Abstract
Hepatocellular carcinoma (HCC) is a threat to public health worldwide. We previously identified the association of a single nucleotide polymorphism (SNP) at the promoter region of the MHC class I polypeptide-related sequence A (MICA) gene with the risk of hepatitis-virus-related HCC. Because this SNP affects MICA expression levels, regulating MICA expression levels may be important in the prevention of HCC. We herein show that the microRNA (miR) 25-93-106b cluster can modulate MICA levels in HCC cells. Overexpression of the miR 25-93-106b cluster significantly suppressed MICA expression. Conversely, silencing of this miR cluster enhanced MICA expression in cells that express substantial amounts of MICA. The changes in MICA expression levels by the miR25-93-106b cluster were biologically significant in an NKG2D-binding assay and an in vivo cell-killing model. These data suggest that the modulation of MICA expression levels by miRNAs may be a useful method to regulate HCCs during hepatitis viral infection.
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related mortality worldwide1. Although multiple major risk factors have been identified, such as genetic factors, environmental toxins, alcohol abuse, obesity, and metabolic disorders2, infection with hepatitis virus B (HBV) or C (HCV) remains the major etiological factor for HCC1.
Disease progression in HBV-induced or HCV-induced HCC is a multistep phenomenon. The clinical outcomes vary among individuals1,3,4 because disease progression is influenced by both environmental and genetic risk factors. In terms of genetic susceptibility factors for HCV-induced HCC, we previously identified a single nucleotide polymorphism (SNP) site in the 5′-flanking region of the MICA gene on 6p21.33 (rs2596452) that is strongly associated with progression from chronic hepatitis C to HCC5. Individuals with the risk allele A of rs2596452 showed lower serum MICA protein levels5. Our subsequent study revealed that the same SNP site was also significantly associated with the risk of HBV-induced HCC6. However, interestingly, the risk allele was G in cases of HBV infection, which differed from HCV infection, and the individuals with the risk allele showed increased MICA protein expression levels6. Despite the different risk alleles at the same SNP site and inverse association between serum MICA levels and HCC risks in these two etiologies, MICA protein expression levels are significantly associated with susceptibility to HCC in chronic hepatitis viral infection.
MICA is highly expressed on viral-infected and cancer cells and acts as a ligand for NKG2D to activate the antitumor effects of natural killer cells and CD8 T cells7,8. This NKG2D-mediated tumor rejection is considered to be effective in the early stages of tumor growth9,10,11. Thus, the expression levels of MICA on the tumor cell surface may determine the antitumor efficacy, and the levels of shedding MICA in serum may act as a decoy of NKG2D to avoid tumor rejection.
Although several stress pathways regulate the transcription of the MICA gene12,13, cellular microRNAs are suggested to control MICA protein expression via post-transcriptional mechanisms14,15. Recently, nucleic-acid-mediated gene therapy has been undergoing clinical trials16. Therefore, to target the clinical application of our GWAS results toward prevention of chronic-hepatitis-infection-induced HCC by nucleic-acid-mediated therapy, we determined the regulatory mechanisms of MICA protein expression using miRNA overexpression and miRNA functional silencing.
Results
HCC cell lines differentially express MICA protein
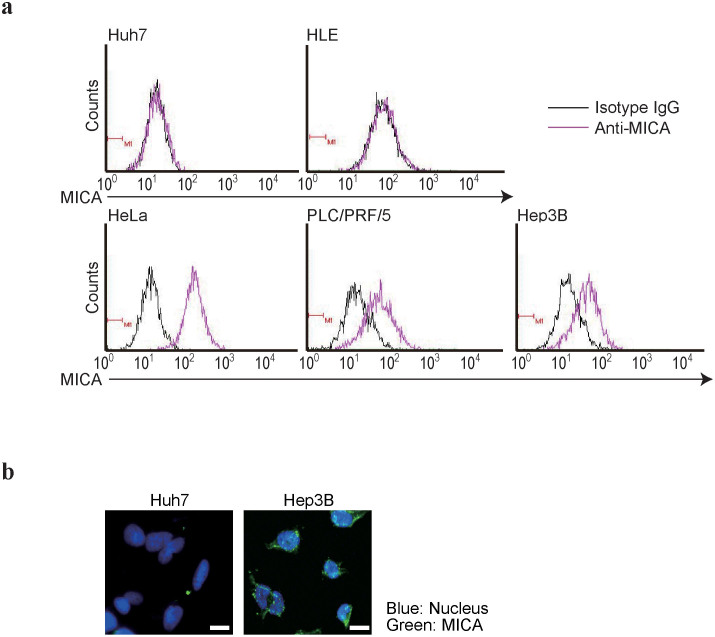
To determine MICA protein levels in HCC cells, four representative HCC cell lines (Huh7, HLE, PLC/PRF/5, and Hep3B cells) underwent flow cytometry to evaluate MICA protein expression because no appropriate antibodies against MICA protein are at present available for western blotting. HeLa cells, which are known to express MICA protein17, were used as a positive control. Hep3B and PLC/PRF/5 cells expressed substantial MICA protein levels, Huh7 and HLE cells expressed no MICA protein (Figure 1a). This was confirmed by immunocytochemistry using Huh7 and Hep3B cells, which showed staining mainly of cell surfaces (Figure 1b). These results suggest that the MICA protein expression status depends on the cell line examined, even those from the same organ.
Figure 1. Expression of MICA protein in HCC cells.
(a), Flow cytometry assessment of MICA protein expression in HCC cells (purple lines). Isotype IgG was used for background staining (black lines). HeLa cells were used as the positive control. Representative results from two independent experiments are shown. (b), Immunofluorescence staining for MICA in Huh7 and Hep3B cells. Representative images from two independent experiments are shown. Scale bar, 25 μm.
The MiR25-93-106b cluster regulates MICA expression
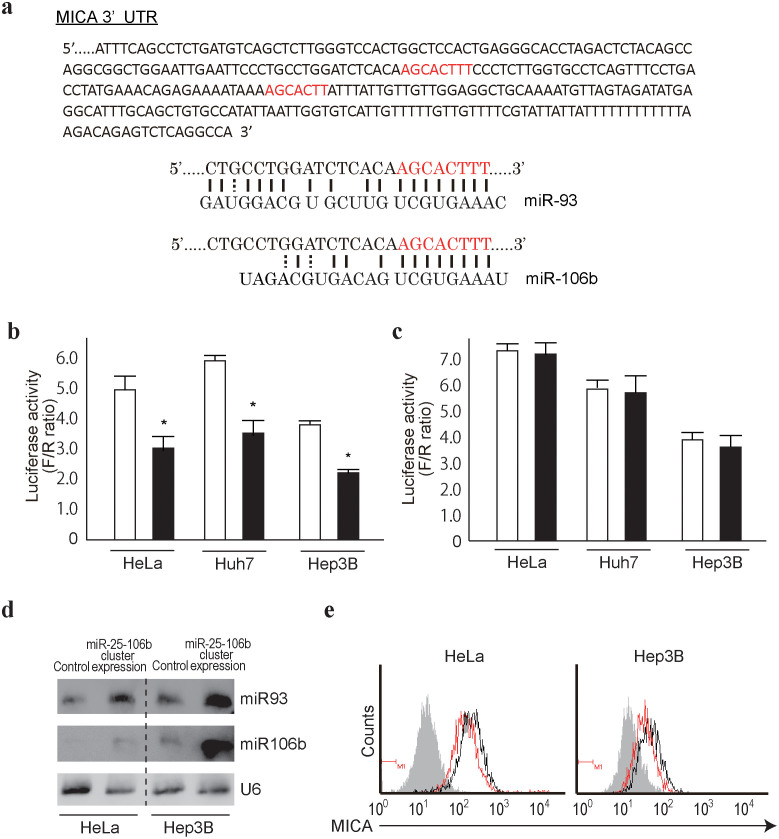
Because upregulation of MICA expression was observed in Dicer-knockdown cells18, we hypothesized that MICA expression levels may be at least partly regulated by miRNAs. We initially tested miRNAs that might affect MICA expression using reporter constructs into which MICA 3′-untranslated region (3′UTR) sequences were cloned and by transiently overexpressing 76 mature synthetic microRNAs, which were selected on the basis of their hepatic expression level, as in our previous studies19,20. Among the microRNAs examined, several may target MICA 3′UTR (Supplementary Figure 1). Among them, we focused on miR93 and miR106b, which were considered to target MICA 3′UTR based partly on the results of our initial miRNA testing described above; in addition, their possible target sequences were identified in the MICA 3′UTR sequences by a computational search using TargetScan 6.0 21. Additional reasons that we focused on these two miRNAs were as follows: 1) these miRNAs share the same seed sequences, to which two perfect-match complementary sequences exist in the 3′UTR of MICA (Figure 2a); 2) the target sequences are highly conserved among mammals and are thus likely to be biologically important sites; and 3) these miRNAs are located as a “miR25-93-106b cluster” on human chromosome 7q22.1, and so they may be expressed together under the same transcriptional control. We introduced mutations in the first possible miRNA target sequences of MICA 3′UTR in the reporter constructs (Supplementary Figure 2a); these sequences have a higher likelihood to be target sites, as determined by TargetScan. Co-transfection experiments revealed that reporter activity was suppressed by overexpression of a miR25-93-106b cluster-expressing plasmid (Figure 2b and Supplementary Figure 2b). The overexpression of an unrelated miR (let-7g)-expressing plasmid did not have any significant effects on the reporter activity (Supplementary Figure 2c) and the suppressive effect was lost using constructs with three point mutations in the seed sequences (Figure 2c), suggesting that miR25-93-106b directly targets these sequences and suppresses gene expression.
Figure 2. miR93 and 106b target MICA 3′UTR.
(a), Sequences of MICA 3′UTR (upper). Letters in red are the sequences completely matched with the seed regions of miR93 and 106b. The complementarities between the first predicted target in the MICA 3′UTR and miRNA sequences are shown below. (b), (c), Cells were co-transfected with pGL4-TK (renilla luciferase as an internal control), Luc-MICA-3′UTRwt (b) or Luc-MICA-3′UTRmut (c), and either an empty control vector (white bar) or miR25-93-106b-cluster expression plasmid (black bar). Data shows the means ± s.d. of the raw ratios (F/R) obtained by dividing firefly luciferase values with renilla luciferase values of three independent experiments. *p < 0.05. (d), miR93 and miR106b expression levels in control and stably miR25-93-106b cluster-expressing cells were determined by northern blotting. U6 levels were used as a loading control. Representative images from two independent experiments are shown. Full-length blot images are available in Supplementary Figure 5. (e), Suppression of MICA expression by overexpression of miRNA93 and 106b. Flow cytometry assessment of MICA protein expression in control (black lines) and stably miR25-93-106b cluster-expressing HeLa and Hep3B cells (red lines). Gray-shaded histograms represent the background staining using isotype IgG. Representative results from two independent experiments are shown.
To confirm these effects, we generated HeLa and Hep3B cell lines that stably expressed the miR25-93-106b-cluster by transducing cells with miR25-93-106b-cluster-expressing lentiviruses (Figure 2d). As expected, the expression of the miR25-93-106b-cluster significantly suppressed MICA protein expression (Figure 2e). However, the expression levels of endogenous miR93 and 106b were not always proportional to the levels of MICA protein expression in the cell lines examined (Supplementary Figure 3). These results suggest that MICA protein expression can be regulated by miR93 and 106b, but that its expression is simultaneously endogenously regulated by other factors (possibly by promoter activities, including epigenetic changes).
Inhibition of miR25-93-106b function increases MICA protein expression
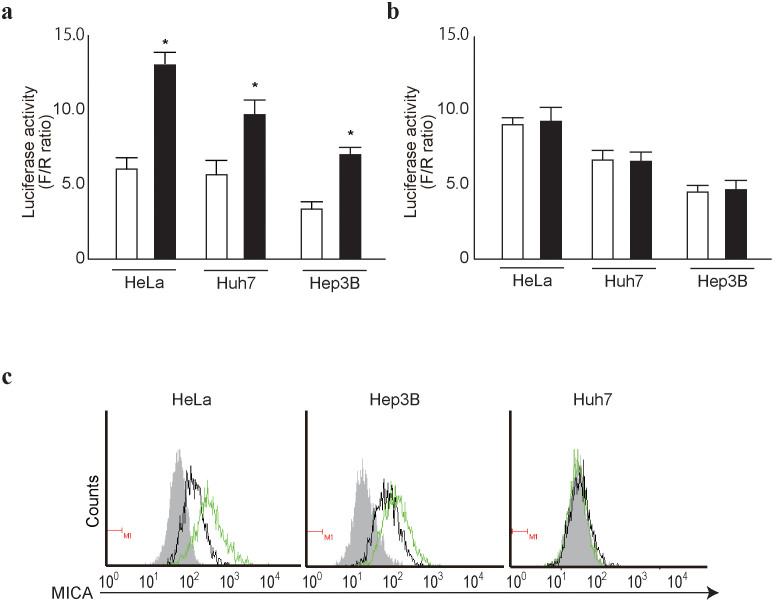
To develop methods of enhancing MICA protein expression levels based on the above results, we examined the effects of functional downregulation of miR25-93-106b on MICA expression. We first performed a reporter assay of transient functional silencing of miR25-93-106b using a construct that produces mature anti-sense RNAs designed to silence miR25-93-106b function. As expected, the reporter activities with MICA 3′UTR sequences were enhanced by the functional silencing of miR25-93-106b in HeLa, Hep3B, and Huh7 cells (Figure 3a). However, such effects were not observed using mutant reporter constructs not targeted by those miRNAs (Figure 3b), suggesting that the enhancing effects of the reporter activities were miRNA-dependent.
Figure 3. Silencing of miR25-93-106b cluster enhances MICA expression.
(a), (b), Cells were co-transfected with pGL4-TK (internal control), Luc-MICA-3′UTRwt (a) or Luc-MICA-3′UTRmut (b), and either an empty control vector (white bar) or plasmid expressing mature anti-sense sequences of miR25-93-106b cluster (black bar). Data shows the means ± s.d. of the raw ratios (F/R) obtained by dividing firefly luciferase values with renilla luciferase values of three independent experiments. *p < 0.05. (c), Enhancement of MICA expression by expression of anti-sense sequences of the miR25-93-106b cluster. Flow cytometry assessment of MICA protein expression in control (black lines) and stably mature anti-sense sequences of miR25-93-106b cluster-expressing cells (green lines). Gray-shaded histograms represent the background staining using isotype IgG. Representative results from three independent experiments are shown.
Next, HeLa, Hep3B, and Huh7 cells were stably transduced with a lentivirus that expresses anti-sense RNAs as described above, and MICA protein expression levels were determined by flow cytometry. Consistent with the reporter assay results, MICA protein expression was increased in HeLa and Hep3B cells by the functional silencing of miR25-93-106b (Figure 3c). However, in Huh7 cells, which express no MICA protein in the normal state, silencing of miR25-93-106b had no effect on MICA protein expression (Figure 3c). These results suggest that MICA protein expression levels can be regulated by modulating miRNA function, albeit only if at least a small quantity of MICA protein is present. In contrast, modulation of miRNA function does not influence MICA protein expression levels when the MICA protein is not expressed, but this could be because there are other forms of regulation at extremely low levels.
MICA protein levels are related to tumor susceptibility to NK cells
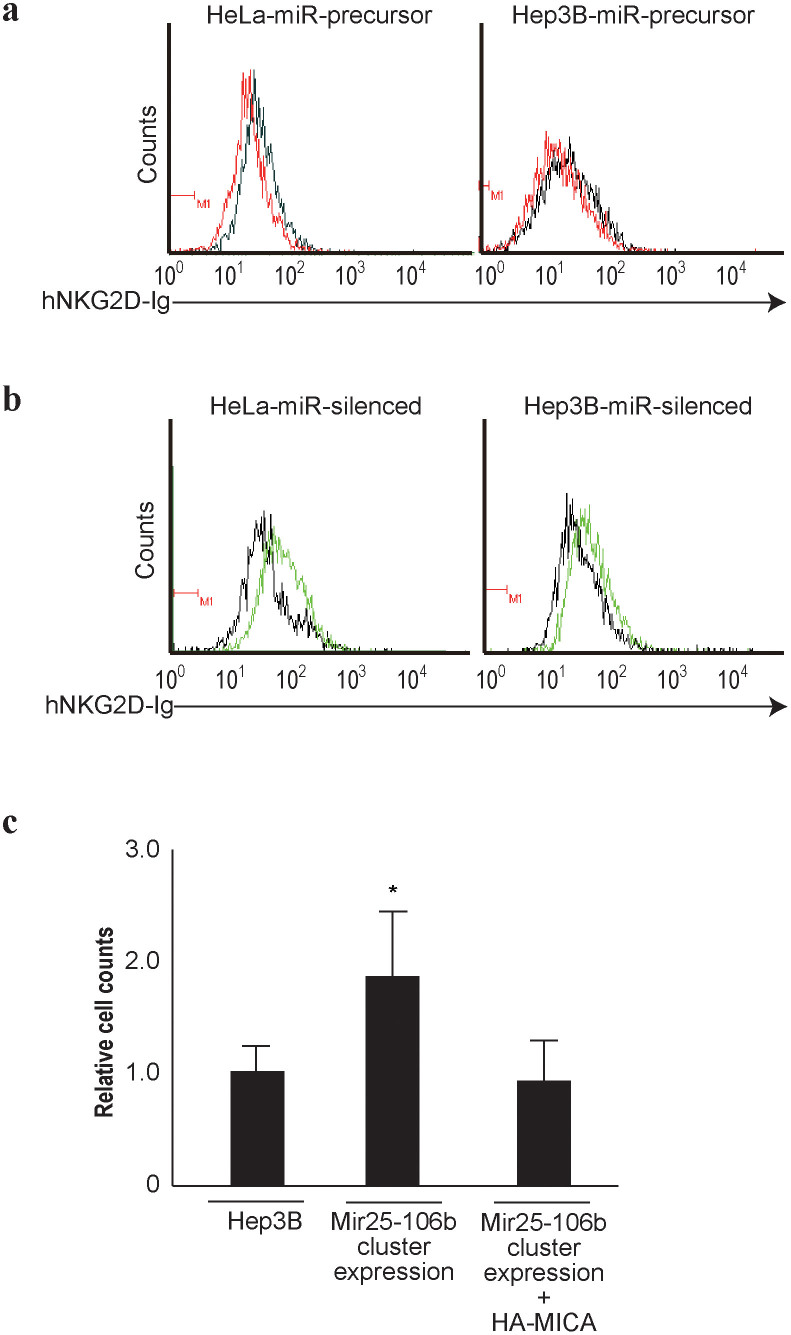
To determine the consequences of the modulation of MICA protein expression levels by miRNAs, we first determined the binding ability of NKG2D, a receptor of MICA, using HeLa and Hep3B cells overexpressing the miR25-93-106b cluster or with silencing of miR25-93-106b function. As expected, the levels of NKG2D binding to the cells, theoretically through binding to MICA, were decreased in HeLa and Hep3B cells overexpressing the miR25-93-106b cluster (Figure 4a). On the contrary, the levels of NKG2D binding to the cells were increased in HeLa and Hep3B cells in which miR25-93-106b function had been silenced (Figure 4b).
Figure 4. NKG2D binding levels change in proportion to MICA expression levels.
(a), (b), Flow cytometry of human IgG-fused NKG2D binding to the control (black lines), miR25-93-106b cluster-expressing cells (red lines) (a), and mature anti-sense sequences of miR25-93-106b cluster-expressing cells (green lines) (b). Representative results from three independent experiments are shown. (c), In vivo killing of DiO-labeled Hep3B and Dil-labeled HeLa cells (internal control cells) injected together into the tail veins of six mice in each group. Fluorescence intensities were quantified by flow cytometry as the ratio of Hep3B to HeLa cells in the lungs. The data from control Hep3B cells were set as 1.0. Data represent the means ± s.d. of three independent experiments. *p < 0.05.
Next, to determine whether tumor cells with different miRNA-induced MICA protein expression levels exhibited differing susceptibilities to NK-cell-mediated killing in vivo, we performed a tumor-clearance assay that measures short-term in vivo killing by NK cells22. Hep3B control cells, Hep3B cells with miR25-93-106b cluster overexpression, or Hep3B cells with miR25-93-106b and HA-tagged MICA overexpression, labeled with fluorescent DiO, were injected into C57Black6/J mouse tail veins together with an equal number of HeLa cells labeled with Dil (internal reference control). After 5 h, surviving Hep3B and HeLa cells in the lungs were enumerated by flow cytometry. The number of Hep3B cells that had survived divided by the number of HeLa cells that had survived represents the relative killing of Hep3B cells in vivo. As shown by the in vitro binding assay using NKG2D, the killing rate of Hep3B cells in which miRNA function had been silenced was higher, and that of cells overexpressing miRNAs was lower, than that of control cells. The effects of miRNA overexpression were similar to those obtained in MICA knocked-down Hep3B cells (supplementary Figure 4). Additionally, the lower cell-killing rate in Hep3B cells overexpressing miRNA was antagonized by the co-expression of exogenous MICA protein (Figure 4c), suggesting that the decreased clearance was mediated by reduced MICA expression levels secondary to overexpression of miRNAs. These results suggest that tumor progression and invasion can be regulated by expression or silencing of miRNAs in at least some cells by regulation of MICA expression levels.
Discussion
In this study, we showed that the miR25-93-106b cluster modulates MICA protein expression by HCC cells. Because our previous GWAS analyses identified that MICA is the critical gene determining HCC susceptibility in patients with chronic hepatitis infection5,6, the herein-described methods of modulating MICA expression may be useful for developing novel methods of prevention and therapeutics against HCCs.
MICA is a membrane protein that acts as a ligand for NKG2D to activate innate anti-tumor effects through natural killer and CD8+ cells7. Our previous GWAS study showed that a risk allele at the SNP in the MICA promoter region was significantly associated with the susceptibility of HCV-induced HCC as well as with lower serum MICA levels. Although polymorphisms at the same SNP site were also associated with HBV-induced HCC, the risk allele determining the susceptibility of HCC was somehow different from that in HCV-induced HCC. While the reason why different MICA gene variations act as risk alleles at the same SNP site between HBV- and HCV-induced HCC has not been elucidated, it is assumed that changes in the membrane-bound MICA and soluble MICA levels due to differences in post-translational processing according to virus type may affect the risk allele results. In any case, because the importance of the regulation of MICA expression levels to prevent development of HCC due to chronic hepatitis viral infection cannot be denied, the regulation of MICA levels by microRNAs as shown here may be useful for the development of preventive methods of preventing HCC development during chronic hepatitis infection.
While several cellular signaling pathways lead to upregulation of MICA12,13, we used microRNAs to regulate the expression levels of MICA in this study. As shown by the results of our GWAS analyses, which found that the polymorphisms in the promoter region of MICA are associated with changes in the sMICA levels5,6, promoter activities of the MICA gene also have significant effects on MICA expression levels23. Our results showed that miR93 and 106b expression levels were not always correlated with those of MICA in HCC cell lines, suggesting that the regulation of MICA expression is not solely dependent on miRNAs. In addition, in cells with no endogenous MICA expression, such as Huh7 cells, modulation of microRNA expression had no effect on the regulation of MICA expression. This suggests that at least low-level endogenous expression, which may be determined by promoter activities, are needed for regulation by miRNA. Therefore, changes in promoter activities and epigenetic changes in the MICA gene should also be determined. This will facilitate application of the regulatory function of miRNAs reported here.
One class of antisense oligonucleotides, namely locked nucleic acids, can be used to sequester microRNAs in the liver of various animals, including humans16,24,25. A clinical trial targeting miR-122 with the anti-miR-122 oligonucleotides miravirsen, the first miRNA-targeted drug, is underway for the treatment of HCV infection16. Thus, nucleic-acid-mediated gene therapy is becoming a realistic option. Modulation of MICA expression levels by such nucleic-acid-mediated therapy based on the results presented herein may also be a promising option for prevention and/or therapy of HCC.
In summary, we have shown that the miR25-93-106b cluster can be used to modulate MICA expression levels in HCC cells. Based on our GWAS results and associated studies, regulation of MICA protein expression levels is crucial to prevent the development of HCC during chronic hepatitis viral infection. It is important to identify the other factors that regulate MICA transcriptional activities as well as the post-translational processes and their association with susceptibility to HCCs. That said, miRNA regulation of MICA expression as shown here may facilitate regulation of the host innate immune system in an HCC-suppressive manner during chronic hepatitis viral infection.
Methods
Cell culture
The human HCC cell lines Huh7, HLE, PLC/PRF/5, and Hep3B were obtained from the Japanese Collection of Research Bioresources (JCRB, Osaka, Japan). The human cervical cancer cell line HeLa was obtained from the American Type Culture Collection (ATCC, Rockville, MD). All cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Mouse
Experimental protocols were approved by the Ethics Committee for Animal Experimentation at the Graduate School of Medicine, the University of Tokyo and the Institute for Adult Disease, Asahi Life Foundation, Japan and conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of the Department of Medicine, the University of Tokyo, and the Institute for Adult Disease, Asahi Life Foundation.
Flow cytometry
Cells were hybridized with anti-MICA (1:500; R&D Systems, Minneapolis, MN) and isotype control IgG (1:500; R&D Systems) in 5% BSA/1% sodium azide/PBS for 1 h at 4°C. After washing, cells were incubated with goat anti-mouse Alexa 488 (1:1000; Molecular Probes, Eugene, OR) for 30 min. Flow cytometry was performed and data analyzed using Guava Easy Cyte Plus (GE Healthcare, Little Chalfont, UK).
Reporter plasmid construction, transient transfections, and luciferase assays
The reporter plasmid for the analysis of the effects of miRNAs on MICA 3′UTR were constructed by subcloning the MICA 3′UTR sequences from pLightSwitch-MICA 3UTR (SwitchGear Genomics, Menlo Park, CA) into the pGL4.50 vector (Promega, Madison, WI) at the FseI site by the In-Fusion method (Clontech, Mountain View, CA) to insert the MICA 3′UTR sequences into the 3′-UTR of the firefly luciferase gene, which was under the control of the CMV promoter. The sequences of the primers were 5′-CTA GAG TCG GGG CGG CG GCC ATT TCA GCC TCT GAT GTC AGC-3′ and 5′-GTC TGC TCG AAG CGG CCG GCC TGG CCT GAG ACT CTG TCT TAA-3′. The resultant plasmid (Luc-MICA 3′UTRwt) was used as a template for the construction of mutant reporter plasmid (Luc-MICA 3′UTRmut), which carries three point mutations in the seed sequences of miR93 and 106b in the MICA 3′UTR, itself generated by a Quik Change II XL Site-directed Mutagenesis Kit (Stratagene, Heidelberg, Germany) according to the manufacturer's instructions. Transient transfection and reporter assays were performed as described previously26.
Lentiviral constructs, viral production, and transduction
To generate a neomycin-resistant miR25-93-106b cluster-expressing lentiviral construct, copGFP in the pmiRNA25-93-106b cluster-expressing plasmid (System Biosciences, Mountain View, CA) was replaced with a neomycin resistant gene, which was subcloned from the pCDH-Neo vector (System Biosciences), at the FseI site. The primers used were 5′-GCT ACC GCT ACG AGG CCG GCC CAT GAT TGA ACA AGA TGG ATT GCA-3′ and 5′-TCG CCG ATC ACG CGG CCG GCC TCA GAA GAA CTC GTC AAG AAG GC-3′. To remove the copGFP region from pmiRZIP25-93-106b (System Biosciences), a construct expressing mature anti-sense sequences of the miR25-93-106b cluster, sequences coding the GFP gene were removed by excision with XbaI and PstI sites followed by connecting the cut ends with annealed oligonucleotides (5′-CTA GAC GCC ACC ATG CTG CA-3′ and 5′-GCA TGG TGG CGT-3′) to maintain the coding frame and the expression of the downstream puromycin-resistance gene. To generate HA-tagged MICA protein overexpressing the lentiviral construct, MICA cDNA was amplified by PCR using a Halo-tag-MICA–expressing plasmid (Promega, Madison, WI) as a template and cloned into a pCDH-puro vector (System Biosciences) at the NotI site. The primer sequences used were 5′-ATC GGA TCC GCG GCC GCA CCA TGT ACC CAT ACG ATG TTC CAG ATT ACG CTA TGG GGC TGG GCC CGG TC-3′ and 5′-AGA TCC TTC GCG GCC GCT TAG GCG CCC TCA GTG GAG C-3′. Let-7g precursor expressing plasmid was generated by inserting about 1,000 bp long PCR product around the let-7g genomic region into pCDH-puro vector using Xba1 and Not1 sites. The production and concentration of lentiviral particles were described previously27. shRNA against MICA-producing lentiviral particles with puromycin resistant gene were purchased from SantaCruz Biotechnology (sc-4924-V, Dallas, TX). Cells were transduced with lentiviruses using polybrene (EMD Millipore, Billerica, MA). The selections were performed with 400 μg/mL G418 and 2 μg/mL (HeLa) or 6 μg/mL (Hep3B) puromycin.
Immunocytochemistry
Cells on two-well chamber slides were fixed with 4% paraformaldehyde. Fixed cells were probed with the primary MICA antibody (R&D Systems) for 1 h after blocking with 5% normal goat serum for 30 min. Cells probed with the MICA antibody were incubated with the secondary Alexa Fluor 488 goat anti-mouse antibody (Molecular Probes) for 30 min. Slides were mounted using VectaShield with DAPI (Vector Labs, Burlingame, CA).
Northern blotting of miRNAs
Northern blotting of miRNAs was performed as described previously27. Briefly, total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Ten micrograms of RNA were resolved in denaturing 15% polyacrylamide gels containing 7 M urea in 1 × TBE and then transferred to a Hybond N+ membrane (GE Healthcare) in 0.25 × TBE. Membranes were UV-crosslinked and prehybridized in hybridization buffer. Hybridization was performed overnight at 42°C in ULTRAhyb-Oligo Buffer (Ambion) containing a biotinylated probe specific for miR93 (cta cct gca cga aca gca ctt tg) and 106b (atc tgc act gtc agc act tta), which had previously been heated to 95°C for 2 min. Membranes were washed at 42°C in 2 × SSC containing 0.1% SDS, and the bound probe was visualized using a BrightStar BioDetect Kit (Ambion). Blots were stripped by boiling in a solution containing 0.1% SDS and 5 mM EDTA for 10 min prior to rehybridization with a U6 probe (cac gaa ttt gcg tgt cat cct t).
miRNA library screening
To screen for miRNAs that target MICA 3′-UTR, synthetic miRNA mimics and reporter constructs were used as described previously19,20. Seventy-six types of synthetic mature miRNAs that are highly expressed in the liver28 were custom-made (B-Bridge, Tokyo, Japan) and transfected by RNAi Max (Life Technologies, Carlsbad, CA) into Huh7 cells in 96-well plates that had been transfected 24 h before with Luc-MICA 3′UTRwt. The cells were then incubated for another 24 h. As negative controls, oligonucleotides of artificial sequences were applied19. The luciferase activities were measured using a GloMax 96 Microplate Luminometer (Promega). The experiments were performed in duplicate.
NKG2D binding assay
Cells were incubated with 4 μg of recombinant human NKG2D fused to human IgG1 Fc chimera protein. After washing, cells were incubated with an Alexa488-conjugated affinity purified F(ab')2 fragment of goat anti-human IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). As a negative control, cells were incubated with only Alexa488 anti-human IgG. The intensity of the fluorescence was determined by flow cytometry.
In vivo cell-killing assay
Hep3B cells and HeLa cells were labeled with the fluorescent dye VybrantDiO and Dil (Molecular Probes), respectively. Cells were mixed at a density of 2 × 107 in 1-ml PBS, and 200 μl was injected into the tail vein. Five hours later, lungs were collected, and single-cell suspensions were collected using a cell strainer. Fluorescence was assayed by flow cytometry, and the ratio of the experimental Hep3B cells to HeLa cells (internal control) was calculated.
Statistical analysis
Statistically significant differences between groups were determined using Student's t-test when variances were equal. When variances were unequal, Welch's t-test was used instead. P-values of < 0.05 were considered to indicate statistical significance.
Author Contributions
T.K., M. Otsuka and K.K. planned the research and wrote the paper. T.K., M. Otsuka, T.Y., M. Ohno, A.T., C.S. and Y.K. performed the majority of the experiments. M.A. and H.Y. supported several experiments and analyzed the data. K.K. supervised the entire project.
Supplementary Material
Supplementary Information (Supplementary Figures S1–5 and Supplementary Figure Legends)
Acknowledgments
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#25293076, #25460979, and #24390183) (to M.Otsuka, Y.K. and K.K.), by Health Sciences Research Grants of The Ministry of Health, Labour and Welfare of Japan (to K.K.), and by grants from the Okinaka Memorial Institute for Medical Research, the Liver Forum in Kyoto, and the Princess Takamatsu Cancer Research Fund (to M.Otsuka).
References
- El-Serag H. B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142, 1264–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. Hepatocellular carcinoma: New and emerging risks. Dig Liver Dis 42, S215–222 (2010). [DOI] [PubMed] [Google Scholar]
- Arzumanyan A., Reis H. M. & Feitelson M. A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 13, 123–135 (2013). [DOI] [PubMed] [Google Scholar]
- Urabe Y. et al. A genome-wide association study of HCV-induced liver cirrhosis in the Japanese population identifies novel susceptibility loci at the MHC region. J Hepatol (2013). [DOI] [PubMed] [Google Scholar]
- Kumar V. et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet 43, 455–458 (2011). [DOI] [PubMed] [Google Scholar]
- Kumar V. et al. Soluble MICA and a MICA variation as possible prognostic biomarkers for HBV-induced hepatocellular carcinoma. PLoS One 7, e44743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccalli C., Scaramuzza S. & Parmiani G. TNK cells (NKG2D+ CD8+ or CD4+ T lymphocytes) in the control of human tumors. Cancer Immunol Immunother 58, 801–808 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinushi M. et al. Impairment of natural killer cell and dendritic cell functions by the soluble form of MHC class I-related chain A in advanced human hepatocellular carcinomas. J Hepatol 43, 1013–1020 (2005). [DOI] [PubMed] [Google Scholar]
- Diefenbach A., Jensen E. R., Jamieson A. M. & Raulet D. H. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature 413, 165–171 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y. Targeting NKG2D in tumor surveillance. Expert Opin Ther Targets 16, 587–599 (2012). [DOI] [PubMed] [Google Scholar]
- Guerra N. et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 28, 571–580 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999). [DOI] [PubMed] [Google Scholar]
- Eleme K. et al. Cell surface organization of stress-inducible proteins ULBP and MICA that stimulate human NK cells and T cells via NKG2D. J Exp Med 199, 1005–1010 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav D., Ngolab J., Lim R. S., Krishnamurthy S. & Bui J. D. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol 182, 39–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N. & Mandelboim O. An integrated view of the regulation of NKG2D ligands. Immunology 128, 1–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen H. L. et al. Treatment of HCV Infection by Targeting MicroRNA. N Engl J Med 368, 1685–94 (2013). [DOI] [PubMed] [Google Scholar]
- Salih H. R., Rammensee H. G. & Steinle A. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J Immunol 169, 4098–4102 (2002). [DOI] [PubMed] [Google Scholar]
- Tang K. F. et al. Decreased Dicer expression elicits DNA damage and up-regulation of MICA and MICB. J Cell Biol 182, 233–239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata A. et al. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem Biophys Res Commun 411, 826–831 (2011). [DOI] [PubMed] [Google Scholar]
- Yoshikawa T. et al. Silencing of microRNA-122 enhances interferon-a signaling in the liver through regulating SOCS3 promoter methylation. Sci. Rep. 2, 637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005). [DOI] [PubMed] [Google Scholar]
- Gazit R. et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol 7, 517–523 (2006). [DOI] [PubMed] [Google Scholar]
- Lo P. H. et al. Identification of a Functional Variant in the MICA Promoter Which Regulates MICA Expression and Increases HCV-Related Hepatocellular Carcinoma Risk. PLoS One 8, e61279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008). [DOI] [PubMed] [Google Scholar]
- Kojima K. et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun 2, 338 (2011). [DOI] [PubMed] [Google Scholar]
- Takata A. et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 57, 162–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J. et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature 438, 685–689 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (Supplementary Figures S1–5 and Supplementary Figure Legends)