SUMMARY
Homeologous recombination between divergent DNA sequences is inhibited by DNA mismatch repair. In Escherichia coli, MutS and MutL respond to DNA mismatches within recombination intermediates and prevent strand exchange by an unknown mechanism. Here, using purified proteins and DNA substrates, we find that in addition to mismatches within the heteroduplex region, secondary structures within the displaced ssDNA formed during branch migration within the recombination intermediate are involved in the inhibition. We present a model that explains how higher-order complex formation of MutS, MutL and DNA blocks branch migration by preventing rotation of the DNA strands within the recombination intermediate. Furthermore, we find that the helicase UvrD is recruited to directionally resolve these trapped intermediates toward DNA substrates. Thus, our results explain on a mechanistic level how the coordinated action between MutS, MutL and UvrD prevents homeologous recombination and maintains genome stability.
INTRODUCTION
DNA mismatch repair (MMR) is one of several important DNA repair processes conserved from bacteria to mammals. MMR repairs base-base mismatches and insertion/deletion loops generated during replication (Jiricny, 2013). In addition, MMR proteins prevent illegitimate recombination between divergent (homeologous) sequences. Thus, inactivation of MMR genes not only results in mutator but also in hyper-recombination phenotypes. The increased frequency (up to 1000 fold) of interspecies conjugation between Escherichia coli and Salmonella typhimurium in mutS, mutL, mutH and uvrD recipients indicates that MMR acts as a barrier to homeologous recombination (Rayssiguier et al., 1989, 1991). Similar findings are obtained for interspecies transduction (Zahrt and Maloy, 1997), transformation (Majewski et al., 2000) and conjugational E. coli-E. coli crosses that involve the creation of mismatches (Feinstein and Low, 1986). Likewise, in eukaryotes, individual MMR proteins have roles of varying magnitude in the prevention of homeologous recombination (Selva et al., 1995; Datta et al., 1996; Nicholson et al., 2000; de Wind et al., 1995). In higher eukaryotes, inhibition of homeologous reactions is thought to protect the genome from recombination events between divergent repeats (de Wind et al., 1999). This is important in humans in which the amount of repetitive DNA is as high as 50%.
An intriguing question is how MMR and recombination pathways are integrated at the molecular level. Common events during MMR and antirecombination include MutS binding to a DNA mismatch, complex formation between MutS and MutL, and MutL-mediated orchestration of MutH (GATC endonuclease) and UvrD (3′–5′ helicase) (Jiricny 2013; Stambuk and Radman, 1998). MutS and MutL are potent suppressors of homeologous recombination in vivo (Rayssiguier et al., 1989). Biochemical studies demonstrate that MutS and MutL block RecA-mediated homeologous strand exchange through prevention of branch migration (Worth et al., 1994). However the molecular mechanism of this inhibition remains unknown. DNA mismatches do not occur within recombination filaments until after strand exchange has taken place, and somehow MMR proteins are able to inhibit the ongoing branch migration into a region where mismatches are not yet present. Although the fate of the trapped homeologous strand exchange intermediates is unknown, cells must disassemble these problematic DNA structures. Initially, genetic data suggested a minor role for the helicase UvrD in this process (Rayssiguier et al., 1989). However, homeologous recombination frequency in a recipient strain lacking both MutH and UvrD is as high as in mutS and mutL strains (Stambuk and Radman, 1998). Based on this, the fate of MutS-MutL-trapped recombination intermediates was hypothesized to depend on two distinct mechanisms One is MutH-independent and involves MutS, MutL and UvrD during the early stage of homeologous recombination. The binding of a mismatch by the MutS-MutL complex is proposed to recruit UvrD to the nearest single strand (ss) / double strand (ds) DNA junction of the recombination intermediate. UvrD then unwinds the heteroduplex from the loading junction toward the mismatch and dissolves the recombination intermediate. Interestingly, in biochemical experiments UvrD can both stimulate and prevent RecA-mediated homologous strand exchange (Morel et al., 1993). Furthermore, UvrD dismantles RecA nucleoprotein filaments, thereby preventing joint molecule formation (Veaute et al, 2005). How these activities are regulated within the context of homeologous recombination remains to be determined. The second mechanism is MutH-dependent and explains events occurring during the late stage of homeologous recombination requiring MutS, MutL, MutH, DNA helicase and de novo DNA synthesis. Both mechanisms have yet to be tested biochemically.
Here, we addressed how MutS and MutL inhibit strand exchange by trapping homeologous recombination intermediates, and how UvrD helicase is directed to resolve the trapped intermediates. Our results provide new insights into the dynamic interplay between these two DNA repair systems with implications for eukaryotic antirecombination.
RESULTS
Characterization of Recombination Intermediates Formed during RecA-Mediated Homeologous Strand Exchange
To mechanistically study MMR-directed inhibition of homeologous recombination, we established and characterized RecA-mediated strand exchange reactions using the 6.4 kb genomes of bacteriophage fd and M13 as DNA substrates. In the homologous reaction, linear dsDNA (designated as species 1 in Figure 1A) and circular ssDNA substrates (species 2) from the bacteriophage M13 were used. The homeologous reaction was performed between fd circular ssDNA and M13 linear dsDNA, whose DNA sequences diverge by approximately 3% (Table S1). Both homologous and homeologous reactions form DNA intermediates (species 3, 4 and 5) before products of nicked circular dsDNA (species 6 in Figure 1A) and linear ssDNA (species 7) are generated.
Figure 1. MutS-MutL Inhibits Progression of DNA Strand Exchange during Homeologous Recombination by Acting on Defined Intermediates.
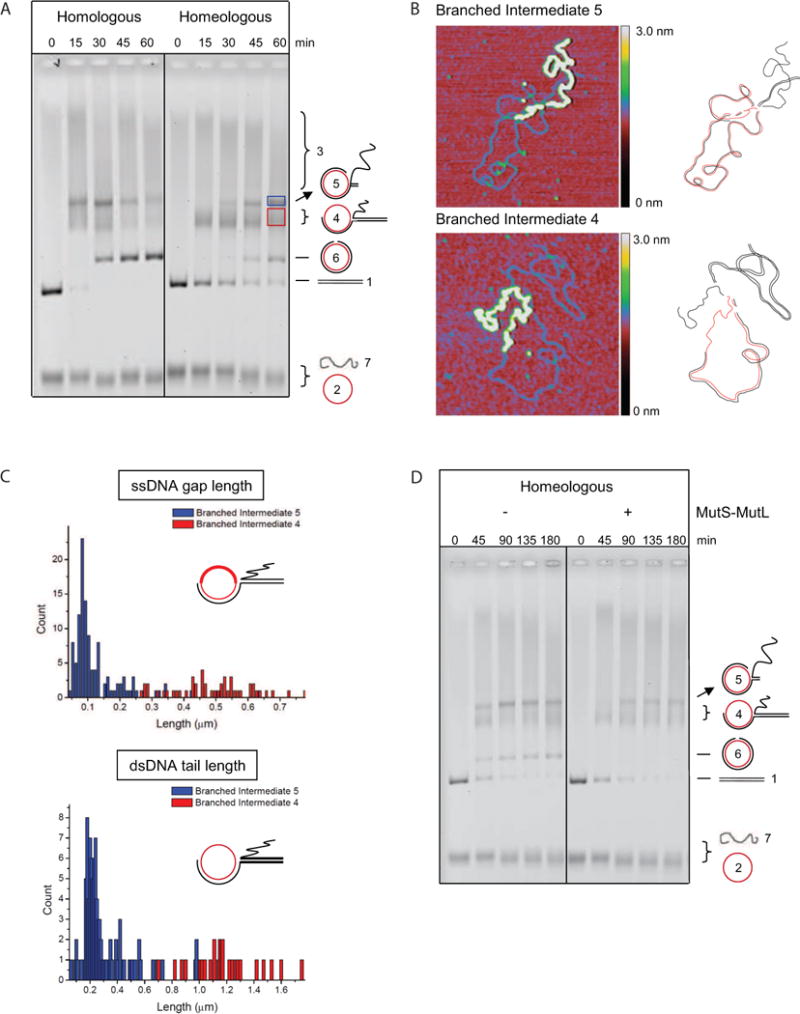
(A) Homologous and homeologous strand exchange using bacteriophage DNA. The numbering system of the DNA species, schematically depicted on the right of the gel, reflects their order of appearance. See also Table S1.
(B) Defined reaction intermediates consist of one ssDNA and one dsDNA molecule. Gel purified intermediates 4 and 5 from the homeologous reaction were incubated with SSB to mark ssDNA regions and were analyzed with scanning force microscopy. Images are 0.6 × 0.6 μm with z dimension indicated by the colored bar. Regions of dsDNA appear as a thin blue line, SSB-coated ssDNA appears as a thick yellow line. Drawings next to the SFM images are interpretations of the intermediates with one circular fd ssDNA (red) and one linear M13 dsDNA (black).
(C) Intermediate 4 has a shorter heteroduplex region than intermediate 5. Upper panel, histogram showing contour length measurements of the SSB-coated ssDNA gap (bold line in cartoon) in intermediate 4 (n = 48, red bars) and 5 (n = 116, blue bars); lower panel, histogram showing the contour length measurement of the linear dsDNA tail (bold line in cartoon) from intermediate 4 (n = 28, red bars) and 5 (n = 96, blue bars).
(D) The effect of MutS (75 nM) and MutL (75 nM) on homeologous strand exchange. See also Figure S1.
Based on structural analysis of DNA intermediates formed during strand exchange with RecA and ATPγS (Menetski et al., 1990) and Rad51 (Holmes et al., 2002), intermediates 3, 4 and 5 observed in our system are expected to be joint molecules. Knowledge of the exact structure and stoichiometry of these intermediates is essential for a mechanistic explanation of the effect of MMR proteins on strand exchange, thus we examined the structures of DNA intermediates 4 and 5 from the homeologous reaction (red and blue box, Figure 1A) with scanning force microscopy (SFM). We incubated purified DNA intermediates with E. coli ssDNA binding protein (SSB) before deposition, allowing us to distinguish ssDNA from dsDNA in the SFM images. Intermediates 4 and 5 are joint molecules consisting of one circular ssDNA and one linear dsDNA substrate (Figure 1B), as was shown for the corresponding joint molecules observed with RecA in the presence of ATPγS (Menetski et al., 1990) and those categorized as JM1 generated with Rad51 (Holmes et al., 2002). Next, we measured the contour length of the protein-free linear dsDNA tail and the SSB-bound ssDNA gap in the circle of the intermediates. Histograms of the measurements (Figure 1C) revealed that DNA intermediate 4 is the precursor of intermediate 5 because it has a longer ssDNA gap in the circle and a longer linear dsDNA tail. Unlike the RecA-ATPγS and Rad51-generated intermediates (Menetski et al., 1990; Holmes et al., 2002), joint molecules containing multiple linear dsDNA molecules are not observed as distinct species. Thus we conclude that DNA intermediates 4 and 5 are sequential on-pathway reaction intermediates that can be used as a read-out for strand exchange progress in addition to the appearance of nicked circular dsDNA product.
MutS and MutS-MutL Block Heteroduplex Formation in Homeologous Strand Exchange
MutS and MutS-MutL inhibit homeologous strand exchange in vitro (Worth et al., 1994, 1998). Here we recapitulated these results. During a one-hour time course we achieved weak inhibition of homeologous recombination by MutS while the homologous reaction remained unaffected (Figure S1A). In addition, we reproduced the observation that MutL functions as an enhancer of MutS-mediated inhibition while affecting neither the homologous nor the homeologous reaction on its own (Figures S1B and S1C). Interestingly, this inhibition depended on ATP hydrolysis by MutL, because ATPase deficient MutL E29A (Ban et al., 1999) was unable to enhance the MutS-mediated inhibition (Figure S4C). To further ensure that MutS-MutL is not just delaying homeologous strand exchange, but really inhibits the reaction, we prolonged the incubation time to 3 hours. We still observed accumulation of DNA intermediates and inhibition of product formation (Figure 1D). Thus, mismatch-specific inhibition of recombination by MutS-MutL suggests that mismatches in the heteroduplex region activate MutS and MutL to prevent DNA intermediates from extensive heteroduplex formation.
MutS and MutS-MutL Prevent Mismatch-Containing D-Loop Molecules from Dissociation
It is at present difficult to envisage how MutS and MutL are able to block strand exchange because mismatches do not arise until after strand exchange has already occurred. It is possible that MMR proteins are able to recognize mismatches within the heteroduplex region directly upon formation, thus within the recombinase filament at the site of synapsis. We therefore decided to test the effect of MutS and MutL on RecA-mediated D-loop formation, a well-established assay to address strand invasion, homologous pairing and strand exchange in the absence of extensive branch migration. We used 5′-labeled linear ssDNA (90 nt), of which 54 nucleotides at the 3′ end are homologous to the supercoiled recipient plasmid DNA, as well as a variant with a single base change that will introduce a DNA mismatch upon invading the supercoiled plasmid. MutS, MutL or MutS-MutL did not affect the efficiency of joint molecule formation in homologous (Figure 2A) or homeologous (Figure 2B) reactions as indicated by the amount of D-loop at the first time point (5 minutes). Thus, MutS and MutS-MutL may not inhibit strand exchange in the absence of extensive branch migration.
Figure 2. MutS and MutS-MutL Prevent Mismatch-Containing D-Loops from Dissociation.
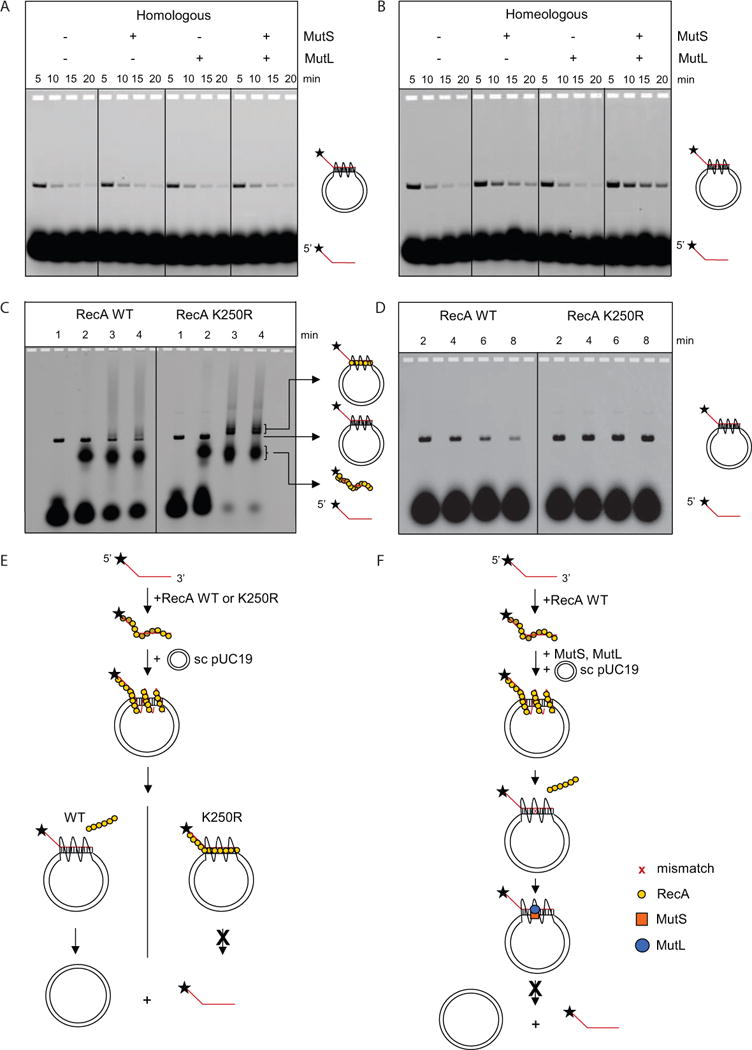
(A) Formation and dissociation of D-loops is not influenced by the presence of 50 nM MutS, 50 nM MutL or both. Diagrams on the right indicate the position of the labeled ssDNA and its joint molecule (D-loop) with a homologous supercoiled plasmid (the helical and supercoiled nature omitted for clarity). See also Figure S2.
(B) Upon formation of a mismatch in the heteroduplex region, MutS and MutS-MutL, but not MutL, inhibit D-loop dissociation.
(C) RecA K250R ATPase mutant is defective in disassembly from the heteroduplex. D-loop samples (time point 2 min) were deproteinized with SDS (lane 1), not treated (lane 2), crosslinked with 0.3% glutaraldehyde (lane 3) or 0.6% glutaraldehyde (lane 4).
(D) D-loop dissociation is correlated to ATP hydrolysis activity by RecA. Homologous D-loop reactions mediated by wild-type RecA (left panel) and RecA ATPase mutant K250R (right panel).
(E) Schematic diagram of D-loop dissociation. RecA forms a filament with ssDNA which invades and pairs with the homologous region within pUC19 supercoiled DNA. At the homologous region, a D-loop structure consisting of a heteroduplex and a displaced ssDNA is formed. After ATP hydrolysis, wild-type RecA proteins disassemble from the DNA, allowing dissociation of the D-loop structure. In contrast, the RecA K250R mutant, which lacks ATPase activity, remains bound to the D-loop structure and prevents its dissociation.
(F) Schematic diagram of the suppression of D-loop dissociation by MutS-MutL. The D-loop structure is formed as in panel E. After ATP hydrolysis and RecA disassembly, the mismatch in the heteroduplex becomes available for MutS binding. MutL forms a complex with MutS and further stabilizes MutS on the heteroduplex DNA. As a result, the dissociation of the D-loop structure is attenuated.
Because we performed the experiments under conditions that permit ATP hydrolysis, we also addressed D-loop dissociation, a well-established phenomenon that occurs possibly via re-invasion of the heteroduplex region by the RecA-bound displaced ssDNA of the D-loop (Shibata et al., 1982). The presence of MMR proteins did not influence the rate of D-loop dissociation in the homologous reaction (Figure 2A). Interestingly, MutS significantly decreased the rate of D-loop dissociation in the homeologous reaction (Figure 2B). In addition, MutL further delayed D-loop dissociation but only in the presence of MutS (Figure 2B) similar to its role in enhancing MutS inhibition during homeologous strand exchange using bacteriophage DNAs (Figure S1). Clearly, MutS and MutL are able to recognize mismatches formed in recombination intermediates, a single mismatch being sufficient to significantly influence progression of the D-loop cycle.
RecA induced D-loops are stable when formed in the presence of a non-hydrolyzable ATP analog (Shibata et al., 1979). Using the ATP hydrolysis-deficient RecA K250R variant (Cox et al., 2008), we tested whether the ATP-hydrolysis-coupled disassembly of RecA from the heteroduplex region allows D-loop dissociation. Upon crosslinking of protein-bound D-loops, we detected more K250R RecA associated with the DNA than wild-type RecA (Figure 2C), confirming that the K250R RecA mutant is defective in disassembly from DNA. Using this mutant RecA, D-loops indeed did not dissociate over the course of the reaction (Figure 2D). These results demonstrate that when RecA is bound to the heteroduplex region of the joint molecule, re-hybridization of the supercoiled dsDNA is prevented (Figure 2E). Interestingly, binding of MutS and MutL to a single mismatch formed in the heteroduplex region also stabilized D-loops even under conditions that permit ATP hydrolysis by RecA (Figure 2F). It is well established that MutS is able to move away from the mismatch as a sliding clamp upon activation by ATP (Acharya et al., 2003). In this case however, because the recombination intermediate consists of multiple intertwined DNA strands, the diffusing complex will be retained on the heteroduplex region, thereby preventing re-invasion by the RecA-bound displaced ssDNA.
MutS and MutL Have No Effect on Homeologous Strand Exchange Using Short DNA Substrates
Because the analysis of D-loop formation suggested that MutS or MutS-MutL do not inhibit strand exchange in the absence of extensive heteroduplex formation, we confirmed this by analyzing early steps of the recombination reaction using short DNA oligonucleotides. This assay addresses DNA pairing and strand exchange without reinvasion of the displaced strand into the heteroduplex region taking place (Bazemore et al., 1997). MutS and MutS-MutL were unable to inhibit homeologous strand exchange between these short substrates (Figure S2) despite a mismatch being formed in the heteroduplex region. This finding is different from MutS and MutS-MutL inhibiting strand exchange using non-identical bacteriophage DNA (Figures 1D, S1A and S1C). A possible explanation for this difference is the shorter lifetime of the intermediates formed by oligonucleotides. Thus, the displaced ssDNA is only transiently available. These considerations prompted us to test whether a long displaced ssDNA in the DNA intermediate might be required for MutS-MutL-mediated antirecombination.
The Displaced ssDNA is Involved in MutS-MutL-Mediated Blocking of Homeologous Strand Exchange
To test the role of the long displaced ssDNA, we used S1 nuclease to specifically degrade the displaced strand of the joint molecules during homeologous strand exchange (Figure S3A–C). Indeed, as shown by Figure 3A, inhibition by MutS and MutL was reduced in the presence of S1 nuclease indicated by the appearance of DNA intermediate 5 at 90 and 120 minutes (panel 4). MutS-MutL in the absence of S1 nuclease blocked DNA intermediate 4 from extensive heteroduplex formation and prevented the formation of DNA intermediate 5 at 90 and 120 minutes (panel 3). S1 nuclease hardly affected the homeologous reaction (panel 2). Therefore, upon removal of the displaced ssDNA, MutS and MutL were unable to efficiently block strand exchange, indicating that the displaced ssDNA is involved mechanistically in the inhibition of homeologous strand exchange.
Figure 3. The Long Displaced ssDNA Tail and MutS tetramerization are Involved in MutS-MutL-Imposed Antirecombination.
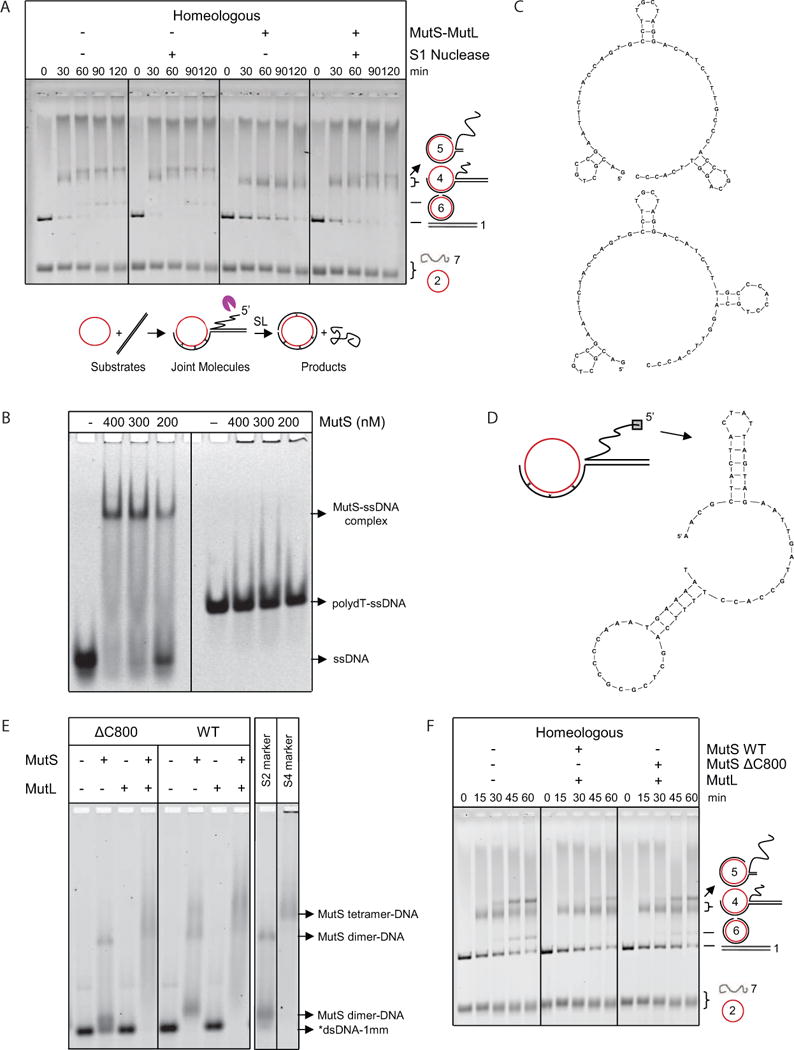
(A) S1 nuclease-mediated degradation of the 5′ displaced ssDNA reduces inhibition of homeologous strand exchange by MutS-MutL. Homeologous strand exchange reactions from left to right: control; with S1 nuclease (0.04 U/μl); with MutS-MutL (100, 100 nM); with S1 nuclease (0.04 U/μl) and MutS-MutL (100, 100 nM). Substrates, intermediates and products are shown below the gel. The purple pacman symbol represents the S1 nuclease. See also Figure S3.
(B) Electrophoretic mobility shift assay of increasing concentrations of MutS binding to ssDNA containing secondary structures and poly-dT.
(C) Predicted secondary structures formed in the ssDNA with arbitrary sequence used in panel B.
(D) Predicted secondary structures formed in the first 60 nucleotides of the 5′ displaced strand of M13.
(E) Binding of wild type MutS, ΔC800 MutS and MutL (400 nM each) alone and together to labeled 90-bp dsDNA with 1 mismatch (*dsDNA-1mm). Markers for DNA-bound MutS dimer (S2) and tetramer (S4) (Groothuizen et al., 2013) are indicated in the panel on the right.
(F) In the presence of MutL, MutSΔC800 only partially blocked homeologous strand exchange as indicated by the formation of higher amounts of DNA intermediate 5 and product compared to the reaction with wild-type MutS-MutL. From left to right, homeologous strand exchange reactions without MutS-MutL, with MutS-MutL (75, 75 nM) and with MutSΔC800-MutL (75, 75 nM).
MutS Binding to the Secondary Structures of ssDNA and Tetramerization are Important for Antirecombination
Because S. typhymurium MutS was reported to cosediment with ssDNA (Pang et al., 1985), we investigated whether E. coli MutS could bind to ssDNA. Using an electrophoretic mobility shift assay (EMSA), we titrated MutS with ssDNA of either arbitrary or poly-dT sequence. This analysis showed that MutS was indeed able to bind ssDNA but only if this ssDNA is capable of forming secondary structures (Figures 3B and C). Similar secondary structures were also predicted to form in the first 60 nucleotides of the 5′ displaced ssDNA of the fd-M13 DNA intermediate (Figure 3D). Because E. coli SSB has high affinity for ssDNA and prevents secondary structure formation, we verified that even in the presence of slightly subsaturating SSB concentrations, as used during strand exchange, MutS is indeed able to bind to ssDNA (Figure S3D). These results suggest that the importance of 5′ displaced ssDNA during MMR-mediated antirecombination is due to the ability of MutS to bind to secondary structures in the displaced strand.
MutS dimers can form tetramers that are able to bind multiple mismatch-containing DNA molecules (Monti et al., 2011) or different segments of DNA creating α-shaped loops (Jiang and Marszalek, 2011). Our findings suggest that simultaneous binding of MutS to mismatches within the heteroduplex region and to secondary structure elements of the displaced ssDNA within recombination intermediates might play a role in blocking strand exchange. Consistent with this notion, in vivo and in vitro evidence indicates that MutSΔC800, which lacks the C-terminal tetramerization domain and therefore can only form monomers and dimers, is compromised in preventing homeologous recombination (Calmann et al., 2005-1, 2005-2). Because of varying reports about mismatch affinity and concentration effects for this mutant (Bjornson et al., 2003; Calmann et al., 2005-1; Lamers et al., 2000), we decided to revisit its mismatch binding and strand exchange inhibition capacities. We were able to demonstrate under our experimental conditions that in the presence of MutL, MutSΔC800 binds to mismatched dsDNA (Figure 3E) and imposes a less stringent block to homeologous recombination than MutS (Figure 3F). This is different from previously reported strand exchange data in which a full block was observed when MutSΔC800 was combined with MutL (Calmann et al., 2005-1). Taken together, these results indicate that the strength of the strand exchange inhibition during homeologous recombination depends on tetramerization of MutS and possibly also on higher order complex formation with MutL.
Bidirectional Unwinding of Homeologous DNA Intermediates by UvrD
We next analyzed the effect of the UvrD helicase on strand exchange using bacteriophage DNA because UvrD dissolves recombination intermediates (Morel et al., 1993). The inclusion of 10 nM UvrD (without MutS and MutL) almost completely inhibited product formation during homologous and homeologous strand exchange (Figure 4A), most likely because the helicase disrupts the RecA-ssDNA filament (Veaute et al., 2005). Reducing UvrD to 1 nM did not affect the homeologous reaction at early time points (45 and 90 min; Figure 4B panel 2), indicating UvrD is no longer dismantling RecA-ssDNA filaments. However, UvrD caused a significant reduction of DNA intermediate 5 at later time points (135 and 180 minutes). Intriguingly, this is coupled with an increase of DNA intermediate 4 and linear dsDNA substrate (indicating that the UvrD helicase is able to drive the homeologous reaction backward), as well as nicked circular dsDNA product (indicating that UvrD at the same time drives the reaction forward) (Figure 4B panel 2; Figure S5). These increases in both substrate and product also occur in the presence of MutL (Figure 4B panel 3; Figure S5) and become more prominent if MutL cannot hydrolyze ATP (Figure S4A). Processing of DNA intermediate 5 required helicase activity because UvrD mutants K35M (Figure S4B) and E221Q (data not shown), which are ATPase deficient but can still bind to DNA (George et al., 1994; Brosh and Matson, 1995), did not affect the homeologous reaction. Because heteroduplex regions containing mismatches cannot spontaneously branch migrate (Biswas et al., 1998), these observations imply that UvrD actively and bidirectionally unwinds strand exchange intermediate 5 (Figure S6).
Figure 4. Bidirectional Unwinding of Homeologous DNA Intermediates by UvrD.
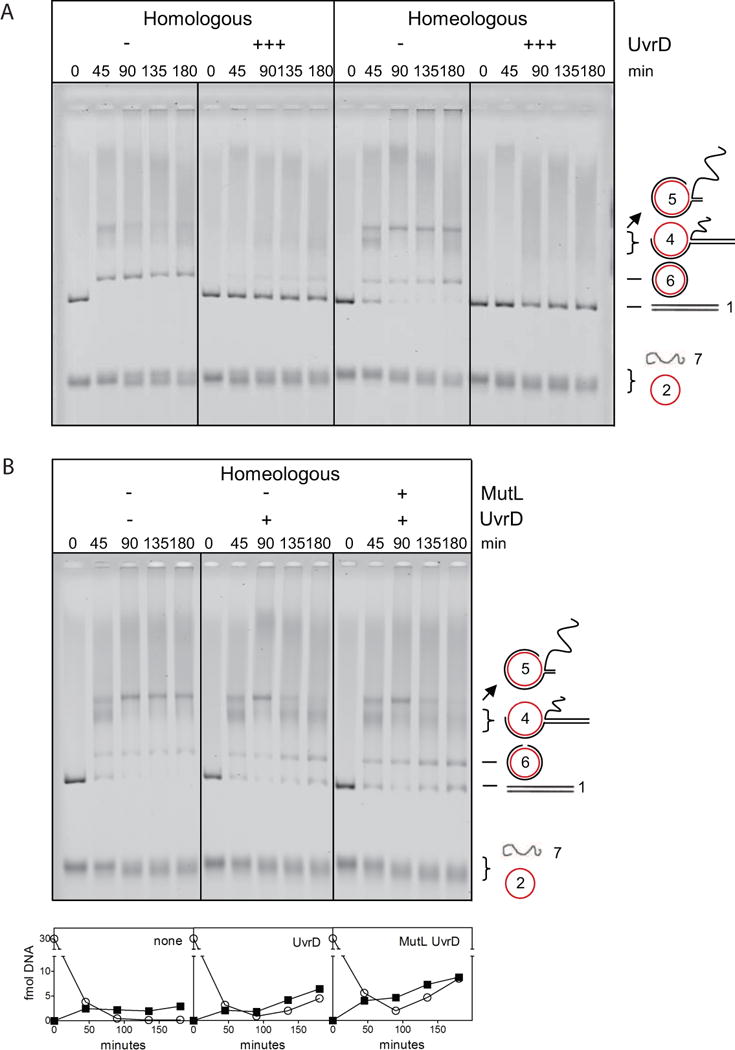
(A) 10 nM UvrD completely inhibits homologous and homeologous strand exchange.
(B) Homeologous strand exchange from left to right: no MMR proteins, with UvrD (1 nM) and with MutL-UvrD (75, 1 nM). Addition of UvrD and of MutL-UvrD significantly increased the amount of substrate and product at 135 and 180 minutes (Figure S5). Panels below the gel show quantified amounts of linear dsDNA substrate (open circles) and nicked circular dsDNA product (closed squares). Also see Figures S4, S5 and S6.
The Mismatch-Activated MutS-MutL Complex Activates UvrD to Resolve Homeologous DNA Intermediates Unidirectionally
Because UvrD can be directed by the mismatch-activated MutS-MutL complex to unwind dsDNA toward the mismatch during MMR (Dao and Modrich, 1998), we tested whether UvrD can likewise be coordinated by the MutS-MutL complex during homeologous recombination (Figure 5A). As was described above, addition of UvrD alone to the homeologous reaction resulted in increased substrate and product formation at the expense of intermediate 5 (panel 2) and addition of MutS-MutL resulted in trapping of strand exchange intermediates and prevented product formation (panel 3). Addition of MutS, MutL and UvrD together (panel 4) again resulted in unwinding of trapped intermediate 5. Interestingly, in this case no additional product was formed but strand exchange intermediates were resolved to substrate (Figure S5). The increase in linear dsDNA substrate is observed at all time points indicating that the MutS-MutL complex coordinates the activity of UvrD early in the strand exchange reaction. Again, the resolution is dependent on helicase activity as UvrD ATPase mutants K35M (Figure S4B) and E221Q (data not shown) did not support this activity. Furthermore, this directed unwinding is largely mismatch-specific, as we did not observe a significant effect in the homologous reaction (Figure 5B). Interestingly, the UvrD-directed unwinding mediated by MutS-MutL seems to depend on ATP hydrolysis by MutL as we failed to detect resolution of the fd-M13 DNA intermediates in the presence of MutS and the MutL E29A ATPase mutant (Figure S4C). Thus the ATP-hydrolysis deficient MutL mutant is not only defective in blocking strand exchange but also unable to confer directionality to UvrD in conjunction with MutS. In conclusion, we demonstrate that the mismatch-activated MutS-MutL complex not only traps the recombination intermediates resulting in inhibition of branch migration but also stimulates UvrD helicase to resolve the DNA intermediates in a directional manner.
Figure 5. MutS and MutL confer directionality to UvrD-mediated unwinding of homeologous recombination intermediates.
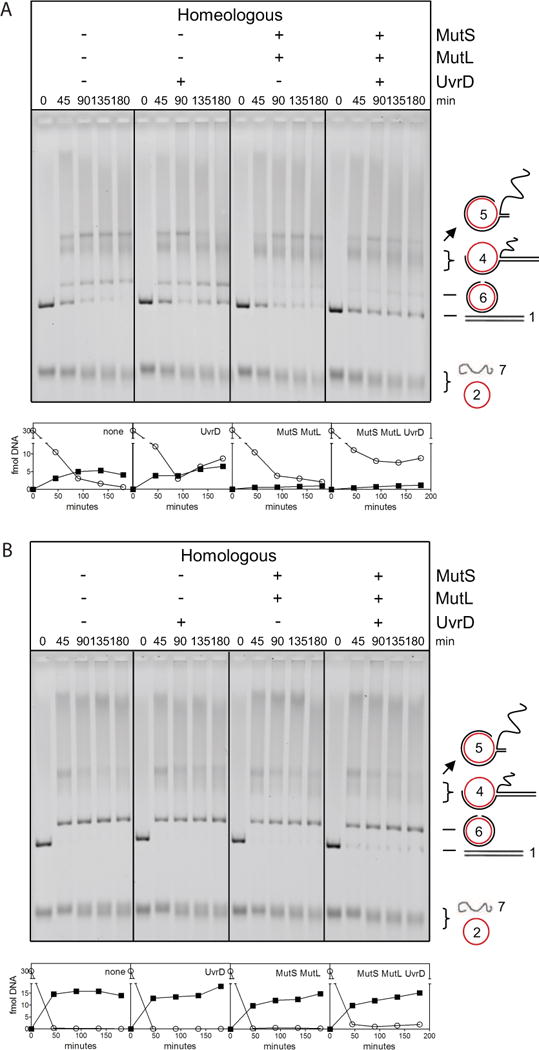
(A) Resolution of homeologous strand exchange intermediates by UvrD is bidirectional, but becomes unidirectional when guided by mismatch-activated MutS-MutL complexes. From left to right, homeologous strand exchange without MMR proteins, with UvrD (1 nM), with MutS-MutL (75, 75 nM) and MutS-MutL-UvrD (75, 75, 1 nM). See also Figures S4, and S5.
(B) MutS, MutL and UvrD do not resolve homologous strand exchange intermediates. From left to right, homologous strand exchange without MMR proteins, with UvrD (1 nM), with MutS-MutL (75, 75 nM) and MutS-MutL-UvrD (75, 75, 1 nM). Panels below the gel show quantified amounts of linear dsDNA substrate (open circles) and nicked circular dsDNA product (closed squares).
DISCUSSION
In this study we found that mismatch repair proteins MutS and MutL (i) trap recombination intermediates using mismatches formed in the heteroduplex region and secondary DNA structures in the displaced ssDNA, and (ii) recruit the UvrD helicase to resolve these trapped intermediates in a directional manner. We thus successfully reconstituted the early-stage antirecombination mediated by MutS, MutL and UvrD, and provided a mechanistic explanation for the observed hyper-recombination phenotypes of E. coli uvrD strains in conjugational crosses involving different species and closely-linked genetic markers (Feinstein and Low, 1986; Stambuk and Radman, 1998).
Binding of MutS and MutL to Recombination Intermediates
It has been postulated that MutS and MutL bind to mismatches within the RecA-bound three-stranded DNA complex (Worth et al., 1994). However, if RecA remains bound, it (i) blocks the access to these mismatches, and (ii) keeps them in a structurally inappropriate conformation for MutS binding because the RecA-dsDNA nucleoprotein filament is stretched and underwound relative to B-form DNA (Di Capua et al., 1982). Because of this, we would not expect MutS-MutL to be able to gain access to the mismatches imbedded within the RecA-heteroduplex filament. We therefore favor a mechanism in which mismatch recognition occurs after RecA disassembly from the heteroduplex region, rather than within the RecA bound three-stranded DNA filament.
Our data indicate that MutS not only becomes activated by mismatches in the heteroduplex formed upon strand exchange, but is also able to bind to secondary structures formed in the displaced ssDNA tail of the joint molecule. The binding of MutS tetramers to two different DNAs may be an important activity as tetramerization-deficient MutS has a strong recombination phenotype in addition to a concentration-dependent mutator phenotype (Calmann et al., 2005-1, 2005-2). MutL is detected in complex with MutS bound to the base of the loop (Allen et al., 1997). We speculate that MutL plays an important role during antirecombination by enhancing the stability of the MutS tetramer and maintaining a more stable higher-order complex of MutS, MutL and DNA strands. This is supported by in vivo studies indicating that low levels of MutL cause a hyper-recombination but not a mutator phenotype (Elez et al., 2007).
A Mechanistic Model for Antirecombination Mediated by MutS, MutL and UvrD Helicase
Taking together our new findings describing the actions of MutS, MutL and UvrD on recombination intermediates, the polarity of RecA-mediated strand exchange (West et al., 1981; Kahn et al., 1981; Cox et al., 1981) and the requirement for rotation during the spooling of DNA into and out of the RecA-bound DNA complex (West, 1992; Honigberg and Radding, 1988; Howard-Flanders et al., 1984), we propose a coherent molecular model for the mechanism of MMR-imposed antirecombination (Figure 6). Initially, the right-handed helical filament formed between RecA and circular ssDNA (Stasiak and Egelman, 1994) pairs with the homologous region in linear dsDNA substrate in the presence of ATP (step 1). Upon intertwining of the incoming dsDNA with the RecA-ssDNA filament, strand exchange is facilitated within the RecA-bound three-stranded DNA complex (step 2) (Menetski et al., 1990; Rosselli and Stasiak, 1990). Upon heteroduplex formation, RecA proteins hydrolyze bound ATP and disassemble from the trailing end of the complex (step 3). RecA disassembly in turn allows the linear displaced ssDNA to spool out from the newly formed heteroduplex. At the same time, the incoming linear dsDNA spools into the groove of the right-handed helical RecA-ssDNA filament at the leading end of the RecA-DNA complex (step 4). These events effectively result in a window of RecA-DNA complex traveling 5′ to 3′ in relation to the displaced ssDNA (van der Heijden et al., 2008) causing rotation of the circular RecA-ssDNA filament around the linear dsDNA (steps 5, 6) (related to Movie S1). ATP hydrolysis and RecA disassembly is therefore important to facilitate the strand exchange reaction of long DNA substrates because the failure of RecA to disassemble would prevent the rotation of circular RecA-ssDNA filament and hinder the formation of RecA-DNA complex at the leading end. In the absence of mismatches, homologous strand exchange proceeds to completion (step 7). In contrast, mismatches formed in the homeologous reaction (step 8) activate MutS-MutL to block extensive branch migration and product formation (step 9). The rotation of RecA-ssDNA filament during strand exchange is inhibited via simultaneous binding of higher-order MutS-MutL complexes to both the heteroduplex DNA and the secondary structures DNA formed by displaced ssDNA (step 9). Throughout the course of heteroduplex formation, the increasing number of mismatches results in multiple loading of MutS-MutL complexes. Eventually, MutS-MutL exerts stronger inhibition on the rotation of the RecA-ssDNA filament circle at later stages of the homeologous strand exchange (step 10).
Figure 6. Model for MMR-Imposed Antirecombination.
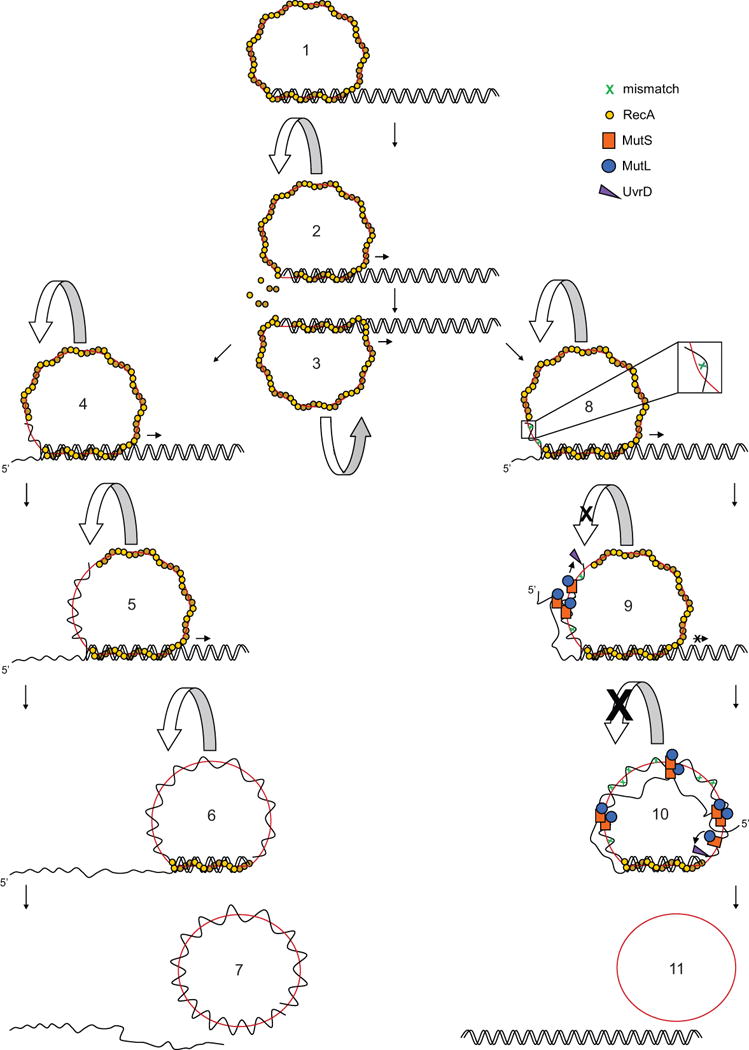
The RecA-ssDNA filament pairs and intertwines with the homologous region of linear dsDNA and strand exchange is catalyzed within the RecA-bound three-stranded DNA complex (step 1). ATP hydrolysis in the RecA-DNA complex allows disassembly of RecA from the heteroduplex, which in turn allows the rotation of RecA-ssDNA nucleoprotein filament. This rotation is promoted by spooling out of displaced ssDNA from the heteroduplex and spooling in of linear dsDNA at the trailing end and the leading end of RecA-DNA complex respectively (steps 2, 3) (related to Movie S1). The rotation of circular RecA-ssDNA filament causes a window of RecA-DNA complex traveling from 5′ to 3′ in relation to the displaced ssDNA throughout the course of strand exchange (steps 4, 5, 6). Upon completion of strand exchange, a nicked circular dsDNA and a linear ssDNA are produced (step 7). The presence of mismatches in the heteroduplex region activates MMR-dependent antirecombination (step 8). MutS and MutL bound to mismatches in the heteroduplex and to secondary structures within the displaced ssDNA form higher-order complexes facilitated by MutS tetramerization. Due to limited freedom of the circular RecA-ssDNA filament during rotation, trapped DNA intermediates are prevented from forming new synapsis at the leading end of the RecA-DNA complex. UvrD is directed by the MutS-MutL complexes on heteroduplex to unwind from the nearest ss/ds DNA junction of heteroduplex and resolve the trapped intermediates (step 9). Stronger strand exchange inhibition is exerted when more mismatches accumulate in the heteroduplex and multiple MutS-MutL complexes are loaded. UvrD is still directed to unwind from the same ss/ds DNA junction by mismatch-activated MutS-MutL complexes (step 10). DNA substrates are re-formed from the MutS-MutL-directed unwinding by UvrD helicase (step 11).
UvrD acts exclusively as an anti-recombinase during RecA-mediated homeologous strand exchange in the presence of MutS and MutL (Figure 6). UvrD is directed by the mismatch-activated MutS-MutL complex to resolve the trapped DNA intermediates specifically toward re-formation of linear dsDNA and circular ssDNA. This is analogous to DNA mismatch repair in which UvrD is directed by the mismatch-activated MutS-MutL complex to unwind DNA toward the mismatch (Dao and Modrich, 1998). The entry site for UvrD unwinding is most likely the ss/ds DNA junction because it is closest to the first mismatch formed in the heteroduplex (steps 9, 10). As a result of the MutS-MutL-directed UvrD unwinding, the initial DNA substrates are re-formed from the trapped DNA intermediates (step 11).
MMR-Dependent Antirecombination in Eukaryotes
Crossover and non-crossover events arise as a consequence of a choice between homologous recombination subpathways in double-strand break repair (DSBR). Synthesis-dependent strand annealing (SDSA) exclusively generates non-crossovers while double Holliday junctions (dHJs) intermediates of the DSBR model generate both crossovers and non-crossovers with the action of appropriate endonucleases (Schwartz and Heyer, 2011). The template within the SDSA recombination intermediate is topologically constrained due to the absence of endonuclease participation. In contrast, the endonucleolytic activities on the Holliday junctions confer some topological freedom to the DNA strands within the dHJs recombination intermediate. Thus, it is possible that the mechanism of the dHJs subpathway intrinsically involves more DNA strand rotation within the recombination intermediates during the process of branch migration than the SDSA subpathway. Interestingly, the yeast MMR machinery impedes crossover events to a much greater extent than non-crossover events in DSBR involving a divergent template (Welz-Voegele and Jinks-Robertson, 2008; Mitchel et al., 2010). This preference may be due to the topological freedom of dHJs recombination intermediate, which could be conferred by endonucleases, targeted and controlled by the MMR machinery. In the light of our finding that MutS-MutL complexes trap the recombination intermediate from branch migration by preventing rotation of the RecA-ssDNA filament and direct UvrD to dissolve the trapped recombination intermediate, the observation by Jinks-Robertson and colleagues supports the possible relevance of our MMR-imposed antirecombination model for eukaryotic crossover inhibition mediated by the MMR system (Welz-Voegele and Jinks-Robertson, 2008).
DNA translocases confer reversibility at several steps of the homologous recombination subpathways by disrupting deadly recombination intermediates (Symington and Heyer, 2006). UvrD and its S. cerevisiae homolog Srs2 both disrupt recombinase-ssDNA filament and prevent strand exchange in vitro (Krejci et al., 2003; Veaute et al., 2003; Veaute et al., 2005 and this study). While a specific role for Srs2 in inhibition of homeologous recombination was not observed using an inverted-repeat assay (Spell and Jinks-Robertson, 2004), recent studies indicate a role for Srs2 during gap repair involving homeologous sequences (Welz-Voegele and Jinks-Robertson, 2008; Mitchel et al., 2013). However, S. cerevisiae Sgs1 (a RecQ homolog) is a potent suppressor of homeologous recombination (Welz-Voegele and Jinks-Robertson, 2008; Myung et al., 2001). sgs1 and msh2 are epistatic in increasing homeologous recombination rates, indicating that Sgs1 may be specifically directed by mismatch-activated Msh2 to heteroduplex DNA (Spell and Jinks-Robertson, 2004). Interestingly, human BLM (also a RecQ homolog) may reverse recombination intermediates (van Brabant et al., 2000) and was shown to interact with human MSH6 both in vivo and in vitro (Pedrazzi et al., 2003). As the mechanistic distinctions between the MMR systems of eukaryotes and bacteria become increasingly understood (Jiricny, 2013), it will be interesting to examine in more detail to what extent the mechanism of antirecombination is conserved throughout evolution.
Conclusion
MutS-MutL complexes inhibit branch migration during homeologous recombination. We propose that the link connecting mismatch binding by MutS or MutS-MutL complexes within the heteroduplex region and the prevention of branch migration is the rotation of RecA-ssDNA nucleoprotein filament around the linear dsDNA. Through the formation of higher-order complexes between MutS-MutL on mismatches and secondary structures of displaced ssDNA at the trailing end of synapsis within the recombination intermediate, filament rotation and branch migration at the leading end of synapsis is prevented. Eventually, UvrD helicase is directed by the mismatch-activated MutS-MutL complexes to reverse the trapped recombination intermediates toward re-formation of DNA substrates. Thus UvrD becomes anti-recombinogenic through specific recruitment by MutS-MutL complexes that interfere with the structural dynamics of recombination intermediates.
EXPERIMENTAL PROCEDURES
Strand exchange using bacteriophage DNA
DNA substrates (M13 and fd ssDNA circles, HpaI-linearized M13 RF DNA) were extensively purified using agarose gel electrophoresis and electroelution. Strand exchange was performed under ATP-hydrolysis-permitting conditions using purified MutS, MutL, RecA, UvrD and variants verified to be free of nuclease contamination. RecA filaments were formed on fd or M13 circular ssDNA and incubated with E.coli SSB. When appropriate, MutS, MutL, UvrD and/or S1 nuclease were added and strand exchange was initiated with linear M13 dsDNA. Deproteinized samples were analyzed using agarose gel electrophoresis, stained using EtBr and intensities for linear dsDNA substrate and products were quantified.
Scanning force microscopy
DNA strand exchange intermediates were purified from agarose gels using electroelution and incubated with E.coli SSB and spermidine prior to deposition on freshly cleaved mica. Surfaces were washed, dried and imaged as specified in supplemental methods. Contour lengths of dsDNA tails and SSB-coated ssDNA gaps were traced manually.
D-loop reaction
RecA filaments were formed on ssDNA oligo’s either fully homologous to pUC19 or containing a single base difference. After incubation with SSB, MutS and MutL were added when appropriate and D-loop formation was initiated by adding supercoiled pUC19. Samples were cross linked with glutaraldehyde when appropriate, deproteinized, analyzed using agarose gel electrophoresis and visualized using the Alexa Fluor 532 label on the ssDNA oligo.
Electrophoretic mobility shift assay
To analyze binding of MutS to ssDNA, the protein was incubated with ssDNA oligonucleotides of arbitrary and poly-dT sequence. Complex formation was analyzed using polyacrylamide gel electrophoresis and visualized using the Alexa Fluor 488 label on the DNA. To analyze binding of MutS and MutL to mismatched DNA, the proteins were incubated with 90 bp dsDNA containing a single mismatch. Complex formation was analyzed on agarose gel and visualized using the Cy5 label on the dsDNA.
Supplementary Material
HIGHLIGHTS.
➢ MutS and MutL trap strand exchange intermediates during homeologous recombination
➢ Trapping involves binding to mismatches in heteroduplex DNA and to displaced ssDNA
➢ Trapping prevents strand exchange by imposing rotational constraints
➢ UvrD is recruited to directionally resolve the trapped recombination intermediates
Acknowledgments
We thank Thomas Holthausen for purified plasmid DNA and Flora Groothuizen for cross-linked MutS. M13 and fd bacteriophages were a gift from Martin G. Marinus, RecA strain GE1710 was a gift from Stephen Kowalczykowski. We thank Stephen Kowalczykowski for discussion and Marcel Reuter, Martin Marinus and Titia Sixma for useful comments on the manuscript. This work was supported by the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement n° HEALTH-F4-2008-223545 and n° HEALTH-F2-2010-259893, a TOP grant and a VIDI grant (700.58.428 to J.L.) from the Netherlands Organization for Scientific Research (NWO) and NIH grant n° GM32335.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Bazemore LR, Folta-Stogniew E, Takahashi M, Radding CM. RecA tests homology at both pairing and strand exchange. PNAS. 1997;94:11863–11868. doi: 10.1073/pnas.94.22.11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Yamamoto A, Hsieh P. Branch migration through DNA sequence heterology. J Mol Biol. 1998;279:795–806. doi: 10.1006/jmbi.1998.1769. 1998. [DOI] [PubMed] [Google Scholar]
- Bjornson KP, Blackwell LJ, Sage H, Baitinger C, Allen D, Modrich P. Assembly and molecular activities of the MutS tetramer. J Biol Chem. 2003;278:34667–34673. doi: 10.1074/jbc.M305513200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Matson SW. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme non-functional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmann MA, Nowosielska A, Marinus MG. Separation of mutation avoidance and antirecombination functions in an Escherichia coli mutS mutant. Nucleic Acids Res. 2005-1;33:1193–1200. doi: 10.1093/nar/gki263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmann MA, Nowosielska A, Marinus MG. The MutS C terminus is essential for mismatch repair activity in vivo. J Bacteriol. 2005-2;187:6577–6579. doi: 10.1128/JB.187.18.6577-6579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Lehman IR. Directionality and polarity in recA protein-promoted branch migration. PNAS. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JM, Li H, Wood EA, Chitteni-Pattu S, Inman RB, Cox MM. Defective dissociation of a “slow” RecA mutant protein imparts an Escherichia coli growth defect. J Biol Chem. 2008;283:24909–24921. doi: 10.1074/jbc.M803934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Adjiri A, New L, Crouse GF, Jinks-Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the Mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- de Wind N, Dekker M, Claij N, Jansen L, van Klink Y, Radman M, Riggins G, van der Valk M, van’t Wout K, te Riele H. HNPCC-like cancer predispostion in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nature Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- Di Capua E, Engel A, Stasiak A, Koller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J Mol Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- Elez M, Radman M, Matic I. The frequency and structure of recombinant products is determined by the cellular level of MutL. PNAS. 2007;104:8935–8940. doi: 10.1073/pnas.0610149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein SI, Low KB. Hyper-recombining recipient strains in bacterial conjugation. Genetics. 1986;113:13–33. doi: 10.1093/genetics/113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JW, Brosh RM, Matson SW. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HHK, Marinus MG, Lebbink JHG, Svergun DI, Friedhoff P, Sixma TK. Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt582. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes VF, Scandellari F, Benjamin KR, Cozzarelli NR. Structure of reaction intermediates formed during Saccharomyces cerevisiae Rad51-catalyzed strand transfer. J Biol Chem. 2002;277:38945–38953. doi: 10.1074/jbc.M206962200. [DOI] [PubMed] [Google Scholar]
- Honigberg SM, Radding CM. The mechanics of winding and unwinding helices in recombination: torsional stress associated with strand transfer promoted by RecA protein. Cell. 1988;54:525–532. doi: 10.1016/0092-8674(88)90074-8. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Marszalek PE. Atomic force microscopy captures MutS tetramers initiating DNA mismatch repair. EMBO J. 2011;30:2881–2893. doi: 10.1038/emboj.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R, Cunningham RP, DasGupta C, Radding CM. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. PNAS. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to G·T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. Barriers to genetics exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol. 2000;182:1016–1023. doi: 10.1128/jb.182.4.1016-1023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Bear DG, Kowalczykowski SC. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. PNAS. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K, Zhang H, Welz-Voegele C, Jinks-Robertson S. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell. 2010;38:211–222. doi: 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. Plos Genetics. 2013;9:e1003340. doi: 10.1371/journal.pgen.1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti MC, Cohen SX, Fish A, Winterwerp HH, Barendregt A, Friedhoff P, Perrakis A, Heck AJ, Sixma TK, van den Heuvel RH, Lebbink JH. Native mass spectrometry provides direct evidence for DNA mismatch-induced regulation of asymmetric nucleotide binding in mismatch repair protein MutS. Nucleic Acids Res. 2011;39:8052–8064. doi: 10.1093/nar/gkr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel P, Hejna JA, Ehrlich SD, Cassuto E. Antipairing and strand transferase activities of E. coli helicase II (UvrD) Nucleic Acids Res. 1993;21:3205–3209. doi: 10.1093/nar/21.14.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nature Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Nicholson A, Hendrix M, Jinks-Robertson S, Crouse GF. Regulation of mitotic homeologous recombination in yeast: functions of mismatch repair and nucleotide excision repair genes. Genetics. 2000;154:133–146. doi: 10.1093/genetics/154.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PP, Lundberg AS, Walker GC. Identification and characterization of the mutL and mutS gene products of Salmonella typhimurium LT2. J Bacteriol. 1985;163:1007–15. doi: 10.1128/jb.163.3.1007-1015.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzi G, Perrera, Bachrati CZ, Selak N, Studer I, Petkovic M, Hickson ID, Jiricny J, Stagljar I. The Bloom’s syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol Chem. 2003;384:1155–1164. doi: 10.1515/BC.2003.128. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Dohet C, Radman M. Interspecific recombination between Escherichia coli and Salmonella typhimurium occurs by the RecABCD pathway. Biochemie. 1991;73:371–374. doi: 10.1016/0300-9084(91)90103-8. [DOI] [PubMed] [Google Scholar]
- Rosselli W, Stasiak A. Energetics of RecA-mediated recombination reactions without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J Mol Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- Schwartz EK, Heyer W. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva EM, New L, Crouse GF, Lahue RS. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, DasGupta C, Cunningham RP, Radding CM. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. PNAS. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Ohtani T, Chang PK, Ando T. Role of superhelicity in homologous pairing of DNA molecules promoted by Escherichia coli recA protein. J Biol Chem. 1982;257:370–376. [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168:1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambuk S, Radman M. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics. 1998;150:533–542. doi: 10.1093/genetics/150.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A, Egelman EH. Structure and function of RecA-DNA complexes. Experientia. 1994;50:192–203. doi: 10.1007/BF01924002. [DOI] [PubMed] [Google Scholar]
- Symington LS, Heyer W. Some disassembly required: role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 2006;20:2479–2486. doi: 10.1101/gad.1477106. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German JL, III, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmes S, Selva M, Jeusset J, Cam EL, Matic I, Fabre F, Petit M. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz-Voegele C, Jinks-Robertson S. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics. 2008;179:1251–1262. doi: 10.1534/genetics.108.090233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC, Cassuto E, Howard-Flanders P. Heteroduplex formation by recA protein: polarity of strand exchanges. PNAS. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- Worth L, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. PNAS. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth L, Bader T, Yang J, Clark S. Role of MutS ATPase activity in MutS, L-dependent block of in vitro strand transfer. J Biol Chem. 1998;273:23176–23182. doi: 10.1074/jbc.273.36.23176. [DOI] [PubMed] [Google Scholar]
- Zahrt TC, Maloy S. Barriers to recombination between closely related bacteria: MutS and RecBCD inhibit recombination between Salmonella typhimurium and Salmonella typhi. PNAS. 1997;94:9786–9791. doi: 10.1073/pnas.94.18.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


