Cystic fibrosis (CF) is the most common inherited disorder of childhood and is caused by autosomal recessive mutations in the CF transmembrane conductance regulator (CFTR) gene (1). CF is most common among Caucasian individuals, with a prevalence of 1 in 2,500 newborns (2). CF can be diagnosed before birth by genetic testing or by a sweat test in early childhood (3,4). Complications of CF include bowel obstruction due to meconium ileus in newborns, poor growth, sinus infections, biliary disorders, infertility, chronic pulmonary infections, and lung disease that requires lung transplantation as CF worsens (2). Exocrine pancreatic insufficiency is observed in 90% of CF patients. Abnormal chloride channel function results in thick viscous secretions that obstruct pancreatic ducts and lead to the retention of digestive enzymes and to pancreas tissue destruction and fibrosis (5). CF-related diabetes (CFRD) is highly prevalent among CF patients and is thought to result from the progressive destruction of islets of Langerhans by pancreatic fibrosis (5). CFRD affects 2% of children, 19% of adolescents, and 40–50% of adults (6). As CFRD is associated with worse pulmonary function (7), failure to maintain weight (8), and higher mortality rates (6), the World Health Organization has recommended annual screening for CFRD using an oral glucose tolerance test. CFRD is considered a clinical entity distinct from that of type 1 diabetes or type 2 diabetes (T2D) and is included in the category “other specific types of diabetes” by the American Diabetes Association (9). Nondiabetic CF patients already display impaired first-phase of insulin secretion in response to intravenous challenges to glucose and delayed and blunted insulin secretion in response to the oral glucose tolerance test (10–12), and these abnormalities are more pronounced when glycemic status worsens (11–13). Insulin therapy is therefore the recommended treatment for CFRD patients (5). While abnormal chloride channel function induced by CFTR mutations is necessary for CFRD to develop, a recent twin study has suggested that genetic modifiers are the primary cause of diabetes in CF subjects (14).
To gain insight into the pathophysiology of CFRD, Blackman et al. (15) report a genome-wide association study (GWAS) of 3,059 individuals with CF (644 with CFRD) in this issue. They found a genome-wide significant association between CFRD and two single nucleotide polymorphisms (SNPs) in complete linkage disequilibrium (rs4077468 and rs4077469) located within and 5′ to the SLC26A9 gene. A directionally consistent association of the polymorphism rs4077468 with CFRD was observed in an independent sample of 409 individuals with CF (124 with CFRD). A joint analysis of discovery and replication samples supports the association of rs4077468 with CFRD (hazard ratio, 1.39 per allele [95% CI 1.25–1.54]; P = 9.8 × 10−10). Beyond the stringent evidence of association observed in this relatively modest sample (ntotal = 3,753 including 768 cases), the biological function of the SLC26A9 gene makes the possibility of a false-positive association signal unlikely (16). The SLC26A9 gene indeed encodes an anion transporter that conducts chloride (17). CFTR physically interacts with SLC26A9 and is required to regulate SLC26A9 chloride anion conductance in human embryonic kidney cells (18). The location of CFRD-associated SNPs in the promoter and first intron region of SLC26A9 suggests a possible role in splicing or expression. Consistent with this hypothesis, in silico analyses predict that SNPs 5′ of SLC26A9 flank a region of DNase I hypersensitivity that binds transcription factors, whereas SNPs in first intron are located near three regions that may represent transcription factor binding sites active in multiple tissues.
Family history of T2D triples the risk of CFRD, indicating that CFRD and T2D may share common molecular determinants (19). To test this hypothesis, Blackman et al. analyzed 13 common variants in 8 loci contributing the most to T2D in their discovery sample (n = 3,059 individuals with CF including 644 CFRD cases). They found significant associations for SNPs at the TCF7L2, CDKN2A/B, CDKAL1, and IGF2BP2 loci, the risk alleles being the same for T2D and CFRD.
Conversely, Blackman et al. investigated the provocative hypothesis that polymorphisms in the CFRD-associated gene SLC26A9 may be associated with common T2D in 9,580 case subjects and 53,810 control subjects of European descent as part of the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium. They found a modest association between SNPs rs4077468 and rs4077469 in/near SLC26A9 and T2D (odds ratio [OR] 1.06; P = 0.003), but the risk alleles for CFRD were protective against T2D.
The original study (15) supports a dual molecular origin for CFRD. Whereas abnormal chloride channel function resulting from CFTR mutations and SNPs at the SLC26A9 locus plays a substantial role in CFRD pathogenesis, at least in part through progressive pancreatic exocrine tissue destruction, impairment of β-cell function (and possibly of insulin sensitivity) conferred by T2D predisposing variants at TCF7L2, CDKN2A/B, CDKAL1, and IGF2BP2 loci worsens the risk of developing diabetes in the high-risk CF population. Furthermore, this study may suggest that the same genetic alterations in SLC26A9 may have the opposite role in CFRD and common T2D predisposition, either by normalizing or exacerbating ion transport abnormalities depending on whether CFTR is functional or not.
The authors may be commended for several reasons. First, the two main hypotheses they tested—genetic modifiers modulate the risk of CFRD and T2D predisposing genes modulate the risk of CFRD—were supported by their previous publications (14,19). They combined with great success two powerful approaches commonly used in gene identification efforts: hypothesis-free GWAS and hypothesis-driven candidate gene study. They used a longitudinal study, the gold standard in genetic epidemiology, instead of a classical cross-sectional case control design and took care to replicate their more promising GWAS signal in a second independent study. Stringent Bonferroni corrections for multiple testing were rigorously applied, lowering the risk of false-positive reporting. The biological relevance of SLC26A9 adds more credit to the statistical evidence of association.
The study also presents several limitations. First, the ethnic composition of the discovery and replication samples is not 100% European, and subjects of non-European ancestry have not been discarded from the analysis, increasing the risk to detect a false-positive association by a population stratification bias. Second, even if we are fully aware of the difficulty to recruit CF patients due to their scarcity, the sample sizes of the discovery and replication studies remain modest and whole-genome and candidate gene association studies for CFRD in larger samples are likely to lead to the discovery of additional CFRD-associated signals. The authors suggest that the unusually large ORs for CFRD conferred by T2D predisposing variants at TCF7L2, CDKN2A/B, CDKAL1, and IGF2BP2 loci may reflect greater disease heterogeneity in T2D compared with CFRD. Another likely explanation omitted by the authors is that the modest sample size used here inflates the OR estimates of CFRD-associated loci. Third, the absence of functional biology experiments is another important limitation of this study. The authors propose that CFRD-associated SNPs in the promoter and first intron region of SLC26A9 may increase the risk of CFRD by modulating splicing or expression of the SLC26A9 gene. They provide in silico data to support this statement, but additional in vitro (e.g., luciferase assay) and in vivo (e.g., genotype/SLC26A9 mRNA level correlations) experiments will ultimately be necessary. Also, further information about the distribution of SLC26A9 transcripts in human tissues relevant for CFRD disease will be valuable. Fourth, 390 glucose-intolerant CF patients have been excluded from this study. Providing evidence of an association between the CFRD-associated SNPs identified in this study and prediabetic status in CF patients would have been highly informative.
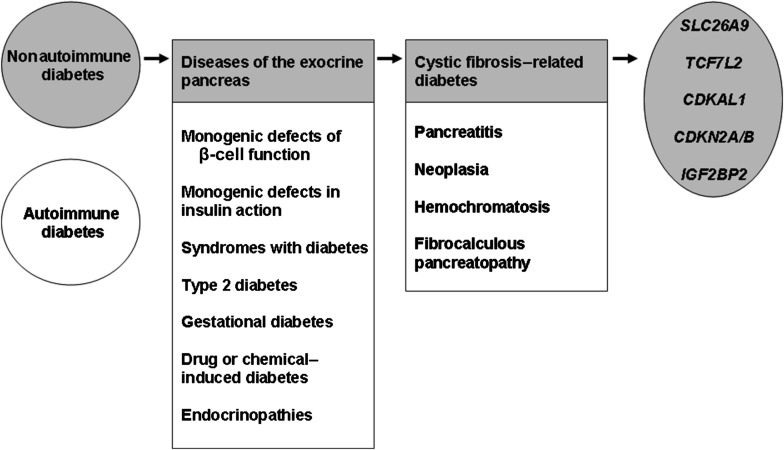
Blackman et al. add new insights into the genetic dissection of diabetes, an exceptionally complex and heterogeneous inherited disorder. First, diabetes can be subclassified as autoimmune (e.g., type 1 diabetes) or nonautoimmune (e.g., T2D) subtypes. The subgroup of nonautoimmune diabetes refers to a mosaic of genetically heterogeneous disease subtypes: syndromes including diabetes as one among many clinical features (e.g., Wolfram syndrome), mitochondrial diabetes, monogenic defects of β-cell function, monogenic defects of insulin action, common T2D, gestational diabetes mellitus, diabetes induced by drugs or chemicals, and diabetes associated with endocrinopathies or diseases of the exocrine pancreas (9). Each nonautoimmune diabetes subtype is complex and can be subgrouped as well. For instance, diseases of the exocrine pancreas resulting in diabetes involve pancreatitis, neoplasia, hemochromatosis, fibrocalculous pancreatopathy, or CF. Genetic dissection of diabetes susceptibility in these subgroups is at best in progress and at worst has not yet started. For instance, the current study identified five SNPs at the SLC26A9, TCF7L2, CDKN2A/B, CDKAL1, and IGF2BP2 loci contributing to CFRD, but many additional CFRD risk gene variants (rare and frequent SNPs or structural variants) still remain to be identified (Fig. 1). The shift from conventional to personalized medicine presupposes that we elucidate the genetic basis of complex inherited disorders before translating our genetic knowledge into clinical benefit (20). The study by Blackman et al. (15) is an additional step in the right direction and encourages us to face and defeat the giant that is diabetes.
FIG. 1.
Diabetes, a heterogeneous complex inherited disorder.
ACKNOWLEDGMENTS
D.M. and G.P. are supported by a Tier 2 Canada Research Chair.
No potential conflicts of interest relevant to this article were reported.
Footnotes
See accompanying original article, p. 3627.
REFERENCES
- 1.Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 1989;245:1066–1073 [DOI] [PubMed] [Google Scholar]
- 2.Ratjen F, Döring G. Cystic fibrosis. Lancet 2003;361:681–689 [DOI] [PubMed] [Google Scholar]
- 3.Mishra A, Greaves R, Massie J. The relevance of sweat testing for the diagnosis of cystic fibrosis in the genomic era. Clin Biochem Rev 2005;26:135–153 [PMC free article] [PubMed] [Google Scholar]
- 4.Langfelder-Schwind E, Kloza E, Sugarman E, et al. National Society of Genetic Counselors Subcommittee on Cystic Fibrosis Carrier Testing Cystic fibrosis prenatal screening in genetic counseling practice: recommendations of the National Society of Genetic Counselors. J Genet Couns 2005;14:1–15 [DOI] [PubMed] [Google Scholar]
- 5.Kelly A, Moran A. Update on cystic fibrosis-related diabetes. J Cyst Fibros 2013;12:318–331 [DOI] [PubMed] [Google Scholar]
- 6.Moran A, Dunitz J, Nathan B, Saeed A, Holme B, Thomas W. Cystic fibrosis–related diabetes: current trends in prevalence, incidence, and mortality. Diabetes Care 2009;32:1626–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milla CE, Warwick WJ, Moran A. Trends in pulmonary function in patients with cystic fibrosis correlate with the degree of glucose intolerance at baseline. Am J Respir Crit Care Med 2000;162:891–895 [DOI] [PubMed] [Google Scholar]
- 8.Koch C, Rainisio M, Madessani U, et al. Investigators of the European Epidemiologic Registry of Cystic Fibrosis Presence of cystic fibrosis-related diabetes mellitus is tightly linked to poor lung function in patients with cystic fibrosis: data from the European Epidemiologic Registry of Cystic Fibrosis. Pediatr Pulmonol 2001;32:343–350 [DOI] [PubMed] [Google Scholar]
- 9.American Diabetes Association Diagnosis and classification of diabetes mellitus (Position Statement). Diabetes Care 2013;36(Suppl. 1):S67–S74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran A, Diem P, Klein DJ, Levitt MD, Robertson RP. Pancreatic endocrine function in cystic fibrosis. J Pediatr 1991;118:715–723 [DOI] [PubMed] [Google Scholar]
- 11.Lanng S, Thorsteinsson B, Røder ME, et al. Pancreas and gut hormone responses to oral glucose and intravenous glucagon in cystic fibrosis patients with normal, impaired, and diabetic glucose tolerance. Acta Endocrinol (Copenh) 1993;128:207–214 [DOI] [PubMed] [Google Scholar]
- 12.Mohan V, Alagappan V, Snehalatha C, Ramachandran A, Thiruvengadam KV, Viswanathan M. Insulin and C-peptide responses to glucose load in cystic fibrosis. Diabete Metab 1985;11:376–379 [PubMed] [Google Scholar]
- 13.Yung B, Noormohamed FH, Kemp M, Hooper J, Lant AF, Hodson ME. Cystic fibrosis-related diabetes: the role of peripheral insulin resistance and β-cell dysfunction. Diabet Med 2002;19:221–226 [DOI] [PubMed] [Google Scholar]
- 14.Blackman SM, Hsu S, Vanscoy LL, et al. Genetic modifiers play a substantial role in diabetes complicating cystic fibrosis. J Clin Endocrinol Metab 2009;94:1302–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackman SM, Commander CW, Watson C, et al. Genetic modifiers of cystic fibrosis–related diabetes. Diabetes 2013;62:3627–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury MJ, Little J, Gwinn M, Ioannidis JP. On the synthesis and interpretation of consistent but weak gene-disease associations in the era of genome-wide association studies. Int J Epidemiol 2007;36:439–445 [DOI] [PubMed] [Google Scholar]
- 17.Loriol C, Dulong S, Avella M, et al. Characterization of SLC26A9, facilitation of Cl(-) transport by bicarbonate. Cell Physiol Biochem 2008;22:15–30 [DOI] [PubMed] [Google Scholar]
- 18.Bertrand CA, Zhang R, Pilewski JM, Frizzell RA. SLC26A9 is a constitutively active, CFTR-regulated anion conductance in human bronchial epithelia. J Gen Physiol 2009;133:421–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackman SM, Hsu S, Ritter SE, et al. A susceptibility gene for type 2 diabetes confers substantial risk for diabetes complicating cystic fibrosis. Diabetologia 2009;52:1858–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet 2013;14:549–558 [DOI] [PubMed] [Google Scholar]