Abstract
All vertebrate nervous systems, except those of agnathans, make extensive use of the myelinated fiber, a structure formed by coordinated interplay between neuronal axons and glial cells. Myelinated fibers, by enhancing the speed and efficiency of nerve cell communication allowed gnathostomes to evolve extensively, forming a broad range of diverse lifestyles in most habitable environments. The axon-covering myelin sheaths are structurally and biochemically novel as they contain high portions of lipid and a few prominent low molecular weight proteins often considered unique to myelin. Here we searched genome and EST databases to identify orthologs and paralogs of the following myelin-related proteins: (1) myelin basic protein (MBP), (2) myelin protein zero (MPZ, formerly P0), (3) proteolipid protein (PLP1, formerly PLP), (4) peripheral myelin protein-2 (PMP2, formerly P2), (5) peripheral myelin protein-22 (PMP22) and (6) stathmin-1 (STMN1). Although widely distributed in gnathostome/vertebrate genomes, neither MBP nor MPZ are present in any of nine invertebrate genomes examined. PLP1, which replaced MPZ in tetrapod CNS myelin sheaths, includes a novel ‘tetrapod-specific’ exon (see also Möbius et al., 2009). Like PLP1, PMP2 first appears in tetrapods and like PLP1 its origins can be traced to invertebrate paralogs. PMP22, with origins in agnathans, and STMN1 with origins in protostomes, existed well before the evolution of gnathostomes. The coordinated appearance of MBP and MPZ with myelin sheaths and of PLP1 with tetrapod CNS myelin suggests interdependence – new proteins giving rise to novel vertebrate structures.
Keywords: Myelin basic protein, myelin protein zero, PMP22, PMP2, stathmin
INTRODUCTION
A prominent feature of the central (CNS) and peripheral (PNS) nervous systems of all extant gnathostomes, with likely origins in placoderms (Zalc et al., 2008) is the myelinated fiber, an entity that speeds, stabilizes and strengthens neuronal communication. Substantial structural and functional alterations in both ancestral glial cells and neurons were required for the initial development of myelinated nervous systems. In both the PNS and CNS, myelin forming glial cells developed mechanisms to: (1) control their proliferation and migration so that appropriate numbers became available to foster and coordinate myelinogenesis; (2) identify axon segments as they become permissive for myelination; (3) form and stabilize appropriately sized spirally wrapped membrane sheets around every ‘large caliber’ axon segment; (4) participate in the development and maintenance of elaborate paranodal junctions that restrict protein movement in the membrane and current leakage at nodes of Ranvier. Concurrent with ensheathment of their axons, neurons developed mechanisms to target and maintain ion channels, adhesive junctions, and scaffold and cytoskeletal proteins in constrained nodal, paranodal, juxtaparanodal and axon initial segment domains. These activities rely on the coordinated activities of many proteins whose histories are traceable with the availability of large and growing numbers of genome and EST databases.
Although there are clearly substantial differences between ancestral myelinogenesis, about which we know little, and myelinogenesis in modern gnathostomes, comparative studies such as this one foster an understanding of the major underlying principles. Using a phylogenetic approach we obtain a historical perspective of a subset of resident myelin proteins, including three proteins that dominate vertebrate myelin sheaths; myelin basic protein, myelin protein zero and proteolipid protein. Studies carried out in species representing each gnathostome taxon have demonstrated that myelin basic protein (MBP) and myelin protein zero (MPZ) are prominent in all PNS myelin sheaths and in CNS sheaths of all fishes while MBP and proteolipid protein (PLP1) are the major proteins in tetrapod CNS myelin (Waehneldt et al., 1986; Waehneldt, 1990; Jeserich et al., 2008). Here we show that all three proteins appeared coordinate with the myelin sheaths that contain them. Peripheral myelin protein-2 (PMP2), derived from an ancient family of fatty acid binding proteins, first appeared in tetrapods. Only peripheral myelin protein-22 (PMP22), with orthologs in agnathans, and stathmin-1 (STMN1), with a broad range of functions in other cell types, appeared prior to the ascension of gnathostomes. However, following their appearances hundreds of millions of years ago, changes, reflected in available sequences from extant species, contain important clues to features that allow these proteins to participate in myelinogenesis.
OBJECTIVE
Vertebrate myelin, a prominent feature of gnathostome central and peripheral nervous system, contains two of three (MBP, MPZ, PLP1) proteins at high levels. With the availability of large numbers of vertebrate and invertebrate genome databases, we documented the phylogenetic distributions of these proteins and compared sequences of orthologs expressed by representatives from different gnathostome taxa. To strengthen our findings that these proteins arose coordinate with their involvement in myelination, similar studies were conducted with three other myelin-associated proteins (PMP2, PMP22 and STMN1). One (PMP2), like PLP1 first appeared in tetrapods whereas the other two existed in agnathans and invertebrate ancestors, respectively.
RESULTS
MBP and MPZ, two proteins that are localized in and define compact PNS myelin (Fig. 1A), are widespread among gnathostome genomes. PLP1, which replaces MPZ in tetrapod CNS myelin sheaths (Fig. 1B), includes a crucial exon that is only found in, and likely originated with, tetrapods. PMP2 and PMP22, located in mammalian PNS myelin, and STMN1, associated with oligodendrocyte cytoskeleton, were included for comparison. Because homologs of MBP and MPZ are absent from invertebrate genome and EST databases (Table 1), their origins are more tightly linked to vertebrate myelination than are PMP2, PMP22 and STMN1, all of which have invertebrate homologs.
Fig. 1. Electron micrographs of rodent (A) PNS and (B) CNS myelin sheaths (originals kindly provided by Dr. Jack Rosenbluth, New York University School of Medicine).
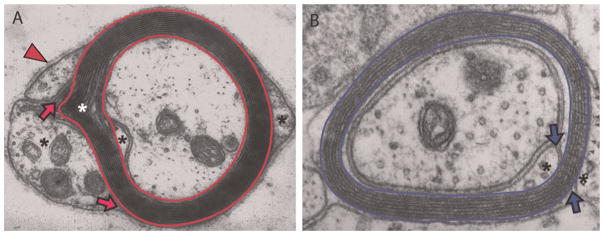
Compact myelin (bordered by red bands in PNS and by blue bands in CNS) is surrounded by glial cell cytoplasm (*), containing organelles where myelin-destined lipids and proteins are synthesized. For peripheral nerves, a supportive basement membrane (arrowhead) is also present and functions to stabilize the fibers and signal myelination (Jessen and Mirsky, 2005). Regions of compact myelin, where MBP, MPZ and PLP1 reside, are separated from regions of non-compacted myelin where MBP, MPZ and PLP1 are made, by tight junctions that form inner and outer mesaxons (arrows). Of many proteins synthesized in glial cell cytoplasm, few accumulate in and help structure myelin. A Schmidt–Lanterman incisure, with cytoplasm included between layers is common in MPZ-based though not PLP1-based myelin (
 ).
).
Table 1.
Distribution of myelin proteins in vertebrate and invertebrate genomes.
| Gene | CNS/PNS myelin | Protein property | Vertebrate | Invertebrate |
|---|---|---|---|---|
| MBP | C/P | Membrane associated | GAX D* H P | – |
| MPZ | P | TM1 | GAX DH P† | – |
| PLP1 | C | TM4 | GAXD* S P | BCSpLCcDmCe N |
| PMP2 | c/P | Membrane associated | GAX DSP | BCSpLCcDmCe N |
| PMP22 | P | TM4 | GAXD* TP | ––‡ |
| STMN1 | – | Cytoplasmic | GAXD* TP | BCSpLCcDm CeN |
Two MBP, DM20, PMP22 and STMN1 genes are present in a number of teleost databases.
Only the N-terminal half of this model corresponds to MPZ (also Fig. 5).
Proteins distantly related to PMP22, LIM2 proteins are identified in lancelet and sea squirt, though no other invertebrate phyla.
Legend: Three genes that encode the major proteins of compact CNS and/or PNS myelin are analyzed as well as one gene that encodes a similar size protein associated with myelinating cells, although not compact myelin. PMP2 is found in some, though not all, mammalian CNS myelin (c). Properties of the proteins are included. Vertebrate orthologs and invertebrate homologs are in black letters, partial orthologs (Methods) are in red, paralogs are in green and absence of a gene is shown in yellow. DM-20 and PLP1 are considered orthologs and GPM6/PLP1, PMP2/FABP and STMN1-4 proteins in invertebrates are all considered homologs. Designations of orthologs and paralogs among gnathostomes are based on reverse BLASTP searches of the mouse NR database (Methods). Letters represent: G – chicken (Gallus gallus), A – lizard (Anolis carolenensis), X – frog (Xenopus sp.), D – zebrafish (Danio rerio), H – horn shark (Heterodontis francisci), S – spiny dogfish (Squalus acanthias), little skate (Leucoraja erinacea), or T – Pacific electric ray (Torpedo californica), P – sea lamprey (Petromyzon marinus), B –lancelet (Branchiostomata floridae), C – sea squirt (Ciona intestinalis), Sp – sea urchin (Strongylocentrotus purpuratus), L – owl limpet (Lottia gigantea), Cc – polychaete worm (Capitella capitata), Dm – fruit fly (Drosophila melanogaster), Ce – roundworm (Caenorhibditis elegans) and N – sea anemone (Nematostella vectensis). Accession numbers of vertebrate and invertebrate homologs are listed in supplementary Tables 1A, 1B and 4 online. Dash (–) indicates that absence of invertebrate models.
MBP – a ubiquitous myelin protein with orthologs limited to gnathostomes
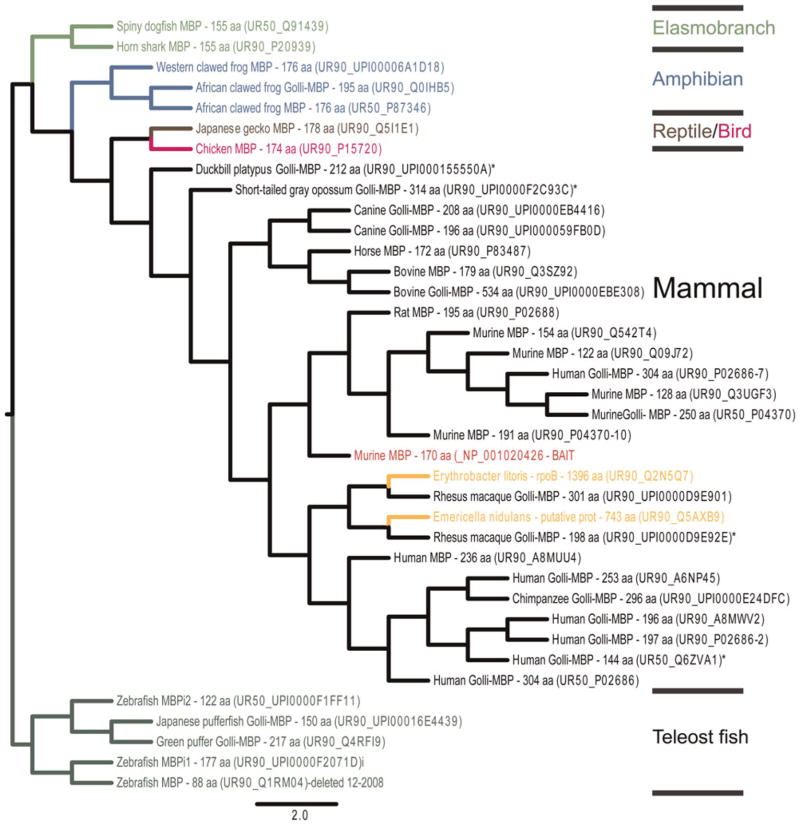
MBP is an intrinsically disordered, highly basic protein prominent in all gnathostome myelin sheaths examined (Boggs, 2006; Harauz and Libich, 2009). It is the product of a large gene with an upstream promoter that generates Golli-MBP proteins. Both neurons and glial cells express Golli-containing proteins, which unlike MBP, are absent from compact myelin (Givogri et al., 2001). Similarity searches performed with a murine MBP primary sequence as ‘BAIT’ (Methods) identified a Swiss-Prot protein set that included Golli-MBP and ‘classic’ MBP (products specific to gnathostome myelin), as well as several weakly related bacterial sequences. Maximum likelihood phylogenetic trees rooted to most divergent teleost MBP sequences (Fig. 2) or to elasmobranch MBPs separate MBPs into taxa-specific clusters. As vertebrate MBP paralogs or invertebrate homologs were not identified, searches were performed with zebrafish and horn shark MBPs as queries and were repeated with all three queries under more relaxed conditions (Methods). Because these additional searches failed to identify vertebrate paralogs and/or invertebrate homologs, and because similarity searches conducted for other myelin proteins: MPZ, PLP1, PMP2, PMP22 and STMN1, identified vertebrate paralogs and/or invertebrate homologs, it seemed unlikely that either vertebrate MBP paralogs or invertebrate MBP homologs exist.
Fig. 2.
Phylogenetic tree constructed with the UniTree program (Methods) with murine MBP (NP_001020426.1) as ‘BAIT’ (red) and with a search specified for 35×UR90 and 7×UR50 genes (3507) and re-rooted to teleost fish clade of MBP using Figtree 2.1 software (http://beast.bio.ed.ac.uk/FigTree). Each Uniprot hit is annotated with common species name, protein name and number of amino acids. In cases in which Uniprot accessions are not annotated, top hits obtained from BLASTP queries of the mouse NR database with FASTA sequences are listed (*). Some accession numbers were inactive (i). The branches, for mammals (black), birds/reptiles (pink/brown), amphibians (blue), teleosts (dark green), elasmobranchs (light green) and interspersed poorly related bacterial sequences (gold) are color coded. Crossbars indicate where branches are shortened to allow the tree to fit the page.
Complementary BLASTP and TBLASTN searches of available invertebrate genome, protein and EST databases (Methods) confirmed the absence of invertebrate MBP (and Golli, not shown) homologs. In contrast, nearly every gnathostome database examined contained MBP (and Golli, not shown) orthologs (Table 1, see supplementary Tables 1A and 1B online). In addition, a prominent myelin-MBP PFAM domain (PF01669), which characterizes MBP isoforms in all gnathostome taxa (Table 2), is not associated with any invertebrate sequence, further suggesting a disjunction between MBP and invertebrate proteins.
Table 2.
PFAM domains in vertebrate myelin proteins
| Gene | PFAM domain | Mouse | Chicken | Lizard | Frog | Zebrafish | Shark | Lamprey |
|---|---|---|---|---|---|---|---|---|
| MBP | Myelin_MBP | 17-170/171 | 16-173/174 | 16-169/170 | 16-175/176 | 2-95/100 | 2-155/155 | – |
| MPZ | V-set | 29-147/248 | 29-147/249 | 6-121/221 | 26-143/243 | 21-139/202 | 27-145/246 | 22-119/240 |
| MPZ | Myelin-P0_C | 190-248 | 190-248 | 164-222 | 186-243 | 176-229/2293 | 193-246 | – |
| PLP1 | Myelin_PLP | 2-277/277 | 2-277/277 | 2-277/277 | 2-280/280 | 2-245/245 | 2-246/246 | 2-108/108 |
| PMP2 | Lipocalin | 6-132/132 | 6-132/132 | 6-132/132 | 6-132/134 | 6-132/132 (FABP7) | 6-132/134 (FABP3) | 5-131/133 (FABP3) |
| PMP22 | PMP22_Claudin | 1-153/161 | 1-153/161 | 1-154/161 | 1-152/159 | 1-150/157 | 1-152/159 | 1-157/163 |
| PMP22 | L_HGMIC_fpl | 6-151 | 6-148 | 13/160 | 13-150 | 1-148 | 6-150 | 9-162 |
| STMN1 | Stathmin | 4-143/148 | 4-143/148 | 4-143/149 | 4-143/145 | 6-145/149 | 6-145/146 | 8-145/148 |
PFAM domain, in trout IP2 has a very low E-value 0.18.
Legend: The following PFAM domains for MBP (Myelin_MBP, PF01669), MPZ (V-set, PF07686 and Myelin-P0_C, PF01570), PLP1 (Myelin_PLP, PF01275), PMP2 (Lipocalin, PF00061), PMP22 (PMP22_Claudin, PF00822 and L_HGMIC_lpf, PF10242) and Stathmin (PF00836) were examined. Sequences (accession numbers are in supplementary Tables 1A, 1B and 4 online) were used as queries for PFAM sequence analyses (http://pfam.sanger.ac.uk/) and for each gene, included; domain ranges are followed after a slash (/) by the total number of amino acids in the search sequence. Proteins with substantially smaller PFAM domains are in red. Dash (–) indicates absences of PFAM domains.
Multi-sequence alignments, pair-wise comparisons and comparisons of known post-translational modifications were used to characterize the relationships of extant gnathostome MBP sequences. Because the Golli portion of MBP is not associated with compact myelin, phylogenetic distributions of Golli exons are not included, although like MBP exons they are widespread among gnathostomes and absent from agnathans and invertebrate databases. Alignments of MBP from species representing each taxon complement similarity search results in showing tetrapod MBPs are far closer to one another than they are to teleost and elasmobranch MBPs. Either when all available full-length sequences (19–24, identities shown in parentheses) or those identified in the same 12 species (see supplementary Tables 1A and 1B online) are aligned, overall sequence identity among mammalian MBPs (65.5%) is much lower than sequence identities for either mammalian MPZ (80%) or PLP1 (83%), although about the same as PMP22 (63%) and higher than for PMP2 (45%) (see supplementary Table 2 online). Furthermore, optimal alignments among mammalian MBPs include eight gaps (not shown), suggesting lower overall conservation compared with mammalian MPZ, PLP1, PMP2 or STMN1 sequence alignments (no gaps) and with mammalian PMP22 (two gaps). When pair-wise comparisons of mammalian MBPs are made with murine MBP, identities are significantly lower than identities of MPZ and PLP1, although higher than for two resident PNS myelin proteins, PMP2 and PMP22 (Fig. 3). Thus, both pair-wise and overall sequence comparisons indicate that even among mammals, MBP sequences are more divergent than the other major myelin proteins, though still higher than for two less abundant proteins in PNS myelin. Exon II is not included in these alignments because it is absent from many mammalian MBP sequences. This exon possibly arose with ancestral mammals, as it is not found in any non-mammalian MBP sequences with the possible exception of zebra finch (184 amino acids, ACH4544).
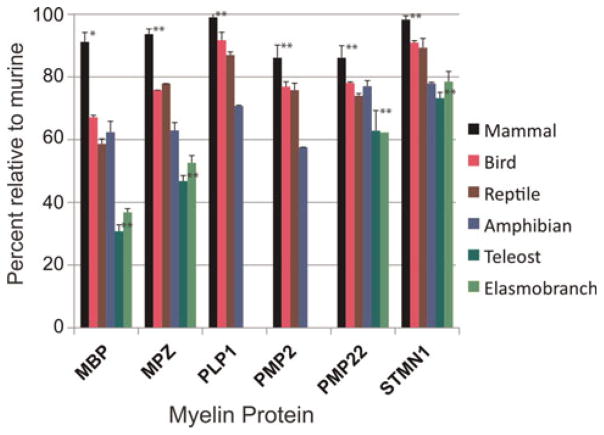
Fig. 3.
Pair-wise amino acid identity comparisons of all full-length sequences (see supplementary Tables 1A and 1B online) from each taxon with its murine counterpart are plotted. Mean and standard deviations are included. Statistical analyses (Prism 4, Graphpad program) were limited to comparisons among mammalian and teleost sequences and significance differences between MBP versus other myelin proteins are displayed (* – P < 0.05, ** – P < 0.0001, all others). Except for mammalian PLP1 and STMN1, comparisons show that conservations of each protein were different from other myelin proteins. Detailed information on comparative studies is presented in supplementary Table 2 online.
Pair-wise comparisons between each avian, diapsid, teleost and elasmobranch MBP sequence with the murine MBP show that for all taxa, with the possible exception of amphibian MPZ and PMP2, MBP sequences are less well conserved than other resident myelin proteins (Fig. 3); too few sequences are available for statistical comparisons. These results suggest that as myelin evolved in the different taxa with the possible exception of amphibians, MBP changes were greater than for both other major and several minor-occurring myelin proteins. It seems plausible that the inherent variability of MBP may stabilize myelin sheaths under the diversified conditions under which they form and are maintained.
A sense of the level of intra-mammalian conservation is reflected in the human-murine MBP sequence comparison (Fig. 4). Exons I, III and VII, which are common to all mammalian MBP isoforms, exhibit foci of high identity as indicated by shading and by identities among PFAM (http://pfam.sanger.ac.uk/) HMM logo amino acids (48% overall and 80% when zebrafish MBP is excluded). The alignment also demonstrates that exon I of teleost and elasmobranch MBPs is smaller than exon I of tetrapod MBPs. In addition, other teleost fish MBP exons are also small, making the sizes of full-length teleost MBPs (132–137 amino acids) only 74–81% the size of tetrapod MBPs and 85–88% the size of elasmobranch MBPs.
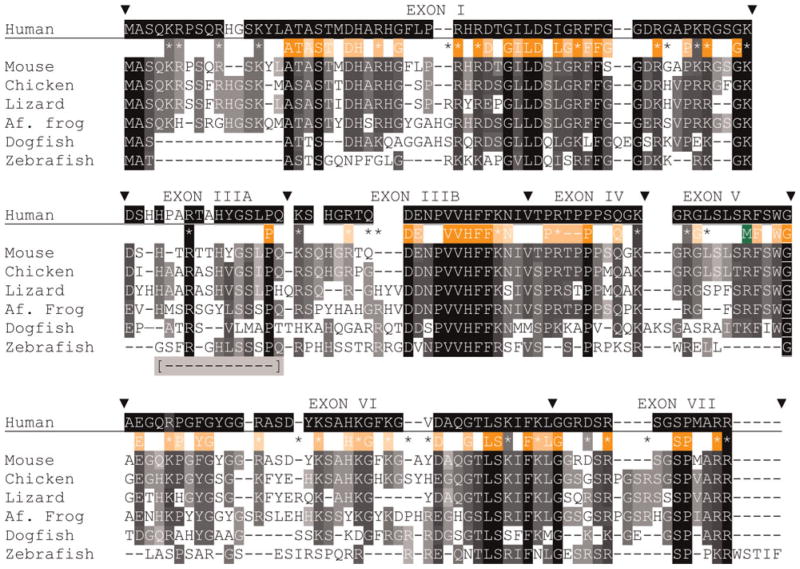
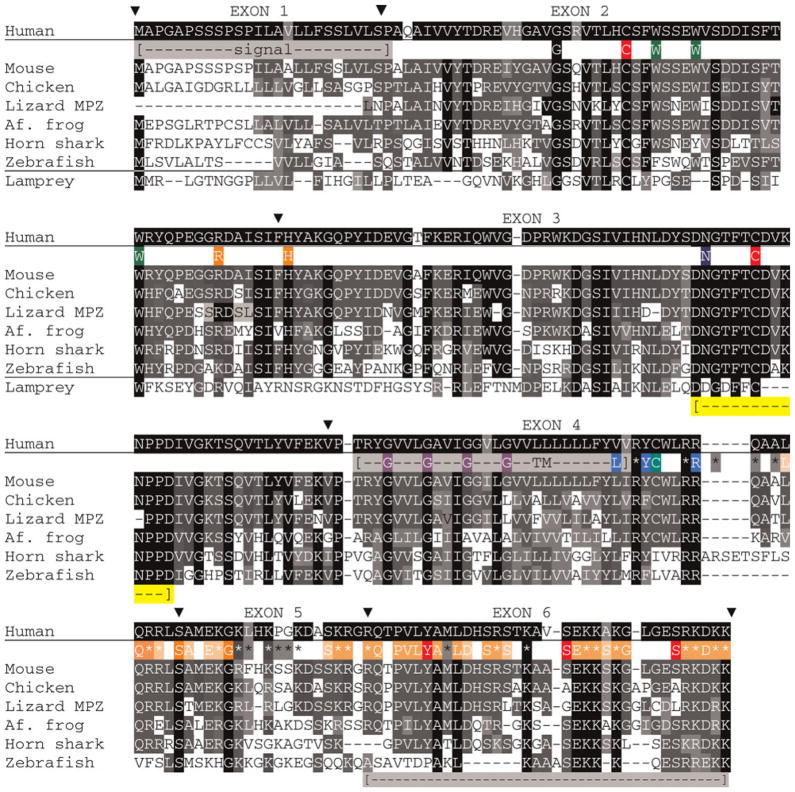
Fig. 4. Alignments of MBP sequences (isoform 3, contains all but exon II) from species that represent each gnathostome taxon.
Accession numbers are listed in supplementary Tables 1A and 1B online. Alignments are made, exon-by-exon with Clustal W (Methods), merged into a single alignment, and finally modified by eye. Comparisons among all listed species, excepting human use white letters on a black background (
 ) for complete identity, white letters on gray background for blocks of identity (>2) and conserved substitutions (
) for complete identity, white letters on gray background for blocks of identity (>2) and conserved substitutions (
 ); the background shading is set to the number of sequences with the same amino acid – darker background are for greater number of sequences. For simplicity, conserved substitutions are considered the same as identical amino acids. Exon boundaries are marked (θ) with exon numbers above the human sequence. Positions of all basic (K, R) amino acids are shown with coloring indicating complete identity (
); the background shading is set to the number of sequences with the same amino acid – darker background are for greater number of sequences. For simplicity, conserved substitutions are considered the same as identical amino acids. Exon boundaries are marked (θ) with exon numbers above the human sequence. Positions of all basic (K, R) amino acids are shown with coloring indicating complete identity (
 ) and (
) and (
 ) conservation and presence in sequences of one or two species (*). Sixty-nine HMM logo (PFAM) amino acids were selected based on individual or conserved substitution contributions >1.0 and shown with white lettering on orange backgrounds. As above, darker background shading indicates greater sequence identity. Letters and basic residue designations are placed between human and murine sequences. A single arginine methylation site is shown below the human sequence (M). A portion of exon III, absent in the common zebrafish 88 amino acid isoform is indicated [
) conservation and presence in sequences of one or two species (*). Sixty-nine HMM logo (PFAM) amino acids were selected based on individual or conserved substitution contributions >1.0 and shown with white lettering on orange backgrounds. As above, darker background shading indicates greater sequence identity. Letters and basic residue designations are placed between human and murine sequences. A single arginine methylation site is shown below the human sequence (M). A portion of exon III, absent in the common zebrafish 88 amino acid isoform is indicated [
 ] and assigned IIIA (see supplementary Table 3 online).
] and assigned IIIA (see supplementary Table 3 online).
Of the 30–32 arginine plus lysine residues in mammalian MBPs (all but two have 31), 26 are identical among all species, four others have conserved (K for R or vice versa) substitutions and only one is absent from MBPs of two species. Of the 35 basic amino acids marked in the alignment (Fig. 4), 12 are totally conserved (two identical and ten conserved substitutions), 11 others are conserved in all but one species, and four others are present in all but two species. The arginine methylation site (M) is not conserved in fish. All 31 basic residues in chicken and zebra finch MBP align. Each of the two non-aligned basic residues in the amphibian sequences is displaced by a single amino acid. Although each of the three reptilian MBP sequences has 32–33 basic residues, six positions do not align. Still, the overall retention of basic amino acid positions, both among taxa and among species within single taxa, suggest that these locations confer function such as binding sites for organizing acidic lipids at the cytoplasmic surfaces of myelin lamellae.
Post-translational modification is another well-known feature of mammalian MBPs with up to 25% of amino acids potentially subject to modification. These post-translational modifications are proposed to represent a ‘recognition’ barcode that modulates lipid–protein interactions and affects myelin stability (Harauz et al., 2004). This barcode appears to be evolutionarily recent. Whereas, 35 of 44 amino acids modified in human MBP are unchanged in mammals, only 6 of 41 are conserved in non-mammalian MBP sequences. Consistent with this notion, relatively few post-translational modifications have been observed in the non-mammalian MBPs examined so far (Zand et al., 2001; Kim et al., 2009).
Based on NR and EST database search results, a single prominent MBP isoform (exons I and III–VII) appears to be expressed by avian, reptilian, amphibian and elasmobranch oligodendrocytes (most ESTs are from brain and not peripheral nerve). A splice variant in horn shark that lacks exon V (Saavedra et al., 1989) also occurs in anole lizard and in three other elasmobranchs; little skate, Pacific electric ray and spiny dogfish (single ESTs). In sharp contrast, many different splice isoforms are observed in mammals (Harauz et al., 2004; Boggs, 2006) and teleosts (see below).
Extensive alternative splicing of MBP in teleost species are derived from two genes, MBP-isoform1 (i1) and MBPi2 (Fig. 2, see supplementary Fig. 1 online). Only a single zebra-fish MBPi2 product, lacking exon V, was identified (see supplementary Table 3 online). It is included in the alignment. The gene products from each species examined are derived from similarly sized exons and exons I, III, IV and V are more highly conserved than exons VI and VII. For example, 16 of 19 basic amino acids are conserved in the first four exons but only 3 of 15 are conserved in the last two exons. Unlike MBPs from all other taxa, teleost express MBPs from i1 and i2 that lack exon VII. Clusters of three or four basic residues occur in both of the teleost MBP gene products but are absent in tetrapod and cartilaginous fish MBPs. Similar basic amino acid clustering occurs in MARCKs, another protein known to associate with acidic lipids (e.g. Dietrich et al., 2009). Whether these amino acid clusters are necessitated by the smaller size of teleost MBPs is currently unknown. The overall amino acid identities between the two isoforms of each teleost species are between 46 and 52%.
Only MBPi1 (88 amino acids) has been identified in zebra-fish myelin (Brösamle and Halpern, 2002) and a single small MBP (73 amino acids) is present in grouper hippocampal myelin (Zhou et al., 2007). To determine the relative abundance of MBP isoforms (mRNAs) expressed by teleosts, counts of ESTs from zebrafish, three-spined stickleback, Japanese medaka and flathead minnow MBP isoforms have been made (see supplementary Table 3 online). Zebrafish and flathead minnow fall into one group; they express eight to nine isoforms, mostly small products derived from i1. The main difference is that zebrafish express two mRNAs that contain a truncated exon III, whereas flathead minnow do not show this truncation. Three-spined stickleback and Japanese medaka fall into another group; they express more products, including many from the second MBP gene, i2. Furthermore, these products tend to be larger. For each of these teleosts, it will be of interest to know the MBP variants present in myelin, whether different sheaths contain different variants and whether patterns of variant expression change during development.
MPZ – a vertebrate-specific protein with three closely related paralogs
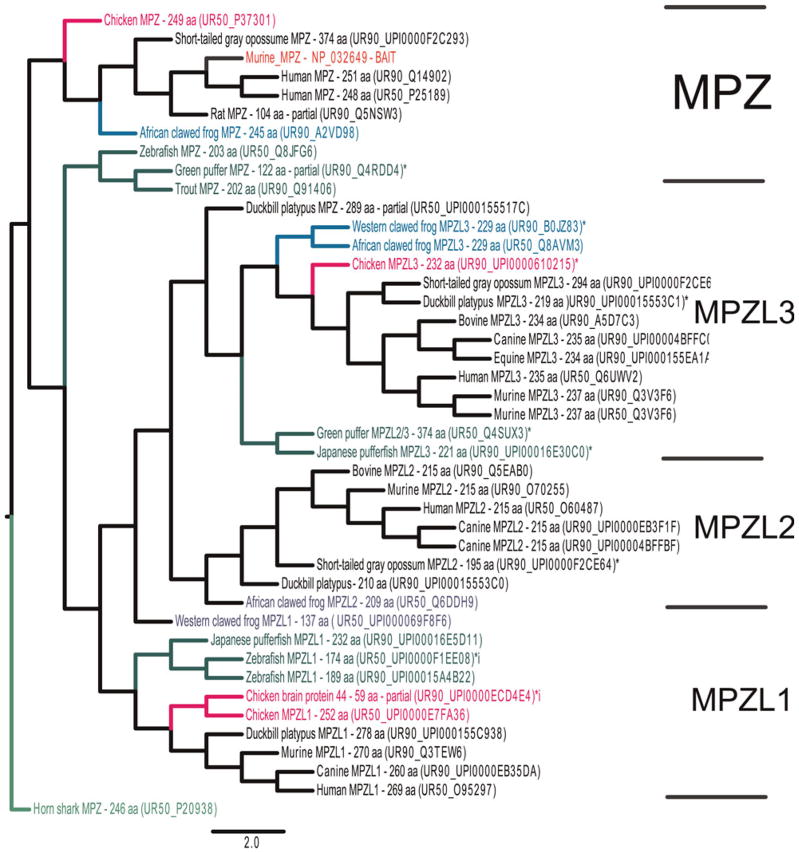
MPZ is the smallest member of the Ig family of proteins. Following a signal peptide, which targets the extracellular V-set containing Ig domain to the external surface, are a single pass transmembrane domain and a unique, highly basic cytoplasmic tail. Originally thought to be a primitive member of a large and abundant family (Williams and Barclay, 1988; Aricescu and Jones, 2007; Shapiro et al., 2007), its phylogenetic distribution suggests that it and other single Ig-domain proteins are recent. MPZ was identified in mammals, aves, diapsids, amphibians, teleost and elasmobranchs using both similarity and database searches (Table 1). Similarity searches queried with murine MPZ identified three paralogs: MPZL1, MPZL2/EVA and MPZL3, all of which are widely expressed in gnathostome taxa (Fig. 5). Queried searches using zebrafish and horn shark MPZs at both normal and relaxed stringency identified additional single Ig-domain families, including the sodium channel beta-subunit (SCN1B-SCN4B) and V-set and immunoglobulin domain containing 1 (VSIG1); however, none of them had invertebrate homologs (not shown). Complementary searches of nine invertebrate genome and EST databases also failed to identify invertebrate MPZ homologs (Table 1). Two MPZ homologs were identified in the current (May 2009) sea lamprey genome, although both gene models are incomplete. Only one has a putative signal peptide and although both have V-set domains, neither has glycosylation sites, transmembrane domains and putative cytoplasmic tail domains. The amino-terminal Ig-containing portion of the most similar lamprey MPZ model is included in the MPZ alignment (Fig. 6). All gnathostome MPZ proteins contain prominent V-set (present in the lamprey models) and carboxyl-terminal myelin-P0_C domains (Table 2). The shorter, more highly expressed teleost MPZ proteins lack the myelin-P0_C domain, as do all three MPZL paralogs.
Fig. 5. Phylogenetic tree constructed with the Unitree program, with murine MPZ (NP_032649) as ‘BAIT’ and with the (40 × 15 stringency, Methods) used in most instances; this tree is re-rooted to horn shark MPZ using Figtree 2.1.
Each hit is annotated, and clusters that include: MPZ, MPZL1, MPZL2/EVA and MPZL3, are separated from one another with brackets. Shading of branches is as described in Fig. 2.
Fig. 6. Alignment of MPZ sequences from species representing different vertebrate taxa.
Alignments are made on complete sequences with Clustal W program and positions of exon boundaries, based on mammalian sequences, are shown (▼). Accession numbers for all sequences are listed in supplementary Tables 1A and 1B online. A horizontal line separates agnathan/lamprey sequence from the gnathostome sequences and only the first 118 amino acids from this model are included as the remainder shows insignificant match. The signal peptide, transmembrane domain and the exon VI missing from many teleost fish MPZ sequences are similarly shaded with the first two regions marked between the human and murine sequences and the third region placed below the zebrafish sequence ([
 ]). Included are the single glycosylation site (N), two cysteine residues (C) that form the disulfide bond, a third (C) that is acylated in mammals. Two tryptophan residues (W) suggested from the X-ray diffraction data (Shapiro et al., 1996) to intercalate the apposing lipid bilayers, a glycine zipper (G) in the transmembrane domain, a tyrosine (Y) and two serine (S) represent phosphorylation sites. The (L), (Y) and (R) are cholesterol recognition motifs are highlighted. Amino acids that form the myelin-PO_C PFAM HMM logo are depicted as in Fig. 4, i.e. they are shaded in orange. A region of high conservation in exon 3 is also marked ([
]). Included are the single glycosylation site (N), two cysteine residues (C) that form the disulfide bond, a third (C) that is acylated in mammals. Two tryptophan residues (W) suggested from the X-ray diffraction data (Shapiro et al., 1996) to intercalate the apposing lipid bilayers, a glycine zipper (G) in the transmembrane domain, a tyrosine (Y) and two serine (S) represent phosphorylation sites. The (L), (Y) and (R) are cholesterol recognition motifs are highlighted. Amino acids that form the myelin-PO_C PFAM HMM logo are depicted as in Fig. 4, i.e. they are shaded in orange. A region of high conservation in exon 3 is also marked ([
 ]).
]).
Overall sequence identity among mammalian MPZs (80%) is substantially higher than for mammalian MBPs (66%) and somewhat lower than for mammalian PLP1 (83%, see supplementary Table 2 online). Pair-wise comparisons suggest less striking differences among mammalian MBP, MPZ and PLP1 orthologs (Fig. 3). When mammalian MPZ domains are analyzed topologically, identity is highest in the extracellular region (84%), followed by cytoplasmic tail (80%) and lastly the transmembrane domain (64%). MPZ alignments, without the amino-terminal signal sequence, show that avian and reptilian MPZ sequences are more highly conserved than MPZs of other taxa and also more highly conserved than their MBP counterparts (Fig. 3, see supplementary Table 2 online). Amphibian MPZs and MBPs show about the same degree of conservation compared with murine counterparts. Fish MPZs are also far closer to mammalian MPZs than corresponding MBPs to mammalian MBPs.
The level of conservation of MPZ across vertebrate taxa is seen in the alignment (Fig. 6). Except for the N-terminal signal peptide (exon 1), high identity is apparent in all exons. Many of the amino acids in the extracellular V-set domain important for structure have been identified by X-ray crystal structure analysis and through point mutations in human peripheral neuropathies (Shy, 2006; Shapiro et al., 2007). Our alignment, which includes the only reptilian sequence from the lizard, Anolis carolinensis, indicates that the most highly conserved region is a 14 amino acid stretch in the 3′-end of exon 3 that includes both the glycosylation site and second of two cysteine residues (C) that form a disulfide bridge. In addition, three amino acids – arginine 45 (R) and histidine 52 (H), that interact at the adhesive interface and – tryptophan 78 (W), proposed to interdigitate in apposing lipid bilayers and stabilize compaction (Shapiro et al., 1996; Luo et al., 2007) – are present in every species. Two others – tryptophan residues (W), one at position 24 and the other at position 28 – are conserved in all but one taxon. Phenylalanine (F) and tyrosine (Y) residues replace these residues in teleost and elasmobranch MPZs, respectively. An additional nearby tryptophan (W) is present in teleost fish MPZs, although not in elasmobranch MPZs.
The importance of the cytoplasmic tail in intracellular and extracellular compaction is well documented (Kirschner et al., 2004), and six of the carboxyl-terminal seven amino acids are either identical or conserved substitutions. The major teleost fish MPZ isoforms expressed in all species (EST database) lack exon 6, which leaves their tails half the size of other MPZs. Variations in elasmobranch MPZs include a slightly larger exon 4 and slightly smaller exon 6 than other MPZs and likely splice variants (see below). Targeting of MPZ to myelin requires a cholesterol recognition/interaction amino acid consensus (CRAC) sequence (-L/V-(X)(1–5)-Y-(X)(1–5)-R/K-) adjacent to the transmembrane domain (Epand, 2006; Saher et al., 2009), conserved in all but avian and teleost fish MPZs. Both have a phenylalanine (F) in place of the tyrosine, a substitution that lowers, though does not eliminate, interactions with cholesterol (Epand, 2006). A cysteine residue in the middle of this domain (C) is fatty acylated in mammals (Bizzozero et al., 1994; Gao et al., 2000) and amphibians (Luo et al., 2008), although the relation between its acylation and cholesterol binding in the CRAC domain is unexplored. A tyrosine phosphorylation site (Y) (Konde and Eichberg, 2006), is present in all gnathostome MPZ sequences. This site and two serine phosphorylation sites are in exon 6, which is not expressed in most teleost MPZs. Neither of two protein kinase substrate sites (S) (Gaboreanu et al., 2007) is conserved in the avian or reptilian sequences. The cytoplasmic tails of all MPZ are highly basic (*) with isoelectric points from 10.7 and 11.9. Amino acids within the Myelin-P0_C motif are less well conserved (only 12 of 39 in all species and 22 of 39 when zebrafish is not included) than those that represent the myelin_MBP domain. A glycine zipper sequence in the TM domain (G), proposed to function in cis-based dimerization (Plotkowski et al., 2007), is conserved in all MPZ sequences except amphibian.
PLP1, a member of the lipophilin gene family, has homologs in fish and many invertebrate genomes
Compared to MBP and MPZ, where the evolutionary relationships among vertebrate and invertebrate orthologs and paralogs (MPZ) and possible invertebrate homologs have received limited attention, studies on the relationships among PLP1 and other lipophilin family members are substantial (Kitagawa et al., 1993; Gow, 1997; Stecca et al., 2000; Gould et al., 2005; Schweitzer et al., 2006). Similarity searches with murine PLP1 identify three invertebrate lipophilins: fruit fly (Q59F95), blood fluke (Q5DHE5) and sea urchin (UPI0000E49D). Complementary searches of genome and EST databases identify a plethora of invertebrate lipophilins in all but sea anemone (Table 1, see supplementary Table 4 online; see also Möbius et al., 2009). Both clustering of invertebrate homologs on phylogenetic trees and reverse BLASTP hits in the murine NR database indicate invertebrate homologs are more closely related to GPM6 sub-family members than to DM-20. Database searches with exon 3B from murine have failed to detect any invertebrate sequences related to this domain. Complementary database searches of the sea lamprey genome have identified a partial (108 amino acid) GPM6 homolog (Table 1), although lipophilins are not represented in either current lamprey or hagfish EST databases. These results support earlier suggestions that whole genome duplication underlie the appearance of DMα from DMγ and that an exonization event in ancestral amphibians that yielded the exon-3B PLP1-specific domain, enabled PLP1 to replace MPZ in tetrapod CNS myelin.
Based both on overall amino acid sequence and pair-wise comparisons, mammalian PLP1 family members are even more highly conserved than MPZ (even than the MPZ extra-cellular domain alone) and far more highly conserved than MBP (Fig. 3). The most divergent PLP1 sequence is that of lesser hedgehog (93% identity, 263 versus 277 amino acids). This sequence has six fewer amino acids in the PLP1-specific domain and a three amino acid deletion in the first extracellular loop (not shown). A duckbill platypus PLP1 (281 amino acids) is nearly as different from murine PLP1 (94% identity). The PLP1-specific domain, even without the lesser hedgehog sequence, has lower identity (78%) than the whole mammalian protein, indicating that while critical for function (Stecca et al., 2000; Sporkel et al., 2002), the primary structure of this domain is not as constrained as the rest of the protein.
Avian and diapsid PLP1 sequences are also well conserved (Fig. 3, see supplementary Table 2 online). Amphibian PLP1 sequences appear to be closer to mammalian PLP1 than amphibian MBP or MPZ orthologs to their mammalian counterparts. Cross-species comparisons among terrestrial sequences indicate levels of conservation in the order from highest to lowest: PLP1 > MPZ > MBP. Pair-wise comparisons demonstrate that DMα sequences in lobe finned, cartilaginous and teleost fishes are also highly conserved both when aligned with mammalian DM-20 (58–66%) and with each other (see supplementary Table 2 online; see also Brösamle, 2009). Structural features, possibly related to higher ordered structural stability, high hydrophobicity and constraints on intracellular trafficking of this tetraspan superfamily member (Southwood et al., 2007a), may prohibit the types of variation seen in the evolution of this lipophilin branch when compared with other myelin proteins, including the PMP22 tetraspan protein.
Like PLP1, PMP22 is well represented in vertebrate and invertebrate databases (see supplementary Tables 1A, 1B and 4 online) and among mammalian myelin proteins, it is even less well conserved than MBP. However, for non-mammalian proteins, avian and reptilian PMP22 although less well conserved than PLP1 counterparts, are better conserved than MBP (Fig. 3).
Other myelin-related proteins have their own unique distribution patterns
In addition to examining compact myelin proteins, we have performed similarity and database searches for 24 other proteins from 20 families. These are from three groups: ones associated with compact myelin (CD9/CD81/TSPAN2, MAL/PLLP, MOBP, PMP2, PMP22), non-compact myelin (CLDN11/CLDN19, CNP, JAM3, MAG, MOG, PADI2, STMN1, TPPP) and enzymes involved with the metabolism of myelin-associated lipids (FA2H, FDFT1, GAL3ST1, HMGCS1, HMGCR, NPC2, UGT8). Most are relatively small and many have invertebrate homologs. Results are outside the scope of this manuscript. With focus on the major myelin proteins, we chose a PMP2 that co-localizes with MBP in mammalian PNS myelin sheaths (Trapp et al., 1984) and STMN1, a cytosolic microtubule binding protein that participates in oligodendrocyte differentiation and in myelination (Liu et al., 2003; Southwood et al., 2007b; Richter-Landsberg, 2008). Unlike other proteins in this study, STMN1 is widely expressed in many cell types. Like MBP, MPZ, PLP1 and PMP22 (included in some comparisons), PMP2 and STMN1 are small, have single domain structures and gnathostome paralogs. Phylogenetic distribution, domain conservation and alignment comparisons are shown (Table 1, Fig. 3, see supplementary Table 2 online).
For PMP2, orthologs are limited to tetrapods (Table 1) although as a member of a large fatty acid binding protein family, paralogs are identified in fishes and in invertebrates (see supplementary Tables 1A, 1B and 4 online). Compared with other myelin proteins, PMP2 is less well conserved among mammals but shows the same or better conservation among avian and diapsid species than MBP, MPZ and PMP22. We were unable to identify orthologs in teleost or cartilaginous fishes, suggesting that like exon-3B-containing PLP1, PMP2 first appeared and had the earliest opportunity to enter myelin in ancestral tetrapods.
Similarity searches using murine and zebrafish STMN1 proteins (74% identical) generate similar maximum likelihood trees that include not only gnathostome homologs: STMN1 (op18), STMN2 (SCG10), STMN3 (SCLIP) and STMN4 (rp3), but also several invertebrate sequences including a fruit fly STMN (Q8IPK0) a silk moth STMN (Q2F5V8) and a sea urchin STMN (UPI0000E47C86). In addition to orthologs in mammals, birds, amphibians and teleosts identified via similarity searching, reptile, elasmobranch and sea lamprey orthologs are identified by database searches (see supplementary Tables 1A and 1B online). In addition to the sea lamprey ortholog are two sea lamprey paralogs (eFD709544 and GENSCAN00000041572). These are 60% identical to STMN1 and 75% identical to one another, suggesting the duplication of an ancestral STMN homolog that coincided with the emergence of agnathans. STMN homologs are also identified in all invertebrate genome databases with the exception of roundworm (Table 1, see supplementary Table 4 online). All vertebrate and invertebrate models include a stathmin PFAM domain (PF00836) that spans each protein (Table 2).
Mammalian STMN1 orthologs are as well conserved as mammalian PLP1 (Fig. 3). Nineteen of 26 mammalian sequences are 100% identical. Also avian and reptilian orthologs show similar levels of conservation to their mammalian counterparts as PLP1, and far higher conservation than MBP, MPZ and PMP22. Three (chicken, wild turkey and zebra finch) of four avian (Patagonia crested duck) STMN1 sequences are completely identical. With the duck sequence included, identity is 98%. Amphibian and elasmobranch STMN1 sequences are >90% identical to one another and a lamprey STMN1 ortholog is 63% identical to mammalian STMN1. Fish STMN1 sequences are far closer to mammalian sequences than any of the fish myelin proteins are to mammalian counterparts. These results indicate that STMN1, with 3D-structure analyzed and sites that interact with tubulin well characterized (Ravelli et al., 2004), is more highly conserved than any myelin-specific protein. However, in contrast to its high conservation among gnathostomes, invertebrate orthologs are far less well conserved with identities to murine ranging from 48% in lancelet to 25% in sea anemone. Unlike the other invertebrates, ESTs from owl limpet STMN1 show variations in both amino- and carboxyl-terminal domains. Two lancelet models (Brafl1: 282926 and Brafl1: 231109) contain a potential N-terminal Golgi targeting sequence specific to STMN paralogs and important for localization to intracellular organelles (Buttmann et al., 2008). In contrast to this extensive sequence identity across vertebrate taxa, the C-terminal domain of the gecko STMN1 gene model is divergent from other STMN1 sequences, which may reflect sequencing errors.
DISCUSSION
Ascensions of MBP and MPZ in early gnathostomes provided a base for the evolution of the myelinated nervous system
Although support cell-derived multi-lamellar membranes that insulate axons have emerged independently at least three times during evolution, vertebrate myelinated fibers are both morphologically and biochemically different from invertebrate myelin (Bullock et al., 1984; Waehneldt, 1990; Schweigreiter et al., 2006; Hartline, 2009; Roots, 2009). We previously addressed the origins of two key proteins in vertebrate myelin evolution, MBP and MPZ by searching the genome database of a basal chordate, Ciona intestinalis (Gould et al., 2005). The absence of these proteins in the Ciona genome indicated that their appearance in basal vertebrates may have coincided with myelinogenesis. The availability of recently sequenced invertebrate genomes and EST libraries has afforded further exploration on the ascension of MBP and MPZ homologs. Their apparent absence in all invertebrate species examined, including ascidians, cephalochordate (lancelet) and echinoderm (sea urchin, Table 1), is consistent with our view that both proteins arose in early vertebrates. Still however, possibilities that MBP and/or MPZ genes existed in invertebrate chordate ancestors and were lost from the extant lineages examined cannot be excluded.
Absence of MBP from the sea lamprey genome (coverage at 5.7×) and agnathan (lamprey and hagfish) EST databases favors the view that MBP arose in early gnathostomes. The possibility of an earlier origin of MPZ is suggested by the presence of two partial MPZ-related gene models in sea lamprey. Both models include V-set domains, which, however, lack the N-glycosylation consensus sequence common among gnathostome MPZ sequences (Fig. 6). An N-terminal signal peptide is present in one model. However, neither model contains transmembrane and cytoplasmic domains. Further sequence information is clearly needed to determine relationships between gnathostome MPZ and these putative agnathan homologs and to find out if they encode secreted or membrane-anchored proteins.
As suggested (Gould et al., 2005), the ascension of MBP and MPZ may have provided glial cells the ability to generate novel, tightly compacted and metabolically active membranes that became a hallmark of vertebrate nervous systems. Although both are small, they are very different from one another. MBP is a peripheral, intrinsically disordered protein (Sedzik and Kirschner, 1992) with positive charges distributed throughout the molecule. MPZ is a type I, membrane-spanning protein that couples an ancient adhesion Ig domain to a cytoplasmic tail that both interacts with cholesterol and via its high content of basic amino acid residues with acidic phospholipids, at least in model in vitro systems (Ding and Brunden, 1994). Whether both proteins were incorporated into the myelinogenesis program at the same time or one was adequate to cause membrane expansion and wrapping is not known. Based on the ability of rodent Schwann cells to generate PNS myelin in the absence of MBP (Privat et al., 1979; Kirschner and Ganser, 1980; Rosenbluth, 1980), it is plausible that MPZ alone may have initiated myelination, and when it entered to myelination program, MBP complemented and enhanced the role of MPZ. Although basic PMP2 may help compensate for the absence of MBP in shiverer mouse PNS myelination, its absence from fish genomes (Table 1) indicates that it could not have participated with MPZ in early PNS myelination. Experimental paradigms to characterize the roles that MBP, MPZ and other candidate proteins play in myelin compaction are needed to obtain knowledge interdependent of their interactions.
A possible link into their recruitment to myelin is the fact that both MBP and MPZ interact with lipids in distinctly different ways. MPZ interacts with cholesterol, through a specific domain adjacent to its transmembrane domain (Saher et al., 2009), a property shared with PLP1 (Simons et al., 2000), and with acidic lipids via its highly basic cytoplasmic tail (Ding and Brunden, 1994). MBP interacts in with phosphatidylinositol-4,5-bis-phosphate and with other acidic lipids that are concentrated in the inner leaflet of plasma membrane bilayer (Boggs, 2006; Musse et al., 2009; Nawaz et al., 2009). Cholesterol, phospholipids, glycosphingolipids, typical of myelin (Quarles et al., 2006), are also constituents of invertebrate membranes and most enzymes responsible for their formation and turnover have homologs in invertebrates (Kishimoto, 1986; Inagaki et al., 2006; Lykidis, 2007; Michell, 2008) (Gould, unpublished). The ability of MBP and MPZ to interact with and organize the lipid structure that makes multilayered myelin such a good insulator may have been key to their initial selection. In this regard, it will be interesting to extend comparisons of interactions of non-mammalian MBP (e.g. Milne et al., 1990; Yamamori et al., 1995) and MPZ proteins with lipids and membranes along the same lines that are being done for mammalian orthologs.
Variability in MBPs residing in myelin sheaths of species from different taxa
Features common among gnathostome MBPs that likely contribute to their ability to enable two cell types, oligodendrocytes and Schwann cells, to myelinate axons in diverse hosts likely include: intrinsic disorder, high portions of basic amino acids (17.5–19% in tetrapods and elasmobranchs and 20–25% in teleost fishes) spread throughout the protein, the ability to bind and cluster acidic phospholipids, particularly polyphosphoinositides (Musse et al., 2009; Nawaz et al., 2009) and finally help with transduction of neuronal signals in the ordering of lipids (Fitzner et al., 2006). Features that are less essential for MBP function, but likely contribute in ways not well appreciated are: (1) alternative splice isoforms as these are mostly lacking in aves, diapsids, amphibians and elasmobranchs; (2) variants that contain exon II, an exon likely introduced with mammals; and (3) most post-translational modifications, including arginine methylation, SH3 binding, some phosphorylation, deimidation and deamidation, as few of the amino acids that undergo these modification are widely conserved in non-mammalian MBPs. Splicing does not appear to be essential even in mammalian systems as the shiverer phenotype (no CNS myelin due to absence of MBP expression) is rescued with single (14 or 17.2 kDa) MBP isoforms (Kimura et al., 1989, 1998). As neither of these studies looked in depth to find out whether development, g-ratios and paranodal specializations were compromised, the need for alternative spice MBP variants is not totally settled.
In addition to interacting with and ordering lipids, particularly phosphatidylinositol 4,5-bisphosphate (PIP2), mammalian MBPs are known to interact with cytoskeletal proteins (actin and tubulin) and (Roth et al., 1993; Boggs et al., 2000; Hill et al., 2005) also with calmodulin (Polverini et al., 2004). Based on the facts that different charge and alternative splice variants as well as different portions of MBP interact with these proteins, it is likely, although not proven, that similar interactions occur with non-mammalian basic proteins, including MBP. The ability of MBPs, especially the divergent forms expressed by fish, to interact with these process-organizing proteins, could solidify the notion of the importance of these interactions in myelination.
Once MBP originated, changes in differences in primary structure seen in extant species comparisons are far greater than primary structural changes in either MPZ or PLP1, although they are similar to changes in PMP22 and less than changes in Golli exons (Fig. 3). Furthermore, unlike MPZ and PLP1, proteins in which numerous point mutation lead to diseases (Shy, 2006; Garbern, 2007), point mutations relating MBP to disease have not been identified. One reason maybe the built-in flexibility/disorder that allows substantial variability in MBP structure. MBPs of elasmobranchs and teleost fishes are smaller and more basic than tetrapod MBPs. In addition, teleosts not only express a variety of splice variants, these are often much smaller than MBPs present in tetrapod myelin sheaths. Furthermore, for teleosts, MBPs work with MPZs having a positively charged tail half the size of MPZs from non-teleosts. These differences likely account for the thinness of the cytoplasmic apposition in teleost myelin sheaths (Avila et al., 2007). How these MBP size variants influence myelin formation and stability is unknown, as is whether differences in MBP composition occur in MPZ-based versus PLP1-based sheaths. The concentration of MBP in mammalian PNS myelin is about half that in mammalian CNS myelin (e.g. Boggs, 2006). An understanding of relative concentrations of MBP and MPZ in sheaths of different species may contain clues as to why different variants are present in mammals and teleosts.
Adaptation of MPZ to myelinogenesis in fishes and tetrapods
MPZ is a member of the Ig superfamily of glycoproteins that functions in cell adhesion by characteristic homophilic and, for some members, heterophilic binding (Lemke and Axel, 1985; Aricescu and Jones, 2007; Shapiro et al., 2007). Compared with other members that are widely distributed in both invertebrates and vertebrates (above references), MPZ is limited to vertebrates and possibly only to gnathostomes. In addition to recognized function in myelin compaction, early embryonic expression (Lee et al., 1997) and expression in Schwann cells at neuromuscular junctions (Georgiou and Charlton, 1999) suggest roles for MPZ different from myelin compaction and a minimum membrane concentration is likely needed for participation in compaction. Limited attention has been paid to possible functions of MPZL1, MPZL2/EVA and MPZL3 paralogs, though, like MPZ, all are widely distributed among gnathostomes. Although the homologs in lamprey are incomplete, once characterized, they may provide clues as to the origins of MPZ and functions that are independent from myelin compaction, but possibly revealing as to structure–function relationships.
Besides the MPZ family, at least two other single-Ig families, SCN1-4B and VSIG1, are known. Both are widespread in gnathostomes and absent in invertebrates and current agnathan databases (Chopra et al., 2007; Fein et al., 2007) (Gould, unpublished). These results suggest that these single Ig-domain families may have appeared in vertebrate/gnathostome ancestors, indicating that a new Ig-topology, introduced with gnathostomes, fostered novel functions. Among these families, MPZ has provided clues that may be applicable to other members. A single Ig-domain was probably best accommodated in the confines of the extracellular apposition, a space lacking cytoplasm and compact so that the space required for multilayered wrap was kept to a minimum. Comparisons of MPZ localization with that of MAG, an immunoglobulin family protein with five Ig-domains highlighted the relationship of Ig-domain size to membrane spacing (Trapp and Quarles, 1984). Inability to identify homologs in invertebrates and possibly in agnathans may be the result of the dramatic structural changes required to support strong and specific interactions that characterize single domain units of MPZ and likely other family members.
Conservation of the intricately woven V-set domain and the requisite disulfide bridge are characteristic of each known MPZ. Both the disulfide bridge and N-glycosylation have been shown to be important in protein–protein interactions as assayed with cell adhesion of expressed proteins (Kirschner et al., 2004). Many potentially important residues have been uncovered through mutagenesis in human diseases (Shy, 2006). It seems unlikely that all of these would be conserved in extracellular domains of other mammalian (84% overall identity) and non-mammalian (56–78% identities) MPZs and more likely 3D structures included many compensatory changes that still enabled the cis- and trans-interactions important for stabilizing the interperiod line. Additional features of the extracellular domain that come from 3D structural determination and modeling includes an arginine–histidine pair that interact in a pH and ionic-strength sensitive manner (Shapiro et al., 1996; Luo et al., 2008). Both are conserved in all available MPZ sequences. Of three tryptophan residues, only one is completely conserved, although hydrophobic substitutions may effectively preserve proposed roles of interacting with lipids in adjacent bilayers.
A novel feature of MPZ compared with other Ig family members is the inclusion of a highly basic cytoplasmic tail captured by the PFAM myelin-P0_C domain and conserved among all known members with MPZ proteins (Table 2). This domain has been suggested to be important for intra-cellular interactions with acidic lipids and compaction in a manner regulated by phosphorylation (Konde and Eichberg, 2006). Not only are most fish MPZs lacking this domain, but also few residues are broadly conserved. In fishes, there may, however, be changes in deployment of MPZs, with larger ones having more elaborate carboxyl-terminal tails being used for early stages of compaction and smaller ones taking over once the process is well underway.
MBP and MPZ are only part of the transition from continuous to saltatory conduction
The emergences of MBP and MPZ alone were unlikely sufficient to foster the evolution of myelinated fibers and saltatory conduction. First, because these proteins primarily interact with lipids, their abilities to influence how initial stages in myelination-competent axons are recognized and sorted are improbable. Their abilities to interact with and organize specific lipid classes may be of utmost importance in their ability to generate myelin sheaths. Second, in the absence of both MBP and MPZ, myelinating cells appear to recognize many axons and carry out initial sorting and wrapping (Martini et al., 1995). Early stages of PNS myelination in particular depend on neuregulin signaling in mammals (Birchmeier and Nave, 2008), zebrafish (Lyons et al., 2005) and likely members of all other gnathostome classes. It is important to gain understanding of the evolution of neuregulin family proteins, of transcription factors (Li and Richardson, 2009; Kucenas et al., 2009), and of the signaling pathways that activate glial cells to separate axon segments from their neighbors and to learn how these induces high MBP, MPZ and lipid synthesis required for elaboration of myelin sheaths. Whether MPZ and/or MBP participate in early roles of myelinogenesis, MPZ is identified in embryonic Schwann cells (Lee et al., 1997), and that with evolution these roles are lost or complemented to such an extent they are no longer distinguishable in animals that do not express them.
Complementing the wrapping of major portions of axons with myelin sheaths in the sequestration of gated sodium channel to nodes of Ranvier and axon initial segments, processes that require multiple protein-protein interactions (Poliak and Peles, 2003; Salzer, 2003; Susuki and Rasband, 2008). Most of these proteins have established invertebrate origins: NRCAM and neurofascin (Hortsch, 2000), spectrin and ankyrin (Baines, 2003), extracellular matrix proteoglycans (Feta et al., 2009), septate junction forming proteins (Banerjee et al., 2006). Little information is available on changes involved in their targeting to initial segments and nodal complexes. One exception is on the characterization of a peptide sequence located between DII and DIII of gnathostome sodium channels (Nav) that bestows binding to ankyrin and, hence, targeting (Garrido et al., 2003). This peptide was traced to lancelet, tunicate, sea lamprey Nav, though not in Nav from any protostome (Hill et al., 2008). A related, but independent evolved, potassium channel motif, specific to the C-terminal tails of KCNQ2 and KCNQ3 and used to target KCNQ channels to axon initial segments and nodes (Pan et al., 2006) is not present in agnathan and invertebrate channels (Hill et al., 2008). These studies indicate that exonization (Schmidt and Davies, 2007) first adapted Nav and subsequently a subset of KCNQ channels to allow them to accumulate at specific axonal locations.
From an evolutionary perspective, the high numbers of tightly packed glial cells that surround large axons of squid (Brown and Abbott, 1993) and lamprey (Schweigreiter et al., 2006) suggest that another important facet of early myelinogenesis was a shift from a situation in which many glial cells were used to contribute to the well being of large axons to a situation in which many fewer glial cells contributed. The reduction in glial number also made their metabolic contribution more limited due to the added responsibility of synthesizing and maintaining myelin sheaths. In other words, did myelinogenesis require a neuronal adjustment due to a need to take a greater role in the metabolic maintenance of their axons?
Nearly 20 years ago, Thomas Waehneldt (1990) discussed the knowledge gap between what we know takes place based on a few laboratory mammals as opposed to what occurs in the rest of the 45,000 species that use myelinated nervous systems. Today, novel microarray (Verheijen et al., 2003; D’Antonio et al., 2006; Dugas et al., 2006; Cahoy et al., 2008; Ogawa and Rasband, 2009) and proteomic (Taylor et al., 2004; Nielsen et al., 2006; Roth et al., 2006; Werner et al., 2007; Ogawa and Rasband, 2009) profiling studies are providing us with a growing list of proteins involved in mammalian myelination and studies to sort out the roles played by some of these are moving rapidly. However, the focus remains primarily on myelination in laboratory rodents, widely used as disease models, and so the knowledge gap is rapidly increasing. It is hoped that this knowledge will be used to help us understand more about the evolutionary history that brought gnathostomes their myelinated nervous systems.
METHODS
The focus of this study is on the proteins listed in Table 1. For each entry, murine protein sequences (single major splice variants) were used as initial queries to search GenBank using BLASTP for closest homolog in both the NR and EST databases. Separate searches were conducted for each of the following species filled in as entry queries: mammal (Homo sapiens, human), bird (Gallus gallus, chicken and Taeniopygia guttata, zebra finch), reptile (Anolis carolinensis, lizard and Gekko japonicus, gecko), amphibian (Xenopus sp., frog) and teleost fishes (Danio rerio, zebrafish; Gasterosteus aculeatus, three-spined stickleback; Ictalurus punctatus, channel catfish; Oryzias latipes, Japanese medaka; Oncorhynchus mykiss, rainbow trout; Pimephales promelas, flathead minnow). In addition, the pre-Ensembl site was used to query genome models in lizard and sea lamprey genomes. EST databases for Squalus acanthias, spiny dogfish; Leucoraja erinacea, little skate; and Torpedo californica, Pacific electric ray were queried, as was the Elephant shark genome database.
Data for vertebrate and invertebrate orthologs/paralogs were obtained from Joint Genome Institute with TBLASTN searches using murine sequences as initial queries. Each of the following databases were searched: Xenopus tropicalis (v4.1), Fugu rubripes (v4.0), Ciona intestinalis (v1.0), Branchiostomata florida (v1.0), Lottia gigantea (v1.0), Capitella sp. 1 (v1.0) and Nematostella vectensis (v1.0) gene models. Separate databases were searched to obtain sequences from an echinoderm, Strongylocentrotus purpuratus (http://www.hgsc.bcm.tmc.edu/blast/blast.cgi?organism=Spurpuratus), Drosophila melanogaster (http://flybase.bio.indiana.edu/blast/) and Caenorhabditis elegans (http://www.wormbase.org/db/searches/blast_blat). Complementing all of these searches were searches for EST sequences in NCBI. Additional mammalian, chicken, frog, fish and sea squirt (Ciona savignyi) sequences were obtained from ENSEMBL and NCBI EST databases. In cases where genes or models were not identified from searches with murine protein sequences, additional searches were performed with frog, zebra-fish and cartilaginous fish protein orthologs or gene models as queries. Accession numbers of all identified sequences are either listed in supplementary Tables 1A and 1B online or see supplementary Table 4 online or in the appropriate table legends.
Similarity searches and phylogenetic analyses
We performed similarity searches and estimated phylogenetic trees for different gene families using an in-house pipeline of shell and perl scripts to merge existing bioinformatics tools. The pipeline involved three steps. First, we chose as ‘BAIT’ sequences known to be expressed in myelin, usually from murine and zebrafish. Each ‘bait’ sequence was used to perform a similarity search using BLASTP of non-redundant protein databases curated by Uniprot. In most cases, we used two blast search strategies for each ‘bait’ gene: 40/15 (where the top 40 blast hits of the uniref90 and the top 15 blast hits of the uniref50 database were retained for further analysis) and 35/25 (where the top 35 and 25 BLASTP hits were retained from uniref90 and uniref50, respectively). In some cases different numbers of top blast hits were retained for subsequent analysis (see text and figure legends). Identical sequences were removed from further analysis, such as identical sequences retained from both uniref90 and uniref50 databases. Second, all retained sequences and bait were aligned using MUSCLE (Edgar, 2004). Third, we estimated maximum likelihood phylogenetic trees using RAxML assuming a JTT (Stamatakis, 2006) model of protein evolution. We visualized resulting phylogenetic trees with TreeView (Page, 2002) or FigTree (http://beast.bio.ed.ac.uk/FigTree).
Multi-sequence alignments
Pair-wise and multi-sequence alignments were made with Vector NTI (versions 10 and 11, Invitrogen, Carlsbad, CA) by sequential profile alignment of increasingly divergent sequences. Incomplete or potentially incorrect sequences were eliminated. The mammalian sequences were mostly obtained from the Ensembl list of orthologs as well as from species searches for ESTs from NCBI.
Supplementary Material
Acknowledgments
We would like to thank Doug Fields for suggesting that I edit this volume of Neuron Glia Biology on the topic of Myelin Evolution and for pushing me to get this contribution completed. We would also like to thank members of the Bay Paul Center at the Marine Biology Laboratory for their input and Dr. Douglas Feinstein for help with statistics. R.M.G. would like to thank the NSF for initial funding that allowed interests in evolutionary biology to blossom and Linda Holland for providing both early access to the lancelet genome database and encouragement to pursue this project. We would like to thank the reviewers for their insightful and comprehensive critique. This work was supported in part by grants to A.G. from the National Multiple Sclerosis Society (RG2891) and the National Institutes of Health (NS043783).
Abbreviations
*Refers to abbreviations from Hugo Gene Nomenclature Committee (http//www.genenames.org/)
- CNS
central nervous system
- GPM6*
glycoprotein M6 with two paralogs GPM6A and GPM6B
- MARCKS
myristoylated alanine-rich C kinase substrate
- MBP*
myelin basic protein
- MPZ*
myelin protein zero
- PLP1*
proteolipid protein
- PMP2*
peripheral myelin protein-2
- PMP22*
peripheral myelin protein-22
- PNS
peripheral nervous system
- STMN1*
stathmin-1
Footnotes
Statement of interest
None.
The supplementary material referred to in this article can be found online at journals.cambridge.org/ngb.
References
- Aricescu AR, Jones EY. Immunoglobulin superfamily cell adhesion molecules: zippers and signals. Current Opinion in Cell Biology. 2007;19:543–550. doi: 10.1016/j.ceb.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Avila RL, Tevlin BR, Lees JP, Inouye H, Kirschner DA. Myelin structure and composition in zebrafish. Neurochemical Research. 2007;32:197–209. doi: 10.1007/s11064-006-9136-5. [DOI] [PubMed] [Google Scholar]
- Baines AJ. Comprehensive analysis of all triple helical repeats in beta-spectrins reveals patterns of selective evolutionary conservation. Cell and Molecular Biology Letters. 2003;8:195–214. [PubMed] [Google Scholar]
- Banerjee S, Pillai AM, Paik R, Li J, Bhat MA. Axonal ensheathment and septate junction formation in the peripheral nervous system of Drosophila. Journal of Neuroscience. 2006;26:3319–3329. doi: 10.1523/JNEUROSCI.5383-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- Bizzozero OA, Fridal K, Pastuszyn A. Identification of the palmitoylation site in rat myelin P 0 glycoprotein. Journal of Neurochemistry. 1994;62:1163–1171. doi: 10.1046/j.1471-4159.1994.62031163.x. [DOI] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cellular and Molecular Life Sciences. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G, Koshy KM, Mueller JP. Adhesion of acidic lipid vesicles by 21.5 kDa (recombinant) and 18.5 kDa isoforms of myelin basic protein. Biochimica et Biophysica Acta: Bio-Membranes. 2000;1463:81–87. doi: 10.1016/s0005-2736(99)00181-9. [DOI] [PubMed] [Google Scholar]
- Brösamle C. The myelin proteolipid DMalpha in fishes. Neuron Glia Biology. 2009 doi: 10.1017/S1740925X09000131. [DOI] [PubMed] [Google Scholar]
- Brösamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Brown ER, Abbott NJ. Ultrastructure and permeability of the Schwann cell layer surrounding the giant axon of the squid. Journal of Neurocytology. 1993;22:283–298. doi: 10.1007/BF01187127. [DOI] [PubMed] [Google Scholar]
- Bullock TH, Moore JK, Fields RD. Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neuroscience Letters. 1984;48:145–148. doi: 10.1016/0304-3940(84)90010-7. [DOI] [PubMed] [Google Scholar]
- Buttmann M, Nowak E, Kroner A, Hemmer B, Lesch KP, Rieckmann P. Analysis of the Stathmin rs182455 single nucleotide promoter polymorphism in patients with multiple sclerosis. Journal of Neurogenetics. 2008;22:181–186. doi: 10.1080/01677060802179287. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. Journal of Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra SS, Watanabe H, Zhong TP, Roden DM. Molecular cloning and analysis of zebrafish voltage-gated sodium channel beta subunit genes: implications for the evolution of electrical signaling in vertebrates. BMC Evolutionary Biology. 2007;7:113. doi: 10.1186/1471-2148-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, Michalovich D, Paterson M, Droggiti A, Woodhoo A, Mirsky R, et al. Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. Glia. 2006;53:501–515. doi: 10.1002/glia.20309. [DOI] [PubMed] [Google Scholar]
- Dietrich U, Kruger P, Gutberlet T, Kas JA. Interaction of the MARCKS peptide with PIP(2) in phospholipid monolayers. Biochimica et Biophysica Acta. 2009;1788:1474–1481. doi: 10.1016/j.bbamem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Ding Y, Brunden KR. The cytoplasmic domain of myelin glycoprotein P 0 interacts with negatively charged phospholipid bilayers. Journal of Biological Chemistry. 1994;269:10764–10770. [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. Journal of Neuroscience. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM. Cholesterol and the interaction of proteins with membrane domains. Progress in Lipid Research. 2006;45:279–294. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Fein AJ, Meadows LS, Chen C, Slat EA, Isom LL. Cloning and expression of a zebrafish SCN1B ortholog and identification of a species-specific splice variant. BMC Genomics. 2007;8:226. doi: 10.1186/1471-2164-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feta A, Do AT, Rentzsch F, Technau U, Kusche-Gullberg M. Molecular analysis of heparan sulfate biosynthetic enzyme machinery and characterization of heparan sulfate structure in Nematostella vectensis. Biochemistry Journal. 2009;419:585–593. doi: 10.1042/BJ20082081. [DOI] [PubMed] [Google Scholar]
- Fitzner D, Schneider A, Kippert A, Mobius W, Willig KI, Hell SW, et al. Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO Journal. 2006;25:5037–5048. doi: 10.1038/sj.emboj.7601376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboreanu AM, Hrstka R, Xu W, Shy M, Kamholz J, Lilien J, et al. Myelin protein zero/P0 phosphorylation and function require an adaptor protein linking it to RACK1 and PKC alpha. Journal of Cell Biology. 2007;177:707–716. doi: 10.1083/jcb.200608060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Li WH, Filbin MT. Acylation of myelin Po protein is required for adhesion. Journal of Neuroscience Research. 2000;60:704–713. doi: 10.1002/1097-4547(20000615)60:6<704::AID-JNR2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Garbern JY. Pelizaeus–Merzbacher disease: genetic and cellular pathogenesis. Cellular and Molecular Life Sciences. 2007;64:50–65. doi: 10.1007/s00018-006-6182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido JJ, Giraud P, Carlier E, Fernandes F, Moussif A, Fache MP, et al. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 2003;300:2091–2094. doi: 10.1126/science.1085167. [DOI] [PubMed] [Google Scholar]
- Georgiou J, Charlton MP. Non-myelin-forming perisynaptic Schwann cells express protein zero and myelin-associated glyco-protein. Glia. 1999;27:101–109. doi: 10.1002/(sici)1098-1136(199908)27:2<101::aid-glia1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Givogri MI, Bongarzone ER, Schonmann V, Campagnoni AT. Expression and regulation of golli products of myelin basic protein gene during in vitro development of oligodendrocytes. Journal of Neuroscience Research. 2001;66:679–690. doi: 10.1002/jnr.10031. [DOI] [PubMed] [Google Scholar]
- Gould RM, Morrison HG, Gilland E, Campbell RK. Myelin tetraspan family proteins but no non-tetraspan family proteins are present in the ascidian (Ciona intestinalis) genome. Biological Bulletin. 2005;209:49–66. doi: 10.2307/3593141. [DOI] [PubMed] [Google Scholar]
- Gow A. Redefining the lipophilin family of proteolipid proteins. Journal of Neuroscience Research. 1997;50:659–664. doi: 10.1002/(SICI)1097-4547(19971201)50:5<659::AID-JNR3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Harauz G, Libich DS. The classic basic protein of myelin –conserved structural motifs and the dynamic molecular barcode involved in membrane adhesion and protein–protein interactions. Current Protein and Peptide Science. 2009;10:196–215. doi: 10.2174/138920309788452218. [DOI] [PubMed] [Google Scholar]
- Hartline DK. What is myelin? Neuron Glia Biology. 2009 doi: 10.1017/S1740925X09990263. [DOI] [PubMed] [Google Scholar]
- Hill AS, Nishino A, Nakajo K, Zhang G, Fineman JR, Selzer ME, et al. Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genetics. 2008;4:e1000317. doi: 10.1371/journal.pgen.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Libich DS, Harauz G. Assembly of tubulin by classic myelin basic protein isoforms and regulation by post-translational modification. Biochemistry. 2005;44:16672–16683. doi: 10.1021/bi050646+. [DOI] [PubMed] [Google Scholar]
- Hortsch M. Structural and functional evolution of the L1 family: are four adhesion molecules better than one? Molecular and Cellular Neuroscience. 2000;15:1–10. doi: 10.1006/mcne.1999.0809. [DOI] [PubMed] [Google Scholar]
- Inagaki M, Nakata T, Higuchi R. Isolation and structure of a galactocerebroside molecular species from the starfish Culcita novae-guineae. Chemical and Pharmaceutical Bulletin (Tokyo) 2006;54:260–261. doi: 10.1248/cpb.54.260. [DOI] [PubMed] [Google Scholar]
- Jeserich G, Klempahn K, Pfeiffer M. Features and functions of oligodendrocytes and myelin proteins of lower vertebrate species. Journal of Molecular Neuroscience. 2008;35:117–126. doi: 10.1007/s12031-008-9035-0. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nature Reviews Neuroscience. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhang R, Strittmatter EF, Smith RD, Zand R. Post-translational modifications of chicken myelin basic protein charge components. Neurochemistry Research. 2009;34:360–372. doi: 10.1007/s11064-008-9788-4. [DOI] [PubMed] [Google Scholar]
- Kimura M, Sato M, Akatsuka A, Nozawa-Kimura S, Takahashi R, Yokoyama M, et al. Restoration of myelin formation by a single type of myelin basic protein in transgenic shiverer mice. Proceedings of the National Academy of Sciences of the USA. 1989;86:5661–5665. doi: 10.1073/pnas.86.14.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Sato M, Akatsuka A, Saito S, Ando K, Yokoyama M, et al. Overexpression of a minor component of myelin basic protein isoform (17.2 kDa) can restore myelinogenesis in transgenic shiverer mice. Brain Research. 1998;785:245–252. doi: 10.1016/s0006-8993(97)01383-8. [DOI] [PubMed] [Google Scholar]
- Kirschner DA, Ganser AL. Compact myelin exists in the absence of basic protein in the shiverer mutant mouse. Nature. 1980;283:207–210. doi: 10.1038/283207a0. [DOI] [PubMed] [Google Scholar]
- Kirschner DA, Wrabetz L, Feltri ML, Lazzarini RA, Griffin JW, Lassmann H, et al. The P0 gene, Myelin Biology and Disorders. San Diego, CA: Elsevier Academic Press; 2004. pp. 523–545. [Google Scholar]
- Kishimoto Y. Phylogenetic development of myelin glycosphingo-lipids. Chemistry and Physics of Lipids. 1986;42:117–128. doi: 10.1016/0009-3084(86)90047-2. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Sinoway MP, Yang C, Gould RM, Colman DR. A proteolipid protein gene family: expression in sharks and rays and possible evolution from an ancestral gene encoding a pore-forming polypeptide. Neuron. 1993;11:433–448. doi: 10.1016/0896-6273(93)90148-k. [DOI] [PubMed] [Google Scholar]
- Konde V, Eichberg J. Myelin protein zero: mutations in the cytoplasmic domain interfere with its cellular trafficking. Journal of Neuroscience Research. 2006;83:957–964. doi: 10.1002/jnr.20793. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Snell H, Appel B. nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biology. 2009 doi: 10.1017/S1740925X09990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Brennan A, Blanchard A, Zoidl G, Dong Z, Tabernero A, et al. P 0 is constitutively expressed in the rat neural crest and embryonic nerves and is negatively and positively regulated by axons to generate non-myelin-forming and myelin-forming Schwann cells, respectively. Molecular and Cellular Neuroscience. 1997;8:336–350. doi: 10.1006/mcne.1996.0589. [DOI] [PubMed] [Google Scholar]
- Lemke G, Axel R. Isolation and sequence of a major cDNA encoding the major structural protein of peripheral nerve. Cell. 1985;40:501–508. doi: 10.1016/0092-8674(85)90198-9. [DOI] [PubMed] [Google Scholar]
- Li H, Richardson WD. The evolution of Olig genes and their roles in myelination. Neuron Glia Biology. 2009 doi: 10.1017/S1740925X09990251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P. Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia. 2003;44:264–274. doi: 10.1002/glia.10290. [DOI] [PubMed] [Google Scholar]
- Luo X, Cerullo J, Dawli T, Priest C, Haddadin Z, Kim A, et al. Peripheral myelin of Xenopus laevis: role of electrostatic and hydrophobic interactions in membrane compaction. Journal of Structural Biology. 2008;162:170–183. doi: 10.1016/j.jsb.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Inouye H, Gross AA, Hidalgo MM, Sharma D, Lee D, et al. Cytoplasmic domain of zebrafish myelin protein zero: adhesive role depends on beta-conformation. Biophysics Journal. 2007;93:3515–3528. doi: 10.1529/biophysj.107.112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykidis A. Comparative genomics and evolution of eukaryotic phospholipid biosynthesis. Progress in Lipid Research. 2007;46:171–199. doi: 10.1016/j.plipres.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, et al. erbb3 and erbb2 are essential for Schwann cell migration and myelination in zebrafish. Current Biology. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Martini R, Mohajeri MH, Kasper S, Giese KP, Schachner M. Mice doubly deficient in the genes for P0 and myelin basic protein show that both proteins contribute to the formation of the major dense line in peripheral nerve myelin. Journal of Neuroscience. 1995;15:4488–4495. doi: 10.1523/JNEUROSCI.15-06-04488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell RH. Inositol derivatives: evolution and functions. Nature Reviews. Molecular Cell Biology. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- Milne TJ, Atkins AR, Warren JA, Auton WP, Smith R. Shark myelin basic protein: amino acid sequence, secondary structure, and self-association. Journal of Neurochemistry. 1990;55:950–955. doi: 10.1111/j.1471-4159.1990.tb04583.x. [DOI] [PubMed] [Google Scholar]
- Möbius W, Patzig J, Nave K-A, Werner HB. Phylogeny of proteolipid proteins: divergence, constraints, and the evolution of novel functions in myelination and neuroprotection. Neuron Glia Biology. 2009 doi: 10.1017/S1740925X0900009X. [DOI] [PubMed] [Google Scholar]
- Musse AA, Gao W, Rangaraj G, Boggs JM, Harauz G. Myelin basic protein co-distributes with other PI(4,5)P2-sequestering proteins in Triton X-100 detergent-resistant membrane micro-domains. Neuroscience Letters. 2009;450:32–36. doi: 10.1016/j.neulet.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Nawaz S, Kippert A, Saab AS, Werner HB, Lang T, Nave KA, et al. Phosphatidylinositol 4,5-bisphosphate-dependent interaction of myelin basic protein with the plasma membrane in oligodendroglial cells and its rapid perturbation by elevated calcium. Journal of Neuroscience. 2009;29:4794–4807. doi: 10.1523/JNEUROSCI.3955-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. Journal of Neuroscience. 2006;26:9881–9891. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Rasband MN. Proteomic analysis of optic nerve lipid rafts reveals new paranodal proteins. Journal of Neuroscience Research. 2009 doi: 10.1002/jnr.21984. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. Current Protocols in Bioinformatics. Unit 6.2. 2002. Visualizing phylogenetic trees using TreeView. [DOI] [PubMed] [Google Scholar]
- Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, et al. A common ankyrin-G-based mechanism retains KCNQ and nav channels at electrically active domains of the axon. Journal of Neuroscience. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkowski ML, Kim S, Phillips ML, Partridge AW, Deber CM, Bowie JU. Transmembrane domain of myelin protein zero can form dimers: possible implications for myelin construction. Biochemistry. 2007;46:12164–12173. doi: 10.1021/bi701066h. [DOI] [PubMed] [Google Scholar]
- Poliak S, Peles E. The local differentiation of myelinated axons at nodes of Ranvier. Nature Reviews. Neuroscience. 2003;4:968–980. doi: 10.1038/nrn1253. [DOI] [PubMed] [Google Scholar]
- Polverini E, Boggs JM, Bates IR, Harauz G, Cavatorta P. Electron paramagnetic resonance spectroscopy and molecular modelling of the interaction of myelin basic protein (MBP) with calmodulin (CaM)-diversity and conformational adaptability of MBP CaM-targets. Journal of Structural Biology. 2004;148:353–369. doi: 10.1016/j.jsb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Privat A, Jacque C, Bourre JM, Dupouey P, Baumann N. Absence of the major dense line in myelin of the mutant mouse shiverer. Neuroscience Letters. 1979;12:107–112. doi: 10.1016/0304-3940(79)91489-7. [DOI] [PubMed] [Google Scholar]
- Quarles RH, Macklin WB, Morell P, Siegel GJ, Albers RW, Brady ST, et al. Myelin Formation, Structure and Biochemistry, Basic Neurochemistry. Amsterdam: Academic Press; 2006. pp. 51–71. [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Richter-Landsberg C. The cytoskeleton in oligodendrocytes. Microtubule dynamics in health and disease. Journal of Molecular Neuroscience. 2008;35:55–63. doi: 10.1007/s12031-007-9017-7. [DOI] [PubMed] [Google Scholar]
- Rosenbluth J. Peripheral nerve in the mouse mutant shiverer. Journal of Comparative Neurology. 1980;193:729–739. doi: 10.1002/cne.901930310. [DOI] [PubMed] [Google Scholar]
- Roth AD, Ivanova A, Colman DR. New observations on the compact myelin proteome. Neuron Glia Biology. 2006;2:15–21. doi: 10.1017/S1740925X06000068. [DOI] [PubMed] [Google Scholar]
- Roth GA, Gonzalez MD, Monferran CG, De Santis ML, Cumar FA. Myelin basic protein domains involved in the interaction with actin. Neurochemistry International. 1993;23:459–465. doi: 10.1016/0197-0186(93)90130-w. [DOI] [PubMed] [Google Scholar]
- Roots BI. The phylogeny of invertebrates and the evolution of myelin. Neuron Glia Biology. 2009 doi: 10.1017/S1740925X0900012X. [DOI] [PubMed] [Google Scholar]
- Saavedra RA, Fors L, Aebersold RH, Arden B, Horvath S, Sanders J, et al. The myelin proteins of the shark brain are similar to the myelin proteins of the mammalian peripheral nervous system. Journal of Molecular Evolution. 1989;29:149–156. doi: 10.1007/BF02100113. [DOI] [PubMed] [Google Scholar]
- Saher G, Quintes S, Mobius W, Wehr MC, Kramer-Albers EM, Brugger B, et al. Cholesterol regulates the endoplasmic reticulum exit of the major membrane protein P0 required for peripheral myelin compaction. Journal of Neuroscience. 2009;29:6094–6104. doi: 10.1523/JNEUROSCI.0686-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer JL. Polarized domains of myelinated axons. Neuron. 2003;40:297–318. doi: 10.1016/s0896-6273(03)00628-7. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Davies CJ. The origins of polypeptide domains. BioEssays. 2007;29:262–270. doi: 10.1002/bies.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweigreiter R, Roots BI, Bandtlow CE, Gould RM. Understanding myelination through studying its evolution. International Review of Neurobiology. 2006;73:219–273. doi: 10.1016/S0074-7742(06)73007-0. [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Becker T, Schachner M, Nave KA, Werner H. Evolution of myelin proteolipid proteins: Gene duplication in teleosts and expression pattern divergence. Molecular and Cellular Neuroscience. 2006;31:161–177. doi: 10.1016/j.mcn.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sedzik J, Kirschner DA. Is myelin basic protein crystallizable. Neurochemical Research. 1992;17:157–166. doi: 10.1007/BF00966794. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P 0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Love J, Colman DR. Adhesion molecules in the nervous system: structural insights into function and diversity. Annual Review of Neuroscience. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- Shy ME. Peripheral neuropathies caused by mutations in the myelin protein zero. Journal of Neurological Science. 2006;242:55–66. doi: 10.1016/j.jns.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Simons M, Krämer EM, Thiele C, Stoffel W, Trotter J. Assembly of myelin by association of proteolipid protein with cholesterol- and galactosylceramiderich membrane domains. Journal of Cell Biology. 2000;151:143–153. doi: 10.1083/jcb.151.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood C, Olson K, Wu CY, Gow A. Novel alternatively spliced endoplasmic reticulum retention signal in the cytoplasmic loop of proteolipid protein-1. Journal of Neuroscience Research. 2007a;85:471–478. doi: 10.1002/jnr.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A. Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochemical Research. 2007b;32:187–195. doi: 10.1007/s11064-006-9127-6. [DOI] [PubMed] [Google Scholar]
- Sporkel O, Uschkureit T, Bussow H, Stoffel W. Oligodendrocytes expressing exclusively the DM20 isoform of the proteolipid protein gene: myelination and development. Glia. 2002;37:19–30. doi: 10.1002/glia.10014. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stecca B, Southwood CM, Gragerov A, Kelley KA, Friedrich VL, Jr, Gow A. The evolution of lipophilin genes from invertebrates to tetrapods: DM20 cannot replace proteolipid protein in CNS myelin. Journal of Neuroscience. 2000;20:4002–4010. doi: 10.1523/JNEUROSCI.20-11-04002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susuki K, Rasband MN. Molecular mechanisms of node of Ranvier formation. Current Opinion in Cell Biology. 2008;20:616–623. doi: 10.1016/j.ceb.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, et al. Proteomic mapping provides powerful insights into functional myelin biology. Proceedings of the National Academy of Sciences of the USA. 2004;101:4643–4648. doi: 10.1073/pnas.0400922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Dubois-Dalcq M, Quarles RH. Ultrastructural localization of P 2 protein in actively myelinating Schwann cells. Journal of Neurochemistry. 1984;43:944–948. doi: 10.1111/j.1471-4159.1984.tb12828.x. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Quarles RH. Immunocytochemical localization of the myelin-associated glycoprotein – fact or artifact? Journal of Neuroimmunology. 1984;6:231–249. doi: 10.1016/0165-5728(84)90011-0. [DOI] [PubMed] [Google Scholar]
- Verheijen MHG, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes and Development. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehneldt TV. Phylogeny of myelin proteins. Annals of the New York Academy of Sciences. 1990;605:15–28. doi: 10.1111/j.1749-6632.1990.tb42377.x. [DOI] [PubMed] [Google Scholar]
- Waehneldt TV, Matthieu JM, Jeserich G. Appearance of myelin proteins during vertebrate evolution. Neurochem Int. 1986;9:463–474. doi: 10.1016/0197-0186(86)90136-1. [DOI] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. Journal of Neuroscience. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily-domains for cell surface recognition. Annual Review of Immunology. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Yamamori C, Terashima M, Ishino H, Shimoyama M. ADP-ribosylation of myelin basic protein and inhibition of phospholipid vesicle aggregation. Enzyme and Protein. 1995;48:202–212. doi: 10.1159/000474990. [DOI] [PubMed] [Google Scholar]
- Zalc B, Goujet D, Colman D. The origin of the myelination program in vertebrates. Current Biology. 2008;18:R511–R512. doi: 10.1016/j.cub.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Zand R, Jin X, Kim J, Wall DB, Gould R, Lubman DM. Studies of posttranslational modifications in spiny dogfish myelin basic protein. Neurochemistry Research. 2001;26:539–547. doi: 10.1023/a:1010921230859. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li CJ, Wang Y, Xia W, Yao B, Jin JY, et al. Identification and characterization of a MBP isoform specific to hypothalamus in orange-spotted grouper (Epinephelus coioides) Journal of Chemical Neuroanatomy. 2007;34:47–59. doi: 10.1016/j.jchemneu.2007.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.