Abstract
Neural stem cells (NSCs) are pluripotent precursors with the ability to proliferate and differentiate into 3 neural cell lineages, neurons, astrocytes and oligodendrocytes. Elucidation of the mechanisms underlying these biologic processes is essential for understanding both physiologic and pathologic neural development and regeneration after injury. Nuclear hormone receptors (NRs) and their transcriptional coregulators also play crucial roles in neural development, functions and fate. To identify key NRs and their transcriptional regulators in NSC differentiation, we examined mRNA expression of 49 NRs and many of their coregulators during differentiation (0–5 days) of mouse embryonic NSCs induced by withdrawal of fibroblast growth factor-2 (FGF2). 37 out of 49 NRs were expressed in NSCs before induction of differentiation, while receptors known to play major roles in neural development, such as THRα, RXRs, RORs, TRs, and COUPTFs, were highly expressed. CAR, which plays important roles in xenobiotic metabolism, was also highly expressed. FGF2 withdrawal induced mRNA expression of RORγ, RXRγ, and MR by over 20-fold. Most of the transcriptional coregulators examined were expressed basally and throughout differentiation without major changes, while FGF2 withdrawal strongly induced mRNA expression of several histone deacetylases (HDACs), including HDAC11. Dexamethasone and aldosterone, respectively a synthetic glucocorticoid and natural mineralocorticoid, increased NSC numbers and induced differentiation into neurons and astrocytes. These results indicate that the NRs and their coregulators are present and/or change their expression during NSC differentiation, suggesting that they may influence development of the central nervous system in the absence or presence of their ligands.
Keywords: central nervous system, histone deacetylase, glucocorticoid, mineralocorticoid, neuron
Introduction
Nuclear hormone receptors (NRs) are intracellular molecules that mediate the actions of their endogenous lipophilic ligands in a broad spectrum of physiological processes [1]. NRs strongly influence the structure and functions of the central nervous system (CNS) by affecting development, growth, differentiation, apoptosis, and death of its component cells. For example, thyroid hormones and their receptors are essential for normal development and function of the CNS, while steroid hormones, such as glucocorticoids, mineralocorticoids, androgens, and estrogens, secreted from the adrenal glands and/or gonadal organs, affect mood, cognitive function and sexual differentiation through their respective receptors expressed in CNS [2 – 4].
Humans and mice respectively express 48 or 49 NRs, and they exert their diverse actions by acting as ligand-dependent transcription factors [1]. NRs efficiently stimulate or repress the transcriptional activity of their target genes either by binding to their cognate DNA sequences (called response elements) located in the regulatory regions of their responsive genes or by interacting with other transcription factors [5]. In the nucleus, DNA- or transcription factor-associated NRs attract their transcriptional coregulators and chromatin-associated molecules [6]. These cofactors influence RNA polymerase II and components of the general transcription machinery, alter the chromatin structure through chemical modification of histones, and ultimately stimulate or repress the transcriptional activity of NR target genes [6]. Currently, over 200 NR cofactor molecules have been reported: These include histone acetyl-transferase coactivators, such as the CREB-binding protein (CBP), p300 and nuclear receptor coactivators (NCoAs), nuclear receptor corepressors (NCoRs), histone deacetylases (HDACs), histone metyltransferases, and further, RNA coregulators, such as the steroid receptor RNA coactivator (SRA) and the growth-specific arrest 5 (Gas5) [6, 7].
The CNS contains populations of neural stem cells (NSCs), defined by their properties of self-renewal and ability to differentiate into multiple neural cell lineages, such as neurons, astrocytes and oligodendrocytes [8]. During development, NSCs provide the cells that will comprise the CNS. In the adult CNS, populations of NSCs still persist, and their localization and biological roles are intensely studied: 2 areas of the adult brain, the dentate gyrus of the hippocampus and the subventricular zone lining the lateral ventricles, are the most established neurogenic zones containing NSCs, while other brain areas comprising NSCs are increasingly being reported [9]. Interestingly, NSCs share some characteristics with the putative progenitor cells found in the adult adrenal medulla, suggesting that NSCs or their related cells may also present in this peripheral neuronal tissue [10, 11]. The roles of adult NSCs may include the supply of new neurons to the olfactory bulb (from the subventricular zone NSC population) and to the hippocampus, thus their self-renewal and differentiation into neural cell lineages potentially affect olfactory function and memory consolidation subserved by these brain areas. More recently, neuroprotective roles of adult NSCs were reported, as NSCs demonstrated powerful neuronal rescue in models of neurodegenerative diseases and neurologic damage induced by pharmacologic treatments [9, 12 – 14].
These pieces of evidence indicate that elucidation of the mechanisms that influence self-renewal and differentiation of NSCs is essential for both understanding the physiology of CNS development and establishing rational therapeutic approaches for neurodegenerative disorders and CNS injury. Recently, primary cell cultures of NSCs have been established, showing functions reminiscent of their in vivo biology [9, 12, 13]; such systems appear useful in testing pertinent hypotheses and rescuing in vivo brain functions [13]. In these in vitro NSCs, maintenance of their characteristics as stem cells, such as self-renewal and differentiation into 3 neural lineages, is absolutely dependent on the presence of fibroblast growth factor-2 (FGF2) in the culture media [13]. Thus, we employed this valuable cellular system – the mouse primary NSCs – and examined mRNA expression of 49 NRs and 35 selected coregulators before and in the course of differentiation induced by withdrawal of FGF2 to identify key NRs and their transcriptional coregulators in the steady state and differentiation process of NSCs. We found that some NRs and coregulators show distinct expression profiles and dramatically altered expression levels during differentiation. Activation of such NRs may offer targets for NSC proliferation and differentiation into neural cells.
Materials and Methods
Preparation of mouse NSCs and treatments
Mouse NSCs were prepared from mouse embryos at embryonic day 13.5, as reported previously [8, 12]. Briefly, brains were obtained from mouse fetus at E13.5 day, and their cortex was harvested, triturated by pipetting, and cultured in N2 medium (Dulbecco’s modified Eagle’s medium/F-12 supplemented with 0.1 mg/ml of apo-transferin, 25 μg/ml of insulin, 0.1 μM of putrescine, 30 nM of selenite, 2 × 10−8 M of progesterone, 10 U/ml of penicillin, 100 μg/ml of streptomycin and 0.25 μg/ml of amphotericin B) in the presence of 30 ng/ml of FGF2 for 4–5 days to isolate mouse NSCs. The purified NSCs were then maintained in N2 medium supplemented with 30 ng/ml of FGF2 daily. NSCs were plated in 10 cm dishes at the concentration of 105 cells/plate. One day after plating, medium was changed to N2 medium without FGF2, and cells were further cultured for another 5 days. During the culture, total RNA was purified daily (Fig. 1). NSCs were also plated in 24-well plates, cultured in the presence or absence of the steroids indicated, the HDAC inhibitor trichostatin- A (TSA), and FGF2 for 2 days, and cell numbers were then counted.
Fig. 1.
Experimental procedures for evaluating NR/coregulator mRNA expression profiles upon differentiation of NSCs. NSCs were plated one day before FGF2 withdrawal. They were then cultured in the absence of FGF2 for 5 days. Total RNA was extracted from the cells daily for examining mRNA expression of NRs and their coregulators using the custom PCR arrays.
PCR arrays
The custom PCR arrays, which have primer pairs for 49 mouse NRs/orphan receptors/membrane receptors, 35 coactivators/corepressors/chromatin modulators and 5 control genes (total 89 genes) listed in Table 1, were produced by SABiosciences Corp. (Frederick, MD), as described previously [15]. Total RNA was purified from mouse NSCs by using the RNeasy Mini kit (Qiagen Inc., Valencia, CA, USA), and was treated with DNase (Promega). The samples were further reverse transcribed into cDNA by using the RT 2 First Strand Kit (SABiosciences Corp.), and real-time PCR reactions were run on the 7 500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA), by following company’s instructions.
Table 1.
Primer pairs used in the SYBR Green-based real-time PCR.
| Gene name | Primer sequence | |
|---|---|---|
| AR | Forward | 5′-CAGAAGATGACTGTATCAC-3′ |
| Reverse | 5′-CTAGATAACAAGGCAGCAAAGG-3′ | |
| COUP-TFI | Forward | 5′-GTCGACTCTGCCGAGTACAG-3′ |
| Reverse | 5′-CAGGGCACACTGTGATTTCTC-3′ | |
| ERRβ | Forward | 5′-GTGGTGGCTTTGGCATTGC-3′ |
| Reverse | 5′-CATGTACTCGCATTTGATG-3′ | |
| GR | Forward | 5′-GTTCCTAAGGAAGGTCTGAAGAG-3′ |
| Reverse | 5′-CAATTCTGACTGGAGTTTCC-3′ | |
| HDAC11 | Forward | 5′-GAGCCACCATCATTGATCTC-3′ |
| Reverse | 5′-GATGGCCTCTTTAGCAAAGC-3′ | |
| LRH1 | Forward | 5′-CGAAGATCATGGCTTACC-3′ |
| Reverse | 5′-GTTCCCTGAAGAAGATACTACTC-3′ | |
| MR | Forward | 5′-GTGTGCTGGAAGAAATGAC-3′ |
| Reverse | 5′-CAGCTTCTTTGACTTTCG-3′ | |
| PMC11 | Forward | 5′-GCTGTAATGCAAATGGTAAAGC-3′ |
| Reverse | 5′-CAGTGTGTGTGCACTTACC-3′ | |
| PR | Forward | 5′-CCAGATAACCCTGATTCAG-3′ |
| Reverse | 5′-CTGCTCATTTAGGATTAGATCAG-3′ | |
| RORγ | Forward | 5′-GAGGAGAGTGGAACATCTG-3′ |
| Reverse | 5′-CTGCAGCTTTTCCACATGTTG-3′ | |
| THRα | Forward | 5′-GTGCTGCTAATGTCAACAG-3′ |
| Reverse | 5′-GTTGTGTTTGCGGTGGTTGAC-3′ | |
| RPLP0 | Forward | 5′-GAGGACCTCACTGAGATTCG-3′ |
| Reverse | 5′-CTGGAAGAAGGAGGTCTTCTC-3′ | |
AR: androgen receptor; CAR: constitutive androstane receptor; COUP-TFI: chicken ovalbumin upstream promoter-transcription factor I; ERRβ: estrogen-related receptor β; GR: glucocorticoid receptor; HDAC11: histone deacetylase 11; LRH1: liver receptor homologue 1; MR: mineralocorticoid receptor; PMC11: progesterone membrane component 11; PR: progesterone receptor; RORγ: RAR-related orphan receptor γ; THRα: thyroid hormone receptor α; RPLP0: acidic ribosomal phosphoprotein P0.
SYBR Green-based real-time PCR
mRNA expression of some NRs and coregulators that demonstrated characteristic expression patterns was further confirmed by using regular SYBR Green-based real-time PCR. Total RNA samples obtained from mouse NSCs were reverse-transcribed to cDNA with the TaqMan reverse transcription reagents (Applied Biosystems), and real-time PCR was performed in triplicate using the SYBR Green PCR Master Mix (Applied Biosystems) in the 7500 Real-time PCR System (Applied Biosystems), as described previously [16]. The primer pairs used for measuring mRNA levels of THRα, RORγ, COUP-TFI, ERRβ, GR, MR, AR, PR, LRH1, PMC11, and HDAC11 in this assay are shown in Table 1. Obtained threshold cycle (C t) values of these molecules were normalized for those of the acidic ribosomal phosphoprotein P0 (RPLP0) and their relative mRNA expressions are demonstrated as fold induction over baseline. The dissociation curves of the primer pairs used showed a single peak and samples after PCR reactions had a single expected DNA band in agarose gel analysis (data not shown).
Indirect immunostaining
Mouse NSCs were plated on 24-well plates or 2-well slide chambers, and were incubated with the indicated steroid(s) or TSA in the absence of FGF2. Cells were then fixed with 4 % paraformal-dehyde and the expression of the glial fibrillary acidic protein (GFAP: astrocyte marker) and neuron-specific class III β-tubulin (TUJ1: neuron marker) was examined by staining with their specific antibodies (purchased from Dako North America, Inc., Carperinteria, CA, USA and R & D Systems, Minneapolis, MN, USA, respectively) followed by appropriate fluorescence-conjugated secondary antibodies. Cells were also stained with the 4′,6-diamidino-2-phenylindole (DAPI). Some cells were cultured in the absence of FGF2 for 5 days, and they were stained with anti-MR (H-300, catalogue # sc-11412, lot # D1709) or -HDAC11 antibody (Q-5, catalogue # sc-101065) purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Staining of these molecules was evaluated under the Leica DMIRB inverted microscope (Leica Microsystems Inc., Bannockburn, IL, USA). Nomarski and fluorescence images were obtained with the C4742-95 CCD camera (Hamamatsu Photonics, Bridgewater, NJ, USA), and were analyzed with the OpenLab v2.2.5 software (Agilent Technologies Inc., Santa Clara, CA, USA).
Statistical analysis
Statistical analyses were carried out by unpaired Student’s t -test with the 2-tailed p-value.
Results
Many NRs and all coregulators examined are expressed in NSCs
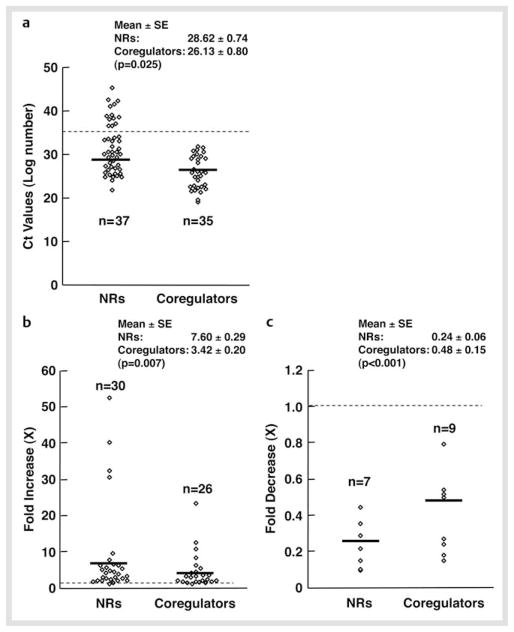
We first examined mRNA expression of 49 NRs and some coregulators in mouse NSCs maintained in the presence of FGF2. C t values of these molecules are shown in Table 2. Thirty seven out of 49 NRs and all (35) coregulators examined were expressed in these cells based on the criterion that the C t value ≤ 35 was the lowest limit for expression. The mean C t value of expressed NRs was 28.62 ± 0.74, while that of coregulators was 26.13 ± 0.74 (p = 0.025), indicating that coregulators tend to be expressed at higher levels than NRs (Fig. 2a).
Table 2.
Ct values of NRs and coregulators in NSCs.
| Subfamilies | Genes | Name | Symbol | Mean | SE | |
|---|---|---|---|---|---|---|
| Nuclear Receptors | ||||||
| 1 | 1A | NR1A1 | Thyroid hormone receptor α | THRα | 25.41 | 0.379 |
| 2 | NR1A2 | Thyroid hormone receptor β | THRβ | 30.97 | 0.645 | |
| 3 | 1B | NR1B1 | Retinoic acid receptor α | RARα | 45.73 | 1.253 |
| 4 | NR1B2 | Retinoic acid receptor β | RARβ | 39.64 | 0.281 | |
| 5 | NR1B3 | Retinoic acid receptor γ | RARγ | 28.58 | 0.142 | |
| 6 | 1C | NR1C1 | Peroxisome proliferator-activated receptor α | PPARα | 30.33 | 0.171 |
| 7 | NR1C2 | Peroxisome proliferator-activated receptor γ | PPARδ | 25.88 | 0.040 | |
| 8 | NR1C3 | Peroxisome proliferator-activated receptor γ | PPARγ | 38.88 | 0.179 | |
| 9 | 1F | NR1F1 | RAR-related orphan receptor α | RORα | 26.28 | 0.126 |
| 10 | NR1F2 | RAR-related orphan receptor β | RORβ | 27.3 | 1.673 | |
| 11 | NR1F3 | RAR-related orphan receptor γ | RORγ | 34.3 | 0.195 | |
| 12 | 1H | NR1H2 | Liver X receptor β | LXRβ | 29.15 | 0.085 |
| 13 | NR1H3 | Liver X receptor α | LXRα | 29.75 | 0.065 | |
| 14 | NR1H4 | Farnesoid X receptor α | FXRα | 39.28 | 0.538 | |
| 15 | NR1H5 | Farnesoid X receptor β | FXRβ | 38.66 | 2.041 | |
| 16 | 1I | NR1I1 | Vitamin D receptor | VDR | 37.05 | 0.404 |
| 17 | NR1I2 | Pregnane X receptor | PXR | 31.16 | 0.220 | |
| 18 | NR1I3 | Constitutive androstane receptor | CAR | 25.46 | 0.081 | |
| 19 | 2A | NR2A1 | Hepatic nuclear receptor 4 α | HNF4α | 33.61 | 0.077 |
| 20 | NR2A2 | Hepatic nuclear receptor 4 γ | HNF4γ | 42.93 | 0.306 | |
| 21 | 2B | NR2B1 | Retinoid X receptor α | RXRα | 27.63 | 1.139 |
| 22 | NR2B2 | Retinoid X receptor β | RXRβ | 24.60 | 0.208 | |
| 23 | NR2B3 | Retinoid X receptor γ | RXRγ | 33.66 | 0.996 | |
| 24 | 2C | NR2C1 | Testicular receptor 2 | TR2 | 26.01 | 0.396 |
| 25 | NR2C2 | Testicular receptor 4 | TR4 | 22.41 | 0.008 | |
| 26 | 2E | NR2E2 | Tailless | TLX | 31.06 | 0.224 |
| 27 | NR3E3 | Photospecific nuclear receptor | PNR | 34.42 | 0.285 | |
| 28 | 2F | NR2F1 | Chicken ovalbumin upstream promoter-TFI | COUP-TFI | 26.85 | 1.530 |
| 29 | NR2F2 | Chicken ovalbumin upstream promoter-TFII | COUP-TFII | 26.37 | 0.053 | |
| 30 | NR2F6 | v-ErbA related 2 | EAR2 | 25.25 | 0.012 | |
| 31 | 3A | NR3A1 | Estrogen receptor α | ERα | 41.43 | 0.706 |
| 32 | NR3A2 | Estrogen receptor β | ERβ | 33.91 | 0.489 | |
| 33 | NR3B1 | Estrogen-related receptor α | ERRα | 42.7 | 2.114 | |
| 34 | 3B | NR3B2 | Estrogen-related receptor β | ERRβ | 32.31 | 0.126 |
| 35 | NR3B3 | Estrogen-related receptor γ | ERRγ | 38.99 | 0.396 | |
| 36 | 3C | NR3C1 | Glucocorticoid receptor | GR | 27.95 | 0.306 |
| 37 | NR3C2 | Mineralocorticoid receptor | MR | 37.07 | 0.130 | |
| 38 | NR3C3 | Progesterone receptor | PR | 30.58 | 0.097 | |
| 39 | NR3C4 | Androgen receptor | AR | 27.00 | 0.106 | |
| 40 | 4A | NR4A1 | NUR77 | NUR77 | 25.29 | 0.097 |
| 41 | NR4A2 | Nuclear receptor-related 1 | NURR1 | 27.78 | 0.073 | |
| 42 | NR4A3 | Neuron-derived orphan receptor 1 | NOR1 | 42.05 | 6.487 | |
| 43 | 5A | NR5A1 | Steroidogenic factor 1 | SF1 | 34.05 | 0.032 |
| 44 | NR5A2 | Liver receptor homologue 1 | LRH1 | 29.91 | 0.093 | |
| 45 | 6A | NR6A1 | Germ cell nuclear factor 1 | GCNF1 | 30.53 | 0.077 |
| 46 | 0B | NR0B1 | Dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1 | DAX1 | 31.83 | 0.220 |
| 47 | NR0B2 | Small heterodimer partner | SHP | 37.48 | 0.167 | |
| Membrane nuclear receptors | ||||||
| 1 | Progesterone membrane component 11 | PMC11 | 26.44 | 0.024 | ||
| 2 | P Protein-coupled estrogen receptor 30 | GOCR30 | 39.69 | 1.686 | ||
| Coregulators | ||||||
| 1 | CREB-binding protein | CBP | 22.77 | 0.110 | ||
| 2 | p300 | p300 | 26.42 | 0.016 | ||
| 3 | Nuclear receptor coactivator 1 | NCoA1 | 25.21 | 0.016 | ||
| 4 | Nuclear receptor coactivator 2 | NCoA2 | 22.73 | 0.008 | ||
| 5 | Nuclear receptor coactivator 3 | NCoA3 | 24.61 | 0.073 | ||
| 6 | p300/CBP-associated factor | p/CAF | 28.45 | 0.318 | ||
| 7 | Nuclear receptor-interacting protein 1 | NRIP1 | 30.95 | 0.016 | ||
| 8 | PPARγ coactivator 1 α | PGC1α | 26.56 | 0.183 | ||
| 9 | Nuclear hormone receptor coregulator 6 | NRC6 | 32.37 | 0.044 | ||
| 10 | Structural maintenance of chromosome protein 1 (SMC) | SMC1 | 29.4 | 0.383 | ||
| 11 | Structural maintenance of chromosomes protein 4 | SMC4 | 29.54 | 0.457 | ||
| 12 | Thyroid receptor-associated proteincomplex 220 kDa component | TRAP220 | 23.07 | 0.089 | ||
| 13 | Thyroid hormone receptor-associated protein complex 170 kDa component | TRAP170 | 23.20 | 0.526 | ||
| 14 | Mediator of RNA polymerase II transcription subunit 6 | MED6 | 27.07 | 1.355 | ||
| 15 | Histone deacetylase 1 | HDAC1 | 22.35 | 0.024 | ||
| 16 | Histone deacetylase 2 | HDAC2 | 30.02 | 0.685 | ||
| 17 | Histone deacetylase 3 | HDAC3 | 22.01 | 0.061 | ||
| 18 | Histone deacetylase 4 | HDAC4 | 30.52 | 0.028 | ||
| 19 | Histone deacetylase 6 | HDAC6 | 25.27 | 0.044 | ||
| 20 | Histone deacetylase 7A | HDAC7 | 25.18 | 0.032 | ||
| 21 | Histone deacetylase 10 | HDAC10 | 31.39 | 0.269 | ||
| 22 | Histone deacetylase 11 | HDAC11 | 31.54 | 0.216 | ||
| 23 | Nuclear receptor corepressor 1 | NCoR1 | 25.56 | 0.069 | ||
| 24 | Nuclear receptor corepressor 2 | NCoR2 | 29.98 | 0.069 | ||
| 25 | SIN3 homolog | Sin3A | 24.64 | 0.469 | ||
| 26 | Set/Template activating factor 1β | Set/TAF-Iβ | 22.93 | 0.004 | ||
| 27 | Coactivator-associated arginine methyltransferase-1 | CARM1 | 21.82 | 0.032 | ||
| 28 | HRMT1-like 2 | HRMT-like 2 | 22.43 | 0.171 | ||
| 29 | Suppressor of variegation 3–9 homolog 1 | SUV39H1 | 32.01 | 1.481 | ||
| 30 | C-terminal tail-binding protein 1 | CtBP1 | 19.63 | 0.036 | ||
| 31 | Growth arrest-specific 5 | Gas5 | 26.27 | 0.881 | ||
| 32 | Steroid receptor RNA coactivator | SRA | 26.35 | 0.351 | ||
| 33 | SNF2 histone linker PHD RING helicase | SHPRH | 23.49 | 0.077 | ||
| 34 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | SMARCA4 | 20.13 | 0.032 | ||
| 35 | Brx (PRKA13) Rho-type GEF | AKAP13 | 29.47 | 0.012 | ||
| Controls | ||||||
| 1 | GUSB | 23.53 | 0.0938 | |||
| 2 | HPT1 | 22.22 | 0.0163 | |||
| 3 | HSP1b | 16.92 | 0.232 | |||
| 4 | GAPDH | 16.97 | 0.191 | |||
| 5 | β-Actin | 15.25 | 0.1633 | |||
Unexpressed molecules (Ct > 35) are shown in grey
Fig. 2.
mRNA expression of 49 NRs and coregulators in NSCs at the baseline and fold changes after FGF2 withdrawal. a The mRNA of 37 NRs receptors and 35 coregulators is expressed in NSCs. Thirty seven NRs and all coregulators examined were expressed in mouse NSCs at baseline, under the criterion that the Ct value ≤ 35 is the lowest limit for expression (shown as a dashed line). Their Ct values are shown as open circles, while mean values are indicated with bold horizontal lines. mRNA expression profiles of NRs and coregulators at the baseline condition were used to create this Figure. b and c Fold increase b and decrease c of mRNA expression of NRs and coregulators in NSCs before and after FGF2 withdrawal. Fold changes in the C t values of expressed NRs and coregulators are shown as open circles, while their mean values are indicated with bold horizontal lines. Dashed line indicates the fold change of “1”. Maximum fold changes found in the mRNA expression profiles of NRs and coregulators upon FGF2 withdrawal were used to create this Figure.
mRNA of the NRs known to have a significant impact on CNS development and function, such as the thyroid hormone receptor α (THRα), retinoid X receptors (RXRs), and chicken ovalbumin upstream promoter-transcription factors (COUP-TFs), NUR77 and v-ErbA related 2 (EAR2) [2, 17 – 19], were highly expressed in NSCs. In addition to these receptors, the peroxisome proliferator-activated receptor δ, testicular receptor (TR) 2 and 4, and the constitutive androstane receptor (CAR), which play important roles in fatty acid, retinoid and xenobiotic metabolism [20 – 22], were abundantly expressed. Among steroid hormone receptors, mRNA of the glucocorticoid (GR), androgen (AR) and progesterone receptor (PR) were moderately expressed in NSCs, while the estrogen receptor (ER) α, ERβ and mineralocorticoid receptor (MR) were poorly expressed or undetectable. Other unexpressed NRs were the retinoic acid receptor (RAR) α and β, PPARγ, farnesoid X receptors (FXRs), vitamin D receptor (VDR), hepatocyte nuclear receptor 4γ (HNF4γ), estrogen-related receptor α (ERRα), neuron-derived orphan receptor 1 (NOR1), and small heterodimer partner (SHP). Among the membrane-associated receptors, the progesterone membrane component 11 (PMC11) was moderately expressed. For coregulators, mRNA of CBP and p300, NCoAs, thyroid hormone receptor-associated protein (TRAP) 220 and 150, HDAC1, 3, 6, and 7, NCoRs, Sin3A, Set/temperature-activating factor (TAF)-Iβ, coactivator-associated arginine methyltransferase 1 (CARM1), HRMT1-like 2, C-terminal tail-binding protein 1 (CtBP1), SNF2 histone linker PHD RING helicase (SHPRH) and the SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4 (SMARCA4) were all highly expressed in NSCs.
Alteration of NR and coregulator mRNA expression upon differentiation of NSCs
We next examined mRNA expression profiles of NRs and coregulators during differentiation of NSCs by culturing them in the absence of FGF2 for up to 5 days (Fig. 2b, c and Suppl. info Table 1). Both fold increase and decrease of NRs after differentiation were greater than changes of coregulator expression (p = 0.007 and 0.001, respectively), indicating that FGF2 withdrawal and subsequent differentiation of NSCs affected more significantly the NR mRNA expression than that of coregulators.
These results further indicate that NR-mediated biological changes observed during differentiation may be regulated more significantly at the levels of NRs than at those of their coregulators. Among NRs, THRα (mean fold expression ± S.E.: 32.98 ± 30.68 on the 2nd day after FGF2 withdrawal), RORγ (40.68 ± 8.28, 4th day), RXRγ (50.94 ± 16.77, 2nd day) and MR (32.73 ± 15.80, 4th day) showed the most significant expression changes, while COUP-TFI, ERRβ and LHR1, which play important roles in the development of CNS [2, 18, 23, 24], demonstrated moderate changes (2.5–5-fold changes).
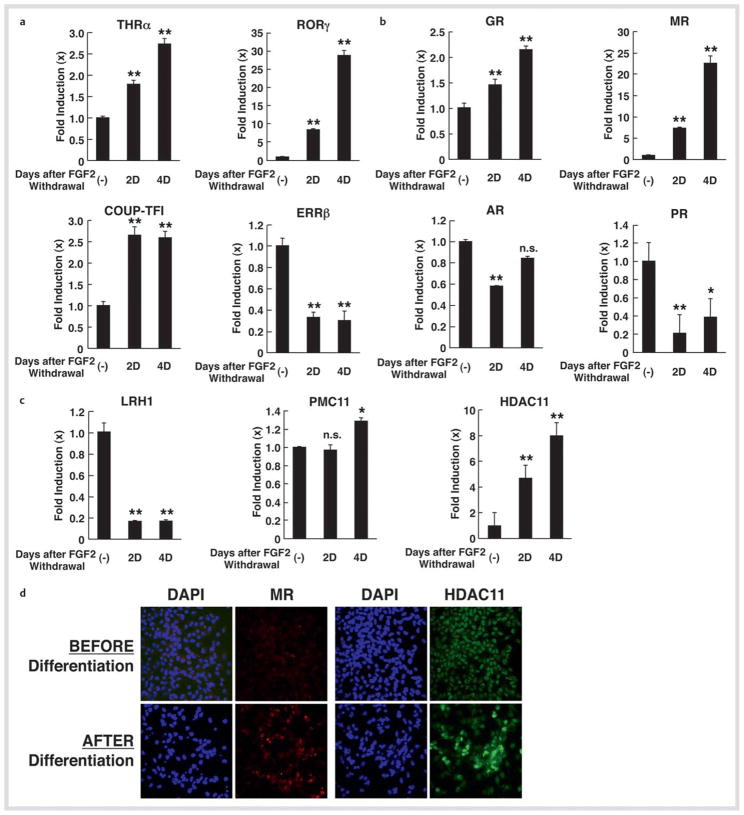
We confirmed mRNA expression of some of these NRs using SYBR Green-based real-time PCR (Fig. 3a, c). Since MR demonstrated a quite dramatic change, we examined its mRNA expression together with that of other steroid hormone receptors in this assay system (Fig. 3b). Significant expression of MR mRNA (over 20-fold) upon differentiation was observed, while GR mRNA expression was elevated by 2-fold. mRNA expression of PR was reduced by ~80 %, whereas that of AR showed minor fluctuations. The progesterone membrane receptor component 11 (PMC11), a membrane steroid hormone receptor expressed in NSCs in the presence of FGF2, did not show any detectable changes in its mRNA expression in the SYBR Green-based realtime PCR (Fig. 3c). Among coregulators, HDAC2, HDAC11, HDAC4 and HDAC10 demonstrated significant mRNA increase (13.23 ± 2.73, 2nd day; 11.35 ± 2.72, 4th day; 8.79 ± 2.70, 2nd day and 6.76 ± 1.87, 2nd day, respectively) in the screening using the PCR arrays, while mRNA expression of HDCA11 demonstrated an ~ 8-fold increase in the SYBR Green-based real-time PCR (Fig. 3c). mRNA expression of p300 (4.66 ± 2.54, 2nd day), nuclear receptor-interacting protein 1 (4.06 ± 1.06, 2nd day), nuclear receptor coregulator 6 (5.04 ± 2.43, 2nd day), nuclear receptor corepressor 1 (5.83 ± 2.59, 2nd day) and SRA (6.92 ± 4.79, 2nd day) demonstrated moderate increases upon differentiation. By using indirect immunostaining, we confirmed that MR and HDCA11 were also increased at the protein level upon differentiation (Fig. 3d).
Fig. 3.
mRNA and protein expression of selected NRs and HDAC11 in NSCs before and after FGF2 withdrawal. a, b, and c mRNA expression of selected NRs and HDAC11 in NSCs upon FGF2 withdrawal. mRNA expression of THRα, RORγ, COUP-TFI and ERRβ a, GR, MR, AR and PR b, and LRH1, PMC11, and HDAC11 c were examined with the SYBR Green-based real-time PCR using their specific primers listed in Table 1. Total RNA samples employed in Fig. 1 were used. Bars represent the mean ± S.E. values of fold mRNA expression upon FGF2 withdrawal compared with baseline (in the absence of FGF2 withdrawal). D: days after GFG2 withdrawal. *p < 0.05, **p < 0.01, n. s.: not significant, compared to baseline. d Protein expression of MR and HDAC11 in NSCs upon FGF2 withdrawal. NSCs before and after incubation for 5 days in the absence of FGF2 were immunostained with anti-MR or -HDAC11 antibody, and subsequently, by appropriate fluorescence-conjugated secondary antibodies. Cells were also stained with DAPI.
Glucocorticoid, mineralocorticoid and TSA alter the numbers of NSCs
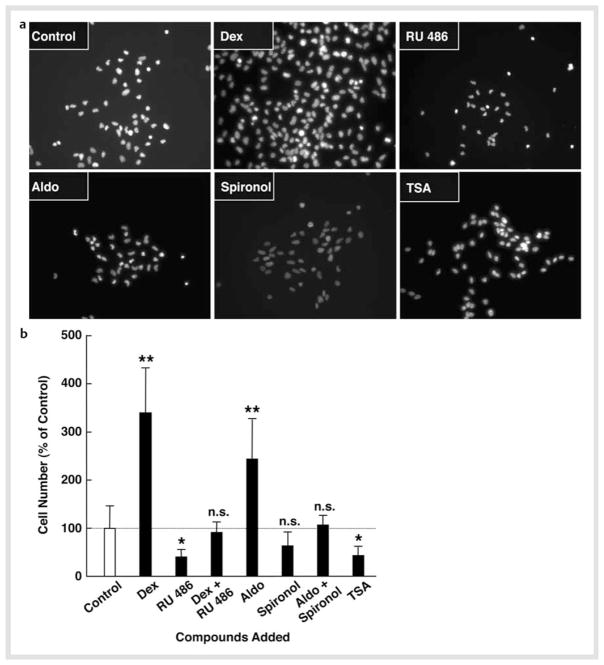
MR was one of the most highly induced NRs upon NSC differentiation. This receptor, together with the classic receptor GR, mediates strong and diverse actions of glucocorticoids in the CNS, as the MR has a higher affinity to glucocorticoids than GR and the CNS does not express 11β-hydroxysteroid dehydrogenase type 2 (HSD2), the enzyme that converts active corticosterone into inactive 11-dehydrocorticosterone [5, 25]. Thus, we focused on these receptors, and examined the effects of the synthetic glucocorticoid dexamethasone and its receptor inhibitor RU 486, and the endogenous mineralocorticoid aldosterone and its inhibitor spironolactone on cell numbers of NSCs by culturing them for 2 days in the presence of the steroid(s) and FGF2. We also tested the general HDAC inhibitor TSA in the same assay system, as mRNA expression of several HDACs was highly regulated upon differentiation. Fig. 4a shows images of DAPI staining of cells cultured with the indicated compounds, while Fig. 4b demonstrates percentage changes of the cell numbers before and after culturing with these compounds. Both dexamethasone and aldosterone significantly increased numbers of NSCs, with the former showing a more pronounced effect. Since these effects were suppressed by co-incubation with their receptor antagonists, we suggest that the observed effects were mediated by the GR and MR, respectively. TSA treatment reduced NSC numbers by ~ 50 % after 2-day culture. Since we did not observe massive cell death during the culture with FGF2, these compounds and/or TSA, it appears that dexamethasone and aldosterone stimulated proliferation of NSCs, while TSA suppressed it. This further suggests that HDACs are necessary for NSCs to maintain their potential for proliferation/self-renewal.
Fig. 4.
Dexamethasone and aldosterone increase NSC numbers, while TSA suppresses them. NSCs were cultured in the presence of 10−6 M of dexamethasone (Dex) and/or RU 486, 10−8 M of aldosterone (Aldo) and/or spironolactone (Spironol), or 10−6 M of trichostatin-A (TSA) in the presence of FGF2 for 2 days. Cells were then fixed and stained with DAPI and their images were taken or the numbers of NSCs were counted under fluorescence microscopy. Images of DAPI staining are shown in a, while percent increases of cell numbers compared to baseline (the cells before FGF2 withdrawal) are shown in b. Bars indicate mean ± S.E. values of % cell numbers compared to control. *p < 0.05, **p < 0.01, n. s.: not significant, compared to control.
Dexamethasone and aldosterone, but not TSA, alter differentiation of NSCs into neurons and astrocytes
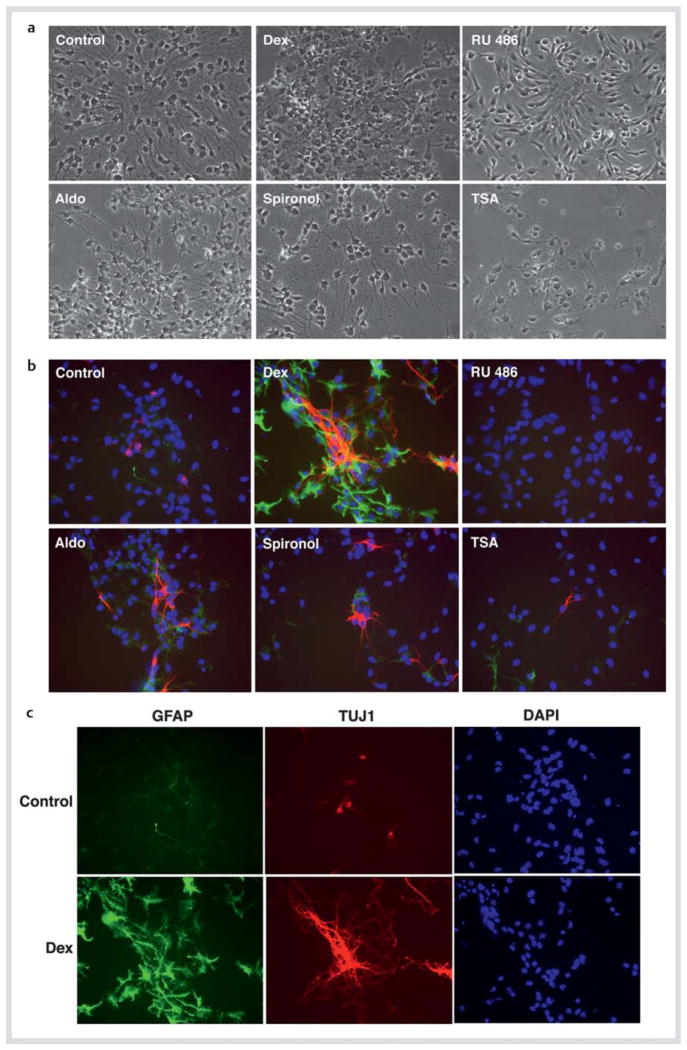
Since GR, MR and HDACs have stimulatory effects on NSC numbers, we next examined dexamethasone, aldosterone, their antagonists and TSA on the differentiation of NSCs into the glial and neuronal lineages, by culturing these cells for 2 days in the presence of these steroid(s) and/or TSA during the differentiation process. We subsequently stained them with the antibodies for GFAP (astrocytes: green) and TUJ1 (neuron: red) for identifying the corresponding neural cells. Given the strong effects of these compounds within the brief treatment period in our experimental setup and slow differentiation of oligodendrocytes and the late expression of their specific cell markers after FGF2 withdrawal, we limited our studies to the glial and neuronal fates, omitting analysis on oligodendrocytes. Fig. 5a demonstrated Nomarski images of the cells treated with the indicated compounds. Dexamethasone significantly increased astrocytes and neurons, while addition of RU 486 strongly suppressed differentiation of NSCs into these cells. Aldosterone increased neuron numbers but not those of astrocytes. TSA had no effect on the differentiation of NSCs (Fig. 5b, c). Percentages of the neurons and astrocytes differentiated from NSCs and those remaining undifferentiated are shown in Table 3.
Fig. 5.
Dexamethasone and aldosterone influence differentiation of NSCs. NSCs were cultured in the presence of 10−6 M of dexamethasone (Dex) or RU 486, 10−8 M of aldosterone (Aldo) or spironolactone (Spironol), or 10−6 M of trichostatin-A (TSA) for 2 days in the absence of FGF2, and were fixed for staining of GFAP (astrocytes: green) and TUJ1 (neurons: red). Nomarski images are shown in a, while images of immunostaining for GFAP and TUJ1 together with DAPI staining are shown in b. c demonstrates results of individual images of GFAP, TUJ1, and DAPI staining for the cells cultured in the presence or absence of dexamethasone.
Table 3.
Percentages of indicated cell lineages in NSC cultured in the absence of FGF2 for 2 days.
| Treatment | Undifferentiated cells | Cell types (means±S.D.) (% of total) Neurons | Astrocytes |
|---|---|---|---|
| No steroids | 77.71±6.67 | 7.86±2.88 | 14.39±5.13 |
| Dexamethasone | 20.12±3.09** | 19.31±3.82* | 60.58±7.46** |
| Aldosterone | 67.15±5.28 | 14.47±0.62* | 18.37±5.45 |
p < 0.05,
p < 0.01, compared to the baseline (no steroids)
Discussion
We showed that many NRs and their coregulators were expressed in NSCs and their expression was altered during differentiation induced by FGF2 withdrawal. These findings indicated that the NR signaling pathways are an integral component of NSC self-renewal and differentiation into 3 neural cell lineages, possibly by mediating the biologic actions of their ligands, such as steroids, lipid metabolites and other compounds available in the environment or produced during cell culturing. Importantly, mRNA expression of many NRs and coregulators were highly elevated or reduced at specific time points after FGF2 withdrawal, suggesting that their effects on NSC differentiation may have specific windows during the differentiation process of these cells.
As expected, THRα, RXRs, COUP-TFs, NUR77 and EAR2, which are known to play important roles in the development/function of the CNS during the fetal life and/or after birth, were highly expressed in NSCs, and their expression levels were relatively constant throughout the differentiation process, consistent with their general and important roles in CNS biology. We also found that PPARδ, TR2 and 4, and CAR were constitutively expressed in NSCs at high levels. Indeed, PPARδ ligands are known to stimulate differentiation of NSCs into oligodendrocytes and to suppress inflammation induced by radiation in microglia [20, 26 – 28]. TR2 and 4, the orphan receptors known to play important roles in spermatogenesis and lipid and lipoprotein metabolism [21], are expressed in the embryonic brain particularly in highly proliferating regions, such as the subventricular zone where embryonic NSCs are accumulated, and are required for normal CNS development [21]. Recent reports indicate that TR2 accumulates in the plaques in the brains of patients with Alzheimer’s disease, while TR4 might function as a receptor for vitamin A [29, 30]. CAR plays a central role in the xenobiotic response by binding to many exogenous compounds like phenobarbital, and by inducing the enzymes for catalyzing their clearance from the body [29]. It would, thus, be quite interesting to examine in future the impact of xenobiotic compounds on NSC activities and subsequent CNS development.
We found that mRNA and protein expression of the MR were highly regulated upon NSC differentiation. In the brain, MR mediates the actions of glucocorticoids in addition to those of mineralocorticoids [31]: These hormones are 2 major steroids produced and secreted from the adrenal glands, and respectively play central roles in the adaptive response to stress by functioning as end products of the hypothalamic-pituitary-adrenal (HPA) axis, and electrolytes and water absorption in the kidney by regulating sodium-potassium exchange in the cortical collecting tubules of this organ [5, 32]. Since corticosterone circulates at 2 log units higher concentrations than the natural mineralocorticoid aldosterone and MR has higher affinity to corticosterone than GR [33], basal physiologic levels of circulating corticosterone fully activate the MR in the CNS, which does not express 11βHSD2, while GR is affected primarily by elevated levels of corticosterone during the circadian surge or during stress [31]. NSCs are found in the dentate gyrus of the hippocampus in humans throughout the lifespan and produce granular cell neurons, which support consolidation of new memories [34]. Interestingly, this portion of the brain expresses high levels of GR and MR, and plays important roles in learning, memory consolidation, emotion and in the control of the HPA axis [31]. Further, increasing cortisol response as occurs in stress is positively associated with encoding of new memories in young subjects, while it correlates negatively with efficiency of their retrieval in older subjects [35]. We found that both dexamethasone and aldosterone increased the numbers of NSCs and induced differentiation of these cells into neurons. Dexamethasone also stimulated their differentiation into astrocytes. We did not examine the effects of these steroids on the differentiation of NSCs into oligodendrocytes. Previous reports indicated that MR activation was protective to granular cell neurons of the hippocampus, while persistent GR stimulation was toxic to these cells [31, 36]. Further, glucocorticoids induce apoptosis of neurons and neural precursor cells in the dentate gyrus of the rat hippocampus as well as the corresponding primary cell cultures, while they inhibit growth of astrocytes in this brain area [37, 38]. These effects of MR and GR on hippocampal neurons may ultimately affect memory consolidation and alterations in mood, cognition and activity of the HPA axis [31]. Thus, our results may provide important information on glucocorticoid-mediated regulation of neurons and astrocytes, underlying in part the known effect of glucocorticoids on neuronal cell fate, and memory consolidation and retrieval. However, it should be kept in mind that our NSCs were obtained from the embryonic brain cortex and were maintained in vitro, in contrast to hippocampal neurons in vivo, which have numerous cellular connections with surrounding and distant cells and are exposed to extracellular bioactive molecules secreted in a paracrine and/or autocrine fashion.
The mRNA and protein expression of several HDACs, including HDAC11, was highly regulated upon NSC differentiation. The HDAC inhibitor TSA decreased numbers of NSCs, but did not influence their differentiation. There are 4 subgroups of HDACs and they influence diverse functions of CNS by deacetylating many molecules localized at different subcellular compartments [39]. Our results are consistent with previous reports that HDAC-mediated transcriptional repression was essential for the proliferation and self-renewal of NSCs through cooperation with the Tailless (TLX) orphan nuclear receptor [40, 41]. Valproic acid, a compound frequently used as a mood stabilizer and as an anticonvulsant, is a HDAC inhibitor [42]. Further, several other HDAC inhibitors alter the fate of neural cells, and are now intensive research targets as potential therapeutic agents for several neurodegenerative diseases, cognitive disorders, multiple sclerosis and CNS trauma/injuries [39, 43 – 45]. Thus, it is quite interesting to elucidate the exact roles of HDAC subtypes on the functions/differentiation of NSCs.
In summary, many NRs and their coregulators are differentially expressed in NSCs and show characteristic changes in their expression during the process of their differentiation. These molecules regulate diverse cellular activities after binding to their ligands, changing the expression of their target genes. Compounds, such as glucocorticoids, gonadal steroids and endocrine disruptors, derived from mothers either by endogenous production or acquisition from the environment, or secreted inside the fetus, could influence epigenetically the development of the CNS and its function after birth by affecting NSC self-renewal and differentiation into neuron and glial cells. These compounds might have potent therapeutic importance for neurodegenerative diseases and CNS injury.
Supplementary Material
Acknowledgments
This study was funded in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD USA; AA-T was supported by a grant from the German Research Foundation (DFG/KFO 252) and a grant from the Wilhelm Sander-Stiftung. We thank Mr. E. K. Zachman and Drs. D. J. Hoeppner and M. G. Pavlatou for their superb technical assistance.
Footnotes
Conflict of Interest
The authors do not have any conflict of interest.
Supporting information available online at http:/www.thieme-connect.de/ejournals/toc/hmr
References
- 1.Burris TP. The nuclear receptor superfamily. In: Burris TP, McCabe ERB, editors. Nuclear receptors and genetic disease. London: Academic Press; 2001. pp. 1–58. [Google Scholar]
- 2.Horn S, Heuer H. Thyroid hormone action during brain development: More questions than answers. Mol Cell Endocrinol. 2010;315:19–26. doi: 10.1016/j.mce.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Wolkowitz OM, Burke H, Epel ES, Reus VI. Glucocorticoids. Mood, memory, and mechanisms. Ann NY Acad Sci. 2009;1179:19–40. doi: 10.1111/j.1749-6632.2009.04980.x. [DOI] [PubMed] [Google Scholar]
- 4.Hughes ZA, Liu F, Marquis K, Muniz L, Pangalos MN, Ring RH, Whiteside GT, Brandon NJ. Estrogen receptor neurobiology and its potential for translation into broad spectrum therapeutics for CNS disorders. Curr Mol Pharmacol. 2009;2:215–236. doi: 10.2174/1874467210902030215. [DOI] [PubMed] [Google Scholar]
- 5.Kino T, Chrousos GP. Glucocorticoid effect on gene expression. In: Steckler T, Kalin NH, Reul JMHM, editors. Handbook on Stress and the Brain. Amsterdam: Elsevier BV; 2005. pp. 295–312. [Google Scholar]
- 6.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 7.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Androutsellis-Theotokis A, Murase S, Boyd JD, Park DM, Hoeppner DJ, Ravin R, McKay RD. Generating neurons from stem cells. Methods Mol Biol. 2008;438:31–38. doi: 10.1007/978-1-59745-133-8_4. [DOI] [PubMed] [Google Scholar]
- 9.Androutsellis-Theotokis A, Rueger MA, Park DM, Mkhikian H, Korb E, Poser SW, Walbridge S, Munasinghe J, Koretsky AP, Lonser RR, McKay RD. Targeting neural precursors in the adult brain rescues injured dopamine neurons. Proc Natl Acad Sci USA. 2009;106:13570–13575. doi: 10.1073/pnas.0905125106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Androutsellis-Theotokis A, Rubin de Celis MF, Ehrhart-Bornstein M, Bornstein SR. Common features between chromaffin and neural progenitor cells. Mol Psychiatry. 2012;17:351. doi: 10.1038/mp.2012.18. [DOI] [PubMed] [Google Scholar]
- 11.Bornstein SR, Ehrhart-Bornstein M, Androutsellis-Theotokis A, Eisenhofer G, Vukicevic V, Licinio J, Wong ML, Calissano P, Nistico G, Preziosi P, Levi-Montalcini R. Chromaffin cells: the peripheral brain. Mol Psychiatry. 2012;17:354–358. doi: 10.1038/mp.2011.176. [DOI] [PubMed] [Google Scholar]
- 12.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 13.Androutsellis-Theotokis A, Rueger MA, Mkhikian H, Korb E, McKay RD. Signaling pathways controlling neural stem cells slow progressive brain disease. Cold Spring Harb Symp Quant Biol. 2008;73:403–410. doi: 10.1101/sqb.2008.73.018. [DOI] [PubMed] [Google Scholar]
- 14.Kittappa R, Bornstein SR. Androutsellis-Theotokis A. The role of eNSCs in neurodegenerative disease. Mol Neurobiol. 2012 doi: 10.1007/s12035-012-8303-8. in press. [DOI] [PubMed] [Google Scholar]
- 15.Ng SS, Chang TH, Tailor P, Ozato K, Kino T. Virus-induced differential expression of nuclear receptors and coregulators in dendritic cells: Implication to interferon production. FEBS Lett. 2011;585:1331–1337. doi: 10.1016/j.febslet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kino T, Ichijo T, Amin ND, Kesavapany S, Wang Y, Kim N, Rao S, Player A, Zheng YL, Garabedian MJ, Kawasaki E, Pant HC, Chrousos GP. Cyclindependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: Clinical Implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol. 2007;21:1552–1568. doi: 10.1210/me.2006-0345. [DOI] [PubMed] [Google Scholar]
- 17.van Neerven S, Kampmann E, Mey J. RAR/RXR and PPAR/RXR signaling in neurological and psychiatric diseases. Prog Neurobiol. 2008;85:433–451. doi: 10.1016/j.pneurobio.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Pereira FA, Qiu Y, Tsai MJ, Tsai SY. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) expression during mouse embryogenesis. J Steroid Biochem Mol Biol. 1995;53:503–508. doi: 10.1016/0960-0760(95)00097-j. [DOI] [PubMed] [Google Scholar]
- 19.Levesque D, Rouillard C. Nur77 and retinoid X receptors: Crucial factors in dopamine-related neuroadaptation. Trends Neurosci. 2007;30:22–30. doi: 10.1016/j.tins.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall MG, Quignodon L, Desvergne B. Peroxisome proliferator-activated receptor β/δ in the brain: Facts and hypothesis. PPAR Res. 2008;2008:780452. doi: 10.1155/2008/780452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YF, Lee HJ, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol. 2002;81:291–308. doi: 10.1016/s0960-0760(02)00118-8. [DOI] [PubMed] [Google Scholar]
- 22.Schnegg CI, Robbins ME. Neuroprotective mechanisms of PPARδ: Modulation of oxidative stress and inflammatory processes. PPAR Res. 2011;2011:373560. doi: 10.1155/2011/373560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Real MA, Heredia R, Davila JC, Guirado S. Efferent retinal projections visualized by immunohistochemical detection of the estrogen-related receptor β in the postnatal and adult mouse brain. Neurosci Lett. 2008;438:48–53. doi: 10.1016/j.neulet.2008.04.044. [DOI] [PubMed] [Google Scholar]
- 24.Grgurevic N, Tobet S, Majdic G. Widespread expression of liver receptor homolog 1 in mouse brain. Neuro Endocrinol Lett. 2005;26:541–547. [PubMed] [Google Scholar]
- 25.Seckl JR, Walker BR. Minireview: 11β-hydroxysteroid dehydrogenase type 1–a tissue-specific amplifier of glucocorticoid action. Endocrinology. 2001;142:1371–1376. doi: 10.1210/endo.142.4.8114. [DOI] [PubMed] [Google Scholar]
- 26.Almad A, McTigue DM. Chronic expression of PPAR-δ by oligodendrocyte lineage cells in the injured rat spinal cord. J Comp Neurol. 2010;518:785–799. doi: 10.1002/cne.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Angelo B, Benedetti E, Di Loreto S, Cristiano L, Laurenti G, Ceru MP, Cimini A. Signal transduction pathways involved in PPARβ/δ-induced neuronal differentiation. J Cell Physiol. 2011;226:2170–2180. doi: 10.1002/jcp.22552. [DOI] [PubMed] [Google Scholar]
- 28.Schnegg CI, Kooshki M, Hsu FC, Sui G, Robbins ME. PPARδ prevents radiation-induced proinflammatory responses in microglia via transrepression of NF-κB and inhibition of the PKCα/MEK1/2/ERK1/2/AP-1 pathway. Free Radic Biol Med. 2012;52:1734–1743. doi: 10.1016/j.freeradbiomed.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel DA, Davies P, Dobrenis K, Huang M. Tomoregulin-2 is found extensively in plaques in Alzheimer’s disease brain. J Neurochem. 2006;98:34–44. doi: 10.1111/j.1471-4159.2006.03801.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhou XE, Suino-Powell KM, Xu Y, Chan CW, Tanabe O, Kruse SW, Reynolds R, Engel JD, Xu HE. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J Biol Chem. 2011;286:2877–2885. doi: 10.1074/jbc.M110.168740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sousa N, Almeida OF. Corticosteroids: Sculptors of the hippocampal formation. Rev Neurosci. 2002;13:59–84. doi: 10.1515/revneuro.2002.13.1.59. [DOI] [PubMed] [Google Scholar]
- 32.Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid resistance/hypersensitivity syndromes. J Endocrinol. 2001;169:437–445. doi: 10.1677/joe.0.1690437. [DOI] [PubMed] [Google Scholar]
- 33.Chrousos GP. Glucocorticoid therapy. In: Felig P, Frohman LA, editors. Endocrinology & Metablism. 4. New York: McGraw-Hill; 2001. pp. 609–632. [Google Scholar]
- 34.Aimone JB, Wiles J, Gage FH. Potential role for adult neurogenesis in the encoding of time in new memories. Nat Neurosci. 2006;9:723–727. doi: 10.1038/nn1707. [DOI] [PubMed] [Google Scholar]
- 35.Kukolja J, Thiel CM, Wolf OT, Fink GR. Increased cortisol levels in cognitively challenging situations are beneficial in young but not older subjects. Psychopharmacology (Berl) 2008;201:293–304. doi: 10.1007/s00213-008-1275-8. [DOI] [PubMed] [Google Scholar]
- 36.Gass P, Kretz O, Wolfer DP, Berger S, Tronche F, Reichardt HM, Kellendonk C, Lipp HP, Schmid W, Schutz G. Genetic disruption of mineralocorticoid receptor leads to impaired neurogenesis and granule cell degeneration in the hippocampus of adult mice. EMBO Rep. 2000;1:447–451. doi: 10.1093/embo-reports/kvd088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S, Yang S, Holsboer F, Sousa N, Almeida OF. Glucocorticoid regulation of astrocytic fate and function. PLoS One. 2011;6:e22419. doi: 10.1371/journal.pone.0022419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu S, Patchev AV, Wu Y, Lu J, Holsboer F, Zhang JZ, Sousa N, Almeida OF. Depletion of the neural precursor cell pool by glucocorticoids. Ann Neurol. 2010;67:21–30. doi: 10.1002/ana.21812. [DOI] [PubMed] [Google Scholar]
- 39.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 40.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun G, Fu C, Shen C, Shi Y. Histone deacetylases in neural stem cells and induced pluripotent stem cells. J Biomed Biotechnol. 2011;2011:835968. doi: 10.1155/2011/835968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chateauvieux S, Morceau F, Dicato M, Diederich M. Molecular and therapeutic potential and toxicity of valproic acid. J Biomed Biotechnol. 2010;2010:479364. doi: 10.1155/2010/479364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faraco G, Cavone L, Chiarugi A. The therapeutic potential of HDAC inhibitors in the treatment of multiple sclerosis. Mol Med. 2011;17:442–447. doi: 10.2119/molmed.2011.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Shein NA, Shohami E. Histone deacetylase inhibitors as therapeutic agents for acute central nervous system injuries. Mol Med. 2011;17:448–456. doi: 10.2119/molmed.2011.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.