Abstract
Bombesin-receptor-subtype-3 (BRS-3) is an orphan G-protein-coupled receptor of the bombesin (Bn) family whose natural ligand is unknown and which does not bind any natural Bn-peptide with high affinity. It is present in the central nervous system, peripheral tissues, and tumors; however, its role in normal physiology/pathophysiology is largely unknown because of the lack of selective ligands. Recently, MK-5046 [(2S)-1,1,1-trifluoro-2-[4-(1H-pyrazol-1-yl)phenyl]-3-(4-{[1-(trifluoromethyl)cyclopropyl]methyl}-1H-imidazol-2-yl)propan-2-ol] and Bantag-1 [Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino) benzylamide N-methylammonium trifluoroacetate], a nonpeptide agonist and a peptide antagonist, respectively, for BRS-3 have been described, but there have been limited studies on their pharmacology. We studied MK-5046 and Bantag-1 interactions with human Bn-receptors—human bombesin receptor subtype-3 (hBRS-3), gastrin-releasing peptide receptor (GRP-R), and neuromedin B receptor (NMB-R)—and compared them with the nonselective, peptide-agonist [d-Tyr6,βAla11,Phe13,Nle14]Bn-(6–14) (peptide #1). Receptor activation was detected by activation of phospholipase C (PLC), mitogen-activated protein kinase (MAPK), focal adhesion kinase (FAK), paxillin, and Akt. In hBRS-3 cells, the relative affinities were Bantag-1 (1.3 nM) > peptide #1 (2 nM) > MK-5046 (37–160 nM) > GRP, NMB (>10 μM), and the binding-dose-inhibition curves were broad (>4 logs), with Hill coefficients differing significantly from unity. Curve-fitting demonstrated high-affinity (MK-5046, Ki = 0.08 nM) and low-affinity (MK-5046, Ki = 11–29 nM) binding sites. For PLC activation in hBRS-3 cells, the relative potencies were MK-5046 (0.02 nM) > peptide #1 (6 nM) > GRP, NMB, Bantag-1 (>10 μM), and MK-5046 had a biphasic dose response, whereas peptide #1 was monophasic. Bantag-1 was a specific hBRS-3-antagonist. In hBRS-3 cells, MK-5046 was a full agonist for activation of MAPK, FAK, Akt, and paxillin; however, it was a partial agonist for phospholipase A2 (PLA2) activation. The kinetics of activation/duration of action for PLC/MAPK activation of MK-5046 and peptide #1 differed, with peptide #1 causing more rapid stimulation; however, MK-5046 had more prolonged activity. Our study finds that MK-5046 and Bantag-1 have high affinity/selectivity for hBRS-3. The nonpeptide MK-5046 and peptide #1 agonists differ markedly in their receptor coupling, ability to activate different signaling cascades, and kinetics/duration of action. These results show that their hBRS-3 receptor activation is not always concordant and could lead to markedly different cellular responses.
Introduction
Bombesin receptor subtype-3 (BRS-3) is a G protein–coupled orphan receptor that is widely distributed in the central nervous system and peripheral tissues (Sano et al., 2004; Jensen et al., 2008; Weber, 2009). Even though it is an orphan receptor, it is classified as a mammalian bombesin (Bn) receptor because of its homology (47–51% amino acid identities) with the two mammalian Bn receptors, gastrin-releasing peptide receptor (GRP-R) and neuromedin B receptor (NMB-R) (Jensen et al., 2008). BRS-3 is putatively involved in a broad range of physiologic and pathophysiological processes (Ohki-Hamazaki et al., 1997; Jensen et al., 2008; Majumdar and Weber, 2012a,b; Sayegh, 2013), including energy homeostasis (Ohki-Hamazaki et al., 1997; Ladenheim et al., 2008; Guan et al., 2010, 2011; Metzger et al., 2010; Feng et al., 2011), feeding behavior (Maekawa et al., 2004; Ladenheim et al., 2008; Majumdar and Weber, 2012a; Sayegh, 2013), glucose metabolism (Ohki-Hamazaki et al., 1997; Nakamichi et al., 2004; Jensen et al., 2008; Feng et al., 2011; Majumdar and Weber, 2012a), gastrointestinal motility (Jensen et al., 2008), and lung cancer development and injury (Shan et al., 2004; Tan et al., 2006). However, exploration of the role of BRS-3 in physiological and pathophysiologic processes had been limited because the natural ligand is unknown, and, in contrast to the other Bn receptors, no selective agonists and antagonists are available (Heinz-Erian et al., 1987; von Schrenck et al., 1990; Wang et al., 1990a,b; Jensen and Coy, 1991; Jensen et al., 2008; Majumdar and Weber, 2012b).
Until recently, the only known high-affinity ligand was the synthetic Bn-related agonist peptide #1 ([d-Tyr6,βAla11,Phe13,Nle14]Bn-(6–14)]), but peptide #1 also has high affinity for GRP-R and NBM-R from all species as well as human bombesin receptor subtype-3 (hBRS-3) (Mantey et al., 1997; Pradhan et al., 1998; Sancho et al., 2010; Uehara et al., 2011). Recent studies have reported that the nonpeptide MK-5046 [(2S)-1,1,1-trifluoro-2-[4-(1H-pyrazol-1-yl)phenyl]-3-(4-{[1-(trifluoromethyl)cyclopropyl]methyl}-1H-imidazol-2-yl)propan-2-ol] is an orally active, potent, and selective BRS-3 agonist (Sebhat et al., 2011; Guan et al., 2011; Reitman et al., 2012), with activity in mice, rats, dogs (Guan et al., 2011), and humans (Reitman et al., 2012). Furthermore, recently a selective peptide antagonist Bantag-1 [Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino) benzylamide N-methylammonium trifluoroacetate] also has been reported (Guan et al., 2010; Feng et al., 2011). There are limited data on the pharmacologic characteristics of these compounds (Guan et al., 2010, 2011; Feng et al., 2011; Sebhat et al., 2011). In addition, for the Bn-receptor family, similar to other gastrointestinal hormone/neurotransmitter G protein–coupled receptors, nonpeptide-agonists are uncommon (Freidinger, 1993; Jensen et al., 2008; Uehara et al., 2011); in fact, MK-5046 is the first high-affinity, widely used nonpeptide agonist for any member of the Bn family of receptors. Therefore, for most of these receptors there are limited data on the comparison of the pharmacological properties of receptor interaction/activation by nonpeptide agonists with peptide-receptor agonists (Freidinger, 1993).
Our study analyses the affinity, selectivity, efficacy, and kinetics of receptor interaction/activation of the nonpeptide agonist MK-5046 compared with a previously reported peptide receptor agonist, peptide #1 (Mantey et al., 2004, 2006; Sancho et al., 2010), for hBRS-3 and for other members of human Bn-receptor family. We also compared the pharmacology of these two agonists with the peptide Bantag-1, which has been described as a selective and potent antagonist of BRS-3 (Guan et al., 2010; Feng et al., 2011).
Materials and Methods
Materials.
Balb 3T3 (mouse fibroblast cells), NCI-N417 (human small-cell lung carcinoma cells), and HuTu-80 (human duodenal cancer cells) were purchased from the American Type Culture Collection (Manassas, VA); NCI-H1299 (non–small-cell lung cancer cells) was obtained from Herb Oie (National Cancer Institute–Navy Medical Oncology Branch, Naval Medical Center, Bethesda, MD). We purchased Dulbecco’s minimum essential medium (DMEM), RPMI 1640 medium, phosphate-buffered saline (PBS), fetal bovine serum (FBS), Dulbecco’s phosphate-buffered saline (DPBS), trypsin-EDTA 1×, penicillin, streptomycin, and Novex 4–20% Tris-glycine gel from Invitrogen (Carlsbad, CA). Geneticin-selective antibiotic (G418 Sulfate) and Tris-buffered saline 10× (TBS) were obtained from CellGro (Mediatech, Inc. Manassas, VA); GRP and NMB were from Bachem (Torrance, CA). Bombesin receptor subtype-3 antagonist (Bantag-1) and MK-5046 were gifts from Merck, Sharp and Dohme (West Point, PA). Iodine-125 radionuclide [125I] (10 mCi, 378 MBq), [5,6,8,9,114,12,14,15-3H(N)]-arachidonic acid (50 mCi, 1.85 MBq), and myo-[2-3H]inositol (5 mCi, 185 MBq) were obtained from PerkinElmer Life and Analytical Sciences (Boston, MA). We purchased AG 1-X8 resin and 10× Tris/glycine/SDS from Bio-Rad Laboratories (Richmond, CA); bacitracin, sodium vanadate, Triton X-100, deoxycholate, Tween 20, phenylmethylsulfonyl fluoride (PMSF), EGTA, EDTA, sodium azide quinacrine dihydrochloride, RHC-80267 [1,6-bis(cyclohexyloximinocarbonylamino)hexane], U-73122 hydrate [1-(6-((17β)-3-methoxyestra-1,3,5[10]-trien-17-yl)amino]hexyl)-1H-pyrrole-2,5-dione]; GF109203X [2-[1-(3-dimethylaminopropyl)-1H-indol-3-yl]-3-(1H-indol-3-yl)-maleimide)], fat-free bovine serum albumin (BSA), and methanol were from Sigma-Aldrich (St. Louis, MO); arachidonyl trifluoromethyl ketone was from Cayman Chemical Company (Ann Arbor, MI); manoalide was from Enzo Life Science (Plymouth Meeting, PA); rabbit anti-α/β-tubulin, rabbit monoclonal p44/42 (Tyr202/204), polyclonal anti-phosphorylated forms p44/42 MAP kinase and paxillin (Tyr118), and rabbit monoclonal antiphosphorylated forms Akt (Ser473) and FAK (Tyr397) were purchased from Cell Signaling Technology (Beverly, MA). The horseradish peroxidase–conjugated secondary antibody (anti-rabbit) and Supersignal Western Pico/Dura were from Thermo Fisher Scientific (Rockford, IL). The nonfat dry milk was from American Bio-analytical (Natick, MA). The protease inhibitor tables came from Roche (Basel, Switzerland). The BSA fraction V and HEPES were from ICN Pharmaceutical (Aurora, OH).
Cell Culture.
Balb 3T3 cells stably expressing human GRP receptor (hGRP-R), human NMB receptor (hNMB-R), and human BRS-3–receptor (hBRS-3) were prepared and used as described previously elsewhere (Benya et al., 1995; Mantey et al., 1997; Ryan et al., 1998a). In addition, NCI-H1299 were transfected to overexpress human NMB receptor (Gonzalez et al., 2009; Uehara et al., 2011). Balb 3T3 cells hGRP-R, hNMB-R, and hBRS-3 were grown in DMEM with 300 mg/l G418 sulfate. The HuTu-80 cells, which contain native hGRP-receptor (Gonzalez et al., 2009; Uehara et al., 2011), were grown in DMEM. The hNMB-R transfected NCI-H1299 cells were grown in RPMI 1640 medium with 300 mg/l G418 sulfate and NCI-N417, which contains native human BRS-3–receptors (Ryan et al., 1998b; Sancho et al., 2010) in RPMI 1640 medium. All of the cells were grown in their respective propagation medium supplemented with 10% FBS and incubated at 37°C in a 5% CO2 atmosphere.
Preparation of Peptides.
The peptides were synthesized using standard solid-phase methods as described previously elsewhere (Mantey et al., 1997; Gonzalez et al., 2009). In brief, solid-phase syntheses of peptide amides were performed using Boc chemistry on methyl benzhydrylamine resin (Advanced ChemTech, Louisville, KY) followed by hydrogen fluoride cleavage of free peptide amides. The crude peptides were purified by preparative high-performance liquid chromatography on columns (2.5 × 50 cm) of Vydac C18 silica (10 µm), which was eluted, with linear gradients of acetonitrile in 0.1% (v/v) trifluoroacetic acid. Homogeneity of the peptides was assessed by analytic reverse-phase high-performance liquid chromatography, and the purity was usually 97% or higher. Amino acid analysis (only amino acids with primary amino acid groups were quantified) gave the expected amino acid ratios. Peptide molecular masses were obtained by matrix-assisted laser desorption mass spectrometry (Thermo Bioanalysis Corp., Hemel, Helmstead, UK), and all corresponded well with calculated values. All peptides/nonpeptides were dissolved as stock solutions in incubation buffer, and all readily dissolved at the concentrations used [peptides–0.01 M, nonpeptides (MK-5046)–0.001 M)].
Preparation of 125I-Labeled Peptides.
125I-peptide #1, with specific activity of 2200 Ci/mmol, was prepared by a modification of methods described elsewhere (Mantey et al., 1997; Sancho et al., 2010). In brief, 0.8 μg of IODO-GEN (in 0.01 μg/ml chloroform) was transferred to a vial, dried under a stream of nitrogen, and washed with 100 μl of 0.5 M KH2PO4, pH 7.4. To the reaction vial 20 μl of 0.5 M KH2PO4, pH 7.4, 8 μg of peptide in 4 μl of water, and 2 mCi (20 μl) Na125I were added, mixed gently, and incubated at room temperature for 6 minutes. The incubation was stopped by the addition of 100 μl of distilled water. Radiolabeled peptide was separated using a Sep-Pak (Waters Associates, Milford, MA) and high-performance liquid chromatography as described previously elsewhere (Mantey et al., 1997; Sancho et al., 2010). The radioligand was stored with 0.5% BSA at 20°C.
Radioligand Binding Assays.
Balb/hBRS-3 (0.5 × 106 cells/ml), Balb/hGRP-R (0.15 × 106), Balb/hNMB-R (0.03 × 106), NCI-N417 (1 × 107), HuTu-80 (0.25 × 106), and NCI-H1299 (1 × 106) cells were incubated for 40–60 minutes at 21°C with 50 pM 125I-labeled ligand in 300 μl of binding buffer as described previously elsewhere (Mantey et al., 1997; Gonzalez et al., 2009; Sancho et al., 2010). The standard binding buffer contained 24.5 mM HEPES, pH 7.4, 98 mM NaCl, 6 mM KCl, 2.5 mM KH2PO4, 5 mM sodium pyruvate, 5 mM sodium fumarate, 5 mM sodium glutamate, 2 mM glutamine, 11.5 mM glucose, 0.5 mM CaCl2, 1.0 mM MgCl2, 0.01% (w/v) soybean trypsin inhibitor, 0.2% (v/v) amino acid mixture, 0.2% (w/v) BSA, and 0.05% (w/v) bacitracin. After the incubation, 100 μl of each sample were removed and added to 300 μl of incubation buffer in 400-μl Microfuge tubes and centrifuged for 1 minute at 10,000g (Microfuge E; Beckman Coulter, Fullerton, CA) to separate the bound radioligand from unbound radioligand. The supernatant was aspirated, and the pelleted cells were rinsed twice with a washing buffer that contained 1% (w/v) BSA in PBS.
The amount of radioactivity bound to the cells was measured in a Cobra II Gamma Counter (Packard Instruments, Meriden, CT). Binding was expressed as the percentage of the total radioactivity that was associated with the cell pellet. Nonsaturable binding was <15% of the total binding in all experiments. Each point was measured in duplicate, and each experiment was replicated at least 4 times. Calculation of affinity was performed by determining the IC50 using the curve-fitting program Prism GraphPad 4.0 (GraphPad Software, Inc., La Jolla, CA).
Measurement of [3H]Inositol Phosphates.
[3H]Inositol phosphates (IP) were measured in the different cells as described previously elsewhere (Rowley et al., 1990; Benya et al., 1992, 1994). In brief, all cells except N417 were subcultured into 24-well plates in regular propagation media and then were incubated for 24 hours at 37°C in a 5% CO2 atmosphere: hGRP-R (0.15 × 106 cells/well), hNMB-R (0.03 × 106), hBRS-3 (5 × 105), HuTu-80 (0.25 × 106), and NCI-H1299 (1 × 106). The cells were then incubated with 3 μCi/ml myo-[2-3H]inositol in growth media supplemented with 2% FBS for an additional 24 hours. After the incubation, the 24-well plates were washed by incubating for 30 minutes at 37°C with 1 ml/well PBS (pH 7.0) containing 20 mM lithium chloride. The wash buffer was aspirated and replaced with 500 μl of IP assay buffer containing 135 mM sodium chloride, 20 mM HEPES, pH 7.4, 2 mM calcium chloride, 1.2 mM magnesium sulfate, 1 mM EGTA, 20 mM lithium chloride, 11.1 mM glucose, and 0.05% BSA (w/v), and was incubated without (control) or with different concentrations of the peptides studied.
The N417 cells (1 × 106 cells/ml), which grow in suspension, were centrifuged to remove the RPMI 1640 medium and were incubated directly with myo-[2-3H]inositol in RPMI 1640 medium with 2% of FBS for 24 hours (Ryan et al., 1998b; Sancho et al., 2010). The N417 cells were centrifuged to remove the wash buffer, and the cells were distributed in 5-ml tubes which were incubated with peptides in 500 μl of IP assay buffer. After 60 minutes of incubation at 37°C, the experiments were terminated by the addition of 1 ml of ice-cold 1% (v/v) hydrochloric acid in methanol.
The total [3H]IP was isolated by anion exchange chromatography as described previously elsewhere (Rowley et al., 1990; Benya et al., 1995; Ryan et al., 1998a; Uehara et al., 2011). Samples were loaded onto Dowex AG1-X8 anion exchange resin columns, washed with 5 ml of distilled water to remove free [3H]IP and then washed with 2 ml of 5 mM disodium tetraborate/60 mM sodium formate solution to remove [3H]glycerophosphorylinositol; after this, 2 ml of 1 mM ammonium formate/100 mM formic acid solution were added to the columns to elute the total [3H]IP. Each eluate was mixed with scintillation cocktail and measured for radioactivity in a scintillation counter.
Western Blot Analysis.
Western blot analysis of the various cells was performed as previously described elsewhere (Berna et al., 2007; Sancho et al., 2010). Balb/hBRS-3 and NCI-N417 cells were washed with PBS and incubated with starvation medium (DMEM or RPMI 1640 medium without FBS) for 2 hours and 3 hours, respectively, at 37°C in a 5% CO2 atmosphere. Afterward, the cells were treated with peptide #1 and MK-5046 for 3 minutes at 37°C in a 5% CO2 atmosphere, and the reaction was stopped at 4°C, and then the cells were washed again with PBS and lysed with lysis buffer containing 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium azide, 1 mM EGTA, 0.4 mM EDTA, 1 mM DTT, 0.4 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and one protease inhibitor tablet per 10 ml. Lysates were sonicated and then centrifuged at 12,200g for 20 minutes at 4°C; the protein concentration was measured using the Bio-Rad protein assay reagent (Hercules, CA).
Equal amounts of protein from whole-cell lysates were loaded on to SDS-PAGE using 4–20% Tris-glycine gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes for 2 hours. The membranes were then washed twice with washing buffer (TBS plus 0.1% Tween 20) and incubated with primary antibody at 1:1000 dilution in washing buffer + 5% BSA overnight at 4°C under constant agitation.
After primary antibody incubation, the membranes were washed twice in blocking buffer (TBS, 0.1% Tween 20, 5% nonfat dry milk) for 5 minutes and then incubated with horseradish peroxidase–conjugated secondary antibody (anti-rabbit) for 1 hour at room temperature under constant agitation. Membranes were washed again twice in blocking buffer for 5 minutes and then twice in washing buffer for 5 minutes. The membranes then were incubated with chemiluminescence detection reagents for 4 minutes and finally were exposed to Kodak Biomax film (maximum resolution, maximum sensitivity; Carestream Health, Rochester, NY). The intensity of the protein bands was measured using Kodak ID Image Analysis (Carestream Health).
Phospholipase A2 Activity.
Phospholipase A2 activity (PLA2) was determined as described previously elsewhere (Moody et al., 1995; Husain and Abdel-Latif, 1998; Xu et al., 2002) in studies demonstrating that activation of Bn-related receptors as well as other G-protein-coupled receptors can stimulate arachidonic acid release through PLA2 activation. In brief, to study the ability of agents to activate PLA2, their effect on [3H-5,6,8,9,11,12,14,15]arachidonic acid ([3H]AA) release from Balb 3T3 cells stably transfected with hBRS-3-receptor (hBRS-3) was assessed. Balb 3T3 cells stably transfected with hBRS-3–receptor (hBRS-3) were subcultured into 24-well plates (5 × 104 cells/well) in regular propagation medium and incubated for 24 hours at 37°C in a 5% CO2 incubator. The medium was aspirated and replaced with DMEM supplemented with 0.2% fatty acid-free BSA (DMEM/BSA) and 1 μCi/4 ml [3H]AA and then incubated for an additional 24 hours. The cells were washed twice with DMEM/BSA and 500 μl of new medium with or without the various agents to be assessed and then was incubated at 37°C. After 40 minutes, the supernatant was pipetted into scintillation vials, scintillation cocktail added, and the samples were counted in a β-counter.
For inhibition experiments with various phospholipase inhibitors, after a 24-hour incubation with [3H]AA (as performed and described previously), the cells were washed twice with DMEM/BSA. The washed cells were treated with 1 μM concentrations of the various inhibitors for 15 minutes before the addition of peptide #1 (0.1 μM) and were incubated for an additional 30 minutes, after which the release of [3H]AA was measured by counting the supernatant in a β-counter.
Statistical Analysis.
All results are expressed as mean ± S.E.M from at least four experiments. P < 0.05 was considered statistically significant, using analysis of variance (ANOVA, nonparametric) or the paired t test when performed with a specifically matched paired control. Prism GraphPad 4.0 was used for all statistical analyses. In addition, binding data and monophasic dose-response curves of cellular activation were curve-fitted using Prism (nonlinear curve-fitting).
Results
Affinities for Human Bn Receptors.
In this study, we compared the ability of the naturally occurring Bn-related peptides GRP and NMB and the universal Bn-receptor agonist ligand peptide #1 (Mantey et al., 1997; Pradhan et al., 1998) to interact with and activate the three human Bn-receptor subtypes—hGRP-R, hNMB-R, and hBRS-3 (Jensen et al., 2008)—with that of MK-5046, a recently reported (Guan et al., 2011; Sebhat et al., 2011) BRS-3-receptor selective nonpeptide agonist, and Bantag-1, a BRS-3 selective peptide antagonist (Guan et al., 2010; Feng et al., 2011). To be certain the results were reflective of the pharmacology of these peptides, two different Bn-receptor–containing cells were used to assess the results with each human Bn-receptor subtype.
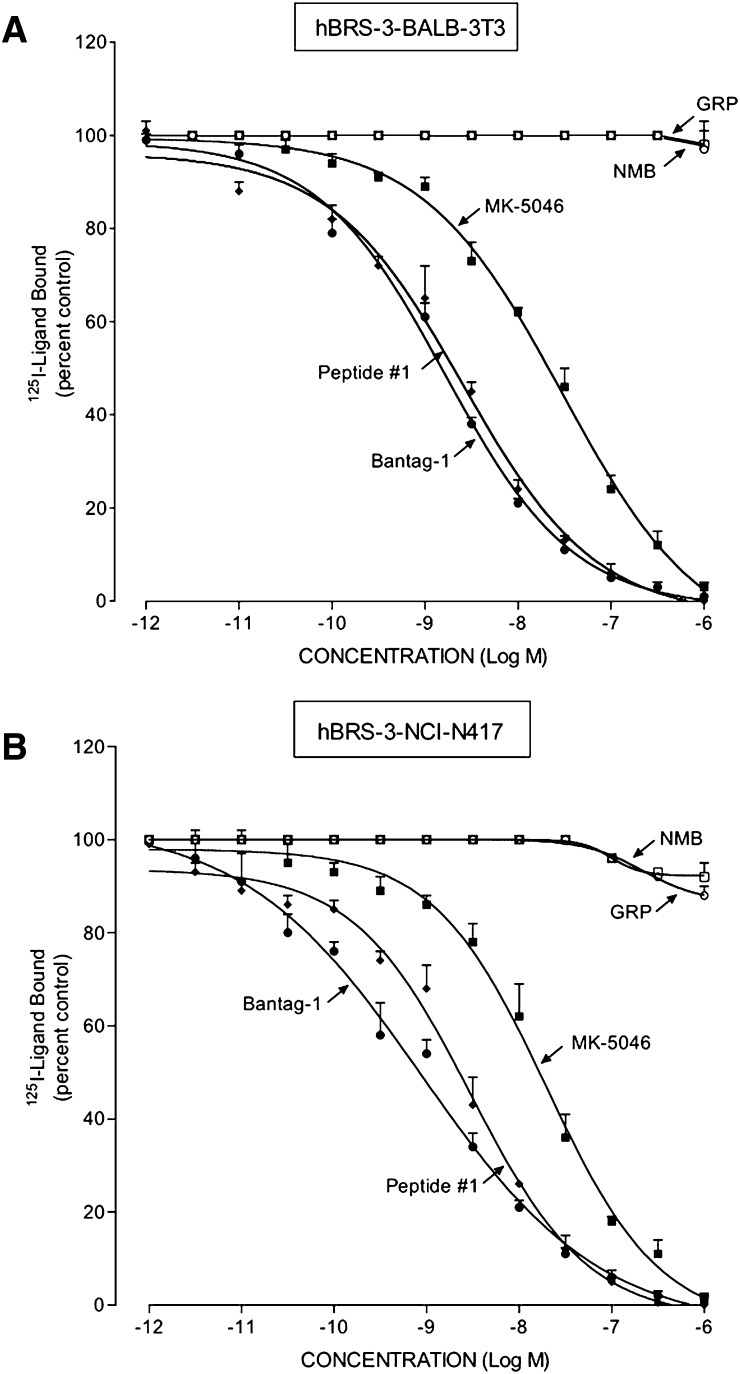
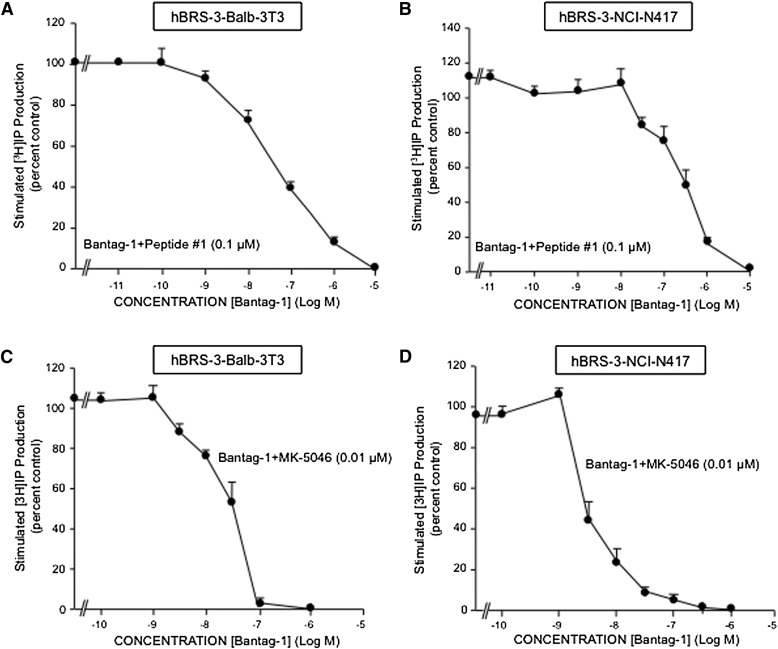
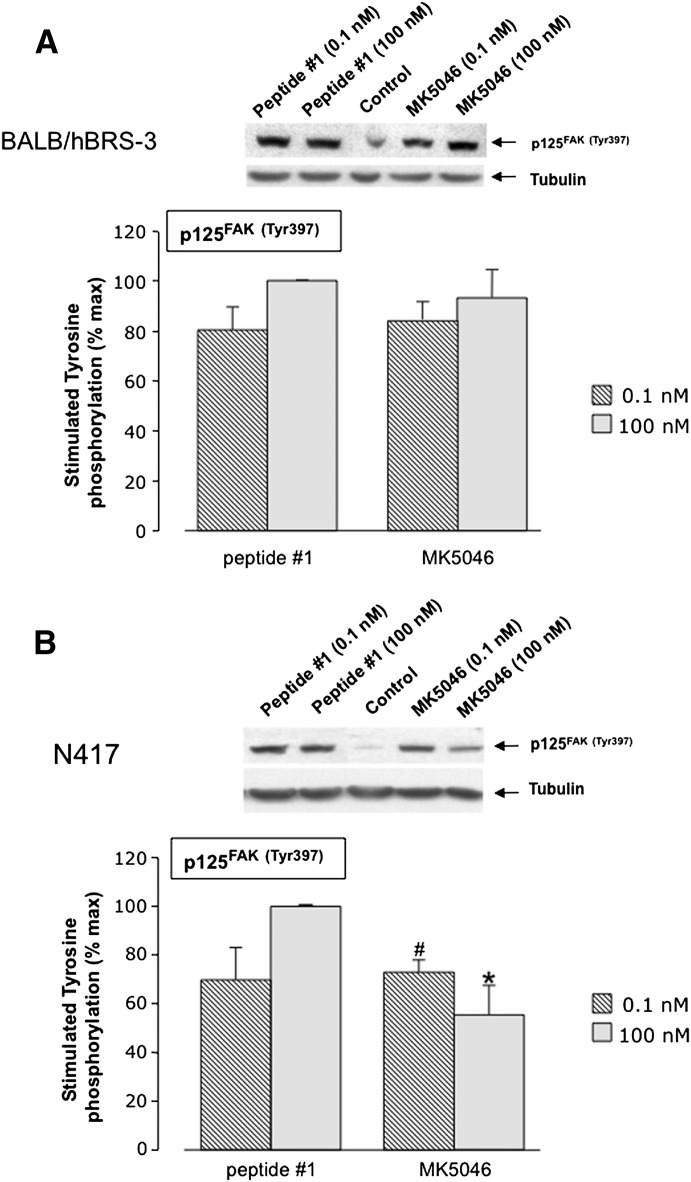
In the cells containing hBRS-3, hBRS-3 transfected Balb 3T3 cells (Benya et al., 1995; Mantey et al., 1997) and NCI-N417 lung cancer cells, which contain a native hBRS-3–receptor (Ryan et al., 1998b; Uehara et al., 2011), the universal ligand peptide #1 had a high affinity for the hBRS-3-receptor (2.5–2.7 nM; Fig. 1, A and B; Table 1), similar to what was reported by others (Gonzalez et al., 2008; Sancho et al., 2010; Uehara et al., 2011); this affinity was 2-fold lower than the affinity observed with the antagonist Bantag-1 (1.0–1.6 nM; Fig. 1; Table 1). However it was 8-fold higher than that for the nonpeptide agonist MK-5046 (17–18 nM; Fig. 1; Table 1), and >10,000-fold higher than the affinity of GRP and NMB for each hBRS-3 cell line (Fig. 1; Table 1).
Fig. 1.
Comparison of the affinities of the peptide agonists GRP, NMB, and peptide #1, nonpeptide agonist MK-5046, and the putative peptide antagonist Bantag-1 for the hBRS-3–receptor in (A) hBRS-3 Balb 3T3 cells and (B) NCI-N417 cells. The peptides were incubated with 50 pM 125I-[d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) for 40–60 minutes at 21°C in 300 μl of binding buffer with hBRS-3 Balb 3T3 cells (0.5 × 106 cells/ml) or NCI-N417 cells (1 × 107 cells/ml), and the saturable binding was determined as described under Materials and Methods. The results are expressed as the percentage of saturable binding without unlabeled peptide added (percentage control). The results are the mean ± S.E.M. from at least four experiments, and in each experiment the data points were determined in duplicate.
TABLE 1.
Binding affinities of different ligands for human BRS-3, GRP, and NMB receptors in various cells
Cells were incubated with 50 pM 125I-[d-Tyr6, βAla11, Phe13, Nle14]Bn-(6–14) for 40–60 minutes at 21°C and binding was determined as described under Materials and Methods. For each ligand, a dose-response curve was performed with concentrations from 1 pM to 1 μM. Data are expressed as the concentration causing half-maximal inhibition of saturable binding. Results were calculated from the dose-response curves shown in Figs. 1 and 2 using a nonlinear regression curve-fitting program (Prism). Results are mean ± S.E.M. from at least four experiments, and >10,000 = IC50 >10,000 nM.
| Ligand | IC50 (nM) |
|||||
|---|---|---|---|---|---|---|
| hBRS-3 |
hGRP-R |
hNMB-R |
||||
| Balb 3T3 | NCI-N417 | Balb 3T3 | HuTu-80 | Balb 3T3 | NCI-H1299 | |
| GRP | >10,000 | >10,000 | 0.23 ± 0.02 | 0.13 ± 0.01 | 246 ± 39 | 127 ± 11 |
| NMB | >10,000 | >10,000 | 57.4 ± 5.9 | 66.4 ± 5.6 | 0.07 ± 0.01 | 0.60 ± 0.11 |
| Peptide #1 | 2.52 ± 0.51 | 2.76 ± 0.41 | 0.26 ± 0.01 | 0.05 ± 0.01 | 1.55 ± 0.15 | 6.00 ± 2.12 |
| MK-5046 | 18.8 ± 0.2 | 17.7 ± 0.2 | >10,000 | >10,000 | >10,000 | >10,000 |
| Bantag-1 | 1.59 ± 0.52 | 1.05 ± 0.55 | >10,000 | >10,000 | >10,000 | >10,000 |
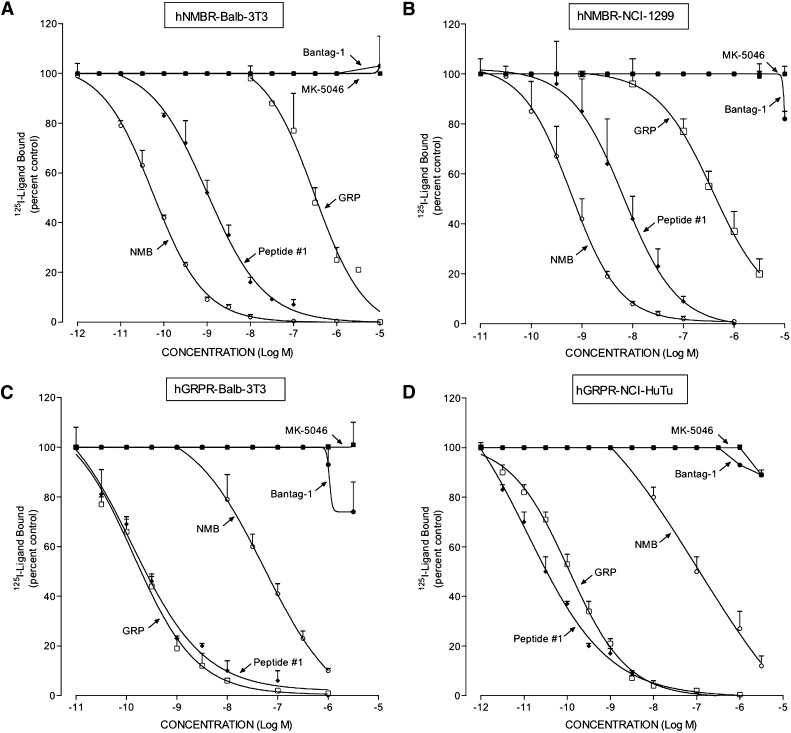
In the two cell lines containing hGRP-R, hGRP-R-transfected Balb 3T3 cells (Benya et al., 1995) and HuTu-80 cancer cells, which natively possess hGRP-R (Gonzalez et al., 2009; Sancho et al., 2010), the universal Bn-receptor ligand peptide #1 had a very high affinity (0.05–0.26; Fig. 2; Table 1). Similarly, the physiologic ligand GRP had a high affinity (0.13–0.23 nM; Fig. 2; Table 1) but GRP-R a lower affinity for NMB-R (57–66 nM; Fig. 2; Table 1), and hGRP-R did not interact with either MK-5046 or Bantag-1, even at concentrations of 10,000 nM (Fig. 2; Table 1).
Fig. 2.
Comparison of affinities of GRP, NMB, peptide #1, MK-5046, and Bantag-1 for cells containing hNMB-R and hGRP-R. Two different cell lines were used to assess human GRP-R interaction: hGRP-R–transfected Balb 3T3 (0.15 × 106 cells/ml) (C) and HuTu-80 native hGRP-R cells (0.25 × 106) (D). For hNMB-R interaction, hNMB-R–transfected Balb 3T3 (0.03 × 106 cells/ml) (A) and NCI-1299 transfected cells (1 × 106) (B) were used. The cells were incubated with 50 pM 125I-[d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) for 40–60 minutes at 21°C in 300 μl of binding buffer either alone or with the indicated peptides, and the saturable binding was determined as described under Materials and Methods. The results are expressed as the percentage of saturable binding without unlabeled peptide added (percentage control). The results are the mean ± S.E.M. from at least four experiments, and in each experiment the data points were determined in duplicate.
In both cell lines containing hNMB-R, hNMB-R-transfected Balb 3T3 cells (Gonzalez et al., 2009; Uehara et al., 2011) and lung carcinoma NCI-H1299 cells (Gonzalez et al., 2009; Uehara et al., 2011), the physiologic ligand NMB had the highest affinity (0.07–0.6 nM; Fig. 2, A and B; Table 1); and peptide #1 also had a high affinity (1.5–6.0 nM; Fig. 2, A and B; Table 1). GRP had a low affinity (127–246 nM; Fig. 2, A and B), and hNMB-R did not interact with MK-5046 or Bantag-1, even at concentrations of 10,000 nM (Fig. 2, A and B; Table 1).
In terms of relative affinities for the different human Bn-receptor subtypes, for the hBRS-3–receptor, the reported antagonist Bantag-1 (Feng et al., 2011) had the highest selectivity of >5000-fold, followed by the nonpeptide BRS-3 agonist MK-5046 (62- to 877-fold). Peptide #1, as reported previously elsewhere (Mantey et al., 1997; Uehara et al., 2011), had a high affinity for all three Bn-receptor subtypes (Table 1). For the hGRP receptor, GRP had the highest selectivity (>2000) over hNMB-R or hBRS-3, whereas peptide #1 had a slight selectivity with a 6- to 31-fold higher affinity than for hNMB-R, and 38- to 74-fold higher than for hBRS-3. Neither MK-5046 nor Bantag-1 interacted with either hGRP-R or hNMB-R. For hNMB-R, only the native peptide NMB interacted selectivity, with a 100–850 higher affinity compared with hGRP-R and >10,000-fold over hBRS-3 (Fig. 2, A and B; Table 1).
In both cell lines containing hBRS-3 receptor, Balb 3T3-transfected cells, and native NCI-N417 lung cancer cells, the binding dose-inhibition curves were broad for peptide #1, Bantag-1, and MK-5046, spanning a >4-fold log range (Fig. 1), which suggests that they could be interacting with more than a single receptor site. This conclusion was supported by the Hill coefficient for each hBRS-3 cell. With peptide #1 in hBRS-3 Balb cells, the Hill coefficient was statistically significantly different from unity at −0.57 ± 0.04 (P < 0.01); in NCI-N417 cells, it was −0.64 ± 0.05 (P < 0.001).
To further analyze this possibility, we examined the dose-inhibition curves of peptide #1 using a least squares curve-fitting program (Prism GraphPad 4.0). For each hBRS-3 cell type, the dose-inhibition curves were statistically significantly (P < 0.001) better fitted by a two-site model compared with a one-site model. The high-affinity site in hBRS-3 Balb 3T3 cells (Kd = 0.0078 ± 0.0027 nM) with a density of 370 ± 6 fmol/106 cells (0.466 ± 0.03 pmol/mg protein) represented 23% of the total, and the low-affinity site (Kd= 4.0 ± 0.5 nM) with 1242 ± 23 fmol/106 cells (1.56 ± 0.08 pmol/mg protein), which was 77% of the total. In NCI-N417 cells, the high-affinity site (Kd = 0.0054 ± 0.007 nM) with a density of 24 ± 1 fmol/106 cells (0.10 ± 0.01 pmol/mg protein) represented 17% of the total, and a low-affinity site (Kd = 3.6 ± 0.4 nM) with 117 ± 6 fmol/106 cells (0.48 ± 0.07 pmol/mg protein), which was 83% of the total.
Likewise, the dose-inhibition curves of both MK-5046 and Bantag-1 were wide, extending over >4-fold log range (Fig. 1). The Schild plot for both was statistically significant different from unity for each cell line containing hBRS-3: hBRS-3 Balb 3T3 cells: MK-5046, n = −0.64 ± 0.03 (P < 0.01 compared with unity); Bantag-1, n = −0.61 ± 0.03 (P < 0.01); and with NCI-N417 cells: MK-5046, n = −0.72 ± 0.05 (P < 0.02); Bantag-1, n = −0.52 ± 0.03 (P < 0.01).
Furthermore, the dose-inhibition curve for MK-5046 and Bantag-1 in both cell lines containing hBRS-3 were better fitted by a two-site model than a one-site model (P < 0.01). In hBRS-3 Balb 3T3 cells, MK-5046 had a Kd of 0.080 ± 0.018 nM for the high-affinity site and Kd of 29.2 ± 0.09 nM for the low-affinity site. In NCI-N417 cells, MK-5046 had an affinity of 0.077 ± 0.005 nM for the high-affinity site, and an affinity of 10.6 ± 2.1 nM for the low-affinity site. On hBRS-3 Balb 3T3 cells, Bantag-1 had an affinity of 0.029 ± 0.008 nM for the high-affinity site, and an affinity of 1.95 ± 0.25 for the low-affinity site. On NCI-417 cells, Bantag-1 had an affinity of 0.017 ± 0.04 nM for the high-affinity site and an affinity of 1.98 ± 0.45 nM for the low-affinity site. These results demonstrate that the hBRS-3–receptor can exist in both high-affinity and low-affinity states.
Activation of PLC in Cells Containing Human Bn Receptors.
The activation of all three Bn-receptor subtypes stimulates PLC and the subsequent generation of IP (Benya et al., 1992, 1994, 1995; Jensen et al., 2008). To determine whether each peptide/nonpeptide functioned as a Bn-receptor agonist or antagonist, its ability to stimulate the generation of [3H]IP was measured in each cell type containing Bn receptors.
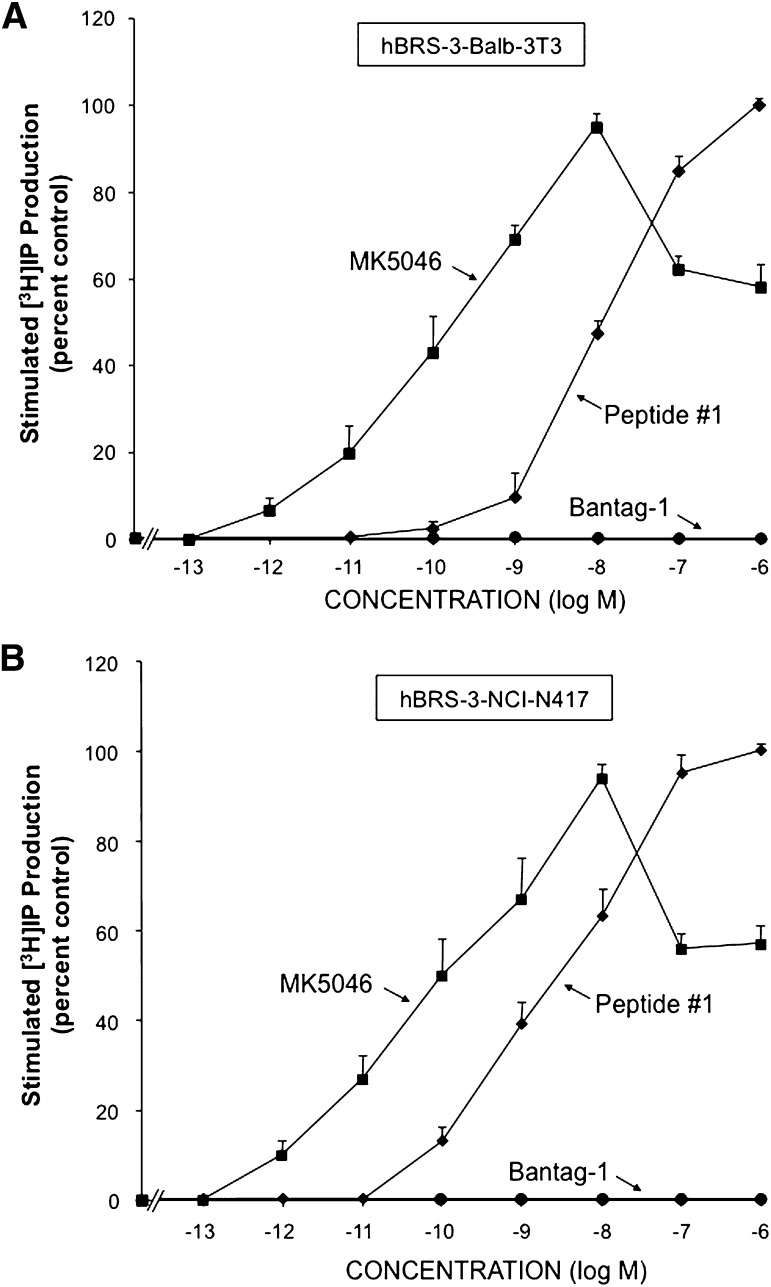
In the two cell lines containing hBRS-3, hBRS-3–transfected Balb 3T3 cells and native NCI-N417 cancer cells, the universal ligand peptide #1 and the nonpeptide MK-5046 each stimulated a dose-dependent response in [3H]IP production and had equal efficacy (Fig. 3). In contrast, even at concentrations up to 1000 nM, Bantag-1 did not activate PLC or alter [3H]IP generation (Fig. 3).
Fig. 3.
Ability of the agonists peptide #1 and MK-5046 or the putative antagonist Bantag-1 to stimulate [3H]IP generation in two different cell types containing hBRS-3: (A) hBRS-3 Balb 3T3 and (B) NCI-N417. After loading the cells with 3 μCi/ml myo-[2-3H]inositol (as described under Materials and Methods), each cell type was incubated with each peptide at the indicated concentration for 60 minutes at 37°C. The [3H]IP measurement was determined as described under Materials and Methods. The results are the mean ± S.E.M. from at least four experiments, and in each experiment the data points were determined in duplicate. The results are expressed as the percentage of stimulation caused by the maximal effective concentration of peptide #1 (1 μM). (A) With hBRS-3 Balb 3T3 cells, the maximal stimulated [3H]IP value by 1 μM peptide #1 was 14,695 ± 1481 dpm, and the control value was 4142 ± 407 dpm (n = 5). (B) With NCI-N417 cells, the maximal stimulation of a 1 μM concentration of peptide #1 was 2897 ± 440 dpm, and the control value was 1251 ± 199 dpm (n = 24).
In the cells containing hBRS-3, peptide #1 caused a detectable increase in [3H]IP at 0.1 nM with a half-maximal effect at 6.5–14.0 nM (Fig. 3; Table 2). In contrast, MK-5046 was 130- to 178-fold more potent, causing a detectable increase at 0.001 nM and having a half-maximal increase at 0.05–0.08 nM (Fig. 3; Table 2). Furthermore, the nonpeptide agonist MK-5046 differed from the Bn-related peptide, peptide #1, in the configuration of their dose-response curves for stimulation of [3H]IP generation (Fig. 3). The dose-response curve with peptide #1 in both cell types containing hBRS-3 (Fig. 3, A and B) was monophasic, with increasing concentrations resulting in increased production of [3H]IP, which reached maximal efficacy at a concentration of 1000 nM. In contrast, the nonpeptide agonist MK-5046 had a biphasic dose-response curve (Fig. 3, A and B), with increasing concentrations increasing the [3H]IP generation, which reached a maximum at 10 nM. With further increases in the concentration of MK-5046, a decrease in the [3H]IP production was seen, with 1 μM MK-5046 causing an 50 ± 4% decrease in the maximal stimulation in hBRS-3 Balb cells (Fig. 3A), and 57 ± 4% decrease in NCI-N417 cells (Fig. 3B).
TABLE 2.
Ability to activate phospholipase C and stimulate [3H]IP production of various Bn-receptor ligands in cells containing human Bn receptors
Cells were incubated with 3 μCi/ml myo-[2-3H]inositol for 24 hours, then washed and incubated with unlabeled ligand for 60 minutes at 37°C. [3H] production was determined as described under Materials and Methods. The EC50 for each ligand was the concentration causing half-maximal increase in [3H] production. Data were calculated from the experiments shown in Figs. 3 and 4 using Prism. Results are mean ± S.E.M. from at least four experiments, and >10,000 = EC50 >10,000 nM.
| Ligand | EC50 (nM) |
|||||
|---|---|---|---|---|---|---|
| hBRS-3 |
hGRP-R |
hNMB-R |
||||
| Balb 3T3 | NCI-N417 | Balb 3T3 | HuTu-80 | Balb 3T3 | NCI-H1299 | |
| GRP | >10,000 | >10,000 | 0.70 ± 0.09 | 4.00 ± 0.50 | 716 ± 13 | 27.1 ± 2.6 |
| NMB | >10,000 | >10,000 | 22.8 ± 2.2 | 220 ± 26 | 12.1 ± 1.7 | 1.01 ± 0.01 |
| Peptide #1 | 14.2 ± 0.9 | 6.51 ± 1.17 | 0.02 ± 0.01 | 0.50 ± 0.03 | 1.00 ± 0.04 | 0.70 ± 0.07 |
| MK-5046 | 0.08 ± 0.02 | 0.05 ± 0.01 | >10,000 | >10,000 | >10,000 | >10,000 |
| Bantag-1 | >10,000 | >10,000 | >10,000 | >10,000 | >10,000 | >10,000 |
Neither GRP nor NMB stimulated the production of [3H]IP in cells containing hBRS-3, even at a concentration of 10,000 nM (Table 2), as was reported by others as well (Ryan et al., 1998a; Gonzalez et al., 2009; Sancho et al., 2010; Uehara et al., 2011).
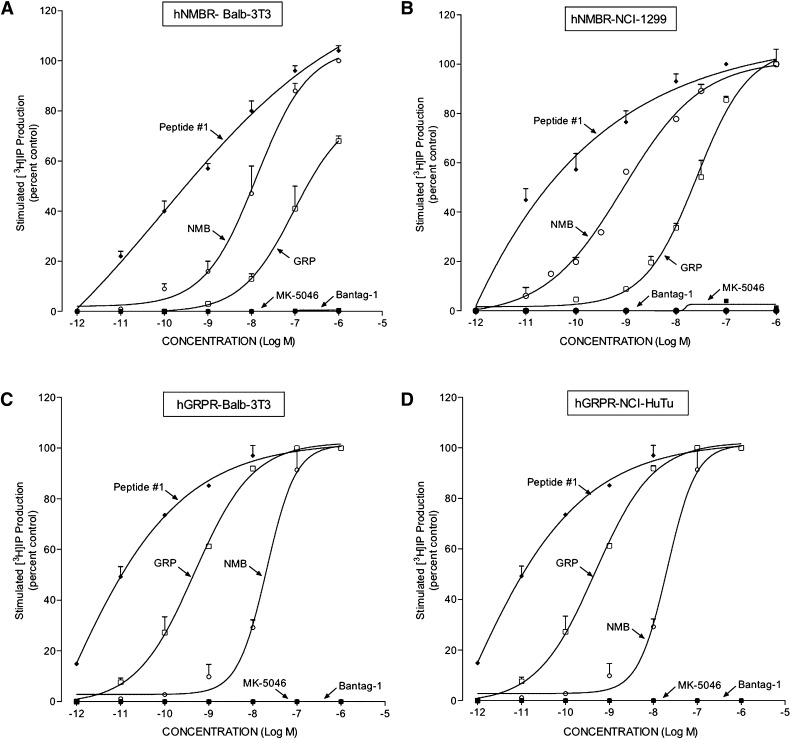
In cells that contained hGRP-R or hNMB-R, the maximally effective concentration of peptide #1, GRP, or NMB had the same efficacy for activating PLC if sufficiently high concentrations were used (Fig. 4, A–D). In the cells containing hGRP-R (hGRP-R-transfected Balb 3T3 cells and the native HuTu-80 duodenal cancer cells), peptide #1 was the most potent at stimulating [3H]IP generation (EC50 0.02–0.05 nM; Fig. 4, C and D; Table 2), as was reported previously elsewhere (Mantey et al., 1997; Pradhan et al., 1998; Uehara et al., 2011). Peptide #1 was 8- to 35-fold more potent than the natural ligand GRP (0.70–4.00 nM; Fig. 4, C and D; Table 1) and 400- to 1150-fold more potent than NMB at stimulating the production of [3H]IP (22–220 nM; Fig. 4, C and D; Table 2) in cells containing hGRP-R. Neither Bantag-1 nor MK-5046 activated hGRP-R, even at concentrations of 1000 nM (Fig. 4, C and D; Table 2).
Fig. 4.
Comparison of the ability of GRP, NMB, peptide #1, MK-5046, and Bantag-1 to stimulate [3H]IP production in cells containing hNMB-R or hGRP-R. The experimental conditions were described in Fig. 3 and under Materials and Methods. The results are the mean ± S.E.M. from at least four experiments, and in each experiment the data points were determined in duplicate. The results are expressed as the percentage of stimulation causing by maximal effective concentration of peptide #1 (1 μM). The control and the maximal stimulated [3H]IP with peptide #1 (1 μM): (A) hNMB-R Balb 3T3 cells, 5336 ± 792 and 47,686 ± 6999 dpm, respectively (n = 6). (B) hNMB-R NCI-1299 cells, 748 ± 109 and 6688 ± 756 dpm, respectively (n = 6). (C) hGRP-R Balb 3T3 cells, 2192 ± 865 and 13,725 ± 4845 dpm, respectively (n = 4). (D) HuTu-80 cells, 1405 ± 219 and 11,266 ± 2973 dpm, respectively (n = 4).
In the two cell types containing hNMB-R (hNMB-R-transfected Balb 3T3 and NCI-H1299 cells), the universal ligand peptide #1 was very potent at activating PLC, causing a detectable increase of [3H]IP production (0.01 nM) and a half-maximal effect at 0.70–1.0 nM (Fig. 4, A and B; Table 2). Peptide #1 was 3- to 12-fold more potent at activating PLC in hNMB-R cells than was the natural ligand NMB (1–12 nM; Fig. 4, A and B; Table 2), and 38- to 700-fold more potent at stimulating the production of [3H]IP than was GRP. Neither Bantag-1 nor MK-5046 activated hGRP-R, even at concentrations of 1000 nM (Fig. 4, A and B; Table 2).
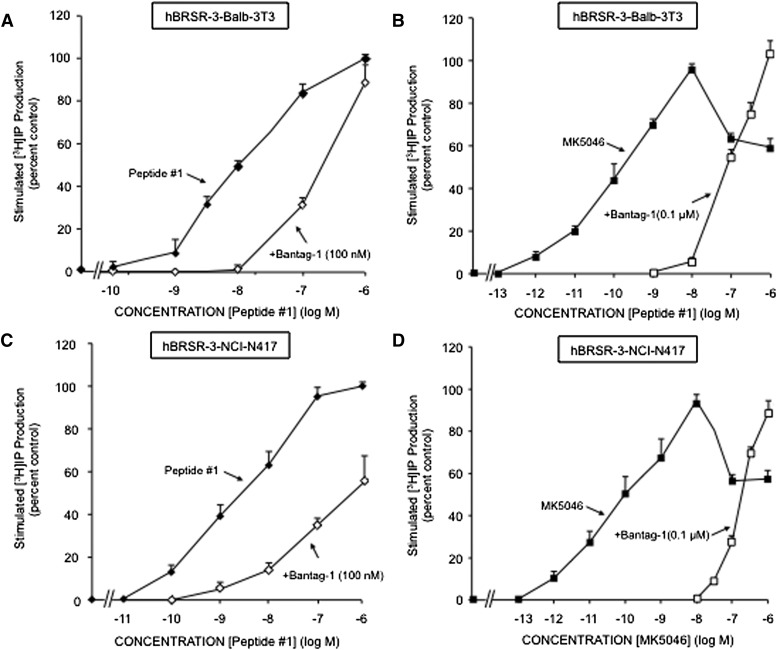
To further explore the ability of Bantag-1 to function as a possible hBRS-3–receptor antagonist, we assessed its ability to inhibit a maximally effective concentration of either peptide #1 (Fig. 5, A and B) or the nonpeptide agonist MK-5046 (Fig. 5, C and D). The antagonist Bantag-1 causes a concentration-dependent inhibition of PLC stimulation caused by both agonists (i.e., peptide #1, MK-5046) in the two hBRS-3 cell types (Fig. 5, A–D). With peptide #1, Bantag-1 caused detectable inhibition at 1 nM, half-maximal inhibition at 52.3 ± 2.7 nM, and complete inhibition at 10 μM in hBRS-3 Balb 3T3 cells (Fig. 5A). In NCI-N417 cells, Bantag-1 caused detectable inhibition of stimulation by peptide #1 at 30 nM, half-maximal inhibition at 137 ± 15 nM, and complete inhibition at 10 μM (Fig. 5B). With MK-5046 (0.01 μM) in hBRS-3 Balb 3T3 cells, Bantag-1 caused half-maximal inhibition at 27.5 ± 2.5 nM (Fig. 5C), whereas in NCI-N417 cells half-maximal inhibition occurred at 3.2 ± 0.56 nM (Fig. 5D). A 100 nM concentration of Bantag-1 caused a parallel rightward shift in the peptide #1 (Fig. 6, A and C) and MK-5046 (Fig. 6, B and D) dose-response curves for generation of [3H]IP. A lesser concentration of Bantag-1 caused a proportional shift in the peptide #1 dose-response curve (data not shown), and when this rightward shift was analyzed by determining the Schild coefficient, it was 1.04 ± 0.09 for peptide #1 in hBRS-3 Balb 3T3 cells (not significantly different than unity), compatible with it functioning as a competitive antagonist with an affinity of 0.57 ± 0.15 nM.
Fig. 5.
Bantag-1 inhibits [3H]IP production in hBRS-3 Balb 3T3 (A and C) and NCI-N417 (B and D) cells. (A and B) Antagonist Bantag-1 alters [3H]IP production stimulated by 0.1 μM peptide #1. The results are expressed as the percentage of stimulation caused by peptide #1 (0.1 μM) alone. (C and D) Antagonist Bantag-1 inhibits [3H]IP generation stimulated by 0.01 μM MK-5046. The results are expressed as the percentage of stimulation caused by MK-5046 (0.01 μM) alone. The experimental conditions were described in Fig. 3 and under Materials and Methods. The results are the mean ± S.E.M. from at least four experiments, and in each experiment the data points were determined in duplicate.
Fig. 6.
Bantag-1 alters the dose-response curves of peptide #1 and MK-5046 stimulated [3H]IP in cells containing hBRS-3. (A and C) Effect of the antagonist Bantag-1 (100 nM) on the dose-response curve of peptide #1 for stimulating [3H] production. The results are expressed as the percentage of stimulation caused by a maximal effective concentration of peptide #1 (100 nM) alone. (B and D) Effect of Bantag-1 (100 nM) on the dose-response curve of MK-5046 for stimulating [3H] production. The results are expressed as the percentage of stimulation caused by the maximal effective concentration of MK-5046 (100 nM) alone.
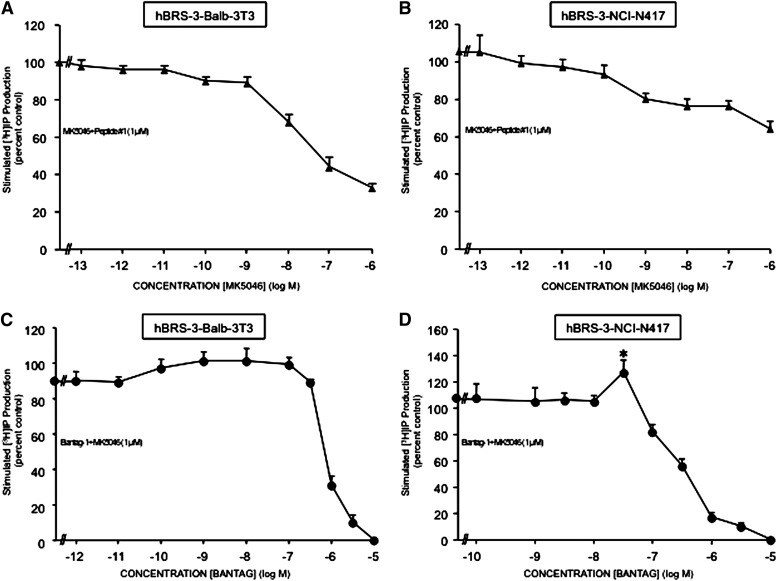
To gain possible insight into the biphasic nature of the MK-5046 dose-response curve for stimulating [3H]IP generation, we examined the effect of adding increasing concentrations of MK-5046 to a maximally effective concentration of peptide #1 (Fig. 7, A and B) or of adding increasing concentrations of the antagonist Bantag-1 to a supramaximal concentration of MK-5046 (Fig. 7, C and D). In both cell types containing hBRS-3-receptor (Fig. 7 A and B), increasing concentrations of MK-5046 resulted in increasing inhibition of the stimulation seen with a maximal effective concentration of peptide #1 (Fig. 7, A and B). Specifically, with hBRS-3 Balb 3T3 cells, MK-5046 caused a detectable inhibition of the maximally stimulated [3H]IP by peptide #1 at 1 nM, half-maximal inhibition at 13.76 ± 0.61 nM, and progressive inhibition with higher doses (Fig. 7A). In NCI-N417 (Fig. 7B), MK-5046 caused progressive inhibition with increasing concentrations >0.1 nM.
Fig. 7.
Increasing concentrations of MK-5046 (A and B) or Bantag-1 (C and D) alter the maximal [3H]IP generation stimulated by peptide #1 (1 μM) (A and B) or by supramaximal MK-5046 (1 μM) (C and D) in hBRS-3 Balb 3T3 and NCI-417 cells. The experimental conditions were described under Materials and Methods. The results are the mean ± S.E.M. of at least four experiments, and in each experiment the data points were determined in duplicate. (A and B) Results expressed as the percentage of stimulation caused by a maximal effective concentration of peptide #1 (1 μM) alone. (C and D) Results expressed as a percentage of the stimulation caused by a supramaximal effective concentration of MK-5046 (1 μM) alone. *P < 0.05 versus 1 μM MK-5046 alone.
These results demonstrate that the supramaximal inhibition caused by high concentrations of MK-5046 can also inhibit maximal stimulation of PLC by peptide #1. To explore further the ability of increasing hBRS-3–receptor occupation to mediate the supramaximal inhibition seen with high concentrations of MK-5046, we assessed the effect of increasing concentrations of the antagonist Bantag-1 when added to a supramaximal concentration of MK-5046, to progressive reduction of the receptor occupation by MK-5046 (Fig. 7, C and D). With increasing concentrations of Bantag-1 in NCI-N417, there was a statistically significant 19 ± 2% increase (P < 0.05) in [3H]IP generation over the stimulation seen by the supramaximal MK-5046 concentration alone (1 μM), demonstrating the reversibility of the biphasic dose-response curve for MK-5046 with decreased hBRS-3–receptor occupation (Fig. 7D).
A similar trend was seen in cells containing hBRS-3 Balb 3T3 (Fig. 7C) over that seen with a supramaximal concentration of MK-5046 alone (1 μM), although the 11 ± 2% increase with increasing Bantag-1 did not reach statistical significance (P < 0.10).
These results demonstrate that increasing occupation of the low-affinity hBRS-3–receptor state mediates the increasing supramaximal inhibition of PLC seen with increasing concentrations of MK-5046, causing the biphasic nature of the MK-5046 dose-response curve.
Ability of the Agonists Peptide #1 and MK-5046 To Stimulate Tyrosine or Serine Phosphorylation in Cells Containing hBRS-3–Receptors.
Activation of Bn-receptor subtypes results in activation of MAPK and stimulation of tyrosine kinases, resulting in phosphorylation of p125FAK, paxillin, and serine/tyrosine phosphorylation of Akt (Tsuda et al., 1997; Ryan et al., 1998b; Jensen et al., 2008; Berna et al., 2009). To assess the ability of the various peptide/nonpeptide agonists to activate various phosphorylation cascades, we determined their abilities to stimulate phosphorylation of MAPK (p42/44), p125FAK, Akt, and paxillin via Western blot analysis in the two cell subtypes containing hBRS-3–receptor, Balb 3T3 and NCI-N417, using specific phosphoantibodies.
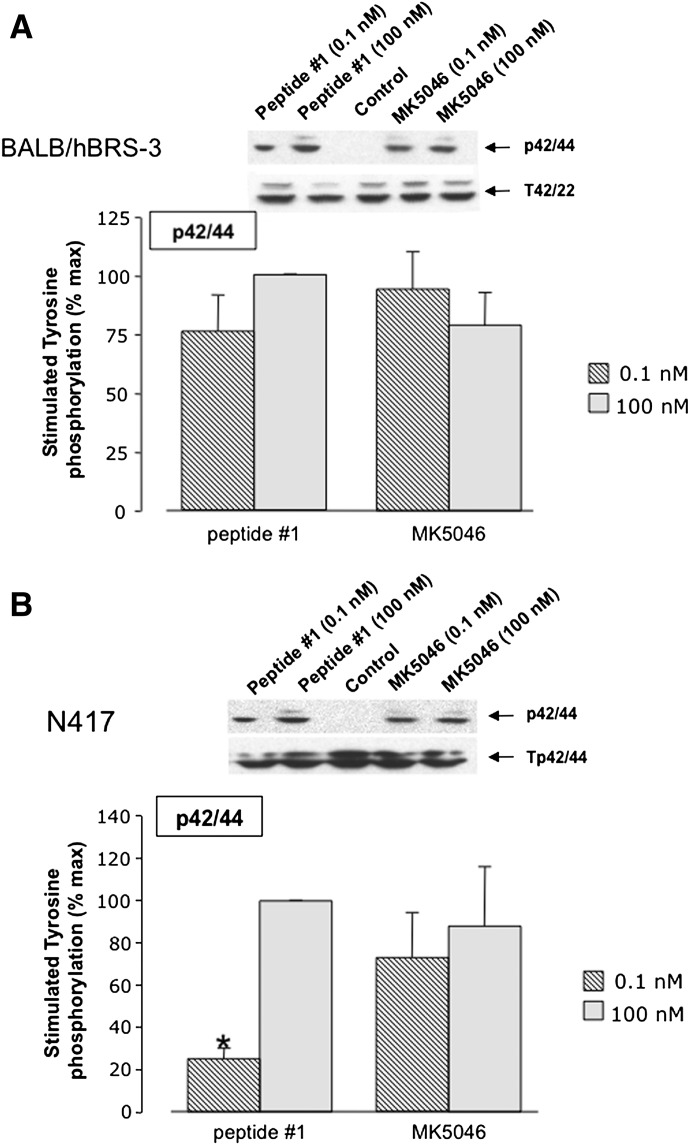
Peptide #1 and MK-5046 stimulated the tyrosine phosphorylation of MAPK in both types of cells containing hBRS-3; in each cell type, they had similar efficacy, but peptide #1 was less potent than MK-5046 in NCI-N417 cells (Fig. 8). A biphasic response was not seen with peptide #1 or MK-5046 in either type of cell with hBRS-3 (Fig. 8, A and B). Likewise, peptide #1 and the nonpeptide agonist MK-5046 stimulated tyrosine phosphorylation of FAK (Tyr397) in hBRS-3-transfected Balb 3T3 cells and native NCI-N417 cells (Fig. 9, A and B). Both peptide #1 and MK-5046 caused similar stimulation of Tyr397 at low concentrations (0.1 nM); at high concentrations (100 nM), peptide #1 was more efficient in NCI-N417 cells (Fig. 9). Furthermore, the two agonists differed in their dose-response curve configuration in NCI-N417 cells where MK-5046 demonstrated a biphasic effect with greater stimulation with the 0.1 nM than the 100 nM dose (Fig. 9B).
Fig. 8.
Peptide #1 and MK-5046 activate MAPK in two different human BRS-3 cell lines: (A) Balb 3T3 or (B) NCI-N417. Above each panel are the representative Western blots of p42/44 MAPK phosphorylation in Balb/hBRS-3 cells. Results are shown of NCI-417 cells treated with 0.1 nM and 100 nM peptide #1 or MK-5046 and incubated for 3 minutes at 37°C. Each panel shows the mean ± S.E.M. from at least four experiments. The results are expressed as the percentage of the maximal concentration of peptide #1 (100 nM). *P < 0.05 versus 100 nM peptide #1.
Fig. 9.
Peptide #1 and MK-5046 stimulate Tyr397 phosphorylation of FAK in (A) hBRS-3-transfected Balb 3T3 cells and (B) NCI-N417 native hBRS-3 cells. Above each panel are representative Western blots of the stimulation of p125FAK(Tyr397) in Balb 3T3 cells (A) and NCI-N417 cells (B) treated with 0.1 nM and 100 nM of peptide #1 or MK-5046 and incubated for 3 minutes at 37°C. Each panel shows the mean ± S.E.M. from at least four experiments. The results are expressed as the percentage of the maximal concentration of peptide #1 (100 nM). *P < 0.05 versus 100 nM peptide #1. #P < 0.05 versus 100 nM MK-5046.
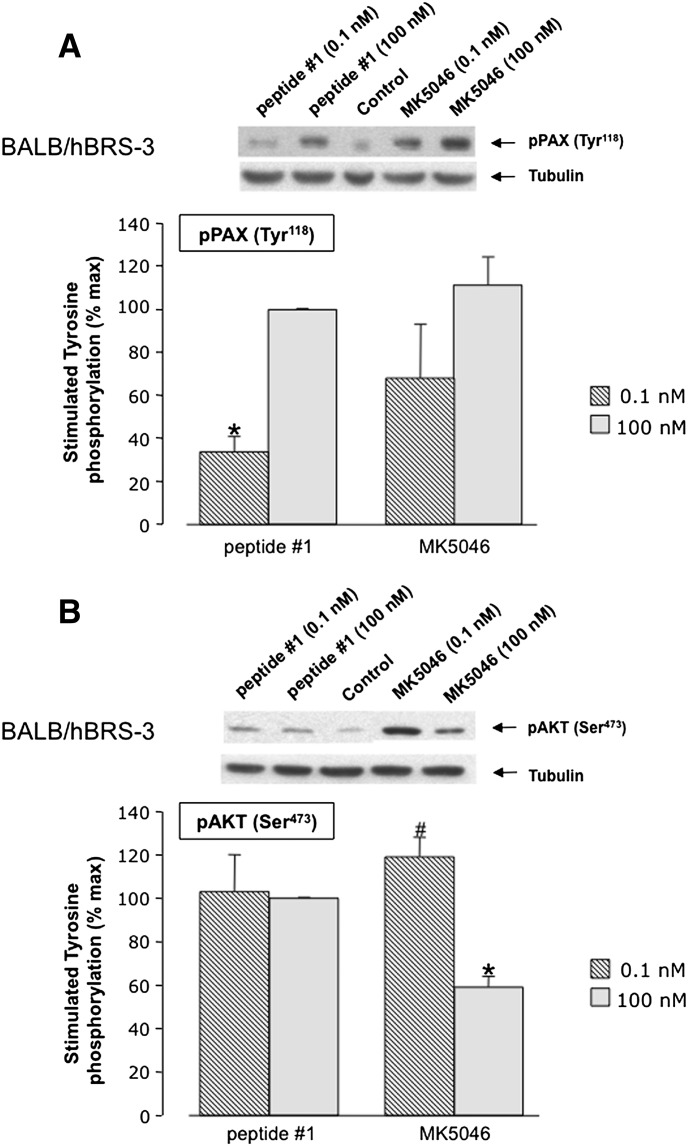
In hBRS-3-transfected 3T3 Balb cells, peptide #1 and MK-5046 stimulated the phosphorylation of paxillin (Tyr118) to a similar extent, but MK-5046 was slightly more potent than peptide #1 (Fig. 10A). Phosphorylation of Akt (Ser473) also was stimulated by peptide #1 and MK-5046 (Fig. 10B); both had a similar maximal efficiency at the 0.1 nM concentration. However, they differed in that MK-5046 showed supramaximal inhibition at a high concentration (i.e., 100 nM)---nM MK-5046 caused only 50% of the maximal stimulation seen with 0.1 nM (P< 0.05) (Fig. 10B).
Fig. 10.
Peptide #1 and MK-5046 stimulate Tyr118 phosphorylation of (A) paxillin and (B) Ser473 phospho-Akt in hBRS-3 Balb 3T3 transfected cells. Above each panel are representative Western blots of the stimulation of paxillin phosphorylation in cells treated with 0.1 nM and 100 nM of peptide #1 or MK-5046 for 3 minutes at 37°C. Each panel provides the results of densitometry analysis with the mean ± S.E.M. from at least four experiments. The results are expressed as the percentage of maximal concentration of peptide #1 (100 nM). *P < 0.05 versus 100 nM peptide #1. #P < 0.05 versus 100 nM MK-5046.
Ability of Peptide #1 and MK-5046 To Stimulate [3H]Arachidonic Acid Release in hBRS Balb 3T3 Cells.
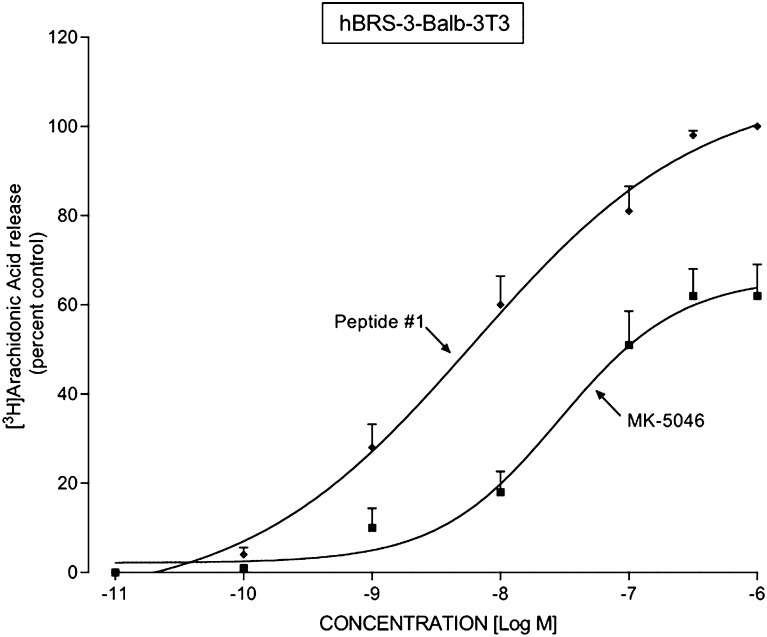
Activation of Bn receptors in a number of cells (Currie et al., 1992) results in activation of PLA2, which stimulates arachidonic acid release. We assessed the ability of peptide #1 and MK-5046 to stimulate PLA2 in hBRS-3 Balb 3T3-transfected cells (Fig. 11). The universal ligand peptide #1 and the nonpeptide MK-5046 each stimulated the release of [3H]AA in a dose-dependent manner. Peptide #1 caused a detectable increase at 1 nM, a half-maximal increase at 4.29 ± 0.31 nM, and a maximal increase at 100 nM (Fig. 11). MK-5046 was 22-fold less potent (EC50 97.2 ± 7.0 nM), and it was less efficacious stimulating a maximal increase of only 62 ± 7.1% of the maximal effect of peptide #1 (Fig. 11).
Fig. 11.
Peptide #1 and MK-5046 stimulate [3H]arachidonic acid release in hBRS-3 Balb 3T3 transfected cells. The hBRS-3 Balb cells (5 × 104 cells/ml) were incubated with [3H-5,6,8,9,11,12,14,15]arachidonic acid (1 μCi/4 ml, [3H]AA) for 24 hours at 37°C, washed, and incubated with the indicated unlabeled concentration of MK-5046 or peptide #1 for 40 minutes at 37°C. [3H]AA release was determined as described under Materials and Methods. The results are expressed as the percentage of the release seen with a maximally effective concentration of peptide #1 (1 μM) and are the mean ± S.E.M. from at least four experiments. In each experiment, the data points were determined in duplicate. The control and the maximal stimulated (1 μM peptide #1) values were 901 ± 94 and 1542 ± 113 dpm, respectively (n = 5).
Arachidonic acid can be released from membrane phospholipids by a number of mechanisms (Husain and Abdel-Latif, 1998). To provide insight into the mechanism of arachidonic acid release in hBRS-3 cells, we preincubated them (Table 3) with quinacrine or manoalide (PLA2 inhibitors); arachidonyltrifluoromethane (AACOCF3), a selective cytosolic PLA2 inhibitor; RHC-80267, a diacylglycerol (DAG) lipase inhibitor; or U-73122, a PLC inhibitor. Only the PLA2 inhibitors decreased peptide #1 stimulation of arachidonic acid release, whereas the PLC (U-73122) or DAG (RHC-80267) inhibitors had no effect (Table 3). Furthermore, Bantag-1 completely inhibited peptide #1 stimulation of arachidonic acid release, demonstrating that the effect of peptide #1 was mediated by hBRS-3 interaction (Table 3).
TABLE 3.
Effect of various inhibitors on the ability of peptide #1 to stimulate phospholipase A2 and release [3H]arachidonic acid in hBRS Balb 3T3 cells
Cells were incubated with 1 μCi/4 ml of [3H-5,6,8,9,11,12,14,15]arachidonic acid for 24 hours, then washed and incubated with the indicated unlabeled compound for 40 minutes at 37°C. [3H]AA release was determined as described under Materials and Methods. Results are mean ± S.E.M. from at least four experiments.
| Additions |
[3H]Arachidonic Acid Released |
|---|---|
| dpm | |
| None | 366 ± 41 |
| Peptide #1 alone (0.1 μM) | 874 ± 121a |
| + Bantag-1 (1 μM) | 432 ± 56b |
| + Manoalide (1 μM) | 553 ± 44b |
| + Quinacrine (1 μM) | 614 ± 32b |
| + AACOCF3 (1 μM) | 574 ± 63b |
| + U-73122 (1 μM) | 950 ± 123 |
| + RHC-80267 (1 μM) | 1067 ± 224 |
P < 0.01 compared with no additions.
P < 0.005 compared with peptide #1 alone.
Time Course of Peptide #1 and MK-5046 Activation of [3H]IP Generation and Stimulation of MAPK Phosphorylation in hBRS Balb 3T3 Cells.
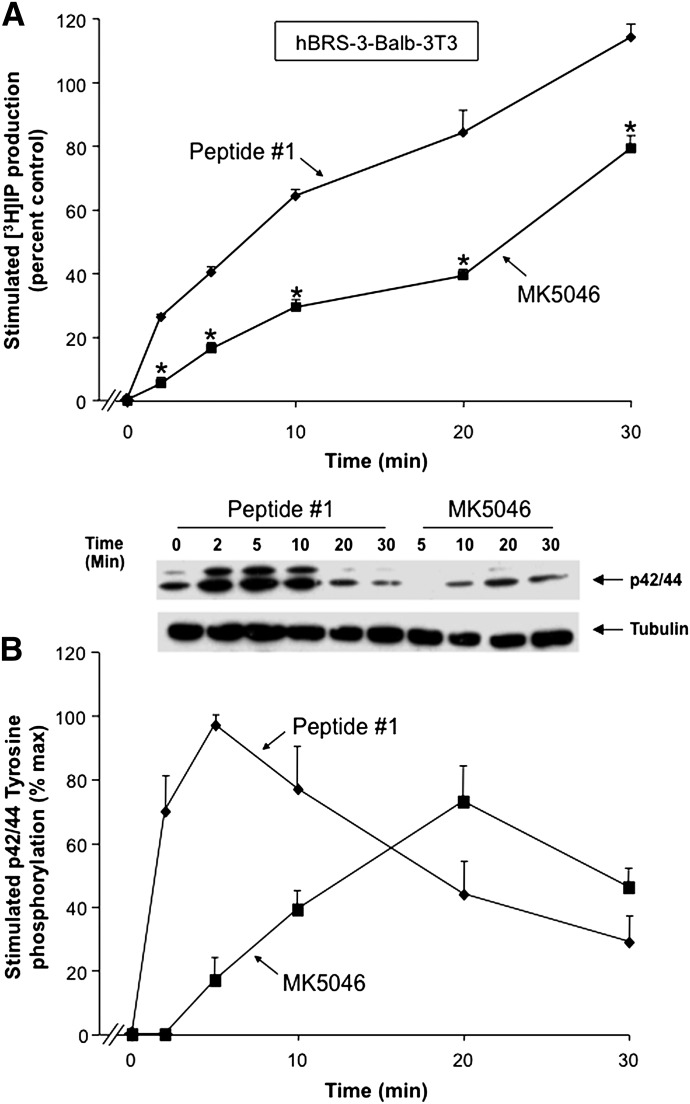
Stimulation of [3H]IP production by either peptide #1 or MK-5046 was time-dependent, reaching an increase after a 30-minute incubation with peptide #1 of 114 ± 14% over control (P < 0.05) and with MK-5046 of 79 ± 4% over control (P < 0.05) (Fig. 12A). Although both hBRS-3-receptor agonists stimulated the generation of [3H]IP, the universal ligand peptide #1 caused a greater increase of [3H]IP generation at all times than was caused by the nonpeptide MK-5046, even though a maximally effective concentration of each was used (Fig. 12A). Specifically, peptide #1 caused a 26 ± 1% increase of [3H]IP production at 2.5 minutes, was 5-fold more efficacious than MK-5046 at this time, and at 9.2 minutes caused a half-maximal increase of that seen at 30 minutes. In contrast, MK-5046 stimulated only 5 ± 0.1% of the control at 2.5 minutes; a half-maximal increase occurred at 20.3 minutes, which was 1.5-fold less than with peptide #1 (Fig. 12A).
Fig. 12.
Time course of peptide #1 and MK-5046 stimulation of [3H]IP production and p42/44 MAPK phosphorylation in hBRS-3 Balb 3T3 transfected cells. (A) Maximal concentration of the agonists peptide #1 (10 nM) and MK-5046 (100 nM) to stimulate [3H]IP generation. The experiment was performed as described in Fig. 3 and under Materials and Methods, and the results are expressed as the percentage of control over basal stimulation. (B) Representative Western blot of p42/44 MAPK phosphorylation in Balb/hBRS-3 cells, and results of the experiment performed as described in Fig. 8 and under Materials and Methods. The concentrations are the same as in A. The results are expressed as the percentage of maximum stimulation caused by a maximal effective concentration of peptide # 1 (10 nM). The results in both panels are the mean ± S.E.M. from at least four experiments, and in each experiment the data points were determined in duplicate. *P < 0.05 versus peptide #1.
In hBRS-3–transfected Balb 3T3 cells, the phosphorylation of MAPK by the universal ligand peptide #1 and the nonpeptide MK-5046 was time-dependent, with a maximal increment after 5 minutes’ incubation by peptide #1 (97 ± 3% over control, P < 0.05), and in the case of MK-5046, after a 20-minute incubation (73 ± 11% over control, P < 0.05) (Fig. 12B). As we show in Fig. 12A, with stimulation of [3H]IP, peptide #1 caused more rapid phosphorylation of p42/44 MAPK with a maximum increase at 5 minutes with a t1/2 of 1.82 minutes (Fig. 12B). In contrast, the maximal increment with MK-5046 occurred at 20 minutes, with t1/2 of 13.5 minutes; the maximal value was 25% less than seen with peptide #1 (Fig. 12B). However, both agonists showed a biphasic time-course curve for stimulation of p42/44 MAPK phosphorylation (Fig. 12B). MK-5046 exhibited a slower rate of p42/44 MAPK phosphorylation, but its duration of action was more sustained than with peptide #1, because its stimulation decreased from maximal by only 27 ± 6% at 30 minutes. In contrast, with peptide #1 at 30 minutes there was a 68 ± 8% decrease in maximal stimulation (Fig. 12B).
Discussion
Our results support the conclusion that the nonpeptide MK-5046 functions as a selective, high-affinity hBRS-3 agonist and that the peptide-Bantag-1 functions as a selective, high-affinity hBRS-3 antagonist. First, our binding data demonstrate that MK-5046 and Bantag-1 have a relatively high affinity for hBRS-3 (Ki 1–19 nM). However, both dose-inhibition curves were broad, extending over >4-log range, and both had Hill coefficients <1 and were best-fitted by a two binding-site model, which demonstrates that hBRS-3 can exist, in both a high- and a low-affinity state. In the present study, in two different hBRS-3-containing cells, MK-5046 had a high affinity for the high-affinity BRS-3-binding site (0.08 nM), as did Bantag-1 (IC50, 0.017–0.029 nM). Second, both MK-5046 and Bantag-1 were highly selective for hBRS-3 (>800-fold) over the other two mammalian Bn receptors (hGRP-R and hNMB-R). In contrast, the agonist peptide #1, as was reported previously by others (Mantey et al., 1997; Pradhan et al., 1998; Sancho et al., 2010), showed a high affinity for all human Bn receptors (IC50 2–6 nM). Third, our results demonstrate that MK-5046 is a hBRS-3 agonist; in two different cell types containing hBRS-3, it activated PLC, stimulated various tyrosine/serine phosphorylation cascades (p125FAK, paxillin, Akt), and activated PLA2. Fourth, that Bantag-1 functioned as a BRS-3 antagonist was demonstrated by its ability to inhibit PLC activation by either peptide #1 or MK-5046. Furthermore, increasing concentrations of Bantag-1 caused a parallel-rightward shift in both the MK-5046/peptide #1 dose-response curves for IP generation. When this rightward shift was analyzed by a Schild plot, the Schild coefficient was not different than unity, compatible with it functioning as a competitive BRS-3 antagonist.
These results show a number of similarities as well as differences from previous studies of hBRS-3 or other Bn receptors. These results demonstrate that the nonpeptide agonist MK-5046 and the peptide antagonist Bantag-1 have as high a selectivity for hBRS-3 over the other Bn receptors, as the natural ligands GRP and NMB have for their receptors (814- to 1069-fold) over the other Bn receptors (Jensen et al., 2008; Uehara et al., 2011). Similarly, the antagonist Bantag-1 has as great a selectivity for hBRS-3, as the best GRP-R/NMB-R antagonist in any of the 10 different GRP/NMB receptor antagonist classes that have been described (Heinz-Erian et al., 1987; Coy et al., 1989; Wang et al., 1990a,b; Jensen and Coy, 1991; Gonzalez et al., 2009). Our results differ from findings with the GRP/NMB receptors whose binding dose-inhibition curves are best-fitted only by a single binding-site model (Mantey et al., 1993; Jensen et al., 2008) because we found that BRS-3–receptor exists in both a high-affinity and low-affinity state. Our results are similar to the previous studies that demonstrated that the nonpeptide agonist MK-5046 and the peptide antagonist–Bantag-1 have high affinity for hBRS-3 and that MK-5046 did not interact with either hGRP-R or hNMB-R even in the micromolar range (Guan et al., 2010; Sebhat et al., 2011). Our results differ from findings previously reported with Bantag-1 in which the antagonist was found to interact with both hGRP-R and hNMB-R in the micromolar range (Guan et al., 2010). Compared with other G protein–coupled receptors, our results with the nonpeptide hBRS-3 agonist MK-5046 are similar to the interaction of the nonpeptide agonist SR146131 (2-[2-[[4-(4-chloro-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]acetic acid) with CCK1-receptors (Gouldson et al., 1999), FR190997 (8-[2,6-dichloro-3-[N-[(E)-4-(N-methylcarbamoyl)cinnamidoacetyl]-N-methylamino]-benzyloxy]-2-methyl-4-(2-pyridylmethoxy)quinoline) with B2 receptors (Aramori et al., 1997), and compound 21 with AT1 receptors (Wan et al., 2004) in that all had a high affinity (IC50 < 10 nM) and also high selectivity (>10,000-fold over the other receptor families).
In addition, our results are similar to the findings of a previous study with a selective CCK1 agonist SR146131, where binding to the CCK1 receptor was best fitted by a two binding-site model with a very high affinity (Ki 0.8 nM) for the high-affinity site (Gouldson et al., 1999). These results demonstrate that nonpeptide agonists differ from many nonpeptide antagonists in their ability to distinguish two receptor sites for some G protein–coupled receptors (CCK1-R and hBRS-3) (Huang et al., 1989, 1994; Gouldson et al., 1999; Jensen et al., 2008).
In this study, we found a number of similarities as well as important differences between nonpeptide-agonist MK-5046 and the peptide agonist peptide #1 in their ability to activate hBRS-3 as well as in their receptor coupling. Our results demonstrate differences in the configuration of their dose-response curves for activating various cell-signaling cascades, their efficacy for activating different signaling cascades, their binding-activation coupling, and their kinetics/duration of cell activation. In regard to the differences in the configuration of their dose-response curves for cell-signaling activation, with both stimulation of PLC and stimulation of Ser/Tyr phosphorylation cascades (i.e., pSer473 Akt, pTyr397p125FAK), the dose-response curves for MK-5046 were biphasic, whereas with peptide agonist peptide #1 the dose-response curves were monophasic. These results raise the possibility that occupation of the high and low hBRS-3 states by MK-5046 could account for the biphasic dose-response curves.
This conclusion was supported by a number of our results from detailed comparative studies of PLC activation by MK-5046 and peptide #1. We found that the antagonist Bantag-1 caused a rightward shift in the MK-5046 dose-response curve for PLC activation, suggesting that both phases of the dose-response curve were mediated by increasing hBRS-3 occupation. This conclusion was supported by the effect of adding increasing concentrations of Bantag-1 to a supramaximal concentration of MK-5046, which demonstrated reversal of the inhibition and changed the MK-5046 dose-response curve from biphasic to monophasic. Furthermore, adding a supramaximal concentration of MK-5046 to a maximal concentration of peptide #1 changed peptide #1’s dose-response curve in a reverse direction, from monophasic to biphasic.
These results show a number of similarities as well as differences from previous studies of nonpeptide agonists of other G protein-coupled receptors. Our results are similar to those obtained with the natural ligand angiotensin II for AT1 receptors (Perlman et al., 1995), which has a monophasic shape for IP accumulation. The nonpeptide agonist L-162,313 (5,7-dimethyl-2-ethyl-3-[[4-[2(n-butyloxycarbonylsulfonamido)-5-isobutyl-3-thienyl]phenyl]methyl]-imadazo[4,5-b]pyridine) has a biphasic dose-response curve. However, our results differ from the majority of the studies with nonpeptide agonists where both the peptide and nonpeptide agonists show monophasic dose-response curves for cell activation (Bignon et al., 1999; Aramori et al., 1997; Asano et al., 1998).
A number of our results lead to the conclusion that there are differences in the ability of peptide (peptide #1) and nonpeptide (MK-5046) agonists to activate different signaling cascades. Whereas both hBRS-3 agonists stimulated the activation of PLA2, MK-5046 was less efficacious than peptide #1. However, MK-5046 functioned as a full agonist compared with peptide #1 for activation of PLC and various Tyr/Ser-phosphorylation cascades (MAPK, paxillin, p125FAK, Akt). Furthermore, the relative potencies of MK-5046 and peptide #1 for activating the PLC, PLA2, or Tyr/Ser-phosphorylation cascades differed. The nonpeptide MK-5046 was 22-fold less potent than peptide #1 for activating PLA2 (EC50, 97.2 nM), but MK-5046 was 130- to 178-fold more potent than peptide #1 for stimulating PLC (EC50 0.05–0.08 nM). Similarly, MK-5046 was more potent than peptide #1 at stimulating the phosphorylation of p42/44 MAPK and paxillin, but the potencies of the two agonists were similar for stimulating the phosphorylation of Tyr397p125FAK or Ser473-Akt. These results demonstrate that nonpeptide MK-5046 and peptide agonist peptide #1 have different abilities to activate different intracellular cascades.
Compared with previous studies with other nonpeptide agonists, our results are similar to those with the nonpeptide CCK1-R agonist SR146131 (Bignon et al., 1999), which is a full agonist compared with the natural peptide ligand CCK-8S for activating PLC but acts as a partial agonist for activation of MAPK. However, our data with MK-5046 differ from those obtained with the CCK1-R nonpeptide agonist SR146131 in other studies, as well as from the majority of other nonpeptide agonists that have been described—L162,313 for AT1-R, FR190997 for B2-R, and OPC-51803 [(5R)-2-[1-(2-chloro-4-(1-pyrrolidinyl)benzoyl)-2,3,4,5-tetrahydro-1H-1-benzazepin-5-yl]-N-isopropylacetamide] for V2-R (Aramori et al., 1997; Perlman et al., 1997; Gouldson et al., 1999; Bignon et al., 1999; Nakamura et al., 2000)—in that MK-5046 activates PLC/Tyr/Ser signaling cascades with a greater potency than does the natural ligand peptide #1. These other nonpeptide agonists are less potent than their respective receptor’s natural peptides for activating PLC.
In this study, we also found differences in the receptor coupling of the two agonists, peptide #1 and nonpeptide MK-5046. Specifically, peptide #1 has a 7-fold higher receptor affinity than MK-5046; however, MK-5046 was 160-fold more potent at activating PLC. Similarly, MK-5046 was more potent than peptide #1 at stimulating MAPK/paxillin phosphorylation, whereas MK-5046 had the same potency as peptide #1 for activating the other signaling cascades (p125FAK, Akt). These results demonstrate that the relationship between receptor occupation and activation differs markedly for a peptide agonist and a nonpeptide agonist. At present, the molecular basis for this difference is unknown. Previous studies have demonstrated for peptide #1 that the molecular receptor determinants of MK-5046 receptor agonism involve amino acids on opposing faces of the extracellular segments of transmembrane (TM) region III, TM-IV, and TM-VII (Gbahou et al., 2010), and binding studies have shown that its affinity depends on interaction with key residues in the extracellular domain 4 (Liu et al., 2002). Unfortunately no comparative data exist for MK-5046.
Our results also differ from those of previous studies with the nonpeptide agonists L-162,313 with the AT1 receptor (Perlman et al., 1997), FR190997 with the B2 receptor (Aramori et al., 1997), and OPC-51803 with the V2 receptor (Nakamura et al., 2000) in that in each case the natural ligand shows a higher affinity for the receptor and was more potent than the nonpeptide agonist at stimulating different signaling cascades.
Our study found differences in the kinetics/duration of action of the two agonist ligands peptide #1 and MK-5046 in stimulating IP generation/p42/44MAPK phosphorylation. Peptide #1 had a more rapid onset and shorter duration of action for each. This difference was not due to a difference in stability with the two agonists, as previous studies had demonstrated that each is stable under incubation conditions at 37°C for at least 1 hour (Mantey et al., 1997; Sebhat et al., 2011; Reitman et al., 2012). Compared with previous studies that investigated the kinetics/duration of action of peptide and nonpeptide agonists, our results are similar to those with the CCK1-receptor nonpeptide agonist SR146131 (Bignon et al., 1999; Schaeffer et al., 2000), where the nonpeptide had a more prolonged duration of the action in stimulating [Ca2+]i when it was compared with an equally effective dose of the natural agonist CCK-8S. Also our results are similar to those of two other studies in which the AT1-receptor nonpeptide agonist L-162,313 (Kivlighn et al., 1995) and the B2-receptor nonpeptide agonist FR190997 (Aramori et al., 1997) were found to have more prolonged action on blood pressure compared with the natural peptide receptor ligand.
In conclusion, our study demonstrates that both nonpeptide MK-5046 and peptide Bantag-1 have high selectivity for hBRS-3 and function as a selective agonist and antagonist, respectively. A detailed comparison of the pharmacologic properties of the nonpeptide agonist MK-5046 versus a peptide agonist for hBRS-3 showed that they differ in their potencies and efficacies for activating various signaling cascades, in their relationship between receptor coupling and cellular activation, and in their kinetics/duration of action. These results demonstrate that although these agonists both bind to the same receptor and activate both its high-affinity and low-affinity states, they differ markedly in pharmacologic properties and ability to activate different signaling cascades. These results suggest that nonpeptide/peptide hBRS-3-receptor activation is not always concordant and could lead to markedly different cellular responses.
Abbreviations
- AACOCF3
arachidonyltrifluoromethane
- β-Ala
β-alanine
- Bantag-1
Boc-Phe-His-4-amino-5-cyclohexyl-2,4,5-trideoxypentonyl-Leu-(3-dimethylamino) benzylamide N-methylammonium trifluoroacetate
- Bn
bombesin
- BRS-3
bombesin receptor subtype-3
- BSA
bovine serum albumin fraction V
- CCK
cholecystokinin
- CCK-R
cholecystokinin receptor
- DMEM
Dulbecco’s minimum essential medium
- DTT
dithiothreitol
- FAK
focal adhesion kinase
- FBS
fetal bovine serum
- FR190997
8-[2,6-dichloro-3-[N-[(E)-4-(N-methylcarbamoyl)cinnamidoacetyl]-N-methylamino]-benzyloxy]-2-methyl-4-(2-pyridylmethoxy)quinoline
- GF109203X
2-[1-(3-dimethylaminopropyl)-1H-indol-3-yl]-3-(1H-indol-3-yl)-maleimide
- GRP
gastrin-releasing peptide
- GRP-R
gastrin-releasing peptide receptor
- [3H]AA
[3H-5,6,8,9,11,12,14,15]arachidonic acid
- hBRS-3
human bombesin receptor subtype-3
- HuTu-80
human duodenal cancer cell line
- IP
inositol phosphate
- L-162,313
5,7-dimethyl-2-ethyl-3-[[4-[2(n-butyloxycarbonylsulfonamido)-5-isobutyl-3-thienyl]phenyl]methyl]-imadazo[4,5-b]pyridine
- MAPK
mitogen-activated protein kinase
- MK-5046
(2S)-1,1,1-trifluoro-2-[4-(1H-pyrazol-1-yl)phenyl]-3-(4-{[1-(trifluoromethyl)cyclopropyl]methyl}-1H-imidazol-2-yl)propan-2-ol
- NMB
neuromedin B
- NCI-H1299
non-small-cell lung cancer cell line
- NCI-N417
human small-cell lung carcinoma cell line
- NMB-R
neuromedin B receptor
- peptide #1
[d-Tyr6, βAla11, Phe13, Nle14]Bn-(6–14)
- OPC-51803
(5R)-2-[1-(2-chloro-4-(1-pyrrolidinyl)benzoyl)-2,3,4,5-tetrahydro-1H-1-benzazepin-5-yl]-N-isopropylacetamide
- PLC
phospholipase C
- PLA2
phospholipase A2
- RHC-80267
1,6-bis(cyclohexyloximinocarbonylamino)hexane, diacylgylcerol lipase inhibitor
- SR146131
2-[2-[[4-(4-chloro-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]acetic acid
- TBS
Tris-buffered saline
- U-73122
1-(6-((17β)-3-methoxyestra-1,3,5[10]-trien-17-yl)amino]hexyl)-1H-pyrrole-2,5-dione
Authorship Contributions
Participated in research design: Moreno, Mantey, Nuche-Berenguer, Reitman, González, Coy, Jensen.
Conducted experiments: Moreno, Mantey, Jensen.
Contributed new reagents or analytic tools: Reitman, Coy, Mantey, Nuche-Berenguer, Jensen.
Performed data analysis: Moreno, Nuche-Berenguer, González, Jensen.
Wrote or contributed to the writing of the manuscript: Moreno, Mantey, Nuche-Berenguer, Reitman, González, Coy, Jensen.
Footnotes
This work was partially supported by the Intramural Research Program of the National Institutes of Health [National Institute of Diabetes and Digestive and Kidney Diseases] [Grants ZIA DK053100-22, ZIA DK053101-22].
References
- Aramori I, Zenkoh J, Morikawa N, Asano M, Hatori C, Sawai H, Kayakiri H, Satoh S, Inoue T, Abe Y, et al. (1997) Nonpeptide mimic of bradykinin with long-acting properties at the bradykinin B2 receptor. Mol Pharmacol 52:16–20 [DOI] [PubMed] [Google Scholar]
- Asano M, Hatori C, Sawai H, Johki S, Inamura N, Kayakiri H, Satoh S, Abe Y, Inoue T, Sawada Y, et al. (1998) Pharmacological characterization of a nonpeptide bradykinin B2 receptor antagonist, FR165649, and agonist, FR190997. Br J Pharmacol 124:441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benya RV, Fathi Z, Kusui T, Pradhan T, Battey JF, Jensen RT. (1994) Gastrin-releasing peptide receptor-induced internalization, down-regulation, desensitization, and growth: possible role for cyclic AMP. Mol Pharmacol 46:235–245 [PubMed] [Google Scholar]
- Benya RV, Kusui T, Pradhan TK, Battey JF, Jensen RT. (1995) Expression and characterization of cloned human bombesin receptors. Mol Pharmacol 47:10–20 [PubMed] [Google Scholar]
- Benya RV, Wada E, Battey JF, Fathi Z, Wang LH, Mantey SA, Coy DH, Jensen RT. (1992) Neuromedin B receptors retain functional expression when transfected into BALB 3T3 fibroblasts: analysis of binding, kinetics, stoichiometry, modulation by guanine nucleotide-binding proteins, and signal transduction and comparison with natively expressed receptors. Mol Pharmacol 42:1058–1068 [PubMed] [Google Scholar]
- Berna MJ, Hoffmann KM, Tapia JA, Thill M, Pace A, Mantey SA, Jensen RT. (2007) CCK causes PKD1 activation in pancreatic acini by signaling through PKC-delta and PKC-independent pathways. Biochim Biophys Acta 1773:483–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna MJ, Tapia JA, Sancho V, Thill M, Pace A, Hoffmann KM, Gonzalez-Fernandez L, Jensen RT. (2009) Gastrointestinal growth factors and hormones have divergent effects on Akt activation. Cell Signal 21:622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignon E, Bachy A, Boigegrain R, Brodin R, Cottineau M, Gully D, Herbert JM, Keane P, Labie C, Molimard JC, et al. (1999) SR146131: a new potent, orally active, and selective nonpeptide cholecystokinin subtype 1 receptor agonist. I. In vitro studies. J Pharmacol Exp Ther 289:742–751 [PubMed] [Google Scholar]
- Coy DH, Taylor JE, Jiang NY, Kim SH, Wang LH, Huang SC, Moreau JP, Gardner JD, Jensen RT. (1989) Short-chain pseudopeptide bombesin receptor antagonists with enhanced binding affinities for pancreatic acinar and Swiss 3T3 cells display strong antimitotic activity. J Biol Chem 264:14691–14697 [PubMed] [Google Scholar]
- Currie S, Smith GL, Crichton CA, Jackson CG, Hallam C, Wakelam MJ. (1992) Bombesin stimulates the rapid activation of phospholipase A2-catalyzed phosphatidylcholine hydrolysis in Swiss 3T3 cells. J Biol Chem 267:6056–6062 [PubMed] [Google Scholar]
- Feng Y, Guan XM, Li J, Metzger JM, Zhu Y, Juhl K, Zhang BB, Thornberry NA, Reitman ML, Zhou YP. (2011) Bombesin receptor subtype-3 (BRS-3) regulates glucose-stimulated insulin secretion in pancreatic islets across multiple species. Endocrinology 152:4106–4115 [DOI] [PubMed] [Google Scholar]
- Freidinger RM. (1993) Toward peptide receptor ligand drugs: progress on nonpeptides. Prog Drug Res 40:33–98 [DOI] [PubMed] [Google Scholar]
- Gbahou F, Holst B, Schwartz TW. (2010) Molecular basis for agonism in the BB3 receptor: an epitope located on the interface of transmembrane-III, -VI, and -VII. J Pharmacol Exp Ther 333:51–59 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Hocart SJ, Portal-Nuñez S, Mantey SA, Nakagawa T, Zudaire E, Coy DH, Jensen RT. (2008) Molecular basis for agonist selectivity and activation of the orphan bombesin receptor subtype 3 receptor. J Pharmacol Exp Ther 324:463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González N, Mantey SA, Pradhan TK, Sancho V, Moody TW, Coy DH, Jensen RT. (2009) Characterization of putative GRP- and NMB-receptor antagonist’s interaction with human receptors. Peptides 30:1473–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouldson P, Legoux P, Carillon C, Delpech B, Le Fur G, Ferrara P, Shire D. (1999) Contrasting roles of leu(356) in the human CCK(1) receptor for antagonist SR 27897 and agonist SR 146131 binding. Eur J Pharmacol 383:339–346 [DOI] [PubMed] [Google Scholar]
- Guan XM, Chen H, Dobbelaar PH, Dong Y, Fong TM, Gagen K, Gorski J, He S, Howard AD, Jian T, et al. (2010) Regulation of energy homeostasis by bombesin receptor subtype-3: selective receptor agonists for the treatment of obesity. Cell Metab 11:101–112 [DOI] [PubMed] [Google Scholar]
- Guan XM, Metzger JM, Yang L, Raustad KA, Wang SP, Spann SK, Kosinski JA, Yu H, Shearman LP, Faidley TD, et al. (2011) Antiobesity effect of MK-5046, a novel bombesin receptor subtype-3 agonist. J Pharmacol Exp Ther 336:356–364 [DOI] [PubMed] [Google Scholar]
- Heinz-Erian P, Coy DH, Tamura M, Jones SW, Gardner JD, Jensen RT. (1987) [D-Phe12]bombesin analogues: a new class of bombesin receptor antagonists. Am J Physiol 252:G439–G442 [DOI] [PubMed] [Google Scholar]
- Huang SC, Fortune KP, Wank SA, Kopin AS, Gardner JD. (1994) Multiple affinity states of different cholecystokinin receptors. J Biol Chem 269:26121–26126 [PubMed] [Google Scholar]
- Huang SC, Zhang L, Chiang HC, Wank SA, Maton PN, Gardner JD, Jensen RT. (1989) Benzodiazepine analogues L365,260 and L364,718 as gastrin and pancreatic CCK receptor antagonists. Am J Physiol 257:G169–G174 [DOI] [PubMed] [Google Scholar]
- Husain S, Abdel-Latif AA. (1998) Role of protein kinase C alpha in endothelin-1 stimulation of cytosolic phospholipase A2 and arachidonic acid release in cultured cat iris sphincter smooth muscle cells. Biochim Biophys Acta 1392:127–144 [DOI] [PubMed] [Google Scholar]
- Jensen RT, Battey JF, Spindel ER, Benya RV. (2008) International Union of Pharmacology. LXVIII. Mammalian bombesin receptors: nomenclature, distribution, pharmacology, signaling, and functions in normal and disease states. Pharmacol Rev 60:1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RT, Coy DH. (1991) Progress in the development of potent bombesin receptor antagonists. Trends Pharmacol Sci 12:13–19 [DOI] [PubMed] [Google Scholar]
- Kivlighn SD, Huckle WR, Zingaro GJ, Rivero RA, Lotti VJ, Chang RS, Schorn TW, Kevin N, Johnson RG, Jr, Greenlee WJ, et al. (1995) Discovery of L-162,313: a nonpeptide that mimics the biological actions of angiotensin II. Am J Physiol 268:R820–R823 [DOI] [PubMed] [Google Scholar]
- Ladenheim EE, Hamilton NL, Behles RR, Bi S, Hampton LL, Battey JF, Moran TH. (2008) Factors contributing to obesity in bombesin receptor subtype-3-deficient mice. Endocrinology 149:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lao ZJ, Zhang J, Schaeffer MT, Jiang MM, Guan XM, Van der Ploeg LH, Fong TM. (2002) Molecular basis of the pharmacological difference between rat and human bombesin receptor subtype-3 (BRS-3). Biochemistry 41:8954–8960 [DOI] [PubMed] [Google Scholar]
- Maekawa F, Quah HM, Tanaka K, Ohki-Hamazaki H. (2004) Leptin resistance and enhancement of feeding facilitation by melanin-concentrating hormone in mice lacking bombesin receptor subtype-3. Diabetes 53:570–576 [DOI] [PubMed] [Google Scholar]
- Majumdar ID, Weber HC. (2012a) Appetite-modifying effects of bombesin receptor subtype-3 agonists. Handb Exp Pharmacol 209:405–432 [DOI] [PubMed] [Google Scholar]
- Majumdar ID, Weber HC. (2012b) Biology and pharmacology of bombesin receptor subtype-3. Curr Opin Endocrinol Diabetes Obes 19:3–7 [DOI] [PubMed] [Google Scholar]
- Mantey S, Frucht H, Coy DH, Jensen RT. (1993) Characterization of bombesin receptors using a novel, potent, radiolabeled antagonist that distinguishes bombesin receptor subtypes. Mol Pharmacol 43:762–774 [PubMed] [Google Scholar]
- Mantey SA, Coy DH, Entsuah LK, Jensen RT. (2004) Development of bombesin analogs with conformationally restricted amino acid substitutions with enhanced selectivity for the orphan receptor human bombesin receptor subtype 3. J Pharmacol Exp Ther 310:1161–1170 [DOI] [PubMed] [Google Scholar]
- Mantey SA, Gonzalez N, Schumann M, Pradhan TK, Shen L, Coy DH, Jensen RT. (2006) Identification of bombesin receptor subtype-specific ligands: effect of N-methyl scanning, truncation, substitution, and evaluation of putative reported selective ligands. J Pharmacol Exp Ther 319:980–989 [DOI] [PubMed] [Google Scholar]
- Mantey SA, Weber HC, Sainz E, Akeson M, Ryan RR, Pradhan TK, Searles RP, Spindel ER, Battey JF, Coy DH, et al. (1997) Discovery of a high affinity radioligand for the human orphan receptor, bombesin receptor subtype 3, which demonstrates that it has a unique pharmacology compared with other mammalian bombesin receptors. J Biol Chem 272:26062–26071 [DOI] [PubMed] [Google Scholar]
- Metzger JM, Gagen K, Raustad KA, Yang L, White A, Wang SP, Craw S, Liu P, Lanza T, Lin LS, et al. (2010) Body temperature as a mouse pharmacodynamic response to bombesin receptor subtype-3 agonists and other potential obesity treatments. Am J Physiol Endocrinol Metab 299:E816–E824 [DOI] [PubMed] [Google Scholar]
- Moody TW, Fagarasan M, Zia F. (1995) Neuromedin B stimulates arachidonic acid release, c-fos gene expression, and the growth of C6 glioma cells. Peptides 16:1133–1140 [DOI] [PubMed] [Google Scholar]
- Nakamichi Y, Wada E, Aoki K, Ohara-Imaizumi M, Kikuta T, Nishiwaki C, Matsushima S, Watanabe T, Wada K, Nagamatsu S. (2004) Functions of pancreatic beta cells and adipocytes in bombesin receptor subtype-3-deficient mice. Biochem Biophys Res Commun 318:698–703 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Yamamura Y, Itoh S, Hirano T, Tsujimae K, Aoyama M, Kondo K, Ogawa H, Shinohara T, Kan K, et al. (2000) Characterization of a novel nonpeptide vasopressin V(2)-agonist, OPC-51803, in cells transfected human vasopressin receptor subtypes. Br J Pharmacol 129:1700–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki-Hamazaki H, Watase K, Yamamoto K, Ogura H, Yamano M, Yamada K, Maeno H, Imaki J, Kikuyama S, Wada E, et al. (1997) Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature 390:165–169 [DOI] [PubMed] [Google Scholar]
- Perlman S, Costa-Neto CM, Miyakawa AA, Schambye HT, Hjorth SA, Paiva AC, Rivero RA, Greenlee WJ, Schwartz TW. (1997) Dual agonistic and antagonistic property of nonpeptide angiotensin AT1 ligands: susceptibility to receptor mutations. Mol Pharmacol 51:301–311 [DOI] [PubMed] [Google Scholar]
- Perlman S, Schambye HT, Rivero RA, Greenlee WJ, Hjorth SA, Schwartz TW. (1995) Non-peptide angiotensin agonist. Functional and molecular interaction with the AT1 receptor. J Biol Chem 270:1493–1496 [DOI] [PubMed] [Google Scholar]
- Pradhan TK, Katsuno T, Taylor JE, Kim SH, Ryan RR, Mantey SA, Donohue PJ, Weber HC, Sainz E, Battey JF, et al. (1998) Identification of a unique ligand which has high affinity for all four bombesin receptor subtypes. Eur J Pharmacol 343:275–287 [DOI] [PubMed] [Google Scholar]
- Reitman ML, Dishy V, Moreau A, Denney WS, Liu C, Kraft WK, Mejia AV, Matson MA, Stoch SA, Wagner JA, et al. (2012) Pharmacokinetics and pharmacodynamics of MK-5046, a bombesin receptor subtype-3 (BRS-3) agonist, in healthy patients. J Clin Pharmacol 52:1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley WH, Sato S, Huang SC, Collado-Escobar DM, Beaven MA, Wang LH, Martinez J, Gardner JD, Jensen RT. (1990) Cholecystokinin-induced formation of inositol phosphates in pancreatic acini. Am J Physiol 259:G655–G665 [DOI] [PubMed] [Google Scholar]
- Ryan RR, Weber HC, Hou W, Sainz E, Mantey SA, Battey JF, Coy DH, Jensen RT. (1998a) Ability of various bombesin receptor agonists and antagonists to alter intracellular signaling of the human orphan receptor BRS-3. J Biol Chem 273:13613–13624 [DOI] [PubMed] [Google Scholar]
- Ryan RR, Weber HC, Mantey SA, Hou W, Hilburger ME, Pradhan TK, Coy DH, Jensen RT. (1998b) Pharmacology and intracellular signaling mechanisms of the native human orphan receptor BRS-3 in lung cancer cells. J Pharmacol Exp Ther 287:366–380 [PubMed] [Google Scholar]
- Sancho V, Moody TW, Mantey SA, Di Florio A, Uehara H, Coy DH, Jensen RT. (2010) Pharmacology of putative selective hBRS-3 receptor agonists for human bombesin receptors (BnR): affinities, potencies and selectivity in multiple native and BnR transfected cells. Peptides 31:1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Feighner SD, Hreniuk DL, Iwaasa H, Sailer AW, Pan J, Reitman ML, Kanatani A, Howard AD, Tan CP. (2004) Characterization of the bombesin-like peptide receptor family in primates. Genomics 84:139–146 [DOI] [PubMed] [Google Scholar]
- Sayegh AI. (2013) The role of bombesin and bombesin-related peptides in the short-term control of food intake. Prog Mol Biol Transl Sci 114:343–370 [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Nestor AL, Prabonnaud V, Bachy A, Laplace MC, Keane PE, Bignon E, Herbert JM. (2000) Characterisation of the effects of SR146131, a novel non-peptide CCK(1) receptor agonist, on IMR-32 human neuroblastoma cells. Eur J Pharmacol 397:303–310 [DOI] [PubMed] [Google Scholar]
- Sebhat IK, Franklin C, Lo MC, Chen D, Jewell JP, Miller R, Pang J, Palyha O, Kan Y, Kelly TM, et al. (2011) Discovery of MK-5046, a potent, selective bombesin recptor subtype-3 agonist for the treatment of obesity. ACS Med Chem Lett 2:43–47 DOI: 10.1021/ml100196d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Emanuel RL, Dewald D, Torday JS, Asokanathan N, Wada K, Wada E, Sunday ME. (2004) Bombesin-like peptide receptor gene expression, regulation, and function in fetal murine lung. Am J Physiol Lung Cell Mol Physiol 286:L165–L173 [DOI] [PubMed] [Google Scholar]
- Tan YR, Qi MM, Qin XQ, Xiang Y, Li X, Wang Y, Qu F, Liu HJ, Zhang JS. (2006) Wound repair and proliferation of bronchial epithelial cells enhanced by bombesin receptor subtype 3 activation. Peptides 27:1852–1858 [DOI] [PubMed] [Google Scholar]
- Tsuda T, Kusui T, Jensen RT. (1997) Neuromedin B receptor activation causes tyrosine phosphorylation of p125FAK by a phospholipase C independent mechanism which requires p21rho and integrity of the actin cytoskeleton. Biochemistry 36:16328–16337 [DOI] [PubMed] [Google Scholar]
- Uehara H, González N, Sancho V, Mantey SA, Nuche-Berenguer B, Pradhan T, Coy DH, Jensen RT. (2011) Pharmacology and selectivity of various natural and synthetic bombesin related peptide agonists for human and rat bombesin receptors differs. Peptides 32:1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schrenck T, Wang LH, Coy DH, Villanueva ML, Mantey S, Jensen RT. (1990) Potent bombesin receptor antagonists distinguish receptor subtypes. Am J Physiol 259:G468–G473 [DOI] [PubMed] [Google Scholar]
- Wan Y, Wallinder C, Plouffe B, Beaudry H, Mahalingam AK, Wu X, Johansson B, Holm M, Botoros M, Karlén A, et al. (2004) Design, synthesis, and biological evaluation of the first selective nonpeptide AT2 receptor agonist. J Med Chem 47:5995–6008 [DOI] [PubMed] [Google Scholar]
- Wang LH, Coy DH, Taylor JE, Jiang NY, Kim SH, Moreau JP, Huang SC, Mantey SA, Frucht H, Jensen RT. (1990a) Desmethionine alkylamide bombesin analogues: a new class of bombesin receptor antagonists with potent antisecretory activity in pancreatic acini and antimitotic activity in Swiss 3T3 cells. Biochemistry 29:616–622 [DOI] [PubMed] [Google Scholar]
- Wang LH, Coy DH, Taylor JE, Jiang NY, Moreau JP, Huang SC, Frucht H, Haffar BM, Jensen RT. (1990b) des-Met carboxyl-terminally modified analogues of bombesin function as potent bombesin receptor antagonists, partial agonists, or agonists. J Biol Chem 265:15695–15703 [PubMed] [Google Scholar]
- Weber HC. (2009) Regulation and signaling of human bombesin receptors and their biological effects. Curr Opin Endocrinol Diabetes Obes 16:66–71 [DOI] [PubMed] [Google Scholar]
- Xu J, Weng YI, Simonyi A, Krugh BW, Liao Z, Weisman GA, Sun GY. (2002) Role of PKC and MAPK in cytosolic PLA2 phosphorylation and arachadonic acid release in primary murine astrocytes. J Neurochem 83:259–270 [DOI] [PubMed] [Google Scholar]