Abstract
microRNAs (miRNAs) are dysregulated in a variety of disease states, suggesting that this newly discovered class of gene expression repressors may be viable therapeutic targets. A microarray of miRNA changes in ALS-model superoxide dismutase 1 (SOD1)G93A rodents identified 12 miRNAs as significantly changed. Six miRNAs tested in human ALS tissues were confirmed increased. Specifically, miR-155 was increased 5-fold in mice and 2-fold in human spinal cords. To test miRNA inhibition in the central nervous system (CNS) as a potential novel therapeutic, we developed oligonucleotide-based miRNA inhibitors (anti-miRs) that could inhibit miRNAs throughout the CNS and in the periphery. Anti-miR-155 caused global derepression of targets in peritoneal macrophages and, following intraventricular delivery, demonstrated widespread functional distribution in the brain and spinal cord. After treating SOD1G93A mice with anti-miR-155, we significantly extended survival by 10 days and disease duration by 15 days (38%) while a scrambled control anti-miR did not significantly improve survival or disease duration. Therefore, antisense oligonucleotides may be used to successfully inhibit miRNAs throughout the brain and spinal cord, and miR-155 is a promising new therapeutic target for human ALS.
INTRODUCTION
Since their discovery in 1993 (1), microRNAs (miRNAs) have emerged as key regulators in numerous physiological and pathological processes. miRNAs are highly conserved, single-stranded, non-coding RNA molecules ∼22 nt in length. miRNAs repress gene expression by inhibiting translation of and/or facilitating the degradation of their target mRNAs via binding to the 3′-untranslated region (UTR). Because only partial complementarity is required for miRNA–mRNA interactions, a single miRNA can potentially regulate hundreds of mRNA transcripts. Testing the potential therapeutic opportunity of dysregulated miRNAs in any particular disease requires not only a careful analysis of the miRNA expression changes in the target tissues but also a method to modulate miRNA function in disease models.
Amyotrophic lateral sclerosis (ALS; also known as Lou Gehrig's Disease) is a fatal adult-onset neurodegenerative disease characterized by the selective loss of motor neurons in the spinal cord and brain leading to stiffness, severe muscle weakness and death because of respiratory failure typically within 3–5 years of disease onset (2). Riluzole, the only FDA approved treatment, prolongs survival by only three to six months. Therefore, discovering novel therapeutic targets is of critical importance.
Amongst the list of dysregulated miRNAs in ALS-model mice and human ALS samples, miR-155 appeared to be an excellent therapeutic target because of its abundance and fold change in ALS, reproducibility across species and various ALS models, and prior work linking miR-155 with immunity and inflammation. miR-155 is highly expressed in hematopoietic cells including T cells and monocytes (3) and can serve to promote pro-inflammatory pathways through targeted repression of anti-inflammatory mediators including Src homology-2 domain-containing inositol 5 phosphatase 1 (SHIP1) (4) and suppressor of cytokine signaling-1 (5). Immune system involvement is important in ALS (6–8), and recent data suggest that miR-155 is increased in peripheral monocytes from ALS-model mice and ALS patients (9). However, as with many changes in the ALS model, whether the miR-155 increase positively or negatively affects ALS remained untested and required development of a method to inhibit miRNAs both in peripheral blood cells as well as in the central nervous system (CNS).
Antisense oligonucleotides can be used to inhibit miRNA function by binding tightly through Watson-Crick base pairing. This miRNA inhibition strategy has been successful in the periphery but has not been readily applied to the CNS. Anti-miRs do not cross the blood brain barrier. To target miRNAs in the CNS, we delivered anti-miRs directly to the cerebral spinal fluid as previously described for mRNA inhibitors (10). We demonstrate here the ability of these anti-miRs to inhibit their cognate miRNA target throughout the CNS. Most importantly, we use these miR-155 inhibitors to test whether the increased miR-155 affects ALS disease phenotype and is thus a viable therapeutic target.
RESULTS
miR-155 is significantly upregulated in rodent and patient ALS spinal cord tissue
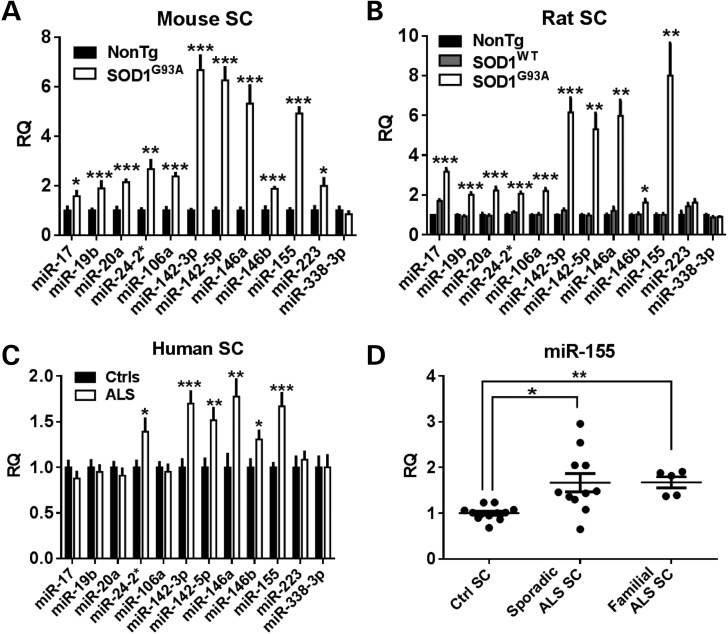
To identify miRNA therapeutic targets for ALS, we measured miRNA changes in both the rodent ALS model and human ALS autopsy samples. Using TaqMan miRNA microarrays that assayed 673 miRNAs, we measured miRNA expression levels in both end-stage mouse and rat spinal cord tissue as compared to their age-matched controls. Twelve miRNAs were identified as significantly increased in both ALS models (Table 1). Using individual miRNA assays, 11 miRNAs were confirmed increased in the mouse, 10 in the rat and 6 in patient ALS autopsy tissues (Fig. 1A–C). Specifically, the most researched amongst these hits, miR-155, was significantly increased in both familial and sporadic patient ALS spinal cord tissue (Fig. 1D). Total RNA integrity from our autopsy spinal cord samples was poor as determined by Agilent Bioanalyzer; however, our miRNA experiments showed well-shaped quantitative polymerase chain reaction (qPCR) amplification curves, high amplification, low noise, and high consistency between samples and even between multiple extractions from the same spinal cord. Furthermore, RNA integrity numbers did not well correlate with miRNA qPCR counts (Supplementary Material, Fig. S1). This is in agreement with published data showing miRNAs to be stable despite extensive degradation of mRNA (11).
Table 1.
Twelve miRNAs are altered in end-stage ALS mouse and rat spinal cords by microarray
| Mouse |
Rat |
|||||
|---|---|---|---|---|---|---|
| RQ: SOD1G93A/non-TG | T-test | RQ: SOD1G93A/non-TG | T-test | RQ: SOD1G93A/SOD1WT | T-test | |
| miR-17 | 2.66 | 0.00 | 1.99 | 0.04 | 11.58 | 0.00 |
| miR-19b | 2.51 | 0.00 | 1.99 | 0.10 | 9.20 | 0.02 |
| miR-20a | 2.33 | 0.01 | 1.79 | 0.10 | 20.60 | 0.01 |
| miR-24-2-5p | 2.44 | 0.02 | 1.78 | 0.06 | 3.22 | 0.00 |
| miR-106a | 2.59 | 0.01 | 2.11 | 0.01 | 11.14 | 0.00 |
| miR-142-3p | 3.36 | 0.04 | 3.81 | 0.06 | 24.84 | 0.02 |
| miR-142-5p | 3.27 | 0.05 | 10.61 | 0.17 | 2.94 | 0.34 |
| miR-146a | 8.86 | 0.00 | 3.89 | 0.02 | 8.79 | 0.01 |
| miR-146b | 3.02 | 0.01 | 1.84 | 0.04 | 2.95 | 0.01 |
| miR-155 | 2.08 | 0.01 | 20.08 | 0.20 | 58.75 | 0.19 |
| miR-223 | 3.52 | 0.05 | 1.89 | 0.14 | 10.25 | 0.02 |
| miR-338-3p | 2.27 | 0.01 | 1.75 | 0.16 | 5.33 | 0.03 |
miRNAs were isolated from end-stage SOD1G93A mouse and rat spinal cords and from age-matched non-transgenic (non-TG) controls (n = 3–4). For rats, miRNAs were also isolated from age-matched SOD1WT control spinal cords (n = 3). A total of 673 miRNAs were assayed using Applied Biosystem's TaqMan Rodent Array A + B Cards v3.0 and 434 miRNAs amplified and were quantified. For mice, 6 miRNAs were decreased and 37 miRNAs were increased greater than 2-fold at P < 0.05. From these 43 miRNAs, 12 were also changed >50% in the rat samples at P < 0.20. RQ, relative quantification.
Figure 1.
miRNA array changes were confirmed in mouse, rat and human spinal cord samples. (A) Using individual TaqMan miRNA assays, 11 out of the 12 miRNA microarray hits were confirmed significantly increased in end-stage mouse spinal cord tissue (n = 5–6). (B) In rat lumbar spinal cord tissue, 10 out of the 12 miRNAs tested were increased in end-stage superoxide dismutase 1 (SOD1)G93A rats over non-transgenic and overexpressing SOD1WT age-matched controls (n = 4,4,4). (C) In autopsy spinal cord samples from 16 ALS patients and 12 non-ALS controls, 6 miRNAs assayed were significantly increased in the ALS tissue. (D) Specifically, both sporadic and familial ALS patients showed a significant increase in miR-155 (n = 12 controls; 1.7 × increase in n = 11 sporadic ALS; 1.7 × increase in n = 5 familial ALS; ANOVA with Tukey's post-hoc test). (SC, spinal cord. *P < 0.05, **P < 0.01, ***P < 0.001.)
Anti-miRs delivered through ventricular osmotic pumps distribute widely in the CNS and derepress target mRNAs
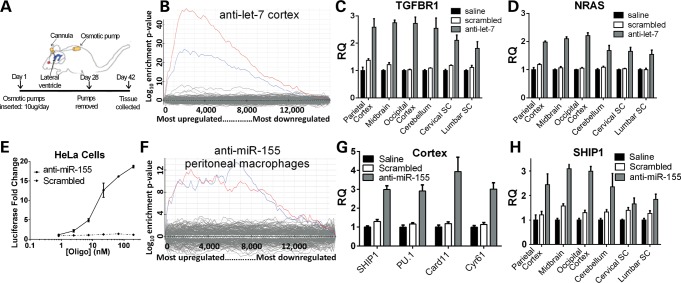
To determine whether anti-miRs could inhibit targets broadly in the CNS, we used a well-characterized miRNA inhibitor of let-7 (12). After treating animals with anti-let-7 via an osmotic pump directed into the lateral ventricles (Fig. 2A), mRNAs with 3′-UTR let-7 binding sites were globally derepressed in cortical tissue (Fig. 2B). After confirming the suitability of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control (Supplementary Material, Fig. S2), analysis of a subset of these mRNAs demonstrated derepression of targets in the parietal cortex, midbrain, occipital cortex, cerebellum, cervical and lumbar spinal cord (Fig. 2C and D).
Figure 2.
Let-7 and miR-155 anti-miRs distribute throughout CNS and derepress target mRNAs. (A) Saline, scrambled anti-miR control or anti-let-7 was infused directly to the lateral ventricle for 28 days with an osmotic pump. At 42 days, RNA was extracted. (B) Using Affymetrix 430 2.0 mouse gene arrays and Sylamer software analysis, cortical mRNA with let-7 binding sites (blue and red lines denoting mRNAs that contain the 1–7 nt or 2–8 nt miRNA seed region compliment, respectively) was enriched amongst the upregulated (derepressed) mRNAs for anti-let-7-treated mice as normalized to scrambled-treated mice. (C and D) mRNA of two confirmed let-7 targets (TGFBR1, NRAS) were significantly increased in anti-let-7-treated mice in all regions assayed (P < 0.05 TGFBR1; P < 0.01 NRAS). (E) Hela cells were transfected with a miR-155 expression plasmid and a luciferase reporter containing two miR-155 binding sites. After 4 h, either anti-miR-155 or a scrambled control anti-miR was transfected into cells (1–200 nM). Quantified 24 h later, luciferase levels were increased in anti-miR-155-treated cells, indicating inhibition of miR-155. (F) After 3 daily IP injections of either anti-miR-155 or scrambled anti-miR control, peritoneal macrophages were isolated from adult mice, stimulated with LPS, and RNA was extracted. An Affymetrix microarray of mRNA followed by Sylamer analysis shows derepression of mRNAs that contain the heptamer seed region compliment to miR-155 (blue line denotes 1–7nt miRNA heptamer compliment, red for 2–8-nt heptamer). (G) anti-miR-155 was infused into the lateral ventricles of adult mice for 4 weeks. Two weeks later, mRNA was extracted. mRNA levels of 4 confirmed and/or predicted miR-155 targets were significantly increased in anti-miR-155-treated mice over both saline- and scrambled-treated mice [Src homology-2 domain-containing inositol 5 phosphatase 1 (SH1P1) (P < 0.001, PU.1 P < 0.001; Card11 P < 0.01, Cyr61 P < 0.001]. (H) SHIP1, a confirmed miR-155 target, was significantly increased in anti-miR-155-treated mice in all regions assayed (P < 0.05).
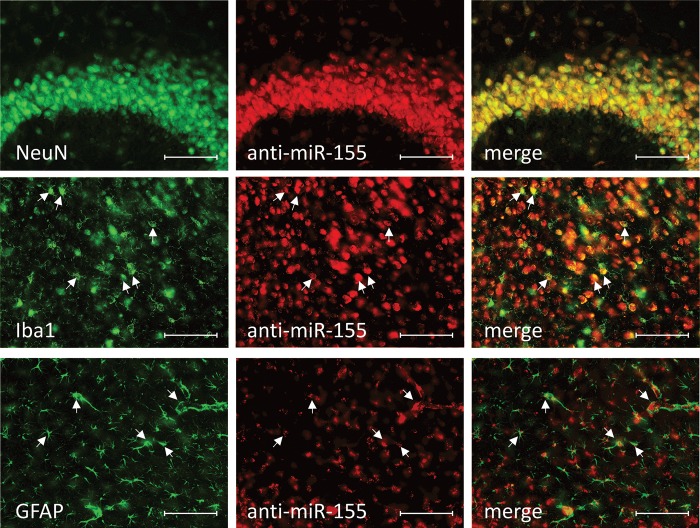
Having established that anti-miRs can target the CNS including the spinal cord, we developed an anti-miR-155 oligonucleotide to test whether this inhibitor would affect the ALS rodent model. In Hela cells transfected with miR-155 and a luciferase reporter containing a 3′-UTR recognized by miR-155, the novel anti-miR-155 derepressed luciferase and led to increased luciferase activity (Fig. 2E). To test efficacy of miR-155 inhibition peripherally in vivo, we administered three daily intraperitoneal (IP) injections of anti-miR-155 in non-transgenic mice. An mRNA profile from lipopolysaccharide (LPS)-stimulated peritoneal macrophages from anti-miR-155-treated mice showed global derepression of mRNAs with a miR-155 3′-UTR binding site (Fig. 2F). Finally, to test efficacy in CNS tissue, we implanted 28-day osmotic pumps with saline, scrambled control or anti-miR-155 that infused directly into the lateral ventricles. We confirmed four known and/or predicted miR-155 mRNA targets (4,13–15) to be derepressed in the cortex following anti-miR-155 treatment. Furthermore, quantification of one of these targets, SHIP1, across the CNS demonstrated a significant increase over saline in all areas assayed (Fig. 2G). In a separate experiment, mice treated with cy3-labeled anti-miR-155 osmotic pumps for two weeks demonstrated clear cellular uptake throughout the brain and spinal cord (Fig. 3). More so, double-label histology showed cy3-anti-miR-155 to be incorporated into distinct cell types including neurons (NeuN+), microglia (Iba1+) and astrocytes (GFAP+) (Fig. 4).
Figure 3.
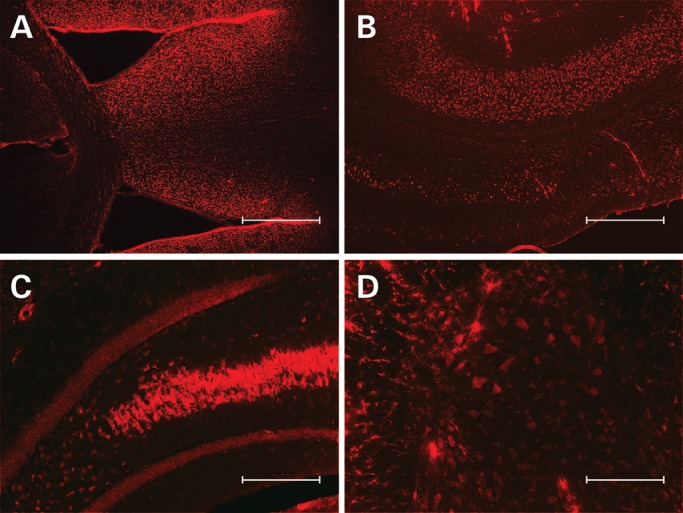
cy3-anti-miR-155 distributes from lateral cerebral ventricle throughout brain and spinal cord. Mice were treated for 2 weeks with an osmotic pump delivering 10 µg/day cy3-labeled anti-miR-155 directly into the lateral ventricle. Anti-miR-155 distributes throughout the (A) subventricular zone (4×, scale bar = 500 µm), (B) cortical layers (4×, scale bar = 500 µm), (C) hippocampus (10×, scale bar = 200 µm) and (D) spinal cord (10×, scale bar = 200 µm).
Figure 4.
cy3-anti-miR-155 distributes into neurons, microglia and astrocytes. Mice were treated for 2 weeks with an osmotic pump delivering 10 µg/day cy3-labeled anti-miR-155 directly into the lateral ventricle (B, E, H). 40-µm brain slices were collected, and cell-specific antibodies were used against NeuN to mark neurons (A), Iba1 to mark microglia (D) and GFAP to mark astrocytes (G). cy3-anti-miR-155 is present in all cell types assayed (C, F, I, arrowheads mark examples of double positive cells) (20×, scale bar = 100 µm).
anti-miR-155 significantly extends survival and disease duration in the SOD1G93A mouse model
To test the therapeutic potential of antagonizing miR-155 in ALS, we treated a large cohort of superoxide dismutase 1 (SOD1)G93A mice at 60 days of age with continuous intraventricular infusion of anti-miR-155 (n = 22), scrambled control anti-miR (n = 21) or saline (n = 20). To account for any peripheral role of miR-155, we supplemented the CNS treatment with weekly anti-miR or saline IP injections.
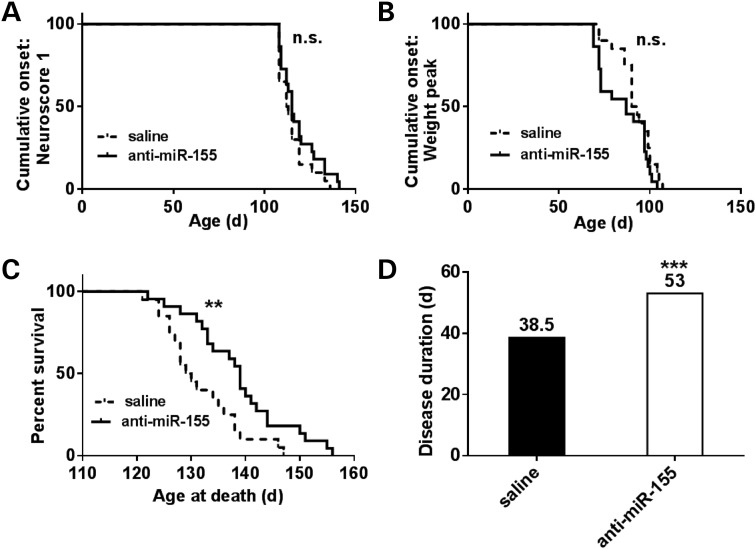
Beginning at 60 days of age, weights and neurological scores (16) were recorded biweekly. Onset was determined in two ways: age when weight peaked and age when a neurological score of 1 was observed marking abnormal hind limb motility. Treatment and monitoring continued until the animal reached end-stage, defined as when the animal was unable to right itself within 30 s after being placed on its side. There was no significant change in disease onset as defined by either a neurological score of 1 (Fig. 5A) or by age at weight peak (Fig. 5B). However, survival was significantly increased in only the anti-miR-155-treated mice (Fig. 5C); these mice had a 9.5-day extension in survival over saline-treated mice (P = 0.006). There was no significant difference in survival between scrambled-treated and saline-treated animals (Table 2). Our controls agreed with published survival data for SOD1G93A B6/SJL animals (129.5 versus 128.9 days16). Furthermore, disease duration was only significantly increased in the anti-miR-155-treated animals (Fig. 5D). These mice had a disease duration 14.5 days longer than saline (38% extension, P = 5.0 × 10−4) and 11 days longer than scrambled (27% extension, P = 2.7 × 10−3). There was no significant difference in disease duration between scrambled-treated and saline-treated animals (Table 2).
Figure 5.
Survival, but not onset, is extended in anti-miR-155-treated ALS mice. SOD1G93A SJL mice were treated with both osmotic pumps directed to the lateral ventricles and with weekly IP injections starting at 60 days of age. Mice were weighed and neurological scores were determined biweekly. A score of 1 marked onset and was characterized by one of the following: (i) failure to fully extend legs when lifted by tail; (ii) failure to spread legs past midline when lifted by tail and (iii) marked tremors when lifted by tail. Survival was determined as when the mouse could not right itself within 30 s of being placed on either side. (A and B) Saline (n = 20) and anti-miR-155-treated mice (n = 22) had no significant difference in onset as determined by neurological score of 1 or by age at peak weight. (C and D) Anti-miR-155-treated mice had a significant extension in survival and a 38% extension in disease duration over saline (median values shown, log-rank test, *P < 0.01, **P < 0.01, ***P < 0.001).
Table 2.
Anti-miR-155-treated mice have extended survival and disease duration over both saline- and scrambled-treated mice
| Days | P-value | Log-rank | Wilcoxon | |
|---|---|---|---|---|
| Onset: Weight Peak | ||||
| Saline | 91.5 | Sal-Scr | 0.10 | 0.26 |
| Scrambled | 90 | Sal-155 | 0.13 | 0.12 |
| anti-miR-155 | 87 | Scr-155 | 0.82 | 0.49 |
| Onset: Neuroscore 1 | ||||
| Saline | 112.5 | Sal-Scr | 0.60 | 0.29 |
| Scrambled | 115 | Sal-155 | 0.21 | 0.19 |
| anti-miR-155 | 115 | Scr-155 | 0.54 | 0.81 |
| Survival | ||||
| Saline | 129.5 | Sal-Scr | 0.51 | 0.24 |
| Scrambled | 133 | Sal-155 | 0.007** | 0.007** |
| anti-miR-155 | 139 | Scr-155 | 0.018* | 0.035* |
| Disease duration | ||||
| Saline | 38.5 | Sal-Scr | 0.081 | 0.15 |
| Scrambled | 42 | Sal-155 | <0.001*** | 0.003** |
| anti-miR-155 | 53 | Scr-155 | 0.038* | 0.047* |
Median onset values are denoted for saline- (n = 20), scrambled- (n = 21) and anti-miR-155- (n = 22) treated mice as determined both by age when the mouse reached weight peak and by a neurological score of 1. There was no significant change between either onset condition by both Mantel–Cox log-rank and Gehan–Breslow–Wilcoxon tests. However, survival and disease duration were significantly extended in only the anti-miR-155-treated mice over both saline and scrambled mice as determined by both log-rank and Wilcoxon tests. (*P, 0.01; **P, 0.01; ***P, 0.001.).
DISCUSSION
To determine the therapeutic potential of targeting miRNAs in ALS, we first generated a list of miRNAs changed in the SOD1G93A rodent model. Six of these miRNA changes are also upregulated in familial and sporadic human ALS spinal cord. However, for each of these miRNAs, whether the change contributes to disease progression or represents a compensatory/beneficial response remained unclear. To test the importance of the miRNA in disease, we developed an antisense oligonucleotide strategy to inhibit miRNAs in the brain and spinal cord. For miR-155, we demonstrated that inhibiting miR-155 slowed disease progression in the SOD1G93A model. Because miR-155 is increased in human sporadic ALS, these data highlight the miR-155 pathway as an exciting, new therapeutic target for ALS.
Antisense oligonucleotides can function as potent inhibitors of miRNAs (anti-miRs), but it remained unclear whether these inhibitors could achieve broad CNS distribution similar to antisense oligonucleotide inhibitors of mRNA (10). We first assayed anti-let-7 because this oligonucleotide is known to be a potent miRNA inhibitor in the periphery (12). let-7 is not known to be important in ALS and is best known for its role in cell cycle regulation, cell differentiation and cancer (17). Following a 28-day infusion of anti-let-7 into the cerebral lateral ventricle in mice, we saw robust derepression of let-7 targets throughout the brain and spinal cord as determined by both global Sylamer analysis and targeted mRNA assays. While other studies have demonstrated anti-miR function in limited sections of cortical tissue (18,19), these data establish a method of widespread inhibition of miRNAs and are the first to show inhibition of miRNAs in the spinal cord. Next, we developed a novel anti-miR against miR-155, previously identified as significantly upregulated in ALS. This anti-miR also showed widespread functional distribution in both the peripheral and central compartments. Anti-miRs have been used safely and effectively in the periphery in recent human clinical trials [NCT00688012 and NCT00979927 (20)], and CSF infusion of antisense oligonucleotides designed to lower mRNA has recently been applied to patients with ALS (21). Thus, CSF delivery of anti-miRs has the real potential to translate to clinical trials.
In order to test the therapeutic potential of inhibiting dysregulated miRNAs in neurodegenerative disease, we treated a large cohort of ALS-model mice with our novel anti-miR-155. Mice treated with anti-miR-155 lived 10 days longer and had a 38% extension in disease duration. Therefore, our data establish that the increased miR-155 negatively contributes to ALS disease progression in the SOD1G93A mouse model. While it is self-evident that patients present after onset of disease, the fact that miR-155 inhibition slowed disease progression may suggest that this preclinical drug trial is particularly relevant for therapies. The majority of drugs tested in the rapidly progressive SOD1G93A model prolong survival by delaying onset of the disease rather than slowing progression of the disease. Of those studies that have reported extended disease duration in the mouse model, the weighted mean difference in a meta-analysis was 10%, while we here report a 38% extension in disease duration (22). For further context, a previous study using antisense oligonucleotides against SOD1 in ALS-model rats did not significantly alter onset but extended survival by 10 days and extended disease duration by 37% (10).
While a number of drugs tested in the SOD1 model have subsequently failed in human clinical trials, we have taken steps to increase the translational potential of our use of this preclinical model. We only considered important those miRNAs increased in both the rodent ALS models and in patient ALS tissue from both familial and sporadic ALS patients. Because miR-155 levels are increased in peripheral blood cells in some ALS patients (9), enrollment for a clinical trial could be determined by elevated miR-155 levels. Similarly, the efficacy of this targeted miR-155 therapy can be read out in peripheral cells throughout treatment. Finally, antisense oligonucleotides have been tested in a Phase I trial in SOD1 human ALS, suggesting this type of strategy to be safe and realistic (21).
Our data clearly identify miR-155 as a promising therapeutic target for ALS and may potentially apply more broadly to neurological disorders where miR-155 is increased including multiple sclerosis (23) and in glioblastoma (24). However, as an ALS therapeutic, some key preclinical issues still need to be addressed. It will be important to test how CNS inhibition alone or peripheral blood cell inhibition alone affects survival in this model since this will have direct implications for which compartment is the most important to target with an anti-miR-155 therapy. In addition, how the timing of miR-155 inhibition will affect survival needs to be examined. Also, our data define other miRNAs that are increased in ALS spinal cord tissue. Interestingly, all six of the patient-confirmed upregulated miRNAs (miR-24, miR-142-3p, miR-142-5p, miR-146a, miR-146b and miR-155) are well studied in the context of immunity and inflammation (25,26). Each of these targets may be potential candidates for therapies for neurodegenerative disease, and testing them will require a similar approach to the one taken here. Another miRNA identified here may have a more potent effect either alone or in concert with other miRNA targets. Finally, to determine mechanism of action and to determine whether neuroinflammation is blunted, a large cohort of mice will need to be treated and sacrificed in age-matched fashion throughout disease for analysis of molecular and cellular targets.
We have demonstrated a therapeutically relevant method to inhibit miRNAs broadly in the brain and spinal cord for the first time. We have also supplied the first published list of altered miRNAs in ALS spinal cord tissue, and we have demonstrated that miRNA inhibition can prolong survival in a neurodegeneration model. miRNA inhibition, and miR-155 specifically, remain exciting avenues for ALS research as potential therapeutic targets.
MATERIALS AND METHODS
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee of Washington University at St. Louis and adhered to NIH guidelines. Animals were provided food and water ad libitum, and cages were changed once a week. All mice were maintained in a 12-hour light/12-hour dark cycle and received routine veterinary monitoring. Post-surgical mice were single-housed as to protect the sutures and tubing. SOD1G93A L26H rats were purchased from Taconic. SOD1WT rats were generously provided by Dr Pak H. Chan (Stanford University School of Medicine). C57Bl/6J SOD1G93A mice were purchased from Jackson Laboratories (stock number: 004435), and B6SJL SOD1G93A mice were gifted from Prize4Life.
Human tissues
Patient tissues were analyzed with approval from the Institutional Review Board from Washington University and Massachusetts General Hospital. Spinal cord autopsy tissue was obtained from 16 ALS patients (nine male, seven female; age range, 34–79, mean 64; from Washington University School of Medicine, Massachusetts General Hospital) and 12 non-ALS control patients (six male, six female; age range, 51–80, mean 70; from National Disease Research Interchange; clinical information in Supplementary Material, Table S1). Postmortem intervals ranged from 1 to 40 h.
miRNA quantification
RNA was extracted from flash-frozen tissue using miRNeasy kits per manufacturer's instructions (Qiagen, Hilden, Germany). Mouse RNA integrity and yield were assessed by Nanodrop before diluting to 150 ng/µl for microarray assay and 2 ng/µl for miRNA individual analysis. miRNA microarrays were performed without amplification using low-density rodent miRNA A + B cards sets 3.0 (Applied Biosystems, Foster City, CA) using a 7900HT quantitative real-time polymerase chain reaction (qRT-PCR) machine for 40 cycles. Analysis was conducted on SDSv2.2 software with automatic thresholding. Microarray hits were confirmed with individual TaqMan miRNA assays (Applied Biosystems) as per the manufacturer's instructions. RNA from patient autopsy samples were also quantified via Nanodrop and diluted to 2 ng/µl. All rodent miRNAs were normalized to endogenous U6, and human to total RNA input. qPCR samples were quantified in technical duplicates on an Applied Biosystems 7500 fast Real-Time PCR System.
mRNA quantification
For mRNA Affymetrix microarray and Sylamer analysis, RNA was diluted to 100 ng/µl and sent to Expression Analysis, Inc (Durham, NC) for running the Affymetrix Mouse Genome 430 2.0 array (Santa Clara, CA). Analysis was then conducted as previously described by others (27). For RT-qPCR of mRNA targets, RNA was normalized to 20 ng/µl using Nanodrop. Primers and probes for mRNA targets were purchased from Integrated DNA Technologies (Coralville, IA) and are detailed in Supplementary Material, Table S2. For let-7 target analysis, 40 ng total RNA was quantified with Express One-Step Superscript Kits (Invitrogen, Carlsbad, CA) as per the manufacturer's protocol. For miR-155 targets, 40 ng total RNA was quantified with Power SYBR Green RNA-to-CT 1-Step Kit (Life Technologies, Carlsbad, CA) as per the manufacturer's protocol. One-step qRT-PCR was conducted on a 7500 fast Real Time PCR System (Applied Biosystems). RNA was quantified as technical duplicates and normalized to GAPDH.
anti-miR sequences
All anti-miRs were synthesized with a full phosphorothioate backbone with alternating blocks of 2′-MOE and 2′fluoro sugar-modified nucleosides. Sequences are denoted in Supplementary Material, Table S2.
anti-miR-155 in vitro luciferase assay
miRNA antisense oligonucleotides were screened for in vitro function as described previously (28). Briefly, Hela cells were co-transfected with a miR-155 expressing plasmid and with a luciferase reporter with perfect 2× miR-155 complementary sequences at the 3′-UTR. Four hours later, anti-miR-155 was added to the media at concentrations ranging from 1 to 200 nM. Twenty-four hours later, luciferase activity was determined and plotted.
Isolation of peritoneal macrophages
Adult C57Bl6 mice were given an IP injection of Thioglycollate followed by three daily IP injections of 25 mg/kg anti-miR or saline. On day 4, peritoneal macrophages were isolated, plated and stimulated with LPS for 24 h (20 ng/ml). RNA was then extracted and analyzed by Sylamer analysis.
Alzet osmotic pump surgeries
Osmotic pumps were prepared per manufacturer protocol (pump model 2004 for 28 day experiments; 2006 for 42 day experiments; Alzet, Cupertino, CA). For implantation, mice were anesthetized in a chamber with 5% isoflurane/oxygen mixture and confirmed to be unconscious before being placed in Kopf Model 940 small animal stereotaxic apparatus, fitted with ear bars (29). The catheter was oriented at 2.00 mm lateral and 1.1 mm posterior to bregma.
Cy3-anti-miR-155 histology
Adult C57Bl6 mice were implanted with a subdermal Alzet osmotic pump to deliver 10 µg/day of cy3-labeled anti-miR-155 (Regulus Therapeutics, San Diego CA) directly into the lateral ventricle. After 2 weeks of treatment, mice were perfused with phosphate buffered saline (PBS) and 4% paraformaldehyde (PFA). Brain and spinal cord tissues were post-fixed in PFA for 48 h and then submerged in 30% sucrose for 2 days. The spinal cord was embedded in O.C.T. (VWR, Radnor, PA) before it and the brain were sliced at 40 µm. Tissue for distribution analysis was then washed in PBS and mounted with Fluoromount G (Southern Biotech, Birmingham, AL) before being coverslipped.
For cell-specific determination, tissues were washed in PBS and blocked in 5% natural horse serum in PBS for 30 min. A dilution of 1:1000 of chicken-anti-GFAP polyclonal antibody (ab4674, Abcam, Cambridge England), 1:1000 goat-anti-Iba1 polyclonal antibody (ab5076, Abcam) or 1:200 mouse anti-NeuN antibody (MAB377, Millipore, MAB377) in blocking buffer was then applied to the tissues overnight at 4°C. Tissues were then washed with PBS and blocked with 5% natural horse serum for 30 min before incubating in their relevant 1:1000 AlexaFluor488-conjugated secondary antibody for 1 h [goat anti-chicken (ab150169, Abcam), donkey anti-goat (ab150129, Abcam) or goat anti-mouse (35502, Thermo Scientific, Waltham, MA)]. All slides were observed at 4×, 10× and 20× objectives using a Nikon Eclipse 80i microscope fitted with a Photometrics CoolSnap EZ camera. All images were taken at ambient temperature with a Cy3 or FITC filter. For image acquisition and formatting, NIS Elements 3.0 (Nikon) and Adobe Photoshop v12.0 were used.
Treatment and analysis of SJL SOD1G93A ALS mice
All B6SJL SOD1G93A were gifted from Prize4Life and were previously confirmed to have high and consistent SOD1G93A copy counts. Mice were implanted with osmotic pumps at 60 days containing 20 µg/day anti-miR-155, 20 µg/day scrambled control or saline. Pumps were removed at 102 days. Weekly IP injections (25 mg/kg) began at 60 days and continued until the mouse reached end-stage. For the first two weeks only, mice received an additional weekly IP injection. Mice were weighed and assessed for neurological score biweekly in blinded fashion starting at 60 days. A neurological score of 1 was given when (i) the mouse was no longer able to fully extend its legs; (ii) the mouse could no longer spread its legs past midline when lifted by its tail or (iii) significant hind limb tremors were present. End-stage was defined as when the mouse was no longer able to right itself after being placed on either side within 30 s. All involved in the administration and monitoring of the animals were blinded with separate people involved in injecting and scoring the mice. Blinding was broken only once all analyses were completed.
Statistical analysis
Data are presented as mean ± standard error of the mean. All statistical tests were conducted with Graphpad Prism 6 Software. All gene expression data were analyzed with a 2-tailed Student t test with the exception of the quantification of miR-155 in patient populations, which was analyzed by ANOVA followed by Tukey's post-hoc test. All survival and onset data were analyzed using both the Mantel–Cox log-rank test and the Gehan–Breslow–Wilcoxon test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Project5 for ALS to T.M.M.; the NIH (R01NS078398 NINDS to T.M.M., K08NS074194 to T.M.M., K08NS062138 to G.F.W., F31NS077781 to E.D.K.); the Robert Packard Center for ALS Research to T.M.M.; the Edward Mallinckrodt, Jr. Foundation to T.M.M., and the Dana Foundation to G.F.W.
Supplementary Material
ACKNOWLEDGEMENTS
SOD1G93A mice were generously provided by ALS Prize for Life. SOD1WT rats were generously provided by Dr Pak H. Chan (Stanford University School of Medicine). We thank National Disease Research Interchange (NDRI) for supplying control patient autopsy spinal cord tissue, and Dr James Berry and Dr Merit Cudkowicz (Massachusetts General Hospital) for supplying ALS patient autopsy spinal cord tissue. We thank ALS patients and families for generously providing research tissues.
Conflict of Interest statement. We disclose that Washington University has applied for a patent on the use of miR-155 inhibitors as a therapy for ALS.
REFERENCES
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein J.D. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann. Neurol. 2009;65(Suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 3.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connell R.M., Chaudhuri A.A., Rao D.S., Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Zhao Q., Matta R., Meng X., Liu X., Liu C.G., Nelin L.D., Liu Y. Inducible nitric-oxide synthase expression is regulated by mitogen-activated protein kinase phosphatase-1. J. Biol. Chem. 2009;284:27123–27134. doi: 10.1074/jbc.M109.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu S., Stavrovskaya I.G., Drozda M., Kim B.Y., Ona V., Li M., Sarang S., Liu A.S., Hartley D.M., Wu D.C., et al. Minocycline inhibits cytochrome c release and delays progression of amyotrophic lateral sclerosis in mice. Nature. 2002;417:74–78. doi: 10.1038/417074a. [DOI] [PubMed] [Google Scholar]
- 7.Boillee S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 8.Beers D.R., Henkel J.S., Zhao W., Wang J., Appel S.H. CD4+ T cells support glial neuroprotection, slow disease progression, and modify glial morphology in an animal model of inherited ALS. Proc. Natl. Acad. Sci. USA. 2008;105:15558–15563. doi: 10.1073/pnas.0807419105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butovsky O., Siddiqui S., Gabriely G., Lanser A.J., Dake B., Murugaiyan G., Doykan C.E., Wu P.M., Gali R.R., Iyer L.K., et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 2012;122:3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H., et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall J.S., Taylor J., Valentine H.R., Irlam J.J., Eustace A., Hoskin P.J., Miller C.J., West C.M. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br. J. Cancer. 2012;107:684–694. doi: 10.1038/bjc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esau C. Keystone Symposia: Diabetes (Z6) Whistler: British Columbia Canada; 2010. [Google Scholar]
- 13.Zhang Y., Diao Z., Su L., Sun H., Li R., Cui H., Hu Y. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am. J. Obstet. Gynecol. 2010;202:466 e461–e467. doi: 10.1016/j.ajog.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 14.Vigorito E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A., et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu G., Fewell C., Taylor C., Deng N., Hedges D., Wang X., Zhang K., Lacey M., Zhang H., Yin Q., et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA. 2010;16:1610–1622. doi: 10.1261/rna.2194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leitner M., Menzies S., Lutz C. Working with ALS mice. 2009 Prize4Life, Inc. [Google Scholar]
- 17.Androsavich J.R., Chau B.N., Bhat B., Linsley P.S., Walter N.G. Disease-linked microRNA-21 exhibits drastically reduced mRNA binding and silencing activity in healthy mouse liver. RNA. 2012;18:1510–1526. doi: 10.1261/rna.033308.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selvamani A., Sathyan P., Miranda R.C., Sohrabji F. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS One. 2012;7:e32662. doi: 10.1371/journal.pone.0032662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S.T., Chu K., Jung K.H., Kim J.H., Huh J.Y., Yoon H., Park D.K., Lim J.Y., Kim J.M., Jeon D., et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012;72:269–277. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 20.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 21.Miller T.M., Pestronk A., David W., Rothstein J., Simpson E., Appel S.H., Andres P.L., Mahoney K., Allred P., Alexander K., et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet. Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benatar M. Lost in translation: treatment trials in the SOD1 mouse and in human ALS. Neurobiol. Dis. 2007;26:1–13. doi: 10.1016/j.nbd.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Junker A., Krumbholz M., Eisele S., Mohan H., Augstein F., Bittner R., Lassmann H., Wekerle H., Hohlfeld R., Meinl E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 24.D'Urso P.I., D'Urso O.F., Storelli C., Mallardo M., Gianfreda C.D., Montinaro A., Cimmino A., Pietro C., Marsigliante S. miR-155 is up-regulated in primary and secondary glioblastoma and promotes tumour growth by inhibiting GABA receptors. Int. J. Oncol. 2012;41:228–234. doi: 10.3892/ijo.2012.1420. [DOI] [PubMed] [Google Scholar]
- 25.Sonkoly E., Stahle M., Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen I., David M. MicroRNAs in the immune response. Cytokine. 2008;43:391–394. doi: 10.1016/j.cyto.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dongen S., Abreu-Goodger C., Enright A.J. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esau C.C. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Devos S.L., Miller T.M. Direct intraventricular delivery of drugs to the rodent central nervous system. J. Vis. Exp. 2013;75:e50326. doi: 10.3791/50326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.