Abstract
The organic cation transporter 1 (OCT1), also known as solute carrier family 22 member 1, is strongly and specifically expressed in the human liver. Here we show that the hepatocyte nuclear factor 1 (HNF1) regulates OCT1 transcription and contributes to the strong, liver-specific expression of OCT1. Bioinformatic analyses revealed strong conservation of HNF1 binding motifs in an evolutionary conserved region (ECR) in intron 1 of the OCT1 gene. Electrophoretic mobility shift and chromatin immunoprecipitation assays confirmed the specific binding of HNF1 to the intron 1 ECR. In reporter gene assays performed in HepG2 cells, the intron 1 ECR increased SV40 promoter activity by 22-fold and OCT1 promoter activity by 13-fold. The increase was reversed when the HNF1 binding sites in the intron 1 ECR were mutated or the endogenous HNF1α expression was downregulated with small interfering RNA. Following HNF1α overexpression in Huh7 cells, the intron 1 ECR increased SV40 promoter activity by 11-fold and OCT1 promoter activity by 6-fold. Without HNF1α overexpression, the increases were only 3- and 2-fold, respectively. Finally, in human liver samples, high HNF1 expression was significantly correlated with high OCT1 expression (r = 0.48, P = 0.002, n = 40). In conclusion, HNF1 is a strong regulator of OCT1 expression. It remains to be determined whether genetic variants, disease conditions, or drugs that affect HNF1 activity may affect the pharmacokinetics and efficacy of OCT1-transported drugs such as morphine, tropisetron, ondansetron, tramadol, and metformin. Beyond OCT1, this study demonstrates the validity and usefulness of interspecies comparisons in the discovery of functionally relevant genomic sequences.
Introduction
The organic cation transporter 1 (OCT1), also known as solute carrier family 22 member 1 (SLC22A1), is strongly expressed in the sinusoidal membrane of the human liver. OCT1 typically accelerates the hepatic uptake of small, hydrophilic, positively charged organic molecules, including both naturally occurring substances and numerous drugs (Koepsell et al., 2007). OCT1 is apparently not essential for life, but it may have substantial medical impact during drug treatment or exposure to certain chemicals (Jonker et al., 2001). A number of clinically relevant drugs have been shown to be substrates or inhibitors of OCT1 (Ahlin et al., 2008; Nies et al., 2011). Although many OCT1 substrates have only been tested in vitro, several studies in humans provide evidence that metformin, tropisetron, ondansetron, tramadol, and morphine are OCT1 substrates (Shu et al., 2007; Tzvetkov et al., 2010, 2011, 2013).
OCT1 is the most strongly expressed drug transporter in the human liver (Hilgendorf et al., 2007; Schaefer et al., 2012). OCT1 mRNA expression is 3-fold greater than the second-most highly expressed human liver influx transporter, OATP1B1 (Hilgendorf et al., 2007). Furthermore, OCT1 mRNA expression is more than 13-fold stronger than the expression of OCT3, the other organic cation transporter that is expressed in the human liver (Hilgendorf et al., 2007; Nies et al., 2009).
In humans, OCT1 is almost exclusively expressed in the liver (Zhang et al., 1997). Although OCT1 expression has also been reported for the human kidney and intestine (Muller et al., 2005; Tzvetkov et al., 2009), the mRNA expression levels in these organs are more than 500-fold lower than in the liver (Nies et al., 2009; Tzvetkov et al., 2009). In addition, de-differentiation of liver hepatocytes to hepatocellular carcinoma cells has been associated with a strong decrease in OCT1 expression (Heise et al., 2012).
OCT1 activity varies greatly among healthy individuals. Approximately 9% of Caucasians completely lack OCT1 activity due to common amino acid polymorphisms (Shu et al., 2003; Tzvetkov et al., 2010). Loss-of-function amino acid substitutions have been associated with altered pharmacokinetics and/or efficacy of metformin, imatinib, tropisetron, ondansetron, tramadol, and morphine (Shu et al., 2007; Tzvetkov et al., 2009, 2010, 2011, 2013; Bazeos et al., 2010).
Loss-of-function amino acid substitutions are not the only factor contributing to the high interindividual variability in OCT1 activity; OCT1 expression also varies widely among individuals. Systematic analyses of OCT1 expression in the human liver showed 113-fold variability in OCT1 mRNA and a corresponding 83-fold variability in OCT1 protein levels (Nies et al., 2009). Only part of this extensive variability could be explained by disease conditions such as cholestasis, or genetic and epigenetic variations in the OCT1 locus (Nies et al., 2009; Schaeffeler et al., 2011). Less is known about the contribution of trans-regulatory factors to the highly variable expression of OCT1.
Two trans-regulatory factors, upstream stimulatory factor (USF) and the hepatocyte nuclear factor 4 (HNF4), are known to regulate OCT1 transcription (Saborowski et al., 2006; Kajiwara et al., 2008). Both factors bind in the 2-kb promoter region of OCT1. USF binds to an E-box element located 90 bp upstream of the translational start site, while HNF4 binds to two tandem direct repeat sequences located 160-bp upstream of the translational start site. However, neither USF nor HNF4 can completely explain the very strong and liver-specific expression of human OCT1. USF1 is a ubiquitously expressed transcriptional factor and thus does not account for the liver specificity of OCT1 expression. On the other hand, HNF4 is a transcription factor highly active in the liver, but it only moderately activates OCT1 transcription. The OCT1 promoter containing the HNF4 binding sites leads to at most a 3-fold increase in luciferase reporter gene activity in HepG2 and Huh7 cells (Kajiwara et al., 2008) and by itself cannot explain the 500-fold stronger expression of OCT1 in the liver compared with the other human organs. Therefore, we hypothesized that other transcription factors may contribute to the liver specificity and high interindividual variability of OCT1 expression.
Here we report that the hepatocyte nuclear factor 1, a major transcriptional regulator in the liver, regulates OCT1 expression via binding to an evolutionary conserved region (ECR) located in intron 1 of the OCT1 gene. We present bioinformatic analyses, electrophoretic mobility shift and chromatin immunoprecipitation assays, reporter gene assays in model cell lines, and HNF1 and OCT1 expression analyses in human liver samples to support this finding.
Materials and Methods
Bioinformatic Analyses.
ECR Browser was used to identify evolutionary conserved regions in the OCT1 gene locus (Ovcharenko et al., 2004). The human locus was compared with rat, mouse, cow, dog, rhesus macaque, and chimpanzee loci. An ECR was defined as a region with a minimum length of 100 bp, showing 70% or higher conservation between the human and tested mammalian genomes. PhyloP software was used for multiple sequence alignment of the 46 vertebrate genomes from the Vertebrate Multiz Alignment Track of the University of California, Santa Cruz Genome Browser database (Rhead et al., 2010). Potential binding sites for the hepatocyte nuclear factor 1 were identified based on the consensus HNF1 recognition sequence GTTAATnATTAAC (Bach et al., 1990).
Isolation of Nuclear Proteins, In Vitro Transcription/Translation, and Electrophoretic Mobility Shift Assay.
Nuclear proteins were extracted by the Dignam method (Dignam et al., 1983) followed by an additional purification with ammonium sulfate (final concentration of ammonium sulfate 0.3 g/ml). Protein concentration was determined using a bicinchoninic acid assay (Smith et al., 1985) with bovine serum albumin as a standard (all reagents were from Sigma-Aldrich, Deisenhofen, Germany).
The electrophoretic mobility shift assays were performed as described previously (Meineke et al., 2008). In brief, 2-pmol annealed oligonucleotide probes were radiolabeled by incubation with 2 U Klenow enzyme (MBI-Fermentas, St. Leon-Roth, Germany) at 30°C for 1 hour in the presence of 20 µCi of [α-32P]dCTP (Hartmann Analytic, Braunschweig, Germany) and a 50 µM concentration of the other three dNTPs. The labeled probes were purified using Mini Quick Spin Oligo Columns (Roche, Mannheim, Germany). The probe sequences are given in Table 1. In the binding reaction, 20-µg nuclear extracts were preincubated for 10 minutes on ice in an 18-µl reaction mixture containing 20 mM HEPES, pH 7.8, 75 mM EDTA, 0.5 mM dithiothreitol, 140 mM KCl, 10% glycerol, and 0.2 pg of poly(dI-dC) as a nonspecific competitor. The radioactive probe (13.5 nCi) was then added and incubated for an additional 15 minutes on ice, and the reaction was run on a 5% native polyacrylamide gel. In the cold competition assays, 3- to 30-fold excesses of unlabeled competitor probes were used. Supershift assays were performed with rabbit polyclonal antibody able to detect both the α and the β isoforms of HNF1 and with rabbit IgG as a control (sc-8986 and sc-2027, respectively; both from Santa Cruz Biotechnology, Heidelberg, Germany). The antibodies were added to the preincubation mixture and incubated for 1 hour instead of 10 minutes before adding the radiolabeled probes.
TABLE 1.
Sequences of the probes and primers used in the study
| Name | Direction | Sequence |
|---|---|---|
| EMSA probes | ||
| GS-ECR | Forward | 5′-gatcCTTAGTTATTCATTTCTGCAGAACTAATTTTTAACCTAG-3′ |
| GS-ECR | Reverse | 5′gatcCTAGGTTAAAAATTAGTTCTGCAGAAATGAATAACTAAG-3′ |
| GS-ECR_mut | Forward | 5′-gatcCTTA-GGGGTTCAGGTCTGCAGAACGGATTTTGGGCCTAG-3′ |
| GS-ECR_mut | Reverse | 5′-gatcCTAGGCCCAAAATCCGTTCTGCAGACCTGAACCCCTAAG-3′ |
| GS-HNF1_cons | Forward | 5′-gatcCCAGGTTAATGATTAACCCA-3′ |
| GS-HNF1_cons | Reverse | 5′-gatcTGGGTTAATCATTAACCTGG-3′ |
| ChIP primers | ||
| ChIP-ECR | Forward | 5′-CTGGCTCGTGGGTAAGAATTGTCTC-3′ |
| ChIP-ECR | Reverse | 5′ATGTCCAAGGCAATGCTAGGTTAAA-3′ |
| ChIP-Intron2 | Forward | 5′-GGCAGCGAGATCGAAGGACAACTGT-3′ |
| ChIP-Intron2 | Reverse | 5′-TCTCCTGCCTTCGGGTTTTCTCCAA-3′ |
| Primers used in the generation of the reporter gene and the HNF1-overexpressing constructs | ||
| OCT1 Promoter | Forward | 5′-TGCACAGAGAGAGAAACCAAAAGTC-3′ |
| OCT1 Promoter | Reverse | 5′-GCCAGCTCGAGATGTCTCCCTCAGAGATCTTTG-3′ |
| ECR | Forward | 5′-TCCTTGGATCCCCAGCTCCTCCTCCAAACT-3′ |
| ECR | Reverse | 5′-CCCGAGTCGACCTCATGATCTTAGCACCTAGCCTT-3′ |
| HNF1A | Forward | 5′-CCTGTGGATCCGAGCCATGGTTTCTAAACTGA-3′ |
| HNF1A | Reverse | 5′-AGCTTATCTAGAGTGGTTACTGGGAGGAAGAGG-3′ |
| HNF1B | Forward | 5′- CTTTTTCCGGATCCTTGGAAAATGGTGTCCAA-3′ |
| HNF1B | Reverse | 5′- GTGGTGTCTAGAGGCATCACCAGGCTTGTAGA-3′ |
| Primers used for site-directed mutagenesis of the ECR1 Binding Sites | ||
| HNF1A(A)_f | Forward | 5′-TTGGGGAATCAATCTTAGGGGTTCAGGTCTGCAGAACTAATTTTT A-3′ |
| HNF1A(A)_r | Reverse | 5′-TAAAAATTAGTTCTGCAGACCTGAACCCCTAAGATTGATTCCCCAA-3′ |
| HNF1A(B)_f | Forward | 5′-GGTTCAGGTCTGCAGAACGGATTTTGGGCCTAGCATTGCCTTGGAC-3′ |
| HNF1A(B)_r | Reverse | 5′-GTCCAAGGCAATGCTAGGCCCAAAATCCGTTCTGCAGACCTGAACC-3′ |
ChIP, chromatin immunoprecipitation. The unspecific sequences used in the radioactive labeling of the EMSA probes are given in lowercase letters. Mutated nucleotides are shown in boldface, and the artificially introduced restriction sites are underlined. The ATG start codons of HNF1α and HNF1β are given in italics.
In vitro transcription-translation was performed using the TNT T7 Quick-Coupled Transcription/Translation System (Promega, Mannheim, Germany) according to the manufacturer’s instructions. In brief, 1 microgram DNA of plasmids pcDNA3.1, pcDNA3.1::HNF1A, or pcDNA3.1::HNF1B (each carrying the T7 promoter) was mixed with 40 µl of TNTT7 Quick Master Mix and 1 µl of 1 mM methionine and incubated for 1 hour at 30°C. Two microliters of the reaction were used instead of nuclear extracts in the electrophoretic mobility shift assays.
Chromatin Immunoprecipitation Assay.
Two million cryopreserved human hepatocytes were rapidly thawed at 37°C, washed once in 10 ml of Dulbecco’s modified Eagle’s medium (Gibco, Life Technologies, Darmstadt, Germany) and one time in 10 ml of phosphate-buffered saline, pH 7.4, and pelleted by centrifugation (300g for 3 minutes at room temperature). The hepatocytes used were from a single male donor and were obtained from Gibco (Life Technologies). The chromatin immunoprecipitation was performed using EpiTect Chromatin Immunoprecipitation OneDay Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The chromatin was sheared in three cycles of 10 minutes at 300 W with 10-second on/off–duty time using a Biorupter (Diagnode, Liege, Belgium). Immunoprecipitation was performed with 4 µg of HNF1 or control IgG antibodies at 4°C overnight. The antibodies used were the same as in the electrophoretic mobility shift assay. The amount of precipitated DNA was analyzed by conventional polymerase chain reaction (PCR) and quantified by real-time PCR. The conventional PCR was performed with KOD Hot Start DNA polymerase (Novagen, Darmstadt, Germany) and the real-time PCR with TaqMan universal master mix (Life Technologies) supplemented with SYBR Green I (Eurogentec, Serang, Belgium). The amplification was carried out under the following conditions: 95°C for 10 minutes, followed by 40 cycles of 94°C for 10 seconds, 60°C for 20 seconds, and 72°C for 50 seconds. Two regions were analyzed: a 149-bp region containing the potential HNF1 binding sites in the intron 1 ECR, and a 141-bp region in intron 2 of the OCT1 gene analyzed as a negative control. The primer sequences are given in Table 1.
Generation of the Luciferase Reporter and the HNF1 Overexpression Plasmids.
Genomic DNA was extracted from an EDTA-preserved venous blood sample from an anonymous healthy Caucasian human donor, using an automated solid phase extraction method according to the manufacturer’s instructions (EZ1 DNA Blood 350-µl Kit used with the BioRobot EZ1; both from Qiagen). The DNA was used for the amplification of the OCT1 core promoter and the intron 1 ECR fragment. All PCR amplifications performed for cloning purposes were performed with a high-fidelity KOD Hot Start DNA polymerase (Novagen, Darmstadt, Germany) and Q-solution (Qiagen).
A 1601-bp promoter fragment spanning 1560 bp upstream to 44 bp downstream of the transcriptional start site of OCT1 was amplified by PCR. The amplification was performed using the OCT1 promoter forward and reverse primers (Table 1) under the following reaction conditions: 95°C for 2 minutes, followed by 35 cycles of 95°C for 30 seconds, 64°C for 30 seconds, 72°C for 90 seconds, and a final elongation of 72°C for 10 minutes. The PCR product was cut with SacI and XhoI restriction enzymes (MBI-Fermentas). The SacI site was present in the template sequence, and the XhoI site was artificially introduced by the reverse primer. The PCR product was cloned into the pGL3basic vector (Promega), which was cut with the same restriction enzymes, to generate the OCT1 promoter reporter construct pGL3b::OCT1promoter.
The 185-bp ECR fragment was amplified using the ECR forward and reverse primers (Table 1) under the following reaction conditions: 95°C for 3 minutes, followed by 35 cycles of 94°C for 15 seconds, 53°C for 30 seconds, 68°C for 1 minute, and a final elongation of 68°C for 7 minutes. The PCR product was cut with BamHI and SalI (MBI-Fermentas) (restriction sites introduced by the primers) and cloned into the pGL3b::OCT1promoter or pGL3-Promoter plasmids (Promega) cut with the same restriction sites.
The two potential HNF1 binding sites in the intron 1 ECR were mutated by fusion PCR followed by classic site-directed mutagenesis. The fusion PCR was used to mutate the upstream HNF1 binding site, while the classic site-directed mutagenesis was used to mutate the downstream HNF1 binding site. In the fusion PCR, the first two DNA fragments, an 88-bp and a 148-bp fragment, were independently amplified using the ECR-containing pGL3b::OCT1promoter plasmid as a template. The 88-bp fragment was amplified using the ECR forward and HNF1A(A) reverse primers (Table 1). The 148-bp fragment was amplified using the HNF1A(A) forward and the ECR reverse primers (Table 1). Both reactions were performed under the following reaction conditions: 95°C for 3 minutes, followed by 35 cycles of 94°C for 30 seconds, 53°C for 30 seconds, 68°C for 2 minutes, and a final elongation of 68°C for 7 minutes.
In the subsequent fusion reaction, the homologous ends of the two mutated ECR fragments were annealed using approximately 1 µg DNA from each initial PCR product and the following conditions: initial denaturing at 95°C for 2 minutes, followed by 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 2 minutes, and a final elongation of 72°C for 7 minutes. Finally, the mutated ECR fragment was amplified by PCR with the flanking primers ECR forward and ECR reverse (Table 1). The PCR product was cloned into the pTOPO vector (Life Technologies), and the mutation was verified by sequencing.
The second potential HNF1 binding site in the ECR fragment was mutated by classic site-directed mutagenesis using the pTOPO plasmid as a template and the forward and reverse mutagenesis primers HNF1A(B). The mutation was verified by sequencing. The 185-bp dual-mutant ECR fragment was cut from the pTOPO plasmid using BamHI and SalI and recloned into the pGL3b::OCT1promoter or the pGL2-Promoter constructs downstream of the firefly luciferase gene.
The HNF1 overexpression vector was constructed using the full-length I.M.A.G.E. cDNA clone IRATp970D11125D (ImaGenes, Berlin, Germany). A 1911-bp fragment, including the complete HNF1A open reading frame, was amplified by PCR from approximately 2 µg of template DNA with the forward and reverse HNF1A primers (Table 1) under the following reaction conditions: 95°C for 3 minutes, followed by 35 cycles of 94°C for 15 seconds, 60°C for 30 seconds, 68°C for 3 minutes, and a final elongation of 68°C for 7 minutes. The PCR product was cloned into the pcDNA3.1 vector (Life Technologies) using the BamHI and XbaI restriction sites downstream of the cytomegalovirus promoter to create the HNF1α overexpression construct pcDNA3.1::HNF1A.
The HNF1β overexpression vector was constructed using a full-length I.M.A.G.E. cDNA clone IRATp970A0421D (ImaGenes). A 1711-bp fragment, including the complete HNF1B open reading frame, was amplified by PCR from approximately 2 µg of template DNA with the forward and reverse HNF1B primers (Table 1) under the following reaction conditions: 95°C for 2 minutes, followed by 35 amplification cycles of 95°C for 30 seconds, 64.6°C for 30 seconds and then 72°C for 2 minutes, and a final elongation of 72°C for 10 minutes. The PCR product was cloned into the pcDNA3.1 vector (Life Technologies) using the BamHI and XbaI restriction sites downstream of the cytomegalovirus promoter to create the HNF1 overexpression construct pcDNA3.1::HNF1B.
Reporter Gene Assays.
HepG2 cells (DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) were cultured in RPMI 1640 GlutaMAX-I supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. Huh7 cells (JCRB Cell Bank, Tokyo, Japan) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. All cell culture media and additives were obtained from Life Technologies. Cells were cultured under standard conditions at 37°C in a humidified atmosphere supplemented with 5% CO2.
For transfection experiments, a total of either 1 × 106 HepG2 or 0.5 × 106 Huh7 cells were plated in a single well of a six-well plate (Nunc, Langenselbold, Germany) and grown for 24 hours to reach approximately 75% confluence.
Linearized reporter gene plasmids were used for the transfection. To linearize the plasmids, approximately 80 µg of plasmid DNA was digested with 15 U AhdI restriction endonuclease (New England Biolabs, Frankfurt am Main, Germany) for 3 hours at 37°C. The digested plasmids were purified through an agarose gel using the QIAquick Gel Extraction Kit from Qiagen. The obtained DNA was quantified spectrophotometrically and used for the transfection.
The HepG2 cells were transfected with 2 µg of reporter gene plasmid and 50 pmol of small interfering RNA (siRNA) per well. For the downregulation of human HNF1A, we used a predesigned Silencer siRNA (Ambion, Life Technologies, Darmstadt, Germany). The sequence of the sense strand of the siRNA was 5′-GGUCUUCACCUCAGACACUtt-3′, and the antisense strand was 5′-AGUGUCUGAGGUGAAGACCtg-3′. This siRNA has been previously shown to efficiently downregulate HNF1A expression as assessed by both mRNA and protein expression (Pelletier et al., 2011). As a control we used nontargeting negative control siRNA #1 (Ambion, Life Technologies). The transfection was performed with 10 µl of Lipofectamine 2000 (Life Technologies) in antibiotic-free medium.
The Huh7 cells were cotransfected with a mixture of 0.9 µg of HNF1 overexpression plasmid and 0.3 µg of reporter plasmid using 3.6 µl of Lipofectamine 2000 (Life Technologies) in antibiotic-free medium. Cells were cotransfected with 9 ng of pRL-cytomegalovirus Renilla luciferase control vector (Promega).
The activity of the firefly and Renilla reniformis luciferase reporter genes was measured with the Dual-Luciferase Reporter 1000 Assay System (Promega) according to the manufacturer’s instructions. In brief, 48 hours after cell transfection, the culture medium was removed and the cells were washed with phosphate-buffered saline. The cells were lysed by a 15-minute incubation in 1× Passive Lysis Buffer (Promega) and then snap-frozen in liquid nitrogen and thawed three times. Cell debris was removed by centrifugation for 5 minutes at 16,000g. The luciferase activity of the supernatant was then directly measured with a GloMax 96 Microplate Luminometer (Promega) using a 10-second integration time. The ratio of firefly to Renilla luciferase signal was calculated for each sample and related to the same ratio from cells transfected with the empty pGL3 basic or pGL3 promoter values to obtain the relative luciferase activity.
Western Blot.
Approximately 2 ×105 cells were lysed in 100 µl of Tris-HCl buffer, pH 7.4, containing 150 mM NaCl, 1 mM EDTA, 1% (v/v) Nonidet P-40, 0.1% (w/v) SDS, 0.25% (w/v) sodium deoxycholate, and 2 mM phenylmethanesulfonyl fluoride. Cell debris was removed by centrifugation (9600g, 10 minutes, 4°C), and the supernatant, which contained the total cellular protein, was used. Ten micrograms of protein were separated on a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (pore size of 0.45 µm; Merck Millipore, Schwalbach, Germany). The membrane was blocked for 1 hour at room temperature with Roti-Block (Carl Roth, Karlsruhe, Germany) and incubated overnight at 4°C with primary antibody diluted in Tris-buffered saline/Tween 20 (TBST) buffer (20 mM Tris, 150 mM NaCl, plus 0.5% Tween 20) containing 5% (w/v) milk powder. We used polyclonal antibodies against HNF1α or HNF1β diluted 1:200 (sc-6547 and sc-7411, respectively; both from Santa Cruz Biotechnology) and a monoclonal antibody against glyceraldehyde-3-phosphate dehydrogenase diluted 1:50,000 (RGM2-6C5; Zytomed Systems, Berlin, Germany). The membrane was washed three times for 5 minutes with TBST buffer. It was incubated for 1 hour with secondary antibodies diluted 1:10,000 in TBST containing 0.5% (w/v) milk powder and washed for 5 minutes in 20 mM Tris supplemented with 150 mM NaCl. We used rabbit-anti-goat (Abcam, Cambridge, UK) or rabbit-anti-mouse (Dianova, Hamburg, Germany) secondary antibodies coupled to horseradish peroxide. The signal was developed using SuperSignal West Pico Kit (Thermo Scientific, Bonn, Germany), according to the manufacturer’s instructions, and detected with the VersaDoc scanner (Bio-Rad, München, Germany). HNF1α migrated at ~78 kDa, and HNF1β migrated at ~65 kDa.
Human Liver Samples.
Forty human liver samples from 10 female and 30 male healthy Caucasian donors were analyzed in the study. The average age of the donors was 37 years (range 2 to 72 years). The human liver samples were provided by the Liver Tissue Procurement and Distribution System and by the Cooperative Human Tissue Network. The origin of the human liver tissue samples and the isolation of DNA and RNA have previously been described in detail (Ramirez et al., 2008). The research on the human liver tissue samples was deemed exempt from ethical review by the Institutional Review Board of the University of Chicago.
Quantitative Reverse Transcription-PCR and Genotyping.
RNA was reverse-transcribed using SuperScript II Reverse Transcriptase (Life Technologies) according to the manufacturer’s instructions. In brief, 0.5 µg of total RNA was incubated at 70°C for 10 minutes in the presence of 0.1 A260 U random hexanucleotide primers (Roche). The samples were allowed to cool to room temperature. Fifty units of reverse transcriptase, 20 units of Recombinant Human Placenta RNase Inhibitor (Affymetrix, High Wycombe, UK), dNTPs to the final concentration of 50 µM, and DTT to the final concentration of 50 µM were added, and the samples were incubated for 1 hour at 42°C. The resulting cDNA was diluted 1:5, and 3 µl (corresponding to 15 ng of the starting RNA) were used for quantitative RT-PCR measurements.
The expression of OCT1, HNF1α, and HNF4 was quantified using predeveloped TaqMan gene expression assays (Life Technologies) according to the manufacturer’s instructions. The OCT1 assay was targeted against the junction of exons 6 and 7 and therefore should detect both the major and the alternative OCT1 transcript variants that were described by Hayer et al. (1999). The expression of the analyzed genes of interest was normalized to the expression of the housekeeping gene TATA-box binding protein (TBP). TBP expression was measured using a predeveloped TaqMan gene expression assay (Life Technologies). The measurements were performed using the 7900HT Fast Real-Time PCR System (Life Technologies).
OCT1 polymorphisms were genotyped using the singe-base primer extension reaction as described previously (Tzvetkov et al., 2009). HNF1A polymorphisms were genotyped using a multiplex single-base primer extension assay. The assay is described in detail in Supplemental Methods and Supplementary Table 1.
Statistical Analyses.
Analyses of variance (ANOVA) followed by post hoc analyses applying Tukey’s honestly significant difference (HSD) were used to compare differences in reporter gene activity. Spearman’s correlation was used to analyze the correlation between OCT1 and HNF1A mRNA expression. Multifactorial effects on OCT1 expression were analyzed using a linear regression model including the logarithm of OCT1 mRNA expression as a dependent variable and the following independent variables: sex; age; HNF1α and HNF4 mRNA expression; the genotypes of the loss-of-function OCT1 polymorphisms Arg61Cys (rs12208357), Cys88Arg (rs55918055), Gly401Ser (rs34130495), Gly465Arg (rs34130495), and Met408Val (rs628031), and a deletion of Met420 (rs72552763). The logarithmic transformation was performed to achieve a normal distribution of OCT1 expression. The effects of HNF1A polymorphisms on OCT1 expression were analyzed using a nonparametric Jonckheere-Terpstra test for comparing three genotype groups and the Mann–Whitney U test for comparing two genotype groups. All statistical analyses were performed with IBM SPSS Statistics version 20.0 (IBM Corporation, Ehningen, Germany).
Results
Evolutionary Conservation of the OCT1 Gene.
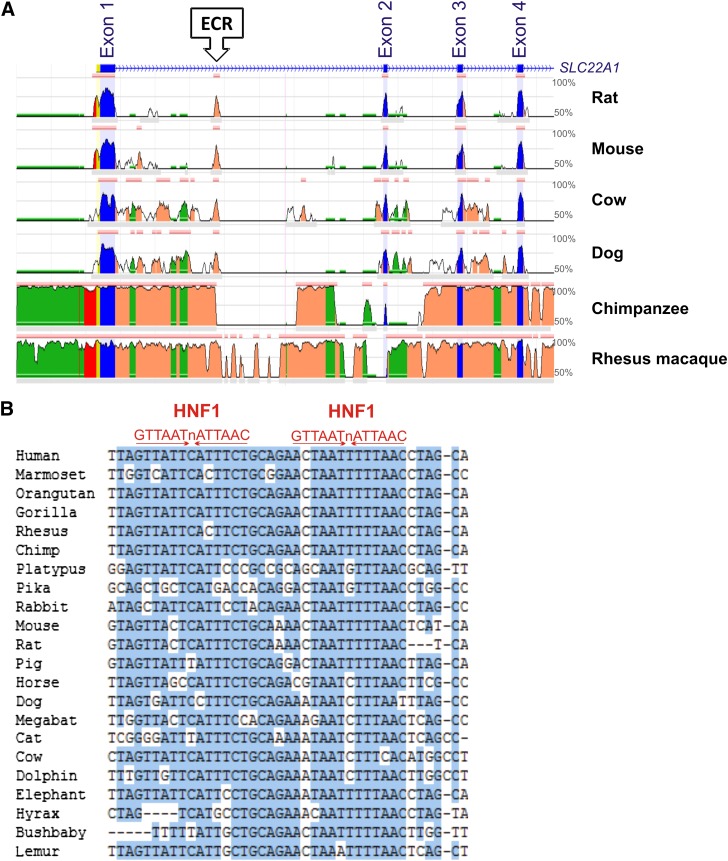
We analyzed evolutionary conservation in the OCT1 locus to identify regions of potential importance for the transcriptional regulation of OCT1. A 185-bp evolutionary conserved region was identified in intron 1 of the gene starting 3362 bp downstream of the translational start site (Fig. 1A). The ECR was highly conserved among the mammals, including human, chimpanzee, rhesus macaque, dog, cow, mouse, and rat. The conservation between humans and any of these species was greater than 70%. No evolutionary conserved regions were identified in the 2-kb promoter region.
Fig. 1.
In silico analyses of evolutionary conservation suggest binding of HNF1 to an evolutionary conserved region in intron 1 of the OCT1 gene. (A) Evolutionary conservation of the organic cation transporter OCT1 gene in mammals visualized using ECR Browser (http://ecrbrowser.dcode.org/). A 15.5-kb region of the OCT1 gene is shown. The region includes the promoter, the first four exons, and three introns. The X-axis represents genomic coordinates. The Y-axis represents the degree of conservation between the human genome (as a reference) and the corresponding mammalian genome. The conserved exon sequences are depicted in blue, the promoter sequences in red, and the intron sequences in orange. The repeated element sequences are depicted in green. The highly evolutionary conserved region in intron 1 is marked with an arrow. (B) Alignment of the highly conserved 40-bp sequence within the intron 1 ECR in 22 mammalian genomes. Conserved bases are highlighted in blue. The consensus binding sequences of HNF1 (shown in red) are given to indicate the two potential HNF1 binding sites.
Next, we analyzed the conservation of the intron 1 ECR in 46 vertebrate genomes. A 40-bp sequence within the intron 1 ECR was highly conserved in 22 mammalian genomes (Fig. 1B). This 40-bp sequence was not conserved in nine mammals (opossum, tree shrew, squirrel, alpaca, microbat, hedgehog, tenrec, armadillo, and sloth) or in the nonmammalian species analyzed. The highly conserved 40-bp region harbored two potential binding sites for the hepatocyte nuclear factor 1. Therefore, we hypothesized that the intron 1 ECR might play a role in the transcriptional regulation of OCT1 by recruiting HNF1, a transcriptional regulatory factor that is predominantly expressed in the liver.
HNF1 Specifically Binds to the ECR in OCT1 Intron 1.
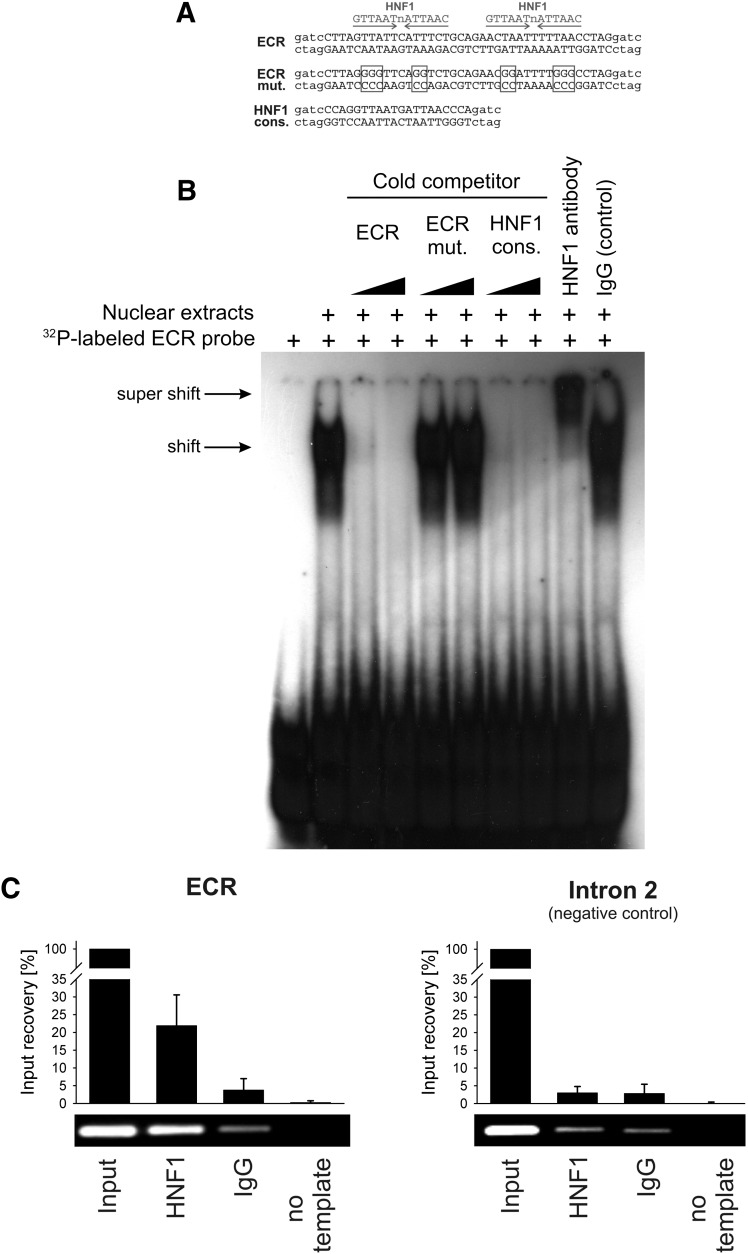
First, we investigated the ability of the transcriptional factor HNF1 to bind the intron 1 ECR in vitro. We performed electrophoretic mobility shift assays using nuclear extracts from HepG2 cells. A clear retention signal was observed for the radiolabeled probe containing the highly conserved 40-bp sequence of the intron 1 ECR (Fig. 2B). The signal was decreased or eliminated by cold competition with the nonlabeled probe, demonstrating binding specificity.
Fig. 2.
Electrophoretic mobility shift and chromatin immunoprecipitation assays demonstrating binding of HNF1 to the evolutionary conserved region in intron 1 of the OCT1 gene. (A) Sequences of the probes used in the electrophoretic mobility shift assay (EMSA). The specific sequences are given in upper case, and the unspecific sequences used in the radioactive labeling of the EMSA probes are given in lower case letters. Positions mutated to disrupt the HNF1 binding sites are indicated by boxes. (B) The 32P-labeled probe containing the 40-bp highly conserved sequence from the intron 1 ECR was incubated with nuclear extracts from HepG2 cells in the absence or presence of unlabeled probes (cold competition) or antibodies (supershift). The unlabeled probes were given in 3- and 30-fold molar excess of the 32P-labeled probe. (C) Chromatin immunoprecipitation assay (ChIP) of isolated human hepatocytes. A representative agarose gel and a real-time PCR-based signal quantification are shown. The intron 1 ECR and a negative control region in intron 2 of the OCT1 gene were analyzed. The quantification results are based on three independent ChIP experiments quantified in duplicate and are shown as means and standard deviations. Identical HNF1-specific and control IgG antibodies were used in the electrophoretic mobility shift and chromatin immunoprecipitation assays. Detailed information about the antibodies used is available in the text.
We next tested whether HNF1 is the nuclear protein that binds to the intron 1 ECR by performing cold competition and supershift assays (Fig. 2B). In the cold competition assay, no competition was observed when the two potential HNF1 binding sites of the ECR probe were mutated, and strong competition was observed when a probe representing the HNF1 consensus binding sequence was used. When the nuclear extracts were preincubated with an antibody against HNF1, the retarded signal was additionally shifted (a so-called supershift). The control reaction with IgG showed no additional shift in the signal.
Finally, we analyzed the binding of HNF1 to the intron 1 ECR in vivo. We performed chromatin immunoprecipitation in human hepatocytes and observed a clear immunoprecipitation signal for the intron 1 ECR (Fig. 2C). The signal was absent if an IgG control was used instead of the HNF1 antibody, or if another region located in intron 2 of the OCT1 gene was analyzed as a negative control. Taken together, these experiments demonstrate that HNF1 binds to the intron 1 ECR of the OCT1 gene.
The Intron 1 ECR Enhances OCT1 Promoter Activity in an HNF1-Dependent Manner.
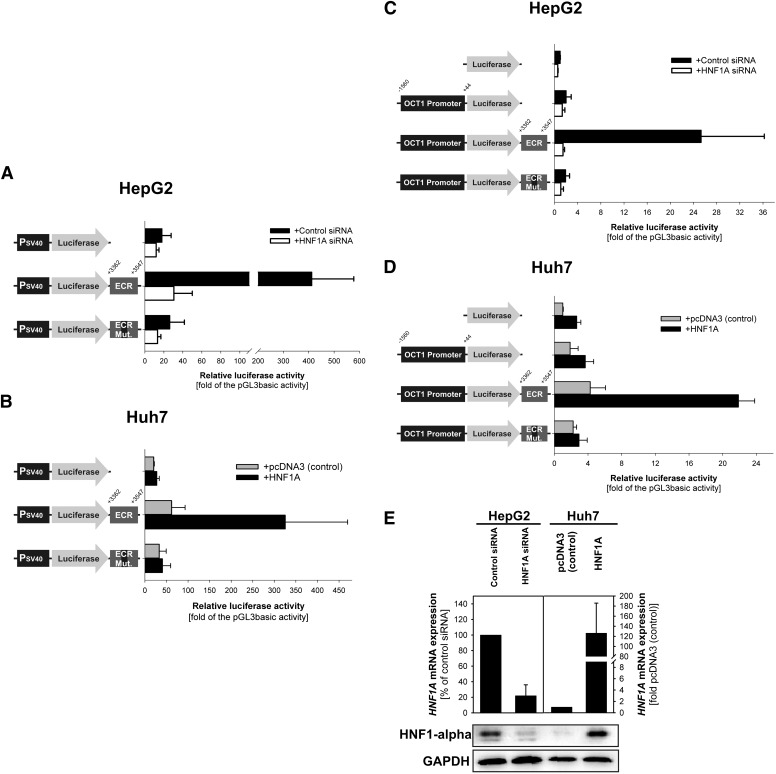
We analyzed whether the binding of HNF1 to the intron 1 ECR affects the transcriptional regulation of OCT1 using luciferase reporter gene assays. The reporter gene assays were performed in the model hepatocarcinoma cell lines HepG2 and Huh7. We used luciferase reporter gene constructs carrying or lacking the intron 1 ECR downstream of the reporter gene. First, we analyzed the effect of the ECR on the promoter activity of the well-defined SV40 promoter. The intron 1 ECR increased the SV40 promoter activity 22-fold in HepG2 cells (P < 0.001, ANOVA F test; P < 0.001, post-hoc Tukey-HSD test for comparison between the promoter activity of SV40 alone or with the intron 1 ECR; Fig. 3A). This increase was absent when the HNF1 binding sites of the ECR were mutated (the same bases were mutated as in the mobility shift experiments; Fig. 2A), or the endogenous HNF1α expression was downregulated with siRNA. The siRNA was specific for HNF1α: HNF1A expression was strongly downregulated (Fig. 3E), whereas HNF1B expression showed a slight but insignificant increase in expression (125 ± 22% of the HNF1B expression in the control transfected cells; data not shown).
Fig. 3.
Effects of HNF1 binding to the intron 1 ECR on the promoter activity of the OCT1 gene. Luciferase reporter gene assay were performed in the model hepatocellular carcinoma cell lines HepG2 (A and C) and Huh7 (B and D). The luciferase gene was cloned under the control of the Simian virus 40 promoter (PSV40) (A and B) or the 1.6-kb promoter fragment of the OCT1 gene (C and D). The constructs included an intact or mutated intron 1 ECR, or no ECR, cloned downstream of the luciferase gene. The coordinates are given in base pairs related to the distance to the transcriptional start site of OCT1. In HepG2 cells, the endogenous HNF1A expression was additionally downregulated using siRNA (white bars) (A and C). In Huh7 cells, HNF1A was additionally overexpressed by cotransfecting the overexpression plasmid pcDNA3::HNF1A with the reporter gene constructs (gray bars) (B and D). Shown are means and standard deviations of at least four independent experiments. (E) Effective downregulation and the overexpression of HNF1A were confirmed at the mRNA level by RT-qPCR (upper part) and at the protein level by Western blot (lower part). The RT-qPCR data represent mean and standard deviation of three independent experiments. The Western blot picture is a single representative experiment.
As with HepG2 cells, the intron 1 ECR increased the SV40 promoter activity 11-fold in Huh7 cells overexpressing HNF1α (P < 0.001, ANOVA F test; P < 0.001, post-hoc Tukey-HSD test for comparison between the promoter activity of SV40 alone or with the intron 1 ECR; Fig. 3B). This increase was absent when the HNF1 binding sites were mutated. In the absence of HNF1 overexpression, the ECR did not significantly increase the SV40 promoter activity in Huh7 cells. These experiments demonstrated that the intron 1 ECR is an enhancer and that its enhancer activity depends on HNF1.
We next investigated whether the intron 1 ECR could also enhance the activity of the original OCT1 promoter. To this end, we performed reporter gene assays using constructs in which the luciferase reporter was under the control of the 1604-bp promoter region of the OCT1 gene. In HepG2 cells, the OCT1 core promoter led only to a nonsignificant 2-fold increase of luciferase expression over the empty pGL3-basic vector control. Importantly, the presence of the ECR fragment downstream of the luciferase gene increased the promoter activity by an additional 13-fold (P < 0.001, ANOVA F test and P < 0.001, post-hoc Tukey-HSD test for comparison between the activity of the OCT1 promoter alone or with the intron 1 ECR; Fig. 3C). This increase was absent when the HNF1 binding sites were mutated or the endogenous HNF1α expression was downregulated using siRNA. Similarly, in HNF1α-overexpressing Huh7 cells, the intron 1 ECR increased the OCT1 promoter activity an additional 6-fold (P < 0.001, ANOVA F test; P < 0.001, post-hoc Tukey-HSD test for comparison between the activity of the OCT1 promoter alone or with the intron 1 ECR; Fig. 3B). This increase was absent when the HNF1 binding sites in intron 1 ECR were mutated, and was limited to 2.3-fold when HNF1A was not overexpressed (P = 0.02, P < 0.001, ANOVA F test; P < 0.001, post-hoc Tukey-HSD test for comparison between the activity of the OCT1 promoter alone or with the intron 1 ECR). From these results we concluded that the intron 1 ECR is an enhancer of the OCT1 promoter and that its enhancer activity depends on HNF1.
Correlation between HNF1α and OCT1 Expression in the Human Liver.
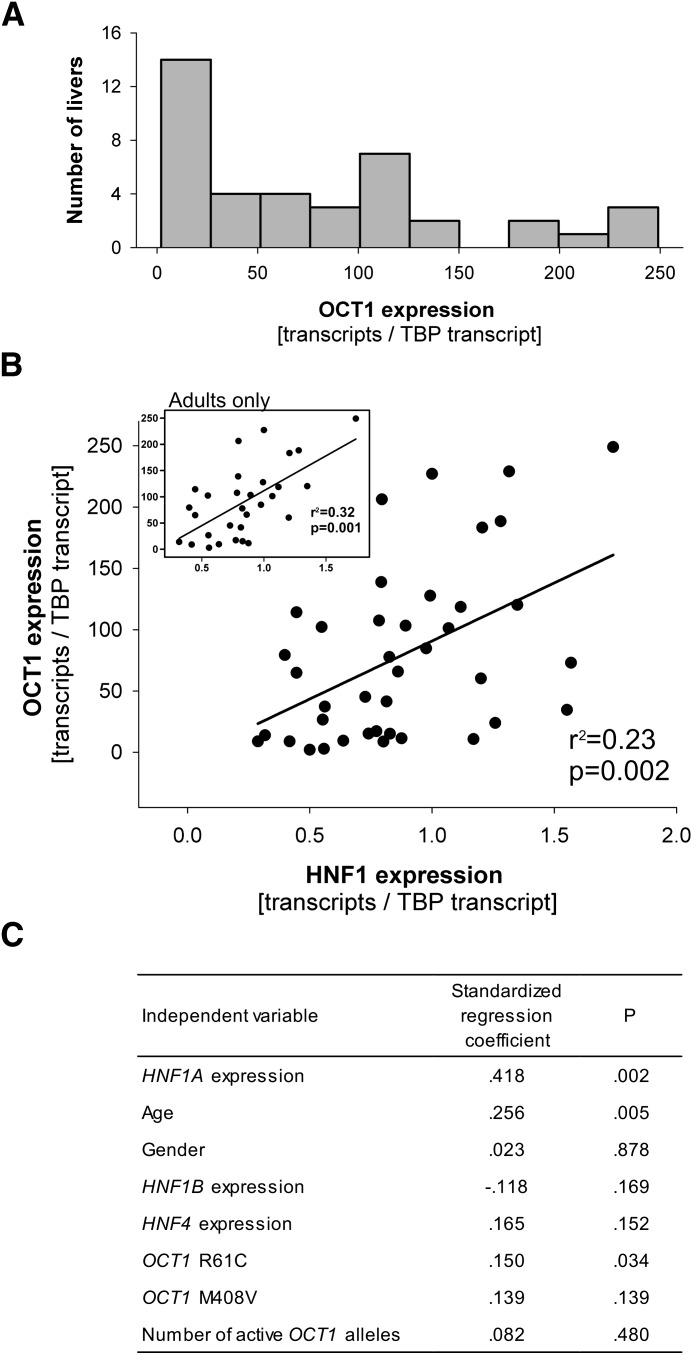
To verify whether the described effects of HNF1 on OCT1 expression play a role in vivo, we analyzed the expression of both HNF1A and OCT1 in 40 human liver samples. OCT1 expression varied 125-fold among the liver samples. The range was 2 to 249 transcripts per TPB transcript (median of 65 transcripts per TBP transcript, 25th and 75th percentile of 15.2 and 117.5 transcripts per TPB transcript, respectively; Fig. 4A). Higher OCT1 expression was correlated with higher HNF1A expression (r = 0.48, P = 0.002; Fig. 4B). This suggests that differences in HNF1A expression accounted for 23% of the variations in OCT1 expression. To rule out possible artifacts due to TBP normalization, we performed correlation analyses of the nonnormalized OCT1 and HNF1A expression. Without normalization to TBP, higher OCT1 expression was still correlated with higher HNF1A expression (r = 0.42, P = 0.007).
Fig. 4.
Variability of OCT1 expression and correlation between OCT1 and HNF1 expression in the human liver. (A) Histogram illustrating the variability in OCT1 expression in 40 human liver samples. (B) Correlation between OCT1 and HNF1A expression in the human liver, based on analysis of 40 samples. The calculated coefficient of determination (r2) and the significance of the correlation (P) are shown. The inset represents the analyses of the subgroup of adult liver tissue donors only (age 19 years or above, n = 30). (C) Multiple linear regression analyses showing the dependence of OCT1 expression on HNF1A expression after adjustment for sex, relevant OCT1 genotypes, and HNF4 expression. Number of active OCT1 alleles is a compound genotype accounting for the loss-of-function amino acid substitutions Arg61Cys (rs12208357), Cys88Arg (rs55918055), Gly401Ser (rs34130495), Gly465Arg (rs34130495), and a deletion of Met420 (rs72552763). We regarded OCT1 alleles carrying any of these polymorphisms as inactive.
We used a multivariate model to assess the combined effects of age and sex of the liver donors, expression of known transcriptional regulatory factors, and genetic polymorphisms in OCT1 on OCT1 expression. We analyzed the following OCT1 polymorphisms: Arg61Cys and Met408Val, which have been reported to affect OCT1 expression (Nies et al., 2009); and Cys88Arg, Gly401Ser, Gly465Arg, and a deletion of Met420, which are known to reduce OCT1 activity and therefore may cause feedback upregulation of OCT1 expression (Shu et al., 2003; Chen et al., 2010; Tzvetkov et al., 2010). HNF1A expression remained the strongest predictor of the variations in OCT1 expression (Fig. 4C). Age and the Arg61Cys polymorphism in OCT1, but not sex, HNF1B expression, HNF4 expression, or other OCT1 polymorphisms were also significantly associated with OCT1 expression. OCT1 expression was significantly lower in the donors 18 years of age or younger (n = 10) than in the adults (n = 30, median expressions of 19.6 and 82.0 transcripts per TBP transcript, respectively, P = 0.02). When analyzed in adults alone, higher OCT1 expression was even more strongly correlated with higher HNF1A expression (r = 0.56, P = 0.001; Fig. 4B).
Five single nucleotide polymorphisms (SNPs) in the HNF1A gene have been reported to associate with the expression levels of HNF1 target genes: two amino acid substitutions (isoleucine-to-leucine in codon 27 and serine-to-asparagine in codon 487), two intronic SNPs (rs1183910 and rs7310409), and an SNP located downstream of the HNF1A gene (rs1169313) (Reiner et al., 2008; Yuan et al., 2008; Dehghan et al., 2011). We analyzed these SNPs in our set of liver samples (Supplemental Methods). Because of the small number of samples, statistical evaluation of the SNP effects on OCT1 expression was not possible. Still, it is interesting to note that homozygous carriers of aspargine 487, and the strongly linked leucine 27, expressed less OCT1 than the isoleucine 27-serine 487 carriers (mean ± S.D. 16 ± 10 compared with 82 ± 71 OCT1 transcripts per TBP transcript, respectively; Supplemental Table 2).
The HNF1α and HNF1β Isoforms Both Interact with the Intron 1 ECR and Enhance OCT1 Promoter Activity.
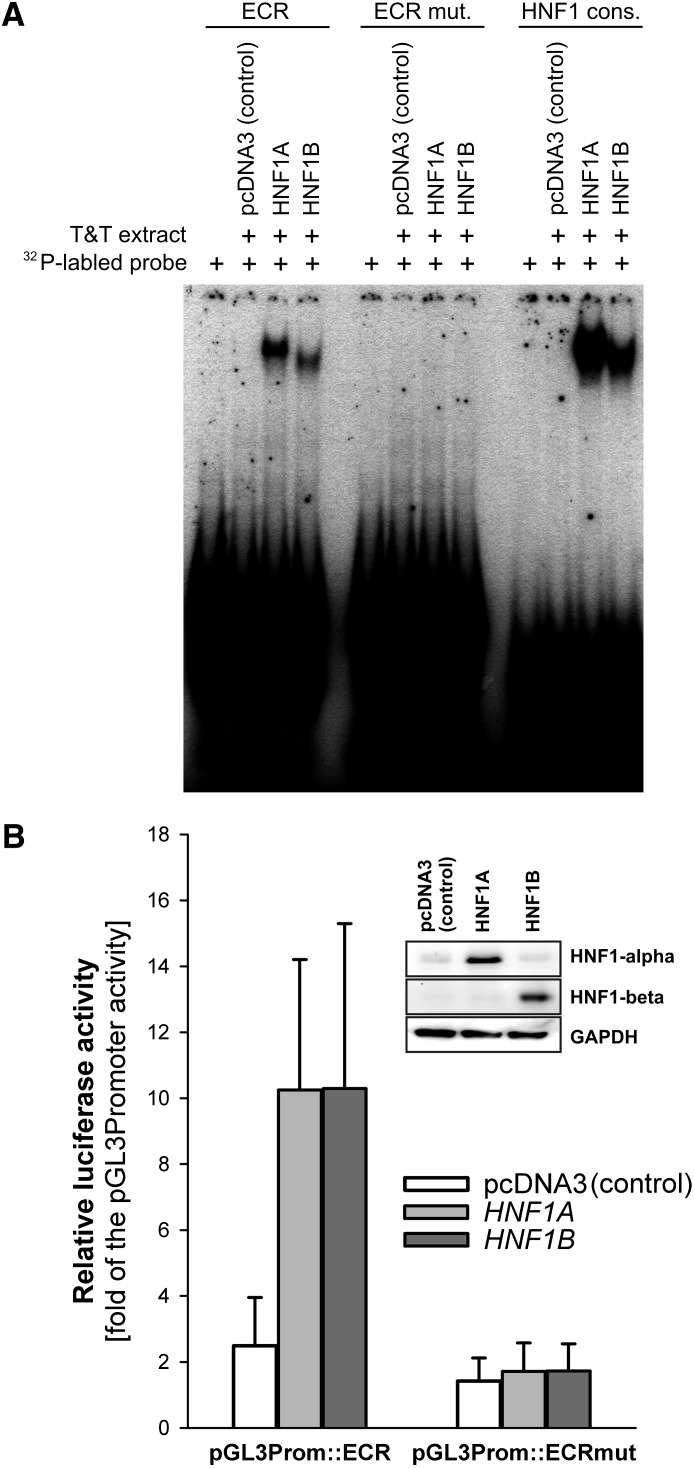
We analyzed whether both α and β isoforms of HNF1 may bind to the intron 1 ECR and enhance the OCT1 promoter activity. First, we expressed the α and β isoforms using an in vitro transcription-translation system. The in vitro expressed proteins were used in an electrophoretic mobility shift assay experiment. Both α and β isoforms bound to the ECR probe (Fig. 5A). The retention signal was similar to those obtained using a consensus recognition sequence for HNF1. No binding was observed when the HNF1 recognition sequences in the ECR probe were mutated.
Fig. 5.
Comparison of the ability of HNF1α and HNF1β isoforms to interact with the intron 1 ECR and to regulate OCT1 promoter activity. (A) Electrophoretic mobility shift assays of in vitro transcribed and translated HNF1α and β. pcDNA3 expression vectors carrying the HNF1A or HNF1B genes, or the empty pcDNA3 vector as a control, were in vitro transcribed and translated using the TNT T7 Quick Coupled Transcription/Translation System (Promega). The in vitro extracts were incubated with 32P-labeled probes containing the 40-bp conserved region from the ECR (ECR), the conserved region with both HNF1 binding sites mutated (ECR mut.), or a consensus binding sequence for HNF1 (HNF1 cons.) (Jain et al., 2007). (B) Luciferase reporter gene assay of Huh7 cells overexpressing the HNF1α or HNF1β isoforms. The overexpression vectors were cotransfected with pGL3 promoter vectors carrying the luciferase reporter gene under the control of the Simian virus 40 promoter and the intact or mutated intron 1 ECR (pGL3Prom::ECR and pGL3Prom::ECR mut., respectively). Shown are means and standard deviation of at least three independent experiments. The overexpression of HNF1α and HNF1β was confirmed by Western blot.
Next, we tested whether the binding of each of the isoforms increased the promoter activity caused by the intron 1 ECR enhancer. To do so, we cotransfected vectors overexpressing either the α or β isoforms, together with SV40 promoter reporter constructs containing the intron 1 ECR. The presence of each of the isoforms increased the reporter gene activity by more than 4-fold in Huh7 cells (Fig. 5B). No increase was observed when the HNF1 recognition sequences in the intron 1 ECR were mutated. From these experiments we concluded that the both the HNF1α and HNF1β isoforms can interact with intron 1 ECR and enhance OCT1 promoter activity.
Discussion
This study shows that the hepatocyte nuclear factor HNF1 is a transcriptional regulator of the organic cation transporter OCT1. By use of the electrophoretic mobility shift, chromatin immunoprecipitation, and reporter gene assays, we demonstrated that HNF1 binds to a highly evolutionary conserved region in intron 1 of the OCT1 gene and that this binding results in a strong enhancement of OCT1 promoter activity. These findings were additionally supported by a significant correlation between HNF1α and OCT1 expression levels in human liver samples.
Our data suggest that the effects of HNF1 on OCT1 expression are stronger than the effects of USF and HNF4, the two transcription factors that were previously known to regulate OCT1 expression (Saborowski et al., 2006; Kajiwara et al., 2008). In vitro, the binding of HNF1 to the intron 1 ECR caused a much larger increase in the luciferase reporter gene activity than the increase caused by the “canonical” OCT1 promoter, which contains the USF and HNF4 binding sites (Fig. 3). In human liver samples, the variations in OCT1 expression were associated with variations in HNF1α, but not with variations in HNF4 expression (Fig. 4).
HepG2 and Huh7 cells lack endogenous OCT1 expression. This is in concordance with the known strong downregulation of OCT1 expression in hepatocellular carcinoma (Schaeffeler et al., 2011; Heise et al., 2012). We demonstrated that HNF1 may regulate promoter activity in HepG2 and Huh7 cells (Figs. 3 and 5). However, the endogenous OCT1 mRNA expression remained low (below 0.1 transcripts per TBP transcript) and did not change after overexpression of HNF1A or HNF1B in Huh7 cells or after HNF1A downregulation in HepG2 cells (data not shown). This suggests an HNF1-independent mechanism of downregulation of OCT1 expression in hepatocellular carcinoma. Still, HNF1 seems to be a relevant regulator of OCT1 expression in the healthy liver, as suggested by the significant correlation between the HNF1A and OCT1 expression in the analyzed human liver samples (Fig. 4).
One potential implication of our findings is that genetic variations in HNF1α may change OCT1 expression and, as a consequence, affect the liver uptake and metabolism of drug substrates of OCT1. Common genetic variants in HNF1A have been reported to affect the expression of a number of HNF1-controlled genes in the liver. In multiple genome-wide association studies, the variants rs7310409, rs2464196, rs1169288, rs1169313, and rs1183910 were associated with changes in the plasma concentrations of liver-expressed C-reactive protein and γ-glutamyltransferase (Reiner et al., 2008; Yuan et al., 2008; Chambers et al., 2011; Dehghan et al., 2011). Although rs2464196 and the highly genetically linked variants showed a trend of reduced OCT1 expression in our study, the reduction was not significant and requires further validation.
Rare mutations in HNF1A are known as the major cause of maturity-onset diabetes of the young type 3 (MODY 3) (Ryffel, 2001). Carriers of these mutations have substantially reduced HNF1α activity, and, based on the results of our study, are thus expected to have a subsequent substantial reduction in OCT1 expression. Reduced OCT1 activity would not be expected to affect the standard drug therapy for MODY 3; these patients are typically treated with sulfonylurea derivatives, which are not known to interact with OCT1 (Ahlin et al., 2008), rather than metformin, which is a well-known OCT1 substrate. However, the potential reduced OCT1 expression in patients with MODY 3 should be considered when developing new therapeutics.
Nongenetic factors also cause changes in HNF1 activity and thus may affect OCT1 expression. Cholestasis was reported to cause a significant decrease in OCT1 expression (Denk et al., 2004; Nies et al., 2009). The decrease was explained by an HNF4-mediated decrease of OCT1 expression induced by the increased bile acid concentration (Saborowski et al., 2006). However, HNF1 is known as the key mediator of cholestatic effects on gene expression in the liver (Jung and Kullak-Ublick, 2003). The major signaling pathways regulated by bile acids, the FXR-SHP pathway, the HNF4 pathway, and the NF-κB pathway, are all known to execute their effects through HNF1. Therefore, based on the direct regulation of OCT1 transcription by HNF1 demonstrated in this study, a substantial contribution of HNF1 to the cholestatic down-regulation of OCT1 expression may be suggested.
The involvement of HNF1 in the regulation of OCT1 reported here may also help predict drug-drug interactions. For example, administering ursodeoxycholic acid (ursofalk), a ursodeoxicholic bile acid used to treat gallstones, may lead to an HNF1-mediated reduction in OCT1 expression because of HNF1’s role in bile acid–induced hepatic gene regulation. Future studies should systematically test whether ursofalk reduces the metabolism of tropisetron, ondansetron, or tramadol, or reduces the efficacy of metformin. A similar interaction has already been demonstrated between ursofalk and rosuvastatin (He et al., 2008). Rosuvastatin is taken up into the liver by OATP1B1, a well-known target of HNF1 regulation. The coadministration of ursofalk resulted in a significant reduction of the oral clearance of rosuvastatin; this result was attributed to HNF1-mediated inhibition of OATP1B1 expression.
HNF1 is a key regulator of drug transport and metabolism in the human liver. Beyond its effects on OCT1 expression suggested here, HNF1 is known to control the expression of the liver uptake transporters OATP1B1, OAT5, and OAT7; the phase II–metabolizing enzymes UGT1A1, 1A3, 1A4, 1A8, 1A9, 1A10, and 2B7; and the efflux transporter MRP2 (Bernard et al., 1999; Ishii et al., 2000; Jung et al., 2001; Gregory et al., 2004; Qadri et al., 2006; Furihata et al., 2007; Gardner-Stephen and Mackenzie, 2007; Klein et al., 2010). Therefore, variations in HNF1 expression or activity may have multiple effects on drug uptake, metabolism, and elimination in the liver. For example, O-desmethyltramadol, the active metabolite of tramadol, depends on OCT1 for its hepatic uptake (Tzvetkov et al., 2011) and UGT2B7 for its metabolism (Lehtonen et al., 2010). Both OCT1 and UGT2B7 are regulated by HNF1A. Therefore variations in HNF1α expression or activity may have complex effects on the hepatic elimination of O-desmethyltramadol and other drugs.
The expression of Oct1 in the liver is unchanged in the Hnf1α knockout mouse, which is a clear contradiction of our findings (Maher et al., 2006). Maher et al. (2006) reported significant decreases in Oat2, Oatp1a1, and Oatp1b2, but not in Oct1 expression in the livers of Hnf1α knockout mice. One explanation for the contradiction between Maher’s and our results may be the ability of the Hnf1β isoform to compensate for the lack of Hnf1α. The expression of Hnf1β is upregulated in the Hnf1α knockout mouse (Pontoglio et al., 1996). Here we demonstrated that the β isoform may also bind to the intron 1 ECR and that this binding results in an enhancement of OCT1 promoter activity similar to that observed in the case of the α isoform binding (Fig. 5). Alternatively, the apparent lack of change in hepatic Oct1 expression in Hnf1α knockout mice may be due to species-specific differences in transcriptional regulation. Our study is focused on the effects in humans and clearly shows the involvement of the human HNF1α in the expression of OCT1 in the human liver both in vitro and in vivo. In contrast, OCT1 expression was not correlated with HNF1β expression in the human liver. Likewise, ECR enhancer activity strongly decreased after downregulation of HNF1A in HepG2 cells, even though HNF1B expression remained unchanged. This suggests that HNF1β may play only a limited role in the regulation of OCT1 expression when active HNF1α is present. Interestingly, Oct1 expression in the duodenum was significantly decreased in Hnf1α knockout mice (Maher et al., 2006), suggesting effects of Hnf1 on extrahepatic Oct1 expression. However, OCT1 extrahepatic expression is very low in humans, and we have not analyzed OCT1 expression in the human duodenum.
Extremely high interindividual variability in the expression of OCT1 was observed in the human liver. The expression of OCT1 varied 125-fold among the liver samples in our study (Fig. 4A). This finding is in complete agreement with the previously reported 113-fold variation in OCT1 expression in the human liver (Nies et al., 2009). Variations in HNF1A expression explained only 23% of the variation in OCT1 expression in our sample (Fig. 4B). Further factors affecting OCT1 expression in our sample were the age of the patient and the genotype of the OCT1 Arg61Cys polymorphism. In the study of Nies et al. (2009), the Arg61Cys variant was also associated with OCT1 expression, but age was not associated and HNF1A expression was not analyzed. The observed discrepant effects of age may be due to differences in the age distribution between the two populations analyzed. While in our study a substantial portion of the liver samples (25%) were obtained from donors younger than 20 years old, in the study of Nies et al. (2009), only two donors (1%) were in this age group. As known from analyses of expression of other pharmacologically relevant genes in the liver and from studying OCT1 expression in mice, the expression may strongly depend on age, with substantial differences reported between adults and younger individuals (Pavlova et al., 2000; Hines, 2008).
Variation in HNF1A expression, age, and OCT1 polymorphisms explained only part of the variability in the OCT1 expression in our study. The remaining variability may be due to variation in the DNA methylation of OCT1 (Schaeffeler et al., 2011), cholestasis (Nies et al., 2009), or amino acid variants affecting the HNF1 activity (Supplemental Table 2). Therefore the high variability in OCT1 expression should be regarded as a multifactorial trait that is controlled by genetic, epigenetic, and nongenetic factors.
In conclusion, HNF1 regulates the expression of OCT1 in humans. The observed strong variation in OCT1 expression in the human liver may be partially explained by variations in HNF1α expression. It remains to be clarified to what extent genetic variations, drugs, and disease conditions that change HNF1 activity may affect the expression of OCT1, and to what extent this may result in changes in the pharmacokinetics or efficacy of drugs taken up in the liver by OCT1.
Supplementary Material
Acknowledgments
The authors thank Alexander Zietlow for contribution to the initial parts of the project and Snezana Mirkov for assistance with preparation of liver samples.
Abbreviations
- ANOVA
analysis of variance
- ECR
evolutionary conserved region
- HNF
hepatocyte nuclear factor
- HSD
honestly significant difference
- MODY 3
maturity-onset diabetes of the young type 3
- OCT1
organic cation transporter 1
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
- SLC22A1
solute carrier family 22 member 1
- SNP
single nucleotide polymorphism
- TBP
TATA-box binding protein
- TBST
Tris-buffered saline/Tween 20
- USF
upstream stimulatory factor
Authorship Contributions
Participated in research design: Tzvetkov, O’Brien, Brockmöller.
Conducted experiments: Bokelmann, O’Brien, Jobst, Tzvetkov.
Contributed new reagents or analytic tools: Ramírez, Ratain.
Performed data analysis: Tzvetkov, O’Brien.
Wrote or contributed to the writing of the manuscript: Tzvetkov, O’Brien, Brockmöller.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft [Grant DFG TZ 74/1-1] (to M.V.T. and J.B.); the National Institutes of Health National Institute of General Medical Sciences [Grant U01-GM061393] (to M.J.R.); and the Fulbright U.S. Student Program (to V.P.O.) The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Ahlin G, Karlsson J, Pedersen JM, Gustavsson L, Larsson R, Matsson P, Norinder U, Bergström CA, Artursson P. (2008) Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J Med Chem 51:5932–5942 [DOI] [PubMed] [Google Scholar]
- Bach I, Galcheva-Gargova Z, Mattei MG, Simon-Chazottes D, Guénet JL, Cereghini S, Yaniv M. (1990) Cloning of human hepatic nuclear factor 1 (HNF1) and chromosomal localization of its gene in man and mouse. Genomics 8:155–164 [DOI] [PubMed] [Google Scholar]
- Bazeos A, Marin D, Reid AG, Gerrard G, Milojkovic D, May PC, de Lavallade H, Garland P, Rezvani K, Apperley JF, et al. (2010) hOCT1 transcript levels and single nucleotide polymorphisms as predictive factors for response to imatinib in chronic myeloid leukemia. Leukemia 24:1243–1245 [DOI] [PubMed] [Google Scholar]
- Bernard P, Goudonnet H, Artur Y, Desvergne B, Wahli W. (1999) Activation of the mouse TATA-less and human TATA-containing UDP-glucuronosyltransferase 1A1 promoters by hepatocyte nuclear factor 1. Mol Pharmacol 56:526–536 [DOI] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, et al. Alcohol Genome-wide Association (AlcGen) Consortium. Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study. Genetic Investigation of Anthropometric Traits (GIANT) Consortium. Global Lipids Genetics Consortium. Genetics of Liver Disease (GOLD) Consortium. International Consortium for Blood Pressure (ICBP-GWAS) Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) (2011) Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 43:1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Takizawa M, Chen E, Schlessinger A, Segenthelar J, Choi JH, Sali A, Kubo M, Nakamura S, Iwamoto Y, et al. (2010) Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther 335:42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, Pellikka N, Wallaschofski H, Kettunen J, Henneman P, et al. (2011) Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation 123:731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk GU, Soroka CJ, Mennone A, Koepsell H, Beuers U, Boyer JL. (2004) Down-regulation of the organic cation transporter 1 of rat liver in obstructive cholestasis. Hepatology 39:1382–1389 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Satoh T, Yamamoto N, Kobayashi K, Chiba K. (2007) Hepatocyte nuclear factor 1 alpha is a factor responsible for the interindividual variation of OATP1B1 mRNA levels in adult Japanese livers. Pharm Res 24:2327–2332 [DOI] [PubMed] [Google Scholar]
- Gardner-Stephen DA, Mackenzie PI. (2007) Isolation of the UDP-glucuronosyltransferase 1A3 and 1A4 proximal promoters and characterization of their dependence on the transcription factor hepatocyte nuclear factor 1alpha. Drug Metab Dispos 35:116–120 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Lewinsky RH, Gardner-Stephen DA, Mackenzie PI. (2004) Coordinate regulation of the human UDP-glucuronosyltransferase 1A8, 1A9, and 1A10 genes by hepatocyte nuclear factor 1alpha and the caudal-related homeodomain protein 2. Mol Pharmacol 65:953–963 [DOI] [PubMed] [Google Scholar]
- Hayer M, Bönisch H, Brüss M. (1999) Molecular cloning, functional characterization and genomic organization of four alternatively spliced isoforms of the human organic cation transporter 1 (hOCT1/SLC22A1). Ann Hum Genet 63:473–482 [DOI] [PubMed] [Google Scholar]
- He YJ, Zhang W, Tu JH, Kirchheiner J, Chen Y, Guo D, Li Q, Li ZY, Chen H, Hu DL, et al. (2008) Hepatic nuclear factor 1alpha inhibitor ursodeoxycholic acid influences pharmacokinetics of the organic anion transporting polypeptide 1B1 substrate rosuvastatin and bilirubin. Drug Metab Dispos 36:1453–1456 [DOI] [PubMed] [Google Scholar]
- Heise M, Lautem A, Knapstein J, Schattenberg JM, Hoppe-Lotichius M, Foltys D, Weiler N, Zimmermann A, Schad A, Gründemann D, et al. (2012) Downregulation of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) in human hepatocellular carcinoma and their prognostic significance. BMC Cancer 12:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf C, Ahlin G, Seithel A, Artursson P, Ungell AL, Karlsson J. (2007) Expression of thirty-six drug transporter genes in human intestine, liver, kidney, and organotypic cell lines. Drug Metab Dispos 35:1333–1340 [DOI] [PubMed] [Google Scholar]
- Hines RN. (2008) The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol Ther 118:250–267 [DOI] [PubMed] [Google Scholar]
- Ishii Y, Hansen AJ, Mackenzie PI. (2000) Octamer transcription factor-1 enhances hepatic nuclear factor-1alpha-mediated activation of the human UDP glucuronosyltransferase 2B7 promoter. Mol Pharmacol 57:940–947 [PubMed] [Google Scholar]
- Jain S, Li Y, Patil S, Kumar A. (2007) HNF-1alpha plays an important role in IL-6-induced expression of the human angiotensinogen gene. Am J Physiol Cell Physiol 293:C401–C410 [DOI] [PubMed] [Google Scholar]
- Jonker JW, Wagenaar E, Mol CA, Buitelaar M, Koepsell H, Smit JW, Schinkel AH. (2001) Reduced hepatic uptake and intestinal excretion of organic cations in mice with a targeted disruption of the organic cation transporter 1 (Oct1 [Slc22a1]) gene. Mol Cell Biol 21:5471–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Hagenbuch B, Gresh L, Pontoglio M, Meier PJ, Kullak-Ublick GA. (2001) Characterization of the human OATP-C (SLC21A6) gene promoter and regulation of liver-specific OATP genes by hepatocyte nuclear factor 1 alpha. J Biol Chem 276:37206–37214 [DOI] [PubMed] [Google Scholar]
- Jung D, Kullak-Ublick GA. (2003) Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology 37:622–631 [DOI] [PubMed] [Google Scholar]
- Kajiwara M, Terada T, Asaka J, Aoki M, Katsura T, Ikai I, Inui K. (2008) Regulation of basal core promoter activity of human organic cation transporter 1 (OCT1/SLC22A1). Am J Physiol Gastrointest Liver Physiol 295:G1211–G1216 [DOI] [PubMed] [Google Scholar]
- Klein K, Jüngst C, Mwinyi J, Stieger B, Krempler F, Patsch W, Eloranta JJ, Kullak-Ublick GA. (2010) The human organic anion transporter genes OAT5 and OAT7 are transactivated by hepatocyte nuclear factor-1α (HNF-1α). Mol Pharmacol 78:1079–1087 [DOI] [PubMed] [Google Scholar]
- Koepsell H, Lips K, Volk C. (2007) Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm Res 24:1227–1251 [DOI] [PubMed] [Google Scholar]
- Lehtonen P, Sten T, Aitio O, Kurkela M, Vuorensola K, Finel M, Kostiainen R. (2010) Glucuronidation of racemic O-desmethyltramadol, the active metabolite of tramadol. Eur J Pharm Sci 41:523–530 [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Callaghan TN, Cheng X, Cheung C, Gonzalez FJ, Klaassen CD. (2006) Alterations in transporter expression in liver, kidney, and duodenum after targeted disruption of the transcription factor HNF1alpha. Biochem Pharmacol 72:512–522 [DOI] [PubMed] [Google Scholar]
- Meineke C, Tzvetkov MV, Bokelmann K, Oetjen E, Hirsch-Ernst K, Kaiser R, Brockmöller J. (2008) Functional characterization of a -100_-102delAAG deletion-insertion polymorphism in the promoter region of the HTR3B gene. Pharmacogenet Genomics 18:219–230 [DOI] [PubMed] [Google Scholar]
- Müller J, Lips KS, Metzner L, Neubert RH, Koepsell H, Brandsch M. (2005) Drug specificity and intestinal membrane localization of human organic cation transporters (OCT). Biochem Pharmacol 70:1851–1860 [DOI] [PubMed] [Google Scholar]
- Nies AT, Koepsell H, Damme K, Schwab M. (2011) Organic cation transporters (OCTs, MATEs), in vitro and in vivo evidence for the importance in drug therapy. Handb Exp Pharmacol 201:105–167 [DOI] [PubMed] [Google Scholar]
- Nies AT, Koepsell H, Winter S, Burk O, Klein K, Kerb R, Zanger UM, Keppler D, Schwab M, Schaeffeler E. (2009) Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology 50:1227–1240 [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. (2004) ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32 (Web Server issue):W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A, Sakurai H, Leclercq B, Beier DR, Yu AS, Nigam SK. (2000) Developmentally regulated expression of organic ion transporters NKT (OAT1), OCT1, NLT (OAT2), and Roct. Am J Physiol Renal Physiol 278:F635–F643 [DOI] [PubMed] [Google Scholar]
- Pelletier L, Rebouissou S, Vignjevic D, Bioulac-Sage P, Zucman-Rossi J. (2011) HNF1α inhibition triggers epithelial-mesenchymal transition in human liver cancer cell lines. BMC Cancer 11:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M. (1996) Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84:575–585 [DOI] [PubMed] [Google Scholar]
- Qadri I, Iwahashi M, Kullak-Ublick GA, Simon FR. (2006) Hepatocyte nuclear factor (HNF) 1 and HNF4 mediate hepatic multidrug resistance protein 2 up-regulation during hepatitis C virus gene expression. Mol Pharmacol 70:627–636 [DOI] [PubMed] [Google Scholar]
- Ramírez J, Mirkov S, Zhang W, Chen P, Das S, Liu W, Ratain MJ, Innocenti F. (2008) Hepatocyte nuclear factor-1 alpha is associated with UGT1A1, UGT1A9 and UGT2B7 mRNA expression in human liver. Pharmacogenomics J 8:152–161 [DOI] [PubMed] [Google Scholar]
- Reiner AP, Barber MJ, Guan Y, Ridker PM, Lange LA, Chasman DI, Walston JD, Cooper GM, Jenny NS, Rieder MJ, et al. (2008) Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet 82:1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. (2010) The UCSC Genome Browser database: update 2010. Nucleic Acids Res 38 (Database issue):D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel GU. (2001) Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol 27:11–29 [DOI] [PubMed] [Google Scholar]
- Saborowski M, Kullak-Ublick GA, Eloranta JJ. (2006) The human organic cation transporter-1 gene is transactivated by hepatocyte nuclear factor-4alpha. J Pharmacol Exp Ther 317:778–785 [DOI] [PubMed] [Google Scholar]
- Schaefer O, Ohtsuki S, Kawakami H, Inoue T, Liehner S, Saito A, Sakamoto A, Ishiguro N, Matsumaru T, Terasaki T, et al. (2012) Absolute quantification and differential expression of drug transporters, cytochrome P450 enzymes, and UDP-glucuronosyltransferases in cultured primary human hepatocytes. Drug Metab Dispos 40:93–103 [DOI] [PubMed] [Google Scholar]
- Schaeffeler E, Hellerbrand C, Nies AT, Winter S, Kruck S, Hofmann U, van der Kuip H, Zanger UM, Koepsell H, Schwab M. (2011) DNA methylation is associated with downregulation of the organic cation transporter OCT1 (SLC22A1) in human hepatocellular carcinoma. Genome Med 3:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D, Kawamoto M, Johns SJ, DeYoung J, Carlson E, et al. Pharmacogenetics Of Membrane Transporters Investigators (2003) Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci USA 100:5902–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, et al. (2007) Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest 117:1422–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85 [DOI] [PubMed] [Google Scholar]
- Tzvetkov MV, Dos Santos Pereira JN, Meineke I, Saadatmand AR, Stingl JC, Brockmöller J. (2013) Morphine is a substrate of the organic cation transporter OCT1 and polymorphisms in OCT1 gene affect morphine pharmacokinetics after codeine administration. Biochem Pharmacol 86:666–678 [DOI] [PubMed] [Google Scholar]
- Tzvetkov MV, Saadatmand AR, Bokelmann K, Meineke I, Kaiser R, Brockmoller J. (2010) Effects of OCT1 polymorphisms on the cellular uptake, plasma concentrations and efficacy of the 5-HT(3) antagonists tropisetron and ondansetron. Pharmacogenomics J 12:22–29 [DOI] [PubMed] [Google Scholar]
- Tzvetkov MV, Saadatmand AR, Lötsch J, Tegeder I, Stingl JC, Brockmöller J. (2011) Genetically polymorphic OCT1: another piece in the puzzle of the variable pharmacokinetics and pharmacodynamics of the opioidergic drug tramadol. Clin Pharmacol Ther 90:143–150 [DOI] [PubMed] [Google Scholar]
- Tzvetkov MV, Vormfelde SV, Balen D, Meineke I, Schmidt T, Sehrt D, Sabolić I, Koepsell H, Brockmöller J. (2009) The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther 86:299–306 [DOI] [PubMed] [Google Scholar]
- Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T, et al. (2008) Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 83:520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Dresser MJ, Gray AT, Yost SC, Terashita S, Giacomini KM. (1997) Cloning and functional expression of a human liver organic cation transporter. Mol Pharmacol 51:913–921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.