Diabetic micro- and macrovascular complications are a major concern for patients with diabetes because they increase morbidity and mortality risk. Diabetes complications are also an economic challenge for both patients and society. Although multifactorial interventions consisting of lifestyle modification and aggressive treatment of hyperglycemia, hypertension, and dyslipidemia reduce progression of microvascular complications, development of cardiovascular events, and mortality by 50% in type 2 diabetes (1), the clinical course and treatment response varies among patients, and some have to halt treatment due to adverse events.
Addressing traditional modifiable risk factors such as blood glucose, blood pressure, cholesterol, and smoking is useful, but is not sufficient to obtain optimal individual treatment or to predict response to treatment. Potential inclusion of genetic information (pharmacogenomics), termed “personalized medicine,” may help fill this gap.
Familial clustering of diabetic nephropathy and cardiovascular disease in diabetes (2–5) has led to a search for genetic risk markers for nephropathy and other complications. Hypothesis-driven evaluations of candidate genes and recent unbiased genome-wide association study analyses have been applied. Although several genetic markers have been identified, they have modest importance and none has been implemented for clinical use (6,7).
Hypothesis-driven research has often focused on genes associated with cardiovascular disease (CVD), with an early focus on an insertion (I)/deletion (D) polymorphism in the ACE gene. Studies initially reported the ACE I/D polymorphism was associated with diabetic nephropathy (8,9). Additional studies suggested a pharmacogenetic effect in which the degree of decline in renal function on ACE inhibitor (ACEi) treatment depended on genotype (10), while no difference was seen for angiotensin receptor blocker (ARB) treatment (11). A post hoc analysis of the Reduction of Endpoints in NIDDM With the Angiotensin II Antagonist Losartan Study of losartan in albuminuric type 2 diabetic patients confirmed that among patients treated with conventional antihypertensive medication plus placebo, DD genotype carriers had 38% increased risk for development of the composite end point of doubling of s-creatinine, end-stage renal disease, or death compared with the II genotype. Similarly, DD genotype also had the greatest effect of losartan with 28% risk reduction, mitigating the effect of genotype on prognosis (12).
The study by Rurali et al. in this issue (13) has applied a comparable, but more comprehensive, hypothesis-driven approach to search for pharmacogenomic factors for the development and treatment of diabetes complications. Endothelial dysfunction is a key feature in diabetes complications that is associated with increased production of the thrombogenic multimers of von Willebrand factor (VWF), as well as reduced production of ADAMTS13. ADAMTS13 is a member of the a disintegrin and metalloproteinase with thrombospondin type 1 motif family that is involved in removal of VWF multimers. In experimental studies, a lack of ADAMTS13 results in uncontrolled VWF-mediated thrombosis. Rurali et al. first demonstrated reduced secretion and activity of ADAMTS13 in cells and blood from healthy control subjects with a specific genetic variation in ADAMTS13. This variant (618Ala) was present in 17% of both patients and healthy control subjects. Second, they analyzed the impact of this gene variant as well as its interaction with ACEi treatment on the development of renal and CVD complications among normoalbuminuric type 2 diabetic patients in the Bergamo Nephrologic Diabetes Complications Trial (BENEDICT). The study included 1,163 patients randomized to ACE inhibition or placebo, and evaluated renal progression (development of persistent microalbuminuria) or CVD events or a combination of the two. Overall, carriers of 618Ala had an equivalent rate of renal and CVD complications as those with the wild-type Pro/Pro genotype. When evaluated by type of treatment, carriers of 618Ala had the highest rate of progression to microalbuminuria (14.6%) compared with wild-type Pro/Pro homozygotes on non-ACEi treatment (10.4%). However, when treated with ACEi—which reduced overall development of the renal end point—the rate among “high risk” 618Ala carriers was reduced to only 2.9%, compared with 6.4% among “low risk” Pro/Pro carriers. Similar findings were observed for the combined CVD and renal end point, whereas for CVD alone the results were less evident. In other words, those who needed the treatment the most experienced the greatest benefit. Based on this observation, the authors suggested that this variant could not only identify patients with excess renal and cardiac risk but also those who may benefit most from the reno- and cardioprotective effects of ACEi.
To add further support to the findings, the authors measured ADAMTS13 activity in a subset of patients and demonstrated less activity in carriers of the Ala variant compared with the low-risk wild-type, but increased activity in Ala carriers on ACEi versus non-ACEi. Finally, ADAMTS13 activity was lower in patients who developed study end points compared with control subjects matched for treatment and when compared with healthy nondiabetic subjects.
The major strengths of the new study are that the effects of the genotype were not only identified on clinical study end points, but a functional link between genotype and phenotype (ADAMTS13 activity) in an experimental setting was also observed. However, as the authors point out, a limitation of this study is that it is a post hoc analysis from the BENEDICT trial, suggesting that confirmation from other studies is needed. Furthermore, the biological rationale for interaction between ACEi and genotype is not known. Whether the same effect would be seen with progression to more advanced stages of diabetic nephropathy or whether it is also relevant for type 1 diabetic patients are unclear. Additionally, results may only apply to Caucasian populations, and whether it is relevant for treatment with other ACEis or ARBs used to block renin-angiotensin system in type 2 diabetes remains uncertain.
If confirmed, ADAMTS13 could pave the way toward the paradise of personalized medicine, as described in the CLIPMERGE PGx program in which a biobank, including genotype information and additional relevant information, is linked to an electronic medical record (14) (Fig. 1). Algorithms then inform decisions on treatment based on the combination of disease and genotype. In future diabetes care, the decision on whether to treat or how to treat could rely on risk profiles built on phenotype information, genomics (or proteomics or other “omics”) (15), as well as other markers for potential treatment side effects. Together, these new approaches may facilitate optimal personalized treatment (16).
FIG. 1.
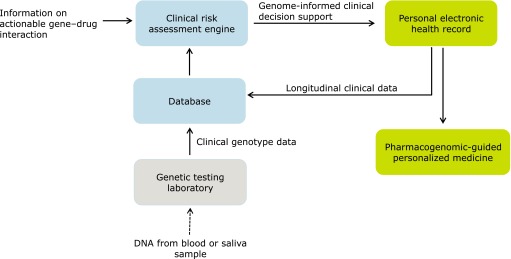
Infrastructure for implementation of genome-informed clinical decision support leading to personalized medicine. Adapted from Gottesman et al. (14).
ACKNOWLEDGMENTS
No potential conflicts of interest relevant to this article were reported.
Footnotes
See accompanying original article, p. 3599.
REFERENCES
- 1.Gaede P, Lund-Andersen H, Parving H-H, Pedersen O. Effect of a multifactorial intervention on mortality in type 2 diabetes. N Engl J Med 2008;358:580–591 [DOI] [PubMed] [Google Scholar]
- 2.Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 1989;320:1161–1165 [DOI] [PubMed] [Google Scholar]
- 3.Canani LH, Gerchman F, Gross JL. Familial clustering of diabetic nephropathy in Brazilian type 2 diabetic patients. Diabetes 1999;48:909–913 [DOI] [PubMed] [Google Scholar]
- 4.Earle K, Walker J, Hill C, Viberti GC. Familial clustering of cardiovascular disease in patients with insulin-dependent diabetes and nephropathy. N Engl J Med 1992;326:673–677 [DOI] [PubMed] [Google Scholar]
- 5.Imperatore G, Knowler WC, Nelson RG, Hanson RL. Genetics of diabetic nephropathy in the Pima Indians. Curr Diab Rep 2001;3:275–81 [DOI] [PubMed] [Google Scholar]
- 6.Alkayyali S, Lajer M, Deshmukh H, et al. Common variant in the HMGA2 gene increases susceptibility to nephropathy in patients with type 2 diabetes. Diabetologia 2013;56:323–329 [DOI] [PubMed] [Google Scholar]
- 7.Schunkert H, König IR, Kathiresan S, et al. Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marre M, Jeunemaitre X, Gallois Y, et al. Contribution of genetic polymorphism in the renin-angiotensin system to the development of renal complications in insulin-dependent diabetes: Genetique de la Nephropathie Diabetique (GENEDIAB) study group. J Clin Invest 1997;99:1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fava S, Azzopardi J, Ellard S, Hattersley AT. ACE gene polymorphism as a prognostic indicator in patients with type 2 diabetes and established renal disease. Diabetes Care 2001;24:2115–2120 [DOI] [PubMed] [Google Scholar]
- 10.Parving HH, Jacobsen P, Tarnow L, et al. Effect of deletion polymorphism of angiotensin converting enzyme gene on progression of diabetic nephropathy during inhibition of angiotensin converting enzyme: observational follow up study. BMJ 1996;313:591–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen S, Tarnow L, Cambien F, et al. Long-term renoprotective effects of losartan in diabetic nephropathy: interaction with ACE insertion/deletion genotype? Diabetes Care 2003;26:1501–1506 [DOI] [PubMed] [Google Scholar]
- 12.Parving HH, de Zeeuw D, Cooper ME, et al. ACE gene polymorphism and losartan treatment in type 2 diabetic patients with nephropathy. J Am Soc Nephrol 2008;19:771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rurali E, Noris M, Chianca A, et al. for the BENEDICT Study Group ADAMTS13 predicts renal and cardiovascular events in type 2 diabetic patients and response to therapy. Diabetes 2013;62:3599–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013;94:214.– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roscioni SS, de Zeeuw D, Hellemons ME, et al. A urinary peptide biomarker set predicts worsening of albuminuria in type 2 diabetes mellitus. Diabetologia 2013;56:259–267 [DOI] [PubMed] [Google Scholar]
- 16.U.S. Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labels [Internet], 2013. Available from http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm Accessed 2 July 2013


