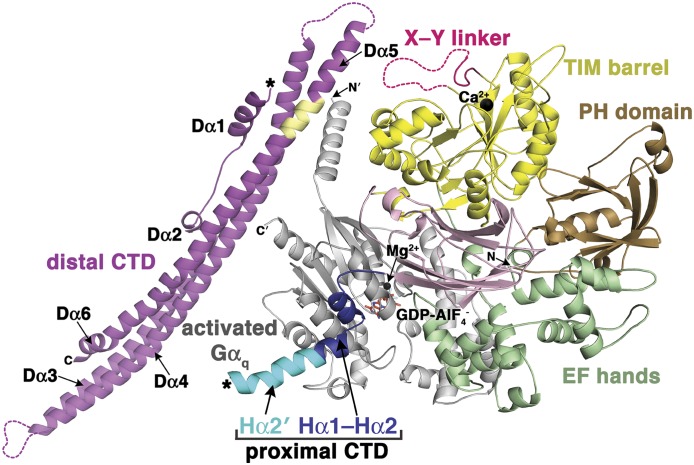
Fig. 2.
Structure of full-length PLCβ3 in complex with activated Gαq. The structure shown is derived from PDB ID 4GNK. The PLCβ3 domains are colored as in Fig. 1, and activated Gαq is colored gray. The hydrophobic surface of the distal CTD that binds the Gαq N-terminal helix is shown in yellow. The observed ends of the proximal and distal CTD are marked with asterisks, and the N and C termini of PLCβ3 and Gαq are labeled N and C or N′ and C′, respectively. The Gαq-bound GDP-AlF4− is shown in orange sticks, and Ca2+ and Mg2+ as black spheres. Disordered regions are shown as dashed lines.
The mechanism by which PLC enzymes hydrolyze PIP2 to generate DAG and IP3 was determined with the help of crystal structures of PLCδ1 (Essen et al., 1996, 1997; Ellis et al., 1998) and is described in greater detail elsewhere (Gresset et al., 2012). Briefly, the catalytic Ca2+ is proposed to decrease the pKa of the inositol 2-hydroxyl group and, with the assistance of the putative catalytic base (Glu341 in PLCβ3), promotes the formation of a 1,2-cyclic monophosphate intermediate and DAG. This cyclic intermediate is stabilized via the 1-phosphate by a histidine (His332 in PLCβ3) and Ca2+. In the next step, another histidine (His379 in PLCβ3) abstracts a proton from water, which attacks the intermediate to release IP3 (Essen et al., 1997; Ellis et al., 1998). A ridge of hydrophobic residues adjacent to the active site also facilitates catalysis (Fig. 3) (Essen et al., 1997). Mutation of these residues within PLCδ (Ellis et al., 1998) or in PLCβ3 (Lyon et al., 2013) decreases basal activity and/or protein expression. Studies of PLCβ1 and PLCβ2 found that increasing surface pressure on lipid bilayers diminishes catalytic activity, suggesting that membrane insertion contributes to activity (James et al., 1997). Taken together, these observations are consistent with the idea that insertion of the hydrophobic ridge into the membrane is required for efficient catalysis.
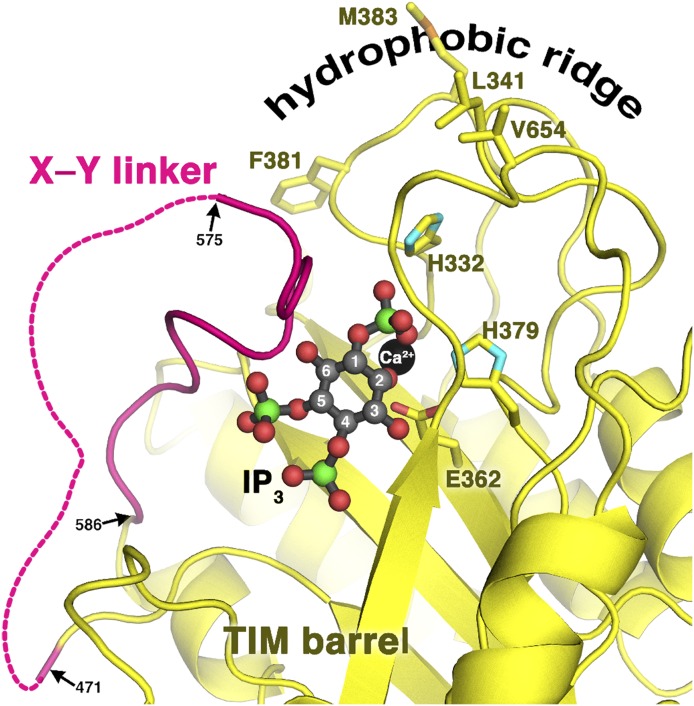
Fig. 3.
The PLCβ X–Y linker blocks the active site. A model of IP3 (derived from PDB ID 1DJX) bound to the PLCβ3 active site reveals a possible mechanism for autoinhibition by the X–Y linker. As observed in six independent structures of PLCβ enzymes, the ordered region of the X–Y linker (PLCβ3 residues 575–586) docks in a position that would prevent PIP2 from entering the enzyme active site. Displacement of this region of the X–Y linker would therefore appear to be a prerequisite for PIP2 binding. The catalytic residues H332, H379, and E362 are shown as sticks, and the active site Ca2+ as a black sphere. Dashed lines indicate the disordered region of the PLCβ3 linker, which contains a span of acidic residues. Side chains of residues that constitute the hydrophobic ridge, which is thought to help anchor the catalytic core to the membrane, are also shown.
Although all of the catalytic machinery is in place, crystal structures suggest that the PLCβ active site cannot readily bind PIP2. The two halves of the PLCβ catalytic TIM barrel-like domain are separated by a poorly conserved X–Y linker that typically bears a stretch of highly acidic residues (Figs. 1 and 2). The C terminus of this linker is ordered in all reported crystal structures (Table 1) and interacts with residues adjacent to the active site cavity in a manner that would sterically prevent the binding of PIP2 (Fig. 3). As discussed below, perturbation of the X–Y linker region may play an important role in the regulation of PLCβ isozymes (Ellis et al., 1993; Schnabel and Camps, 1998; Zhang and Neer, 2001; Hicks et al., 2008).
TABLE 1.
Crystal structures of PLCβ domains and complexes
| Structure | PDB ID | Residue Range(s) Used | Species | Resolution |
|---|---|---|---|---|
| Å | ||||
| Distal CTD | 1JAD | 878–1158 | Turkey | 2.4 |
| PLCβ2 | 2ZKM | 1–799 | Human | 1.6 |
| Sepia PLC21 | 3QR0 | 1–816 | Cuttlefish | 2.0 |
| Loligo PLC21 | 3QR1 | 1–813 | Squid | 3.2 |
| Rac1-PLCβ2 | 2FJU | Rac1:1–89 | Human | 2.2 |
| PLCβ2: 1–799 | Human | |||
| Gαq-PLCβ3 | 3OHM | Gαq: 35–359 | Mouse | 2.7 |
| PLCβ3: 1–887 | Human | |||
| Gαq-PLCβ3 | 4GNK | Gαq: 7–359 | Mouse | 4.0 |
| PLCβ3: 1–1234 | Human |
The defining element of the PLCβ subfamily is an approximately 400 amino acid C-terminal extension that contains highly conserved segments at its N terminus [the proximal C-terminal domain (CTD)] and an elongated approximately 300 amino acid coiled-coil domain (the distal CTD) separated by a 28–61 residue flexible linker region (the CTD linker). Numerous studies have shown that the C-terminal extension is required for membrane and/or particulate fraction binding, Gαq binding, and maximum basal and Gαq-stimulated activity (Park et al., 1993; Schnabel et al., 1993; Kim et al., 1996; Jenco et al., 1997; Adjobo-Hermans et al., 2008, 2013), yet it is dispensable for Rac and Gβγ activation (Lee et al., 1993b; Wu et al., 1993a; Illenberger et al., 2003a; Waldo et al., 2010). The proximal CTD is composed of the first approximately 40 amino acids immediately following the C2 domain and contains the primary Gαq binding site (Waldo et al., 2010), followed by an autoinhibitory helix designated Hα2′ (Lyon et al., 2011) (Figs. 2 and 5). The role of these structural elements in regulation of activity is discussed in later sections.
Fig. 5.
The proximal CTD is an allosteric site for Gαq activation. (A) PLCβ3 is colored as in Fig. 1, and activated Gαq is shown as a gray surface with the switch regions colored orange. In the absence of Gαq, the Hα2′ helix (cyan) is bound to the PLCβ catalytic core (right), and is connected to the C terminus of the C2 domain by an approximately 25-amino acid disordered loop (dashed line). Gαq binds the disordered loop via its switch regions, ordering the Hα1/Hα2 element (dark blue). Additional interactions between switch regions of Gαq, the EF hands, and the C2 domain displace Hα2′ from the catalytic core (left). The interactions between Gαq and the Hα1/Hα2 element are largely hydrophobic, and mutation of Leu859 eliminates Gαq binding and activation. The intrinsic GAP activity of PLCβ relies on Asn260, positioned in a loop between two EF hand domains, which interacts with the catalytic glutamine of Gαq. (B) In the resting cell (left), PLCβ is in an autoinhibited state, wherein the Hα2′ and the X–Y linker are bound to the catalytic core. The distal CTD interacts with the cell membrane or the hydrophobic ridge of the catalytic core, which may help dictate the distribution of the enzyme between the membrane and cytosol. Gαq binding leads to allosteric activation through displacement of Hα2′ and recruitment of the PLCβ catalytic core to the membrane surface. Anionic phospholipids in the inner leaflet eject the acidic X–Y linker. The orientation of the active site at the membrane is further optimized by interactions between the membrane, the palmitoylated N terminus of Gαq, and the distal CTD (right).
The distal CTD is believed to be the primary membrane binding determinant in PLCβ isozymes and is required for maximal basal and stimulated activity (Lee et al., 1993b; Kim et al., 1996; Waldo et al., 2010; Lyon et al., 2011, 2013). One mystery concerning the distal CTD is its lack of strong sequence conservation (approximately 30–35% identity across PLCβ isoforms) despite its importance in activity and regulation by Gαq. Structural insights into the distal CTD were first obtained from a crystal structure of an isolated engineered domain derived from turkey PLCβ (Singer et al., 2002), revealing an unusual approximately 140 Å–long helical bundle composed primarily of three long, kinked helical spans and several shorter bridging helices (Figs. 2 and 4). The “core” of the domain, which contains some of the most highly conserved residues, is found where the Dα2 helix crosses one face of the helical bundle. The entire distal CTD is stabilized primarily through coiled-coil interactions, which may have relatively low stringency for amino acid side chains, and thus could account for the low sequence conservation (Singer et al., 2002; Zhang et al., 2006; Lyon et al., 2013). The tertiary structure of the distal CTD was confirmed in the crystal structure of full-length PLCβ3 in complex with Gαq (Figs. 2 and 4) (Lyon et al., 2013). The PLCβ3 distal CTD has a greater degree of curvature compared with the turkey structure, likely due to the inherent flexibility of the domain and to differences in sequence and crystal contacts. Comparisons of the PLCβ distal CTD to other structures identified the Bin-Amphiphysin-Rvs domains as distant structural homologs. These domains are also extended helical bundles that interact with negatively charged phospholipids (Peter et al., 2004; Qualmann et al., 2011). Although an intriguing possibility, it is unknown whether the PLCβ distal CTD can sense and/or induce membrane curvature, as do some Bin-Amphiphysin-Rvs domains.
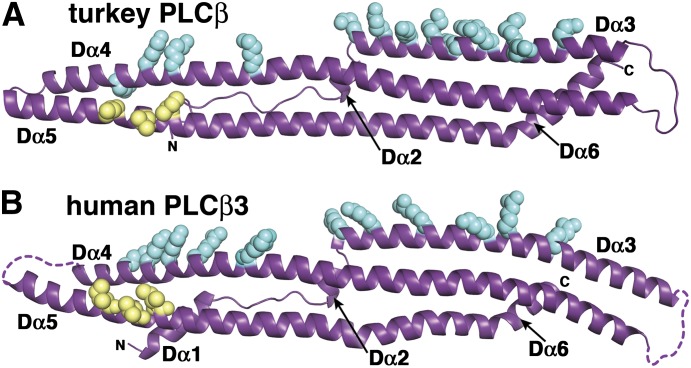
Fig. 4.
The structure and surface of the distal CTD are conserved. The isolated turkey distal CTD (PDB ID 1JAD) (A) and human PLCβ3 distal CTD (PDB ID 4GNK) (B) have the same fold and similar conserved surfaces. Basic residues within Dα3 and Dα4 (blue spheres) form an extended conserved surface along one face of the domain, which likely functions as a membrane binding site. The conserved hydrophobic patch on Dα5, which interacts with the N-terminal helix of Gαq in the 4GNK structure (Fig. 2), is shown as yellow spheres. The turkey PLCβ CTD was engineered to facilitate crystallization by deletion of 32 residues from the Dα3–Dα4 loop.
The turkey distal CTD crystallized as a dimer, burying approximately 3100 Å2 of accessible surface area. Mutation of conserved hydrophobic residues within the analogous dimer interface of PLCβ1 were shown to impair activation by Gαq (Ilkaeva et al., 2002), and size exclusion analysis of both purified PLCβ proteins and isolated distal CTDs, as well as cell-based studies, suggested the existence of dimers (Singer et al., 2002; Zhang et al., 2006). Conversely, studies of full-length human PLCβ3 found no evidence of oligomerization as assessed by size exclusion chromatography, multiangle light scattering, cryo-electron microscopy, or X-ray crystallography (Lyon et al., 2013). Instead, many of the conserved residues that contributed to the dimer interface in the turkey distal CTD structure instead form an intermolecular contact with the N terminus of Gαq (Figs. 2 and 4).
PLCβ Isoforms, Splice Variants, and Function
There are four PLCβ isoforms (PLCβ1−4), three of which are expressed as splice variants (Fig. 1). The sites of variation are typically localized within the C-terminal extension and alter the total length of the enzyme, potentially effecting membrane association and/or the ability to interact with scaffolding proteins or activators (Suh et al., 2008). Now that a full-length PLCβ enzyme has been structurally characterized, the sites of variation can be more accurately mapped and functional differences resulting from these changes can be considered.
PLCβ1 is expressed in the cerebral cortex and hippocampus, where the enzyme regulates neuronal activity (Kim et al., 1997; Böhm et al., 2002), and in the cardiovascular system (Ushio-Fukai et al., 1998; Mende et al., 1999; Arthur et al., 2001; Descorbeth and Anand-Srivastava, 2010). In vascular smooth muscle cells exposed to high glucose concentrations, Gαq and PLCβ1 expression increases, resulting in higher intracellular Ca2+. This Ca2+ increase is thought to be an underlying mechanism in vascular complications of diabetes (Descorbeth and Anand-Srivastava, 2010). There are two PLCβ1 splice variants, each of which has been assigned a specific role (Faenza et al., 2000; Grubb et al., 2008; Filtz et al., 2009; Woodcock et al., 2009b). PLCβ1a and PLCβ1b differ at the extreme C terminus, beyond the last residue observed in the reported crystal structures. PLCβ1a is longer and contains a consensus postsynaptic density protein/Drosophila disc large tumor suppressor/zona occludens-1 protein motif at its C terminus, whereas PLCβ1b contains a proline-rich region (Bahk et al., 1994, 1998; Grubb et al., 2008). Both variants are reported to interact with the membrane, suggesting full function of the distal CTD, although there may be variation between cell lines (Adjobo-Hermans et al., 2008, 2013; Grubb et al., 2008). PLCβ1a and PLCβ1b have been detected in the nucleus, where they contribute to the regulation of cell cycle progression, in particular the G1/S transition (Bahk et al., 1998; Faenza et al., 2000; O'Carroll et al., 2009; Fiume et al., 2012). The PLCβ1 variants have been reported to have unique functions with the cardiac sarcolemma. PLCβ1b is membrane associated and interacts with the scaffold proteins Homer1b/c and Shank3, enabling its rapid activation upon Gq-coupled receptor stimulation (Shin et al., 2003; Grubb et al., 2008, 2011, 2012). In contrast, PLCβ1a is cytosolic and does not interact with these scaffold proteins. Upregulation of the Gαq-PLCβ1b pathway results in increased cell size and expression of hypertrophic markers (Filtz et al., 2009; Descorbeth and Anand-Srivastava, 2010).
PLCβ2 is expressed in hematopoietic cells and platelets, where it is involved in chemotaxis (Mao et al., 2000; Sun et al., 2007; Suh et al., 2008; Tang et al., 2011). Paradoxically, loss of PLCβ2 in neutrophils increased their sensitivity to inflammatory agents and chemoattractants, despite a requirement for Ca2+ and IP3 during the early stages of chemotaxis. It may be that in later stages of chemotaxis, PLCβ2 has an inhibitory role (Jiang et al., 1997; Li et al., 2000). PLCβ2 is also required for thrombin-induced Ca2+ release in platelets through a Gαq-dependent mechanism (Vaidyula and Rao, 2003). PLCβ2 is found as two splice variants, PLCβ2a and PLCβ2b, where PLCβ2b is missing 19 internal residues that span the C terminus of the CTD linker and the Dα1 helix of the distal CTD (human PLCβ3 residues 930–948). Based on the structure of full-length PLCβ3, this deletion is expected to unmask a hydrophobic patch on the surface of the PLCβ2b distal CTD, but it is unclear whether this would significantly alter known functions of the domain (Fig. 4).
Only one variant of PLCβ3 has been characterized in humans, where it is expressed in the brain, liver, parotid gland (Jhon et al., 1993; Han et al., 2006; Bianchi et al., 2009), hematopoietic cells (Li et al., 2000; Cai et al., 2005; Xiao et al., 2009), and the cardiovascular system (Mende et al., 1999; Arthur et al., 2001). Within the nervous system, PLCβ3 is required for opioid-induced Ca2+ release through a Gβγ-dependent pathway, and it also mediates Ca2+ release in response to noxious stimuli (Xie et al., 1999; Han et al., 2006; Mathews et al., 2008; Bianchi et al., 2009). In the hematopoietic system, PLCβ3 inhibits proliferation by preventing differentiation through interactions with the transcription factor Stat5 and its regulator SHP1 (Xiao et al., 2009), and it also contributes to regulation of chemotaxis in neutrophils (Li et al., 2000). Lastly, in mouse models of Gq-mediated cardiac hypertrophy, increased PLCβ3 expression and activity have been reported (Mende et al., 1998, 1999).
PLCβ4 is most similar to NorpA, the invertebrate PLCβ homolog required for phototransduction, and is highly expressed in the retina and the cerebellum (Lee et al., 1993a; Jiang et al., 1996; Adamski et al., 1999; Suh et al., 2008). Within the retina, PLCβ4 is required for visual processing events after phototransduction (Jiang et al., 1996), and loss of PLCβ4 in mice also results in motor defects (Kim et al., 1997). Two splice variants of PLCβ4 have been identified in humans. PLCβ4a is the full-length protein, whereas PLCβ4b is truncated at the extreme C terminus after the end of the structurally characterized distal CTD in PLCβ3 (Fig. 1) (Adamski et al., 1999), which has been proposed to alter the efficacy of Gαq-dependent activation. An interesting splice variant of PLCβ4 has been identified in rat retina, in which the protein is truncated at the beginning of helix Dα4 (human PLCβ3 residue 1040) (Kim et al., 1998). This variant would clearly disrupt the fold of the distal CTD, likely explaining its loss of membrane association and Gαq responsiveness. As Kim et al. (1998) conjectured, even though the remaining portion of the distal CTD in this variant contain some of the most significant stretches of basic charge, their spatial localization, as dictated by a properly folded domain, seems to be essential for association with the particulate fraction of cells.
Regulation of PLCβ Basal Activity
PLCβ Membrane–Binding Determinants.
To prevent aberrant signaling and retain sensitivity to extracellular signals, PLCβ isozymes must have very low intrinsic activity. Because they interact with phospholipid bilayers to hydrolyze PIP2, control of membrane localization provides a straightforward mechanism for regulation of basal enzymatic activity (Romoser et al., 1996; Runnels et al., 1996; Jenco et al., 1997; Scarlata, 2002). Each PLCβ isoform has a unique subcellular distribution, despite sharing a conserved structure and membrane binding determinants. Therefore, relatively subtle differences in their amino acid sequences, and potentially their interactions with scaffolding proteins, likely dictate their cellular location. In general, PLCβ1 and PLCβ4 variants seem to be primarily membrane associated, whereas PLCβ2 and PLCβ3 seem primarily cytosolic (Illenberger et al., 2003b; Adjobo-Hermans et al., 2008, 2013; Grubb et al., 2008), although it is likely that all of these isoforms are in equilibrium between the membrane and cytoplasm.
The PLCβ PH domain has been proposed to contribute to membrane binding, in part because some PH domains have high specificity and affinity for certain phospholipids (Philip et al., 2002; Lemmon, 2004). The PLCδ PH domain specifically binds PIP2 and flexibly tethers the rest of enzyme to the membrane (Cifuentes et al., 1993; Ferguson et al., 1995; Garcia et al., 1995; Essen et al., 1996). However, most of the residues that coordinate the inositol head group in PLCδ are absent in PLCβ, and the PLCβ PH domain has micromolar affinity and little specificity for negatively charged phospholipids (Tall et al., 1997; Wang et al., 1999b). In addition, in all reported structures the PLCβ PH domain forms an extended interface with the EF hands and the X domain of the TIM barrel, burying an approximately 3000 Å2 of accessible surface area (Jezyk et al., 2006; Hicks et al., 2008; Waldo et al., 2010; Lyon et al., 2011, 2013). Therefore, lipid interactions with the PH domain could, in principle, directly influence the orientation of the entire catalytic core at the membrane.
However, the primary membrane binding element within PLCβ enzymes seems to be the distal CTD. Truncation of the C-terminal extension, internal deletions, and mutations within the distal CTD are sufficient to abrogate association with the particulate fraction of cells, membranes, and liposomes (Lee et al., 1993b; Schnabel et al., 1993; Wu et al., 1993a; Kim et al., 1996, 1998; Jenco et al., 1997), and the overexpressed C-terminal extensions of PLCβ associate with membranes in cells (Adjobo-Hermans et al., 2013). Initial studies led to the identification of the “P box,” a 127-residue region (human PLCβ3 residues 947–1057) essential for membrane association (Wu et al., 1993a). Additional studies of the C-terminal extension identified three highly conserved basic clusters whose mutation significantly decreased particulate association and lowered basal activity. However, only deletion of the entire C-terminal extension completely eliminated particulate association (Lee et al., 1993b; Wu et al., 1993a; Kim et al., 1996; Ilkaeva et al., 2002). Crystallographic studies confirmed that these basic residue clusters fall on the same face of the distal CTD formed by the Dα3 and Dα4 helices, generating a long and highly polarized electrostatic surface that likely forms the primary membrane interaction site (Fig. 4) (Singer et al., 2002; Lyon et al., 2013). Sequence variation among the PLCβ isoforms in the distal CTD may result in different degrees of membrane association, or lead to distinct modes of autoinhibition via protein–protein interactions in cis. For example, in both the crystal structure and single particle cryoelectron microscopy three-dimensional reconstruction of full-length PLCβ3, the distal CTD interacts with the hydrophobic ridge of the catalytic core, sequestering the basic surface of the distal CTD and preventing the hydrophobic ridge from accessing the membrane (Lyon et al., 2013). These observations may help partially explain the cytosolic localization of PLCβ3 and its lower basal activity compared with that of the other isoforms (Smrcka and Sternweis, 1993; Philip et al., 2010; Adjobo-Hermans et al., 2013). The PLCβ2 C-terminal extension has also been shown to influence the equilibrium between membrane-bound and cytosolic populations of this enzyme (Illenberger et al., 2003b).
Autoinhibition by the X–Y Linker.
In PLCβ, the TIM barrel-like domain is split into X and Y halves connected by a poorly conserved linker, which contains highly acidic stretches in mammalian enzymes. An autoinhibitory role for this X–Y linker was identified in reconstitution studies of PLCβ2, wherein fragments containing the PH, EF hands, and X domain were combined with fragments containing the Y and C2 domains, and exhibited an approximately 10-fold increase in basal activity relative to the intact protein (Zhang and Neer, 2001). Treatment of the PLCβ2 catalytic core with trypsin or the V8 protease, both of which cleave the linker, also increased basal activity compared with the intact PLCβ2 catalytic core (Schnabel and Camps, 1998).
In the six reported structures of PLCβ enzymes (Table 1), the X–Y linker varies in length and degree of order, from 28 observed residues (of 38) in cuttlefish PLC21, to 13 (of 116) in the structure of PLCβ3 in complex with Gαq. However, in each structure the C-terminal 12 amino acids of the linker adopt a similar structure and, based on ligand-bound structures of PLCδ (Essen et al., 1996, 1997), would block access of the phosphoinositide head group to the active site, thereby providing a molecular basis for autoinhibition by the linker (Fig. 3). Selective deletions in the PLCβ2 X–Y linker or single amino acid point mutations to disrupt its interaction with the TIM barrel increased basal activity up to 20-fold over wild-type PLCβ2 (Hicks et al., 2008). Confusingly, the consistently ordered C-terminal portion of the linker in PLCβ enzymes is not conserved in other PLC families, and was disordered in the PLCδ structures, which allowed cocrystallization with various ligands (Essen et al., 1996, 1997). Nonetheless, deletion of this linker in PLCδ increased basal activity 10-fold (Hicks et al., 2008). The disordered regions of the X–Y linker contain highly acidic stretches in many PLC enzymes, and this may hinder basal interactions between the catalytic core and the negatively charged inner leaflet of the plasma membrane (Hicks et al., 2008; Waldo et al., 2010). PLC21, an invertebrate homolog of PLCβ, does not contain an acidic stretch, and instead features a well ordered helix, which is stabilized by an internal series of i, i+4 salt bridges (Lyon et al., 2011). Therefore, the mechanisms underlying autoinhibition by the X–Y linker may be different for each PLC enzyme, and may include electrostatic repulsion with the membrane, well ordered structural elements that occlude the active site, or both, as seems to be the case for mammalian PLCβ enzymes.
Autoinhibition by the Proximal CTD.
In crystal structures, the C-terminal ∼25 amino acids of the PLC21 proximal CTD form a well-ordered helical hairpin. The Hα2′ helix of the hairpin binds to a cleft on the catalytic core formed at the interface of the TIM barrel and C2 domains. The cleft contains residues that are uniquely conserved in the PLCβ subfamily, and places the helical hairpin in close proximity to the active site and the X–Y linker, suggesting a role for this region in regulating PLCβ activity (Lyon et al., 2011). This interaction is recapitulated in two unique structures of human PLCβ3, albeit via in trans crystal contacts (Waldo et al., 2010; Lyon et al., 2013), suggesting that the interaction is evolutionarily conserved (Koyanagi et al., 1998). The role of the Hα2′-catalytic core interaction was assessed in human PLCβ3, where point mutations in Hα2′ or its binding site on the catalytic core decreased the thermal stability of the enzyme and increased basal activity up to 50-fold over wild-type PLCβ3 (Lyon et al., 2011). Although other mechanisms are possible, one possible model based on these observations is that in the inactive state, Hα2′ binds to and stabilizes the PLCβ catalytic core in a catalytically quiescent state that could hinder displacement of the ordered portion of the X–Y linker. Differences in the affinity of the Hα2′ interaction may contribute to differences in basal activity among PLCβ isoforms.
Mechanisms of Activation
Multiple mechanisms of autoinhibition can beget multiple modes of activation. On the basis of biochemical and structural data, it is clear that Gαq has a distinct binding site and activation mechanism from Gβγ and the Rho GTPases. Indeed, for some isoforms, regulation by these molecules has been shown to be synergistic (Roach et al., 2008; Philip et al., 2010; Rebres et al., 2011). Below we discuss the current state of knowledge regarding the molecular basis of PLCβ activation by four key regulators: the phospholipid bilayer, Gαq, the Gβγ heterodimer, and Rho GTPases.
Interfacial Activation.
All known activating proteins for PLCβ are lipid modified, and these groups are required for maximum efficacy of PLCβ activation (Dietrich et al., 1994, 1996; Hepler et al., 1996; Illenberger et al., 1998; Lyon et al., 2013). Although this might imply that membrane recruitment serves as the dominant activation mechanism, it has been shown these activators do not dramatically alter the membrane or particulate association of full-length PLCβ in vitro (Romoser et al., 1996; Runnels et al., 1996; Jenco et al., 1997; Scarlata, 2002). Cell-based assays have shown colocalization between PLCβ isoforms and activators, but it is not clear whether these interactions also lead to increased membrane affinity (Illenberger et al., 2003b; Adjobo-Hermans et al., 2013). Because deletions within the X–Y linker increase basal activity (Schnabel and Camps, 1998; Zhang and Neer, 2001; Hicks et al., 2008), it has been proposed that activating proteins serve to orient the active site of the enzyme near the surface of the negatively charged membrane. This would electrostatically repel the acidic regions of the X–Y linker, which in turn would destabilize the ordered region of the PLCβ linker and allow free access of substrate into the active site (Hicks et al., 2008; Waldo et al., 2010). However, such interfacial activation is clearly not the entire story, as Gαq, Gβγ, and Rac still significantly activate PLCβ proteins when the ordered or acidic portions of the X–Y linker are deleted (Hicks et al., 2008). This additional increase in activity may reflect either the contribution of optimizing the orientation of the catalytic core (e.g., facilitating insertion of the hydrophobic ridge) or other allosteric effects, as discussed below.
Regulation by Activated Gαq.
Gαq activates each PLCβ enzyme to a different extent. PLCβ3 is the most sensitive, with reported approximately 20- to 80-fold increases over basal activity upon interactions with Gαq. PLCβ1 is activated to a similar extent, whereas PLCβ2 and PLCβ4 are typically activated approximately 2- to 10-fold over basal, depending on the experimental method (Smrcka and Sternweis, 1993; Jiang et al., 1994; Lee et al., 1994; Paterson et al., 1995; Biddlecome et al., 1996; Philip et al., 2010). The Gαq interaction is also of high affinity, with EC50 values of 1–400 nM depending on the experimental approach (Smrcka et al., 1991; Runnels and Scarlata, 1999; Waldo et al., 2010; Lyon et al., 2011, 2013). PLCβ enzymes are also able to rapidly terminate their own activation by Gαq by serving as a GTPase activating protein (GAP). PLCβ3 and PLCβ1 increase the rate of GTP hydrolysis by Gαq approximately 100- to 1000-fold, respectively (Berstein et al., 1992; Chidiac and Ross, 1999; Waldo et al., 2010), providing an additional level of temporal control in downstream signaling events (Berstein et al., 1992; Chidiac and Ross, 1999; Cook et al., 2000; Waldo et al., 2010).
Gαq binding, activation, and GAP activity were long attributed to various regions within the C-terminal extension, as its presence increases basal and Gq-saturated PLCβ3 activity by approximately 3- and 40-fold, respectively (Lee et al., 1993b; Park et al., 1993; Wu et al., 1993a; Kim et al., 1996; Paulssen et al., 1996; Jenco et al., 1997; Ilkaeva et al., 2002; Lyon et al., 2011, 2013). However, it was not entirely clear whether these results were due to defects in Gαq binding, or structural changes within the C-terminal extension that altered its ability to interact with membranes, which would also lower activity by decreasing membrane association. Gαq does not seem to alter the subcellular distribution of PLCβ or increase its affinity for membranes (Runnels et al., 1996; Jenco et al., 1997; Scarlata, 2002; Gutman et al., 2010), supporting a mechanism of Gαq activation that is independent of increased membrane association, despite being palmitoylated at its amino terminus (Hepler et al., 1996).
The structure of a C-terminal truncation of human PLCβ3 (PLCβ3-Δ887) in complex with activated Gαq provided the first glimpse into the molecular basis for recognition of activated Gαq and for GAP activity (Fig. 5A). The interface between Gαq and PLCβ3-Δ887 buries approximately 3100 Å2 of accessible surface area and involves multiple domains of PLCβ. The most important interaction is formed by a helix-turn-helix (Hα1/Hα2) in the first 25 residues of the proximal CTD (Waldo et al., 2010). This region is disordered in the absence of Gαq and precedes the autoinhibitory Hα2′ helix (Lyon et al., 2011). Hα1/Hα2 binds to the canonical effector binding site on Gαq, burying approximately 1650 Å2 of accessible surface area, in a manner highly analogous to the interaction made by a helix-turn-helix in p63RhoGEF (Lutz et al., 2007). Both utilize an ALXXPI binding motif (residues 858–863 in human PLCβ3). Single amino acid substitutions (e.g., L859A; Fig. 5A) are sufficient to abolish Gαq binding and activation (Waldo et al., 2010; Adjobo-Hermans et al., 2013). Furthermore, fusing the PLCβ3 Hα1/Hα2 element to the C terminus of PLCδ conferred some Gαq-dependent activation on this otherwise insensitive enzyme (Waldo et al., 2010). The C2 domain and the loop connecting it to the TIM barrel also contribute to the Gαq–PLCβ3 interface through interactions with the switch 1 and 2 regions of Gαq, burying approximately 1100 Å2 of accessible surface area. Mutations within this interface decreased Gαq-dependent activation, but did not eliminate it, further confirming Hα1/Hα2 as the primary Gαq binding site (Waldo et al., 2010). The isolated C2 domains from PLCβ1 and PLCβ2 were previously reported to bind Gαq (Wang et al., 1999a), but the interface between the C2 domain alone and Gαq only buries approximately 400 Å2. Thus, it is unclear whether this interaction would persist in the absence of the other PLCβ binding surfaces. A third set of interactions between Gαq and the PLCβ3 core is mediated by residues 260–264 in the loop between the third and fourth EF hands, which buries approximately 900 Å2 of accessible surface area (Fig. 5A). This loop is highly conserved and unique to PLCβ isozymes. Asn260 forms a hydrogen bond with the side chain of Gαq-Gln209, whose side chain in turn coordinates the hydrolytic water during GTP hydrolysis, an interaction essentially identical to that observed in Gαi/q subunits in complex with regulator of G protein signaling proteins (Tesmer et al., 1997; Slep et al., 2001; Nance et al., 2013), indicating a conserved GAP mechanism. Mutation of Asn260 eliminated GAP activity, as did exchange of the EF3–EF4 loop with that of PLCδ (Cook et al., 2000; Waldo et al., 2010).
The structure of full-length human PLCβ3 in complex with activated Gαq provided additional insights regarding the distal CTD, previously reported to be required for maximum activity and high affinity binding to Gαq (Lee et al., 1993b; Park et al., 1993; Schnabel et al., 1993; Wu et al., 1993a; Kim et al., 1996; Lyon et al., 2011). In this structure, a conserved hydrophobic patch of the distal CTD interacts the N-terminal helix of Gαq, burying approximately 850 Å2 of accessible surface area (Figs. 2 and 4). Cryoelectron microscopy three-dimensional reconstructions of the Gαq–PLCβ3 complex confirmed that this interaction also occurs in solution. Mutation of residues in the hydrophobic patch or deletion of the N-terminal helix of Gαq decreased the efficacy of Gαq activation approximately 2-fold, but had no effect on basal activity or affinity for Gαq. Loss or mutation of the palmitoyl groups of Gαq (Hepler et al., 1996) also decreased maximum Gαq-stimulated activity, but only in the context of full-length PLCβ3. Thus, the N terminus of Gαq appears to play a role in activation, likely by virtue of its coordinate interaction with the distal CTD and with the membrane via its palmitoyl groups (Lyon et al., 2013). However, the relative importance of this interaction in a physiological context remains to be determined.
Unexpectedly, the CTD linker, which is disordered in the Gαq–PLCβ3 complex, also seems important for Gαq activation. Deletion of the linker in PLCβ3 eliminated Gαq-dependent activation at all concentrations tested and modestly increased basal activity, but did not alter the binding affinity of Gαq. Thus, the length and conformational flexibility of the linker may be essential for activation by Gαq (Lyon et al., 2013). Whether the relative length of the CTD linker is a determinant of isoform sensitivity to Gαq-dependent activation is unknown. However, of the human PLCβs, PLCβ2 has the shortest linker (28 residues) and is most weakly activated by Gαq, whereas PLCβ1 and PLCβ3 have longer linker regions (61 and 56 residues, respectively) and are robustly activated by Gαq (Smrcka and Sternweis, 1993; Biddlecome et al., 1996; Philip et al., 2010).
In light of the two Gαq-PLCβ3 crystal structures and associated biochemical data, we propose the following molecular mechanism for PLCβ3 activation by Gαq (Fig. 5). In the resting cell, the Hα2′ helix of the proximal CTD is bound to the catalytic core, inhibiting basal activity, and the preceding Hα1/Hα2 element is disordered and freely accessible to Gαq. The X–Y linker and the interactions between the distal CTD and the ridge of the catalytic core also likely repress basal activity. Upon Gq-coupled receptor activation, Gαq binds to Hα1/Hα2 and displaces the Hα2′ element from the catalytic core by approximately 50 Å, leading to allosteric activation of PLCβ. The interactions between the membrane, the palmitoylated N terminus of Gαq, and the distal CTD help bring the catalytic core into close proximity with the membrane. The conformational flexibility provided by the CTD linker is required for this optimization. The repulsion between the negatively charged residues in the X–Y linker and the membrane facilitates ejection of the ordered portion of the linker through interfacial activation, facilitating membrane insertion of the hydrophobic ridge and substrate binding (Hicks et al., 2008; Waldo et al., 2010; Lyon et al., 2011, 2013).
There are several outstanding questions regarding Gαq activation of PLCβ that remain to be addressed. The first is that we can only conjecture what a PLCβ enzyme looks like in a fully activated state. Neither of the two Gαq-PLCβ3 crystal structures likely represent the “fully activated” conformation of PLCβ3, as they both preserve the Hα2′-catalytic core interaction via in trans crystal contacts. However, crystal structures of PLCβ2, which are truncated immediately after the C2 domain, do not exhibit large conformational differences compared with the PLCβ3 structures, implying that any allosteric change that occurs upon displacement of Hα2′ is subtle. Second, the mechanism by which the Hα2′-catalytic core interaction regulates activity remains unknown. Finally, the relative importance of allosteric versus interfacial activation is not understood. In fact, they could be intimately linked: displacement of the X–Y linker upon interaction with the plasma membrane may promote displacement of the Hα2′ helix, or vice versa.
Regulation by the Gβγ Heterodimer.
As in Gαq activation, each PLCβ isoform is differentially activated upon binding to Gβγ. PLCβ3 and PLCβ1 show the greatest increase in activity (approximately 10-fold over basal), whereas PLCβ2 is activated approximately 5- to 20-fold over basal and PLCβ4 is unresponsive (Lee et al., 1994). However, PLCβ2 is most sensitive to Gβγ, with an EC50 of approximately 30 nM, compared with the approximately 90–200 nM EC50 values reported for PLCβ1 and PLCβ3 (Camps et al., 1992; Katz et al., 1992; Smrcka and Sternweis, 1993; Lee et al., 1994; Hicks et al., 2008). The source of Gβγ in cells is thought to be generated by Gi-coupled receptors, such as the δ- and μ-opioid receptors, as activation by Gβγ can be inhibited by treatment with pertussis toxin (Camps et al., 1992; Katz et al., 1992; Wu et al., 1998; Xie et al., 1999) and because Gi-coupled receptors are more abundant than Gq-coupled receptors in cells in which Gβγ-dependent activation occurs (Kadamur and Ross, 2013).
Gβγ activation of PLCβ does not require the proximal and distal CTDs (Lee et al., 1993b; Kim et al., 1996; Waldo et al., 2010), and Gβγ only activates PLCβ when the Gγ subunit is prenylated (Katz et al., 1992; Dietrich et al., 1994, 1996). These observations suggest that Gβγ simply recruits PLCβ enzymes to the membrane. However, as reported for Gαq, there is no evidence that Gβγ changes the affinity for membranes or liposomes or the cellular distribution of PLCβ (Schnabel et al., 1993; Romoser et al., 1996; Runnels et al., 1996; Jenco et al., 1997; Wang et al., 1999b; Scarlata, 2002). If this is so, then Gβγ must instead impart an allosteric change or help orient the PLCβ catalytic core in a manner that optimizes its function.
Although there are currently no reported structures of a Gβγ-PLCβ complex that could help shed light on the molecular basis for their interaction and for activation, many studies have sought to map their protein–protein interface. GDP-bound Gαi subunits can inhibit PLCβ activation, suggesting a common protein interaction surface on Gβγ, which was confirmed by mutagenesis studies (Ford et al., 1998; Li et al., 1998; Panchenko et al., 1998; Buck et al., 1999; Scott et al., 2001). The outer strands of Gβ1 blades 2, 6, and 7 (Panchenko et al., 1998) and the N terminus of Gβ1 (Bonacci et al., 2005; Friedman et al., 2009) have also been implicated in PLCβ binding. These regions of Gβγ may contribute to differences in the sensitivity of PLCβ isoforms to activation (Li et al., 1998; Panchenko et al., 1998; Chen et al., 2005; Friedman et al., 2009). It has also been hypothesized that Gγ or its prenyl group may be directly involved (Katz et al., 1992; Dietrich et al., 1994, 1996). Loss of prenylation eliminated interactions between PLCβ2 and PLCβ3 with Gβγ (Fogg et al., 2001), but these defects could simply reflect impaired targeting of Gβγ to the membrane. Interestingly, movement of the prenyl group with respect to the Gβ subunit via deletions at the C terminus of Gγ also reduced PLCβ activation (Akgoz et al., 2002). This could imply that either the C terminus of Gγ is part of the interface, or that shortening this loop effects the orientation of Gβγ at the membrane such that its ability to productively interact with PLCβ or to orient the catalytic core of the enzyme at the membrane is impaired.
The location of the Gβγ binding site on PLCβ is less well defined, although the PH domain has emerged as a strong candidate (Feng et al., 2005; Drin et al., 2006; Han et al., 2011), and PH domains in other proteins, such as G protein–coupled receptor kinase 2, interact with Gβγ (Lodowski et al., 2003). One approach suggesting that the PLCβ PH domain is the site of Gβγ binding takes advantage of the similarity between PLCβ and PLCδ enzymes. A PLCδ chimera, in which its PH domain was replaced with that of PLCβ2, could interact with and be stimulated by Gβγ (Runnels and Scarlata, 1999; Wang et al., 2000; Guo et al., 2003; Drin et al., 2006), whereas the reverse chimera lost responsiveness as determined through activity and fluoresence resonance energy transfer–based assays (Guo et al., 2003). Furthermore, the isolated PH domains of PLCβ2 and PLCβ3 have been shown to directly bind to Gβγ by fluoresence resonance energy transfer methods (Wang et al., 1999b).
Another candidate Gβγ binding site lies within the Y domain of the TIM barrel. A chimera in which the PLCβ2 PH, EF hands, and TIM barrel were fused to the PLCβ1 C2 domain and C-terminal extension retained the ability to be activated by Gβγ. Replacing the PLCβ2 PH and EF hands with those of PLCβ1 had no effect on Gβγ activation (Wu et al., 1993b). Subsequently, 20 amino acid peptides corresponding to PLCβ2 Y domain (residues 564–583 and 575–594) were identified that inhibited Gβγ-dependent activation of PLCβ2 and PLCβ3, impaired association between Gβγ and inactive Gαi, and directly interacted with Gβγ in crosslinking studies (Kuang et al., 1996; Sankaran et al., 1998; Bonacci et al., 2005). These peptides correspond to the Tβ5-Tβ6 loop, Tβ6, Tα5, and Tα5′, with the area of overlap between the peptides centered on the Tα5 helix. Point mutants within Tα5 decreased Gβγ-dependent activation (Bonacci et al., 2005; Rebres et al., 2011). Interestingly, this helix interacts with both the X–Y linker and Hα2′ in the PLCβ structures, raising the possibility that it could contribute to regulation.
The Tα5 helix and the PH domain reside on opposite faces of the catalytic core, and a single Gβγ molecule cannot simultaneously interact with both sites (Fig. 6A). Thus, clarification of the Gβγ binding site on PLCβ awaits further structural and biochemical characterization. Different activation mechanisms can be envisioned for each putative binding site. If Gβγ binds to the PH domain in a manner overlapping or adjacent to the Rac1 binding site on PLCβ2, then their activation mechanism are likely very similar: they may simply interact with the catalytic core of PLCβ at the membrane and optimize its orientation (Dietrich et al., 1994, 1996; Drin et al., 2006; Han et al., 2011), which could promote interfacial activation by ejection of the X–Y linker. If Gβγ binds to the Tα5 helix of the TIM barrel-like domain, the same orientation effects could occur, but there may also be a significant allosteric component to activation because this helix contacts two autoinhibitory elements, the X–Y linker and Hα2′ helix (Fig. 6B), and thus could contribute to their displacement.
Fig. 6.
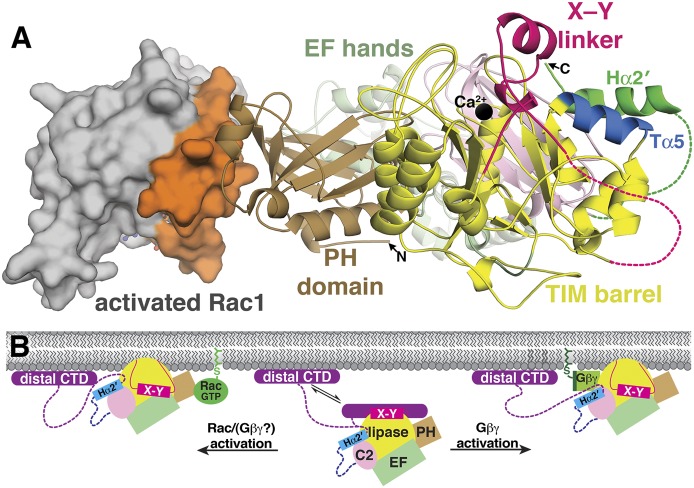
Gβγ and Rac1 bind the PLCβ catalytic core. (A) Rac1 (gray surface) binds exclusively to the PH domain via its switch regions (orange surface), which enables PLCβ to detect the activation state of Rac1. PLCβ domains are colored as in Fig. 1. Current biochemical data predict that Gβγ binds to either the PH domain or a helix within the Y domain of the TIM barrel (Tα5, in blue), which is in close proximity to the X–Y linker and the Hα2′ helix. The active site Ca2+ is shown as a black sphere, disordered regions as dashed lines, and GTPγS bound to Rac1 as ball and sticks. (B) Rac1/Gβγ likely share similar PLCβ activation mechanisms. In the resting state (center), PLCβ is in an autoinhibited state, as described in Fig. 5B. Rac1/Gβγ binding to the PLCβ catalytic core is dictated in part by the geometry imposed by the cell membrane, which likely increases the affinity between these activators and PLCβ. The interaction between Rac1/Gβγ and the PH domain (left) or between Gβγ and the catalytic core (right) likely optimize the orientation of the active site at the membrane surface, overcoming repulsion between the membrane and the acidic region in the X–Y linker, and thereby opening access to the active site.
Synergistic Activation by Gαq and Gβγ.
On the basis of current evidence, the Gαq and Gβγ binding sites within PLCβ are likely spatially separated and involve some independent steps leading to activation. Early evidence for synergistic activation of PLCβ enzymes came from macrophages, where treatment with Gi- and Gq-coupled receptor agonists resulted in superadditive Ca2+ increases over what either agonist could induce alone. This synergistic Ca2+ release required the activity of PLCβ3 (Roach et al., 2008). In the presence of excess activated Gαq and Gβγ, PLCβ3 activity is stimulated approximately 19-fold over what either Gαq or Gβγ can induce, which appears to depend on the very low basal activity of PLCβ3 (Philip et al., 2010). PLCβ2 can also be synergistically activated, but over a much narrower range of Gαq and Gβγ concentrations and to a lesser degree than PLCβ3 (Rebres et al., 2011). At this time, it is unclear how widespread synergistic activation of PLCβ enzymes is and how robust synergistic activation is among cell types.
Regulation by Small G Proteins.
Rho-dependent activation of PLCβ was first identified in cytosolic preparations from granulocytes, where treatment with the nonhydrolyzable GTP analog GTPγS resulted in increased rates of PIP2 hydrolysis (Camps et al., 1990). Subsequent studies identified Cdc42, Rac1, and Rac2, but not RhoA, as the activators. These G proteins were subsequently shown to directly bind and stimulate PLCβ2 and PLCβ3, but not PLCβ1 or PLCβ4 (Illenberger et al., 1997, 2003a). As with Gαq, only the GTP-bound conformation of the small G proteins can productively engage PLCβ (Illenberger et al., 1998; Snyder et al., 2003), and as with Gβγ, the C terminus of the GTPase must be prenylated for activation and does not require the PLCβ C-terminal extension (Illenberger et al., 1997, 1998, 2003a).
The binding site for small G proteins on PLCβ was first identified through chimeras between PLCβ1 and PLCβ2. Replacement of the PLCβ2 PH domain with that of PLCβ1 eliminated GTPase binding and activation (Illenberger et al., 2003a,b). The G protein binding site is localized entirely within the PH domain, as the isolated domains from PLCβ2 and PLCβ3 were able to bind activated Rac1 with affinity comparable to the full-length enzymes (Kd approximately 25 μM) (Snyder et al., 2003). This interaction is relatively weak compared with the affinities measured for Gαq and Gβγ, suggesting that colocalization at the membrane is essential for Rac-dependent activation. In support of this mechanism, Rac1 has been shown to increase the membrane association time of PLCβ2 (Illenberger et al., 2003b; Gutman et al., 2010).
The crystal structure of the Rac1–PLCβ2 catalytic core complex confirmed the PH domain as the sole Rac1 binding site, burying approximately 1200 Å2 of total accessible surface area (Fig. 6A) (Jezyk et al., 2006). Rac1 contacts PLCβ2 via its switch 1 and 2 regions, which undergo conformational changes upon GTP binding. Accordingly, point mutations within the switch regions of Cdc42 or Rac1 eliminated its ability to activate PLCβ (Illenberger et al., 1998; Jezyk et al., 2006). Point mutations within the PLCβ2 PH domain decreased Rac1-dependent activation, but had little to no effect on Gβγ-mediated activation (Jezyk et al., 2006). Thus, if the PH domain is the binding site for both Rac1 and Gβγ, they interact with distinct sites, or the residues involved have different degrees of importance for each activator. Indeed, in one instance, Gβγ and Rac2 have been reported to additively increase PLCβ2 activity (Illenberger et al., 2003a). Comparison of the Rac1-PLCβ2 structure with the apo-PLCβ2 structure did not reveal any large conformational changes occurring upon complex formation (Jezyk et al., 2006; Hicks et al., 2008), suggesting that the mechanism of activation does not have an allosteric component. However, as all of the PLCβ crystal structures have been determined in the absence of phospholipid bilayers, it remains possible that such conformational changes are also dependent on a membrane environment.
Overall, Rac1-dependent activation likely shares similarities with Gβγ activation in that both proteins must be prenylated and activate PLCβ via interactions with the catalytic core of the enzyme, and the PLCβ C-terminal extension is not required. Although Rac1 binding does not appear to elicit a conformational change, this is not yet clear if this is also the case for Gβγ. The prenylated C terminus of activated Rac1 restricts the orientation of the protein at the membrane and may promote higher affinity binding to the PLCβ PH domain. As a result, the Rac1–catalytic core complex is brought in close proximity to the membrane, possibly promoting interfacial activation (Fig. 6B) (Illenberger et al., 2003b; Hicks et al., 2008). An interesting question is whether the Hα2′ helix remains associated with the PLCβ catalytic core during Rac-dependent activation. If the X–Y linker and Hα2′ are allosterically coupled, then displacement of one element could influence the other.
Small Molecule Modulators of PLCβ Activity
Selective small molecule probes can aid in elucidating the roles of specific proteins in cells and whole organisms, and, importantly, serve as leads for future therapeutic agents. Development of PIP2-based chemical probes has been difficult, as modification of the inositol group and/or the acyl chains decreases PLC binding, hydrolysis, and catalytic efficiency (Bruzik and Tsai, 1994; Essen et al., 1997; Wu et al., 1997). The lipid analog edelfosine [1-0-octadecyl-2-0-methyl-glycerol-3-phosphocholine (ET-18-OCH3)] was one of the first molecules identified that selectively decreased Ca2+ release and inositol phosphate accumulation in tumor cells (Berkovic, 1998). Its lipid-like structure allows for incorporation into cell membranes, where it can disrupt membrane integrity, protein–membrane interactions, and the catalytic activity of membrane-associated enzymes, such as PLC. As such, it is difficult to directly associate the effects of edelfosine treatment strictly with PLC inhibition (Seewald et al., 1990; Powis et al., 1995; Arthur and Bittman, 1998). Another PLC inhibitor is the aminosteroid 1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione (U73122) (Bleasdale et al., 1989; Wu et al., 1998; Kobrinsky et al., 2000; Ward et al., 2003; Hou et al., 2004; Oh et al., 2004; Tanski et al., 2004; Horowitz et al., 2005; Suire et al., 2012). Accumulating reports of off-target effects (Hughes et al., 2000; Klose et al., 2008; Burgdorf et al., 2010; Macmillan and McCarron, 2010) prompted efforts to identify its mechanism of action. Purified PLC isoforms treated with U73122 and assayed for activity in vitro showed diverse effects, including increased activity for some PLC isoforms. The maleimide group in U73122, which is required for its inhibitory action, reacts with exposed sulfhydryl groups on the protein surface. For PLCβ, several of the modified sulfhydryl groups are on the same face of the catalytic core as the active site, and these hydrophobic adducts are proposed to increase membrane association and activity (Klein et al., 2011).
A renewed effort to develop PIP2-based chemical probes for PLC is underway. The C6 hydroxyl group of the inositol head group was found to be amenable to chemical modifications, with little effect on PLC activity (Wang et al., 2012). A soluble PIP2 analog with a cleavable fluorescent tag (WH-15) has also been synthesized, and is hydrolyzed at a rate comparable to that of PIP2 (Huang et al., 2011). Although high selectivity among PLC isozymes is unlikely to be exhibited by compounds that bind in the active site, such soluble analogs will greatly facilitate high-throughput screening efforts to identify more potent PLC probes (Huang et al., 2013).
In light of the recent structural and functional findings, is there a rational approach to developing PLCβ-specific modulators? Selectivity would arguably best be achieved by targeting known allosteric and/or regulatory sites. An interesting possibility is the Hα2′ binding site on the catalytic core. This cleft contains residues unique to the PLCβ family; thus, small molecules that target this site would likely be PLCβ-specific. However, the effect of chemical probes that would bind at this site is not clear. Such molecules would likely displace the Hα2′ helix. However, if they do not fully reproduce the autoinhibition mediated by Hα2′, they would serve as activators. On the other hand, if they did repress activity, such molecules could likely inhibit PLCβ even in the face of persistent Gαq activation. Intermolecular protein–protein interaction sites within PLCβ are also potential targets. For example, molecules that target the Rac1 binding surface of the PH domain would enable the selective study of PLCβ function downstream of pathways that activate small molecular weight GTPases. A similar strategy was recently proposed as a treatment in PLCβ-mediated cardiac hypertrophy and heart failure, as peptides or small molecules that disrupt membrane association of PLCβ1b in the sarcolemma are of therapeutic interest (Woodcock et al., 2010). Despite the fact that protein–protein interfaces can be very difficult to “drug,” there are proofs of principle that compounds disrupting the interactions between PLCβ and its protein regulators can be identified (Bonacci et al., 2006). The small molecule M119 has already been used to demonstrate the involvement of Gβγ-activated PLCβ in antinociception induced by opioid receptor activation (Mathews et al., 2008).
Future Directions
Recent structural studies of PLCβ enzymes and their activation complexes have provided atomic-level insight into mechanisms of PLCβ regulation and activation. In particular, tremendous progress has been made in elucidating the molecular mechanisms of Gαq and Rac-dependent activation. Structural insights into how Gβγ interacts with and stimulates PLCβ remain lacking. An unexpected consequence of the most recent structural studies is recognition that the membrane itself is an active player in PLCβ regulation. The membrane serves to increase the local concentration of the enzyme and its activators, and may also alter the structure of the PLCβ core, leading to increased activity. An intriguing possibility is that the distal CTD could also influence the membrane association of the catalytic core by inducing differences in local membrane curvature. An additional layer of regulatory complexity arises from the observation that PLCβ isozymes interact with numerous scaffolding proteins to form signaling complexes (Cai et al., 2005; Cartier et al., 2011; Sun et al., 2013). How these higher order complexes contribute to PLCβ regulation and whether they alter activation by Gαq, Gβγ, and small GTPases are not understood, and represent the next frontier for structure/function analyses of PLCβ enzymes.
Abbreviations
- CTD
C-terminal domain
- DAG
diacylglycerol
- ET-18-OCH3
1-0-octadecyl-2-0-methyl-glycerol-3-phosphocholine
- GAP
GTPase activating protein
- GPCR
G protein–coupled receptor
- IP3
inositol-1,4,5-triphosphate
- PH
pleckstrin homology
- PIP2
phosphatidylinositol-4,5-bisphosphate
- PLC
phospholipase C
- TIM
triose phosphate isomerase
- U73122
1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Lyon, Tesmer.
Footnotes
This work was supported by the American Heart Association [Grant 11POST7620083]; the National Institutes of Health National Heart, Lung, and Blood Institute [Grants R01 HL086865 and R01 HL071818]; and the National Institutes of Health National Institute of General Medical Sciences [Grant GM081655].
References
- Adamski FM, Timms KM, Shieh BH. (1999) A unique isoform of phospholipase Cβ4 highly expressed in the cerebellum and eye. Biochim Biophys Acta 1444:55–60 [DOI] [PubMed] [Google Scholar]
- Adjobo-Hermans MJ, Crosby KC, Putyrski M, Bhageloe A, van Weeren L, Schultz C, Goedhart J, Gadella TW., Jr (2013) PLCβ isoforms differ in their subcellular location and their CT-domain dependent interaction with Gαq. Cell Signal 25:255–263 [DOI] [PubMed] [Google Scholar]
- Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr (2008) Regulation of PLCβ1a membrane anchoring by its substrate phosphatidylinositol (4,5)-bisphosphate. J Cell Sci 121:3770–3777 [DOI] [PubMed] [Google Scholar]
- Akgoz M, Azpiazu I, Kalyanaraman V, Gautam N. (2002) Role of the G protein γ subunit in β γ complex modulation of phospholipase Cβ function. J Biol Chem 277:19573–19578 [DOI] [PubMed] [Google Scholar]
- Arthur G, Bittman R. (1998) The inhibition of cell signaling pathways by antitumor ether lipids. Biochim Biophys Acta 1390:85–102 [DOI] [PubMed] [Google Scholar]
- Arthur JF, Matkovich SJ, Mitchell CJ, Biden TJ, Woodcock EA. (2001) Evidence for selective coupling of alpha 1-adrenergic receptors to phospholipase C-β 1 in rat neonatal cardiomyocytes. J Biol Chem 276:37341–37346 [DOI] [PubMed] [Google Scholar]
- Bahk YY, Lee YH, Lee TG, Seo J, Ryu SH, Suh PG. (1994) Two forms of phospholipase C-β 1 generated by alternative splicing. J Biol Chem 269:8240–8245 [PubMed] [Google Scholar]
- Bahk YY, Song H, Baek SH, Park BY, Kim H, Ryu SH, Suh PG. (1998) Localization of two forms of phospholipase C-β1, a and b, in C6Bu-1 cells. Biochim Biophys Acta 1389:76–80 [DOI] [PubMed] [Google Scholar]
- Berkovic D. (1998) Cytotoxic etherphospholipid analogues. Gen Pharmacol 31:511–517 [DOI] [PubMed] [Google Scholar]
- Berridge MJ. (2003) Cardiac calcium signalling. Biochem Soc Trans 31:930–933 [DOI] [PubMed] [Google Scholar]
- Berstein G, Blank JL, Jhon DY, Exton JH, Rhee SG, Ross EM. (1992) Phospholipase C-β 1 is a GTPase-activating protein for Gq/11, its physiologic regulator. Cell 70:411–418 [DOI] [PubMed] [Google Scholar]
- Bianchi E, Norcini M, Smrcka A, Ghelardini C. (2009) Supraspinal Gbetagamma-dependent stimulation of PLCbeta originating from G inhibitory protein-μ opioid receptor-coupling is necessary for morphine induced acute hyperalgesia. J Neurochem 111:171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlecome GH, Berstein G, Ross EM. (1996) Regulation of phospholipase C-β1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J Biol Chem 271:7999–8007 [DOI] [PubMed] [Google Scholar]
- Bleasdale JE, Bundy GL, Bunting S, Fitzpatrick FA, Huff RM, Sun FF, Pike JE. (1989) Inhibition of phospholipase C dependent processes by U-73, 122. Adv Prostaglandin Thromboxane Leukot Res 19:590–593 [PubMed] [Google Scholar]
- Böhm D, Schwegler H, Kotthaus L, Nayernia K, Rickmann M, Köhler M, Rosenbusch J, Engel W, Flügge G, Burfeind P. (2002) Disruption of PLC-β 1-mediated signal transduction in mutant mice causes age-dependent hippocampal mossy fiber sprouting and neurodegeneration. Mol Cell Neurosci 21:584–601 [DOI] [PubMed] [Google Scholar]
- Bonacci TM, Ghosh M, Malik S, Smrcka AV. (2005) Regulatory interactions between the amino terminus of G-protein βγ subunits and the catalytic domain of phospholipase Cβ2. J Biol Chem 280:10174–10181 [DOI] [PubMed] [Google Scholar]
- Bonacci TM, Mathews JL, Yuan C, Lehmann DM, Malik S, Wu D, Font JL, Bidlack JM, Smrcka AV. (2006) Differential targeting of Gβγ-subunit signaling with small molecules. Science 312:443–446 [DOI] [PubMed] [Google Scholar]
- Braz JC, Gregory K, Pathak A, Zhao W, Sahin B, Klevitsky R, Kimball TF, Lorenz JN, Nairn AC, Liggett SB, et al. (2004) PKC-α regulates cardiac contractility and propensity toward heart failure. Nat Med 10:248–254 [DOI] [PubMed] [Google Scholar]
- Bruzik KS, Tsai MD. (1994) Toward the mechanism of phosphoinositide-specific phospholipases C. Bioorg Med Chem 2:49–72 [DOI] [PubMed] [Google Scholar]
- Buck E, Li J, Chen Y, Weng G, Scarlata S, Iyengar R. (1999) Resolution of a signal transfer region from a general binding domain in gβ for stimulation of phospholipase C-β2. Science 283:1332–1335 [DOI] [PubMed] [Google Scholar]
- Burgdorf C, Schäfer U, Richardt G, Kurz T. (2010) U73122, an aminosteroid phospholipase C inhibitor, is a potent inhibitor of cardiac phospholipase D by a PIP2-dependent mechanism. J Cardiovasc Pharmacol 55:555–559 [DOI] [PubMed] [Google Scholar]
- Cai Y, Stafford LJ, Bryan BA, Mitchell D, Liu M. (2005) G-protein-activated phospholipase C-β, new partners for cell polarity proteins Par3 and Par6. Oncogene 24:4293–4300 [DOI] [PubMed] [Google Scholar]
- Camps M, Carozzi A, Schnabel P, Scheer A, Parker PJ, Gierschik P. (1992) Isozyme-selective stimulation of phospholipase C-β 2 by G protein β γ-subunits. Nature 360:684–686 [DOI] [PubMed] [Google Scholar]
- Camps M, Hou CF, Jakobs KH, Gierschik P. (1990) Guanosine 5′-[gamma-thio]triphosphate-stimulated hydrolysis of phosphatidylinositol 4,5-bisphosphate in HL-60 granulocytes. Evidence that the guanine nucleotide acts by relieving phospholipase C from an inhibitory constraint. Biochem J 271:743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier A, Parent A, Labrecque P, Laroche G, Parent JL. (2011) WDR36 acts as a scaffold protein tethering a G-protein-coupled receptor, Gαq and phospholipase Cβ in a signalling complex. J Cell Sci 124:3292–3304 [DOI] [PubMed] [Google Scholar]
- Chen S, Lin F, Hamm HE. (2005) RACK1 binds to a signal transfer region of G βγ and inhibits phospholipase C β2 activation. J Biol Chem 280:33445–33452 [DOI] [PubMed] [Google Scholar]
- Chidiac P, Ross EM. (1999) Phospholipase C-β1 directly accelerates GTP hydrolysis by Gαq and acceleration is inhibited by Gβ γ subunits. J Biol Chem 274:19639–19643 [DOI] [PubMed] [Google Scholar]
- Cifuentes ME, Honkanen L, Rebecchi MJ. (1993) Proteolytic fragments of phosphoinositide-specific phospholipase C-δ 1. Catalytic and membrane binding properties. J Biol Chem 268:11586–11593 [PubMed] [Google Scholar]
- Cook B, Bar-Yaacov M, Cohen Ben-Ami H, Goldstein RE, Paroush Z, Selinger Z, Minke B. (2000) Phospholipase C and termination of G-protein-mediated signalling in vivo. Nat Cell Biol 2:296–301 [DOI] [PubMed] [Google Scholar]
- Descorbeth M, Anand-Srivastava MB. (2010) Role of growth factor receptor transactivation in high glucose-induced increased levels of Gq/11α and signaling in vascular smooth muscle cells. J Mol Cell Cardiol 49:221–233 [DOI] [PubMed] [Google Scholar]
- Dietrich A, Brazil D, Jensen ON, Meister M, Schrader M, Moomaw JF, Mann M, Illenberger D, Gierschik P. (1996) Isoprenylation of the G protein γ subunit is both necessary and sufficient for β γ dimer-mediated stimulation of phospholipase C. Biochemistry 35:15174–15182 [DOI] [PubMed] [Google Scholar]
- Dietrich A, Meister M, Brazil D, Camps M, Gierschik P. (1994) Stimulation of phospholipase C-β 2 by recombinant guanine-nucleotide-binding protein β γ dimers produced in a baculovirus/insect cell expression system. Requirement of γ-subunit isoprenylation for stimulation of phospholipase C. Eur J Biochem 219:171–178 [DOI] [PubMed] [Google Scholar]
- Drin G, Douguet D, Scarlata S. (2006) The pleckstrin homology domain of phospholipase Cβ transmits enzymatic activation through modulation of the membrane-domain orientation. Biochemistry 45:5712–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MV, Carne A, Katan M. (1993) Structural requirements of phosphatidylinositol-specific phospholipase C δ 1 for enzyme activity. Eur J Biochem 213:339–347 [DOI] [PubMed] [Google Scholar]
- Ellis MV, James SR, Perisic O, Downes CP, Williams RL, Katan M. (1998) Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of plcδ1. J Biol Chem 273:11650–11659 [DOI] [PubMed] [Google Scholar]
- Essen LO, Perisic O, Cheung R, Katan M, Williams RL. (1996) Crystal structure of a mammalian phosphoinositide-specific phospholipase C δ. Nature 380:595–602 [DOI] [PubMed] [Google Scholar]
- Essen LO, Perisic O, Katan M, Wu Y, Roberts MF, Williams RL. (1997) Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry 36:1704–1718 [DOI] [PubMed] [Google Scholar]
- Faenza I, Matteucci A, Manzoli L, Billi AM, Aluigi M, Peruzzi D, Vitale M, Castorina S, Suh PG, Cocco L. (2000) A role for nuclear phospholipase Cβ 1 in cell cycle control. J Biol Chem 275:30520–30524 [DOI] [PubMed] [Google Scholar]
- Feng J, Roberts MF, Drin G, Scarlata S. (2005) Dissection of the steps of phospholipase C β 2 activity that are enhanced by G β γ subunits. Biochemistry 44:2577–2584 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83:1037–1046 [DOI] [PubMed] [Google Scholar]
- Filtz TM, Grubb DR, McLeod-Dryden TJ, Luo J, Woodcock EA. (2009) Gq-initiated cardiomyocyte hypertrophy is mediated by phospholipase Cβ1b. FASEB J 23:3564–3570 [DOI] [PubMed] [Google Scholar]
- Fiume R, Keune WJ, Faenza I, Bultsma Y, Ramazzotti G, Jones DR, Martelli AM, Somner L, Follo MY, Divecha N, et al. (2012) Nuclear phosphoinositides: location, regulation and function. Subcell Biochem 59:335–361 [DOI] [PubMed] [Google Scholar]
- Fogg VC, Azpiazu I, Linder ME, Smrcka A, Scarlata S, Gautam N. (2001) Role of the γ subunit prenyl moiety in G protein β γ complex interaction with phospholipase Cbeta. J Biol Chem 276:41797–41802 [DOI] [PubMed] [Google Scholar]
- Ford CE, Skiba NP, Bae H, Daaka Y, Reuveny E, Shekter LR, Rosal R, Weng G, Yang CS, Iyengar R, et al. (1998) Molecular basis for interactions of G protein βγ subunits with effectors. Science 280:1271–1274 [DOI] [PubMed] [Google Scholar]
- Friedman EJ, Temple BR, Hicks SN, Sondek J, Jones CD, Jones AM. (2009) Prediction of protein-protein interfaces on G-protein β subunits reveals a novel phospholipase C β2 binding domain. J Mol Biol 392:1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Gupta R, Shah S, Morris AJ, Rudge SA, Scarlata S, Petrova V, McLaughlin S, Rebecchi MJ. (1995) The pleckstrin homology domain of phospholipase C-δ 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry 34:16228–16234 [DOI] [PubMed] [Google Scholar]
- Gresset A, Sondek J, Harden TK. (2012) The phospholipase C isozymes and their regulation. Subcell Biochem 58:61–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb DR, Iliades P, Cooley N, Yu YL, Luo J, Filtz TM, Woodcock EA. (2011) Phospholipase Cβ1b associates with a Shank3 complex at the cardiac sarcolemma. FASEB J 25:1040–1047 [DOI] [PubMed] [Google Scholar]
- Grubb DR, Luo J, Yu YL, Woodcock EA. (2012) Scaffolding protein Homer 1c mediates hypertrophic responses downstream of Gq in cardiomyocytes. FASEB J 26:596–603 [DOI] [PubMed] [Google Scholar]
- Grubb DR, Vasilevski O, Huynh H, Woodcock EA. (2008) The extreme C-terminal region of phospholipase Cβ1 determines subcellular localization and function; the “b” splice variant mediates α1-adrenergic receptor responses in cardiomyocytes. FASEB J 22:2768–2774 [DOI] [PubMed] [Google Scholar]
- Guo Y, Philip F, Scarlata S. (2003) The Pleckstrin homology domains of phospholipases C-β and -δ confer activation through a common site. J Biol Chem 278:29995–30004 [DOI] [PubMed] [Google Scholar]
- Gutman O, Walliser C, Piechulek T, Gierschik P, Henis YI. (2010) Differential regulation of phospholipase C-β2 activity and membrane interaction by Galphaq, Gβ1γ2, and Rac2. J Biol Chem 285:3905–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DS, Golebiewska U, Stolzenberg S, Scarlata SF, Weinstein H. (2011) A dynamic model of membrane-bound phospholipase Cβ2 activation by Gβγ subunits. Mol Pharmacol 80:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Mancino V, Simon MI. (2006) Phospholipase Cβ 3 mediates the scratching response activated by the histamine H1 receptor on C-fiber nociceptive neurons. Neuron 52:691–703 [DOI] [PubMed] [Google Scholar]
- Hepler JR, Biddlecome GH, Kleuss C, Camp LA, Hofmann SL, Ross EM, Gilman AG. (1996) Functional importance of the amino terminus of Gq α. J Biol Chem 271:496–504 [DOI] [PubMed] [Google Scholar]
- Hicks SN, Jezyk MR, Gershburg S, Seifert JP, Harden TK, Sondek J. (2008) General and versatile autoinhibition of PLC isozymes. Mol Cell 31:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz LF, Hirdes W, Suh BC, Hilgemann DW, Mackie K, Hille B. (2005) Phospholipase C in living cells: activation, inhibition, Ca2+ requirement, and regulation of M current. J Gen Physiol 126:243–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Kirchner T, Singer M, Matheis M, Argentieri D, Cavender D. (2004) In vivo activity of a phospholipase C inhibitor, 1-(6-((17β-3-methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl)-1H-pyrrole-2,5-dione (U73122), in acute and chronic inflammatory reactions. J Pharmacol Exp Ther 309:697–704 [DOI] [PubMed] [Google Scholar]
- Huang W, Barrett M, Hajicek N, Hicks S, Harden TK, Sondek J, Zhang Q. (2013) Small molecule inhibitors of phospholipase C from a novel high-throughput screen. J Biol Chem 288:5840–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Hicks SN, Sondek J, Zhang Q. (2011) A fluorogenic, small molecule reporter for mammalian phospholipase C isozymes. ACS Chem Biol 6:223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SA, Gibson WJ, Young JM. (2000) The interaction of U-73122 with the histamine H1 receptor: implications for the use of U-73122 in defining H1 receptor-coupled signalling pathways. Naunyn Schmiedebergs Arch Pharmacol 362:555–558 [DOI] [PubMed] [Google Scholar]
- Ilkaeva O, Kinch LN, Paulssen RH, Ross EM. (2002) Mutations in the carboxyl-terminal domain of phospholipase C-β 1 delineate the dimer interface and a potential Gαq interaction site. J Biol Chem 277:4294–4300 [DOI] [PubMed] [Google Scholar]
- Illenberger D, Schwald F, Gierschik P. (1997) Characterization and purification from bovine neutrophils of a soluble guanine-nucleotide-binding protein that mediates isozyme-specific stimulation of phospholipase C β2. Eur J Biochem 246:71–77 [DOI] [PubMed] [Google Scholar]
- Illenberger D, Schwald F, Pimmer D, Binder W, Maier G, Dietrich A, Gierschik P. (1998) Stimulation of phospholipase C-β2 by the Rho GTPases Cdc42Hs and Rac1. EMBO J 17:6241–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illenberger D, Walliser C, Nurnberg B, Diaz Lorente M, Gierschik P. (2003a) Specificity and structural requirements of phospholipase C-β stimulation by Rho GTPases versus G protein β γ dimers. J Biol Chem 278:3006–3014 [DOI] [PubMed] [Google Scholar]
- Illenberger D, Walliser C, Strobel J, Gutman O, Niv H, Gaidzik V, Kloog Y, Gierschik P, Henis YI. (2003b) Rac2 regulation of phospholipase C-β 2 activity and mode of membrane interactions in intact cells. J Biol Chem 278:8645–8652 [DOI] [PubMed] [Google Scholar]
- James SR, Paterson A, Harden TK, Demel RA, Downes CP. (1997) Dependence of the activity of phospholipase C β on surface pressure and surface composition in phospholipid monolayers and its implications for their regulation. Biochemistry 36:848–855 [DOI] [PubMed] [Google Scholar]
- Jenco JM, Becker KP, Morris AJ. (1997) Membrane-binding properties of phospholipase C-β1 and phospholipaseC-β2: role of the C-terminus and effects of polyphosphoinositides, G-proteins and Ca2+. Biochem J 327:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezyk MR, Snyder JT, Gershberg S, Worthylake DK, Harden TK, Sondek J. (2006) Crystal structure of Rac1 bound to its effector phospholipase C-β2. Nat Struct Mol Biol 13:1135–1140 [DOI] [PubMed] [Google Scholar]
- Jhon DY, Lee HH, Park D, Lee CW, Lee KH, Yoo OJ, Rhee SG. (1993) Cloning, sequencing, purification, and Gq-dependent activation of phospholipase C-β 3. J Biol Chem 268:6654–6661 [PubMed] [Google Scholar]
- Jiang H, Kuang Y, Wu Y, Xie W, Simon MI, Wu D. (1997) Roles of phospholipase C β2 in chemoattractant-elicited responses. Proc Natl Acad Sci USA 94:7971–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lyubarsky A, Dodd R, Vardi N, Pugh E, Baylor D, Simon MI, Wu D. (1996) Phospholipase C β 4 is involved in modulating the visual response in mice. Proc Natl Acad Sci USA 93:14598–14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wu D, Simon MI. (1994) Activation of phospholipase C β 4 by heterotrimeric GTP-binding proteins. J Biol Chem 269:7593–7596 [PubMed] [Google Scholar]
- Kadamur G and Ross EM (2013) Mammalian phospholipase C. Annu Rev Physiol 75:127–154. [DOI] [PubMed]
- Katz A, Wu D, Simon MI. (1992) Subunits β γ of heterotrimeric G protein activate β 2 isoform of phospholipase C. Nature 360:686–689 [DOI] [PubMed] [Google Scholar]
- Kim CG, Park D, Rhee SG. (1996) The role of carboxyl-terminal basic amino acids in Gqα-dependent activation, particulate association, and nuclear localization of phospholipase C-β1. J Biol Chem 271:21187–21192 [DOI] [PubMed] [Google Scholar]
- Kim D, Jun KS, Lee SB, Kang NG, Min DS, Kim YH, Ryu SH, Suh PG, Shin HS. (1997) Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature 389:290–293 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Min DS, Ryu SH, Suh PG. (1998) A cytosolic, gαq- and βγ-insensitive splice variant of phospholipase C-β4. J Biol Chem 273:3618–3624 [DOI] [PubMed] [Google Scholar]
- Klein RR, Bourdon DM, Costales CL, Wagner CD, White WL, Williams JD, Hicks SN, Sondek J, Thakker DR. (2011) Direct activation of human phospholipase C by its well known inhibitor u73122. J Biol Chem 286:12407–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose A, Huth T, Alzheimer C. (2008) 1-[6-[[(17β)-3-methoxyestra-1,3,5(10)-trien-17-yl]amino]hexyl]-1H-pyrrole-2,5-dione (U73122) selectively inhibits Kir3 and BK channels in a phospholipase C-independent fashion. Mol Pharmacol 74:1203–1214 [DOI] [PubMed] [Google Scholar]
- Kobrinsky E, Mirshahi T, Zhang H, Jin T, Logothetis DE. (2000) Receptor-mediated hydrolysis of plasma membrane messenger PIP2 leads to K+-current desensitization. Nat Cell Biol 2:507–514 [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Ono K, Suga H, Iwabe N, Miyata T. (1998) Phospholipase C cDNAs from sponge and hydra: antiquity of genes involved in the inositol phospholipid signaling pathway. FEBS Lett 439:66–70 [DOI] [PubMed] [Google Scholar]
- Kuang Y, Wu Y, Smrcka A, Jiang H, Wu D. (1996) Identification of a phospholipase C β2 region that interacts with Gβ-γ. Proc Natl Acad Sci USA 93:2964–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Lee KH, Lee SB, Park D, Rhee SG. (1994) Regulation of phospholipase C-β 4 by ribonucleotides and the α subunit of Gq. J Biol Chem 269:25335–25338 [PubMed] [Google Scholar]
- Lee CW, Park DJ, Lee KH, Kim CG, Rhee SG. (1993a) Purification, molecular cloning, and sequencing of phospholipase C-β 4. J Biol Chem 268:21318–21327 [PubMed] [Google Scholar]
- Lee SB, Shin SH, Hepler JR, Gilman AG, Rhee SG. (1993b) Activation of phospholipase C-β 2 mutants by G protein α q and β γ subunits. J Biol Chem 268:25952–25957 [PubMed] [Google Scholar]
- Lemmon MA. (2004) Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans 32:707–711 [DOI] [PubMed] [Google Scholar]
- Li Y, Sternweis PM, Charnecki S, Smith TF, Gilman AG, Neer EJ, Kozasa T. (1998) Sites for Galpha binding on the G protein β subunit overlap with sites for regulation of phospholipase Cβ and adenylyl cyclase. J Biol Chem 273:16265–16272 [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. (2000) Roles of PLC-β2 and -β3 and PI3Kγ in chemoattractant-mediated signal transduction. Science 287:1046–1049 [DOI] [PubMed] [Google Scholar]
- Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. (2003) Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gβγ. Science 300:1256–1262 [DOI] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, et al. (2007) Structure of Gαq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318:1923–1927 [DOI] [PubMed] [Google Scholar]
- Lyon AM, Dutta S, Boguth CA, Skiniotis G, and Tesmer JJ (2013) Full-length Gαq-phospholipase C-β3 structure reveals interfaces of the C-terminal coiled-coil domain. Nat Struct Mol Biol 20:355–362. [DOI] [PMC free article] [PubMed]
- Lyon AM, Tesmer VM, Dhamsania VD, Thal DM, Gutierrez J, Chowdhury S, Suddala KC, Northup JK, Tesmer JJ. (2011) An autoinhibitory helix in the C-terminal region of phospholipase C-β mediates Gαq activation. Nat Struct Mol Biol 18:999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan D, McCarron JG. (2010) The phospholipase C inhibitor U-73122 inhibits Ca(2+) release from the intracellular sarcoplasmic reticulum Ca(2+) store by inhibiting Ca(2+) pumps in smooth muscle. Br J Pharmacol 160:1295–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao GF, Kunapuli SP, Koneti Rao A. (2000) Evidence for two alternatively spliced forms of phospholipase C-β2 in haematopoietic cells. Br J Haematol 110:402–408 [DOI] [PubMed] [Google Scholar]
- Mathews JL, Smrcka AV, Bidlack JM. (2008) A novel Gbetagamma-subunit inhibitor selectively modulates μ-opioid-dependent antinociception and attenuates acute morphine-induced antinociceptive tolerance and dependence. J Neurosci 28:12183–12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende U, Kagen A, Cohen A, Aramburu J, Schoen FJ, Neer EJ. (1998) Transient cardiac expression of constitutively active Gαq leads to hypertrophy and dilated cardiomyopathy by calcineurin-dependent and independent pathways. Proc Natl Acad Sci USA 95:13893–13898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende U, Kagen A, Meister M, Neer EJ. (1999) Signal transduction in atria and ventricles of mice with transient cardiac expression of activated G protein α(q). Circ Res 85:1085–1091 [DOI] [PubMed] [Google Scholar]
- Murthy KS, Makhlouf GM. (1996) Opioid μ, δ, and κ receptor-induced activation of phospholipase C-β 3 and inhibition of adenylyl cyclase is mediated by Gi2 and G(o) in smooth muscle. Mol Pharmacol 50:870–877 [PubMed] [Google Scholar]
- Nance MR, Kreutz B, Tesmer VM, Sterne-Marr R, Kozasa T, Tesmer JJ. (2013) Structural and functional analysis of the regulator of g protein signaling 2-gαq complex. Structure 21:438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC. (2010) Protein kinase C: poised to signal. Am J Physiol Endocrinol Metab 298:E395–E402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll SJ, Mitchell MD, Faenza I, Cocco L, Gilmour RS. (2009) Nuclear PLCβ1 is required for 3T3-L1 adipocyte differentiation and regulates expression of the cyclin D3-cdk4 complex. Cell Signal 21:926–935 [DOI] [PubMed] [Google Scholar]
- Oh YS, Jo NW, Choi JW, Kim HS, Seo SW, Kang KO, Hwang JI, Heo K, Kim SH, Kim YH, et al. (2004) NHERF2 specifically interacts with LPA2 receptor and defines the specificity and efficiency of receptor-mediated phospholipase C-β3 activation. Mol Cell Biol 24:5069–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniyandi SS, Sun L, Ferreira JC, Mochly-Rosen D. (2009) Protein kinase C in heart failure: a therapeutic target? Cardiovasc Res 82:229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchenko MP, Saxena K, Li Y, Charnecki S, Sternweis PM, Smith TF, Gilman AG, Kozasa T, Neer EJ. (1998) Sites important for PLCβ2 activation by the G protein βγ subunit map to the sides of the β propeller structure. J Biol Chem 273:28298–28304 [DOI] [PubMed] [Google Scholar]
- Park D, Jhon DY, Lee CW, Ryu SH, Rhee SG. (1993) Removal of the carboxyl-terminal region of phospholipase C-β 1 by calpain abolishes activation by G α q. J Biol Chem 268:3710–3714 [PubMed] [Google Scholar]
- Paterson A, Boyer JL, Watts VJ, Morris AJ, Price EM, Harden TK. (1995) Concentration of enzyme-dependent activation of PLC-β 1 and PLC-β 2 by G α 11 and β γ-subunits. Cell Signal 7:709–720 [DOI] [PubMed] [Google Scholar]
- Paulssen RH, Woodson J, Liu Z, Ross EM. (1996) Carboxyl-terminal fragments of phospholipase C-β1 with intrinsic Gq GTPase-activating protein (GAP) activity. J Biol Chem 271:26622–26629 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499 [DOI] [PubMed] [Google Scholar]
- Philip F, Guo Y, Scarlata S. (2002) Multiple roles of pleckstrin homology domains in phospholipase Cbeta function. FEBS Lett 531:28–32 [DOI] [PubMed] [Google Scholar]
- Philip F, Kadamur G, Silos RG, Woodson J, Ross EM. (2010) Synergistic activation of phospholipase C-β3 by Gα(q) and Gβγ describes a simple two-state coincidence detector. Curr Biol 20:1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis G, Hill SR, Frew TJ, Sherrill KW. (1995) Inhibitors of phospholipid intracellular signaling as antiproliferative agents. Med Res Rev 15:121–138 [DOI] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. (2011) Let’s go bananas: revisiting the endocytic BAR code. EMBO J 30:3501–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebres RA, Roach TI, Fraser ID, Philip F, Moon C, Lin KM, Liu J, Santat L, Cheadle L, Ross EM, et al. (2011) Synergistic Ca2+ responses by Gαi- and Gαq-coupled G-protein-coupled receptors require a single PLCβ isoform that is sensitive to both Gβγ and Gαq. J Biol Chem 286:942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach TI, Rebres RA, Fraser ID, Decamp DL, Lin KM, Sternweis PC, Simon MI, Seaman WE. (2008) Signaling and cross-talk by C5a and UDP in macrophages selectively use PLCβ3 to regulate intracellular free calcium. J Biol Chem 283:17351–17361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romoser V, Ball R, Smrcka AV. (1996) Phospholipase C β2 association with phospholipid interfaces assessed by fluorescence resonance energy transfer. G protein βγ subunit-mediated translocation is not required for enzyme activation. J Biol Chem 271:25071–25078 [DOI] [PubMed] [Google Scholar]
- Runnels LW, Jenco J, Morris A, Scarlata S. (1996) Membrane binding of phospholipases C-β 1 and C-β 2 is independent of phosphatidylinositol 4,5-bisphosphate and the α and βγ subunits of G proteins. Biochemistry 35:16824–16832 [DOI] [PubMed] [Google Scholar]
- Runnels LW, Scarlata SF. (1999) Determination of the affinities between heterotrimeric G protein subunits and their phospholipase C-β effectors. Biochemistry 38:1488–1496 [DOI] [PubMed] [Google Scholar]
- Sankaran B, Osterhout J, Wu D, Smrcka AV. (1998) Identification of a structural element in phospholipase C β2 that interacts with G protein βγ subunits. J Biol Chem 273:7148–7154 [DOI] [PubMed] [Google Scholar]
- Scarlata S. (2002) Regulation of the lateral association of phospholipase Cβ2 and G protein subunits by lipid rafts. Biochemistry 41:7092–7099 [DOI] [PubMed] [Google Scholar]
- Schnabel P, Camps M. (1998) Activation of a phospholipase Cβ2 deletion mutant by limited proteolysis. Biochem J 330:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel P, Camps M, Carozzi A, Parker PJ, Gierschik P. (1993) Mutational analysis of phospholipase C-β 2. Identification of regions required for membrane association and stimulation by guanine-nucleotide-binding protein β γ subunits. Eur J Biochem 217:1109–1115 [DOI] [PubMed] [Google Scholar]
- Scott JK, Huang SF, Gangadhar BP, Samoriski GM, Clapp P, Gross RA, Taussig R, Smrcka AV. (2001) Evidence that a protein-protein interaction ‘hot spot’ on heterotrimeric G protein βγ subunits is used for recognition of a subclass of effectors. EMBO J 20:767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seewald MJ, Olsen RA, Sehgal I, Melder DC, Modest EJ, Powis G. (1990) Inhibition of growth factor-dependent inositol phosphate Ca2+ signaling by antitumor ether lipid analogues. Cancer Res 50:4458–4463 [PubMed] [Google Scholar]
- Shin DM, Dehoff M, Luo X, Kang SH, Tu J, Nayak SK, Ross EM, Worley PF, Muallem S. (2003) Homer 2 tunes G protein-coupled receptors stimulus intensity by regulating RGS proteins and PLCβ GAP activities. J Cell Biol 162:293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AU, Waldo GL, Harden TK, Sondek J. (2002) A unique fold of phospholipase C-β mediates dimerization and interaction with G α q. Nat Struct Biol 9:32–36 [DOI] [PubMed] [Google Scholar]
- Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. (2001) Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A. Nature 409:1071–1077 [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Hepler JR, Brown KO, Sternweis PC. (1991) Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science 251:804–807 [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Sternweis PC. (1993) Regulation of purified subtypes of phosphatidylinositol-specific phospholipase C β by G protein α and β γ subunits. J Biol Chem 268:9667–9674 [PubMed] [Google Scholar]
- Snyder JT, Singer AU, Wing MR, Harden TK, Sondek J. (2003) The pleckstrin homology domain of phospholipase C-β2 as an effector site for Rac. J Biol Chem 278:21099–21104 [DOI] [PubMed] [Google Scholar]
- Suh PG, Park JI, Manzoli L, Cocco L, Peak JC, Katan M, Fukami K, Kataoka T, Yun S, Ryu SH. (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep 41:415–434 [DOI] [PubMed] [Google Scholar]
- Suire S, Lécureuil C, Anderson KE, Damoulakis G, Niewczas I, Davidson K, Guillou H, Pan D, Clark J, Hawkins PT, andStephens L. (2012) GPCR activation of Ras and PI3Kc in neutrophils depends on PLCβ2/β3 and the RasGEF RasGRP4. EMBO J 31:3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Mao G, Kunapuli SP, Dhanasekaran DN, Rao AK. (2007) Alternative splice variants of phospholipase C-β2 are expressed in platelets: effect on Gαq-dependent activation and localization. Platelets 18:217–223 [DOI] [PubMed] [Google Scholar]
- Sun Z, Smrcka AV, Chen S. (2013) WDR26 functions as a scaffolding protein to promote Gβγ-mediated PLCβ2 activation in leukocytes. J Biol Chem. 288:16715–16725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall E, Dormán G, Garcia P, Runnels L, Shah S, Chen J, Profit A, Gu QM, Chaudhary A, Prestwich GD, et al. (1997) Phosphoinositide binding specificity among phospholipase C isozymes as determined by photo-cross-linking to novel substrate and product analogs. Biochemistry 36:7239–7248 [DOI] [PubMed] [Google Scholar]
- Tang W, Zhang Y, Xu W, Harden TK, Sondek J, Sun L, Li L, Wu D. (2011) A PLCβ/PI3Kγ-GSK3 signaling pathway regulates cofilin phosphatase slingshot2 and neutrophil polarization and chemotaxis. Dev Cell 21:1038–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanski WJ, Roztocil E, Hemady EA, Williams JA, Davies MG. (2004) Role of Gαq in smooth muscle cell proliferation. J Vasc Surg 39:639–644 [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. (1997) Structure of RGS4 bound to AlF4—activated G(i α1): stabilization of the transition state for GTP hydrolysis. Cell 89:251–261 [DOI] [PubMed] [Google Scholar]
- Ushio-Fukai M, Griendling KK, Akers M, Lyons PR, Alexander RW. (1998) Temporal dispersion of activation of phospholipase C-β1 and -γ isoforms by angiotensin II in vascular smooth muscle cells. Role of alphaq/11, α12, and β γ G protein subunits. J Biol Chem 273:19772–19777 [DOI] [PubMed] [Google Scholar]
- Vaidyula VR, Rao AK. (2003) Role of Gαq and phospholipase C-β2 in human platelets activation by thrombin receptors PAR1 and PAR4: studies in human platelets deficient in Gαq and phospholipase C-β2. Br J Haematol 121:491–496 [DOI] [PubMed] [Google Scholar]
- Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, Wang X, Montell C, Kozasa T, Sondek J, et al. (2010) Kinetic scaffolding mediated by a phospholipase C-β and Gq signaling complex. Science 330:974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Dowal L, El-Maghrabi MR, Rebecchi M, Scarlata S. (2000) The pleckstrin homology domain of phospholipase C-β(2) links the binding of gβγ to activation of the catalytic core. J Biol Chem 275:7466–7469 [DOI] [PubMed] [Google Scholar]
- Wang T, Pentyala S, Elliott JT, Dowal L, Gupta E, Rebecchi MJ, Scarlata S. (1999a) Selective interaction of the C2 domains of phospholipase C-β1 and -β2 with activated Gαq subunits: an alternative function for C2-signaling modules. Proc Natl Acad Sci USA 96:7843–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pentyala S, Rebecchi MJ, Scarlata S. (1999b) Differential association of the pleckstrin homology domains of phospholipases C-β 1, C-β 2, and C-δ 1 with lipid bilayers and the β γ subunits of heterotrimeric G proteins. Biochemistry 38:1517–1524 [DOI] [PubMed] [Google Scholar]
- Wang X, Barrett M, Sondek J, Harden TK, Zhang Q. (2012) Fluorescent phosphatidylinositol 4,5-bisphosphate derivatives with modified 6-hydroxy group as novel substrates for phospholipase C. Biochemistry 51:5300–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PD, Ouyang H, Thakker DR. (2003) Role of phospholipase C-β in the modulation of epithelial tight junction permeability. J Pharmacol Exp Ther 304:689–698 [DOI] [PubMed] [Google Scholar]
- Woodcock EA, Grubb DR, Filtz TM, Marasco S, Luo J, McLeod-Dryden TJ, Kaye DM, Sadoshima J, Du XJ, Wong C, et al. (2009a) Selective activation of the “b” splice variant of phospholipase Cβ1 in chronically dilated human and mouse atria. J Mol Cell Cardiol 47:676–683 [DOI] [PubMed] [Google Scholar]
- Woodcock EA, Grubb DR, Iliades P. (2010) Potential treatment of cardiac hypertrophy and heart failure by inhibiting the sarcolemmal binding of phospholipase Cβ1b. Curr Drug Targets 11:1032–1040 [DOI] [PubMed] [Google Scholar]
- Woodcock EA, Kistler PM, Ju Y-K. (2009b) Phosphoinositide signalling and cardiac arrhythmias. Cardiovasc Res 82:286–295 [DOI] [PubMed] [Google Scholar]
- Wu D, Jiang H, Katz A, Simon MI. (1993a) Identification of critical regions on phospholipase C-β 1 required for activation by G-proteins. J Biol Chem 268:3704–3709 [PubMed] [Google Scholar]
- Wu D, Katz A, Simon MI. (1993b) Activation of phospholipase C β 2 by the α and β γ subunits of trimeric GTP-binding protein. Proc Natl Acad Sci USA 90:5297–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Perisic O, Williams RL, Katan M, Roberts MF. (1997) Phosphoinositide-specific phospholipase C δ1 activity toward micellar substrates, inositol 1,2-cyclic phosphate, and other water-soluble substrates: a sequential mechanism and allosteric activation. Biochemistry 36:11223–11233 [DOI] [PubMed] [Google Scholar]
- Wu YL, Pei G, Fan GH. (1998) Inhibition of phospholipase C blocks opioid receptor-mediated activation of Gi proteins. Neuroreport 9:99–103 [DOI] [PubMed] [Google Scholar]
- Xiao W, Hong H, Kawakami Y, Kato Y, Wu D, Yasudo H, Kimura A, Kubagawa H, Bertoli LF, Davis RS, et al. (2009) Tumor suppression by phospholipase C-β3 via SHP-1-mediated dephosphorylation of Stat5. Cancer Cell 16:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Samoriski GM, McLaughlin JP, Romoser VA, Smrcka A, Hinkle PM, Bidlack JM, Gross RA, Jiang H, Wu D. (1999) Genetic alteration of phospholipase C β3 expression modulates behavioral and cellular responses to μ opioids. Proc Natl Acad Sci USA 96:10385–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Neer EJ. (2001) Reassembly of phospholipase C-β2 from separated domains: analysis of basal and G protein-stimulated activities. J Biol Chem 276:2503–2508 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Vogel WK, McCullar JS, Greenwood JA, Filtz TM. (2006) Phospholipase C-β3 and -β1 form homodimers, but not heterodimers, through catalytic and carboxyl-terminal domains. Mol Pharmacol 70:860–868 [DOI] [PubMed] [Google Scholar]